Abstract
The prediction of pairing between microRNAs (miRNAs) and the miRNA recognition elements (MREs) on mRNAs is expected to be an important tool for understanding gene regulation. Here, we show that mRNAs that contain Pumilio recognition elements (PRE) in the proximity of predicted miRNA-binding sites are more likely to form stable secondary structures within their 3′-UTR, and we demonstrated using a PUM1 and PUM2 double knockdown that Pumilio proteins are general regulators of miRNA accessibility. On the basis of these findings, we developed a computational method for predicting miRNA targets that accounts for the presence of PRE in the proximity of seed-match sequences within poorly accessible structures. Moreover, we implement the miRNA-MRE duplex pairing as a two-step model, which better fits the available structural data. This algorithm, called MREdictor, allows for the identification of miRNA targets in poorly accessible regions and is not restricted to a perfect seed-match; these features are not present in other computational prediction methods.
INTRODUCTION
MicroRNAs (miRNAs) are ∼21–22 nt endogenous RNAs that direct the post-transcriptional repression of protein-coding genes by imperfect pairing to miRNA recognition elements (MREs) within their transcripts. Their deep involvement in physiological and pathological processes makes the understanding of the mechanisms by which miRNA select their targets a major challenge.
A conserved Watson–Crick pairing to the bases 2–8 of the miRNA’s 5′ region, which is also called the miRNA seed (1–6), is crucial for miRNA targeting. This relationship is confirmed by the strong conservation that is observed for 7mer that are complementary to the seed region within protein-coding genes’ 3′-UTRs (7–9) as well from motif-enrichment analysis performed on top downregulated genes on ectopic expression of miRNAs (10–12). Crystallographic analysis of Argonaute (Ago) proteins in complex with miRNAs provided a structural explanation by showing that the bases of the seed are uniquely constrained in a conformation that makes them solvent accessible and primed for miRNA pairing nucleation (1,2–6,13). However, a perfect match of the seed sequence is not always functional nor does it lead to similar repression activity. Other determinants that must be involved in the effectiveness of miRNA-mediated regulation have been described, such as functional miRNA-MRE pairing in the absence of perfect seed complementarity as well as non-functional pairing in the presence of a perfect seed match (7–9,14,15).
Local target accessibility appears to play a key role because a large fraction of validated MREs preferentially reside outside of a 3′-UTR’s stable secondary structure (10–12,16,17), which is reflected by the local nucleotide composition being skewed toward a higher AU content (2,4,13). However, the prediction of the local accessibility is a difficult task because the RNA secondary structure as well as the formation of the duplex between miRNA and mRNA are multifactorial events. Moreover, RNA-binding proteins can regulate positively or negatively the function of miRNA on specific mRNA by altering MRE accessibility. HuR (ELAVL1) and DND1 have been proven to antagonize miRNA binding, respectively, to CAT-1 and p27 mRNAs (8,14,15). The human PUM1 has been shown to be required for the repression that is mediated by miR-221/222 on p27 mRNA and to enhance the activity of multiple E2F3 targeting miRNAs (11,16,17). This feature appears to be conserved because the Caenorhabditis elegans Pumilio homolog puf-9 is required for the repression that is mediated by let-7 on the Hunchback homolog hbl-1 (2,4,18). PUF proteins represent a highly conserved family of ubiquitously expressed RNA-binding proteins that play an important role in stem cell maintenance, development and differentiation by binding to conserved elements within target mRNA 3′-UTR (8,19). An important feature of the PUF family is the highly conserved C-terminal RNA-binding domain termed the Pumilio homology domain (11,20), which binds to a conserved 8 nt sequence UGUANAUA, called the Pumilio Recognition Element (PRE) (1,3,5,6,21–23). A genome-wide analysis has shown that the PRE is highly enriched around predicted miRNA-binding sites (7,9,24). Moreover, it has been recently shown that Pumilio proteins can form a complex with Ago proteins and the core elongation factor eEF1A to repress translational elongation (10,12,25).
Here, we developed a highly sensitive computational method for miRNA target prediction that accounts for the role of PRE in the accessibility of miRNA as well as the dynamics of the miRNA-MRE pairing, and the sites that were predicted were validated experimentally.
MATERIALS AND METHODS
Sequences and validated miRNA targets
The 3′-UTRs sequences were obtained from UTRdb (26) (Release 2010). To enable automatic retrieval of up-to-date sequences, an object-oriented Perl module was developed (available on request). miRNA sequences were obtained from miRBase (27) (version 19), whereas seed sequences for conserved miRNA families were taken from a previous study (13).
Validated miRNA–MRE interactions (Supplementary Tables S1 and S3) were obtained from previously published data. The positive data set includes sites from miRecords database(28), TarBase (29) and from individual studies (see references in Supplementary Tables S1 and S3). The negative data set was obtained from previously published data (30) and from individual studies (see references in Supplementary Tables S1 and S3).
Comparison with state-of-the-art algorithms
Whole-genome predictions were downloaded from the respective sites for TargetScan (http://www.targetscan.org/vert_61/vert_61_data_download), miRanda (http://cbio.mskcc.org/microrna_data), PITA (http://genie.weizmann.ac.il/pubs/mir07/catalogs for PITA 0/0 or http://genie.weizmann.ac.il/pubs/mir07/catalogs for PITA 3/15), TargetMiner (http://www.isical.ac.in/∼bioinfo_miu/Genome_Wide_Target(Human).rar), MultiMiTar (http://www.isical.ac.in/∼bioinfo_miu/multimitar-genomewide-prediction.zip), MirTarget2 (http://mirdb.org/miRDB/download/MirTarget2_v4.0_prediction_result.txt.gz) and TargetSpy (http://www.targetspy.org/data/hsa_refseq_all.gz). For each method, we calculated the numbers of predicted true positives (TP), true negatives (TN), false positives (FP) and false negatives (FN). We then calculated the standard performances comparison measures of sensitivity [SN = TP/(TP + FN)], specificity [SPC = TN/(TN + FP)], accuracy [ACC = (TP + TN)/(TP + TN + FP + FN)] and Matthews Correlation Coefficient {MCC = [TP*TN – FP*FN]/sqrt[(TP + FP)*(TP + FN)*(TN + FP)*(TN + FN)]}.
miRNAs targeting determinants computation
For each validated MRE in our training data set (Supplementary Table S2), we computed duplex free energy (ΔGduplex), duplex loop position and class, target site accessibility (ΔGaccess), seed-match accessibility (ΔGseed), free energy gain (ΔΔG) and the local AU content.
All of the energy-related features were calculated using the ViennaRNA Package 2.0 (31) API.
ΔGaccess is calculated as the difference between the ΔG of the ensemble of 3′-UTR structures and the ΔG of the structures ensemble in which a constraint is imposed to make the nucleotide positions between 15 nt upstream and 3 nt downstream of the seed-match unpaired, whereas ΔΔG corresponds to the difference between ΔGduplex and ΔGaccess (10).
Duplex loop position and class were computed as previously described (32).
Constructs design and cloning
A window of 200 bases centered on REST and Sirt1, and DDX58-predicted MREs were amplified from HEK293 FT genomic DNA and cloned into a pMIR-Report (Invitrogen, cat. AM5795) vector, within SpeI and HindIII restriction sites. Each construct was verified by sequencing.
Cell cultures, transfection and Luciferase assay
HEK293 cells (Invitrogen) were maintained in Dulbecco Modified Eagle Medium, supplemented with 10% Fetal Bovine Serum, 1% Pen-Strep and 1% Sodium Pyruvate. For the luciferase assays, 3 × 104 cells were seeded per well in a 96-well plate the day before transfection. The cells were transfected with Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions, with 100 ng of pMIR-Report (Invitrogen, cat. AM5795) vector bearing the tested MRE, 5 ng of SV40-Renilla and either 105 nM of AllStars negative control siRNA (Qiagen, cat.1027280) or 50 nM of miRNA mimic, plus 55 nM of control siRNA or 50 nM of miRNA mimic, plus 45 nM of Pum1 siRNA (Ambion, cat.138317) and 10 nM of Pum2 siRNA (Ambion, cat. s23672). Firefly luciferase activity was assayed after 24 h using the Dual Luciferase Reporter Assay System (Promega, cat.E1910) and was normalized over the Renilla intensity.
Protein extraction and western blot
Approximately 1 × 106 cells were scraped in 1 ml of cold phosphate buffered saline (PBS) and centrifuged for 5′ at 1000g; then, cell pellets were resuspended in 200 μl of cold F-Buffer [10 mM Tris–HCl (pH 7.0), 50 mM NaCl, 30 mM Sodium Pyrophosphate, 50 mM Sodium Fluoride and 5 μM ZnCl2]. Cells were subjected to three cycles of sonication (30′′ ON, 30′′ OFF, High) and then stored on ice for 10′. Cell extract was then centrifuged for 10′ at 14 000g, and the pellet was discarded. PUM1 and PUM2 were revealed by western blot using Pumilio AbVantage Pack antibodies (A310-056A, Bethyl Laboratories).
RNA immunoprecipitation
Approximately 3 × 107 cells were scraped in 5 ml of cold PBS and were centrifuged for 5′ at 1000g. Cells were resuspended in cold Polysomal Lysis Buffer [10 mM HEPES (pH 7.0), 100 mM KCl, 5 mM MgCl2, 25 mM EDTA, 0.5% NP-40, 2 mM DTT, 50 U/ml Rnase OUT and 50 U/ml Superase IN] and stored on wheel at 4°C for 30′. Cells were then centrifuged for 10′ at 14 000g, and the supernatant was recovered. A total of 60 μl of protein G beads were added per ml of extract and were stored on wheel at 4°C for 1 h to perform pre-clearing. The lysate was then recovered from the beads and incubated with 5 μg of IgG, PUM1 or PUM2 antibodies (A310-056A, Bethyl Laboratories) for 16 h on wheel at 4°C. The next day, 25 μl of protein G beads saturated for 16 h in PBS + BSA (1%) was added to the lysate and stored on wheel for 2 h at 4°C. Once recovered, the beads were washed four times with cold PLB. To elute the RNA, 1 ml of TRIzol reagent (Invitrogen) was added directly to the beads and left on wheel at RT for 20′.
Microarray
HEK293 cells were transfected with either 105 nM of AllStars negative control siRNA (Qiagen, cat.1027280) or 50 nM of miR-297 mimic (Qiagen, cat. MSY0004450) plus 55 nM of control siRNA or 50 nM of miR-297 mimic, plus 45 nM of Pum1 siRNA (Ambion, cat.138317) and 10 nM of Pum2 siRNA (Ambion, cat. s23672). A total RNA extraction was performed using Trizol reagent (Invitrogen), and sample quality control was performed with Agilent Bioanalyzer 2100. Detection was performed using Human HT-12 v4 Expression BeadChip (Illumina) on an Illumina HiScanSQ platform.
Microarray analysis was performed with Illumina GenomeStudio software. Briefly, signals were quantile normalized and background adjusted; then, probes with P < 0.05 were classified as present (P), probes with P-values between 0.05 and 0.065 were classified as marginal (M), and probes with a P-value under 0.065 were classified as absent (A). For downstream analysis, we kept only the genes that were classified as present in at least one condition or those that were classified as marginal in at least two conditions. Moreover, we selected only those genes that had a perfect 7–8mer seed-match for miR-297, which were considered to avoid side effects and to somehow allow a reasonable comparison with other prediction algorithms. Moreover, three analysis data sets were defined after clustering, as follows: [i] PUM-independent cluster composed of genes with FC ≤ −0.5 in both miR-297 and miR-297 plus PUM1/2 double knockdown transfections versus control; [ii] PUM-dependent cluster composed of genes with FC ≤ −0.5 in miR-297 transfection versus control and FC ≥ 0.5 in miR-297 plus PUM1/2 double knockdown versus miR-297 alone; and [iii] Non-regulated cluster composed of expressed genes that had at least one miR-297 perfect seed-match, which did not exhibit significant expression change.
Heatmap and clustering of microarray data was performed using the R package (v2.14.1).
RT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen). Real-time PCR was performed using the SuperScript III Platinum One-Step Quantitative RT-PCR System (Invitrogen, cat.11732-020) following the manufacturer’s instructions. The complete list of the primers that were used is provided in the Supplementary Table S7.
Analysis of miR-297 target accessibilities
For each cluster of genes from the microarray, we computed an overall accessibility as follows. First, we scanned the entire 3′-UTR sequence of each gene for possible 7–8mer matches; then, the accessibility for each seed-match was computed as the difference between the free energy 3′-UTR’s structures ensemble and the free energy of the 3′-UTR in which a constraint was imposed to make the positions 15 nt upstream and 3 nt downstream of the seed-match unpaired. For genes that had more than one seed-match, we averaged the accessibility of each possible target. The same calculation was iterated using different window sizes (100, 250, 500 and 750 nt around the predicted seed-match), and the overall accessibility was obtained by averaging the ΔGaccess calculated over the different windows. This approximation allows us to minimize the error that is introduced by the employment of arbitrary window sizes.
Algorithm description and implementation
Given a miRNA sequence, the algorithm extracts the first 8 nt positions at the 5′ side (seed ± 1 nt) and computes the reverse complement. Starting from this template, the following possible seed-matches are generated (Supplementary Figure S4): (i) 8-mer (perfect pairing to positions 1–8); (ii) 8mer-A1 (perfect pairing to positions 2–8, plus an unpaired A at position 1 of the MRE); (iii) 7mer-m8 (perfect pairing to positions 2–8); (iv)7mer-A1 (perfect pairing to positions 2–7, plus an unpaired A at position 1 of the MRE); (v) 6mer (6 contiguous bases paired starting either at positions 1, 2 or 3); (vi) 5mer (5 contiguous bases paired starting either at positions 1, 2 or 3); (vii) 4mer (4 contiguous bases paired starting either at positions 2 or 3); (viii) G:U wobble (7 contiguous bases paired, allowing for 1 or 2 G:U wobbles); (ix) Target bulge (1 nt of the MRE forms a bulge and does not take part in the duplex nucleation), (x) Seed bulge (1 nt of the seed forms a bulge and does not take part in the duplex nucleation); and (xi) Mismatch (1 nt of the seed is not complementary to its corresponding base in the MRE).
Once identified, the algorithm extracts a window of 200 nt (where possible) that are centered on the seed-match and computes the regional ΔGaccess. If the accessibility cost exceeds the threshold value (default: −10 kcal/mol), a positional weight matrix (PWM) is used to locate possible PRE motifs within the same window. Any site that resides within an inaccessible region that lacks a nearby PRE motif is discarded. For each of the remaining sites, the algorithm performs a thermodynamic simulation of the miRNA–MRE pairing using a two-step model composed of a first nucleation step at the seed level followed by propagation of the duplex pairing toward its 3′-end. First, it extracts a region of n – m nucleotides upstream of the seed-match, where n is the length of the miRNA and m is the length of the seed match, and then, it computes the duplex minimum free energy under the constraints imposed by the seed. Any structure that is not compliant with these constraints is discarded. The same calculation is then iterated 35 – n times, and the structure with both the minimum ΔGduplex and the overall system’s free energy is selected. The value of 35 nt was chosen because the longest mirRNA–MRE interaction identified so far extends for 35 nt (33,34).
Next, miRNA-mRNA duplexes are filtered according to different ΔGduplex cut-off values on the basis of their seed-match class (ΔGperfect for perfect 7–8mer sites, and ΔGimperfect for imperfect sites with fewer than 7 bases paired in the seed). Whole genome prediction for a given miRNA takes about 15 min.
Implementation
MREdictor can be run online on user-provided data on the HuGeF website (http://mredictor.hugef-research.org).
RESULTS
MREs with nearby PREs tend to reside within inaccessible contexts
To determine the features that significantly contribute to miRNA-mediated repression, we first created a data set of literature-validated miRNA–MRE interactions (Supplementary Table S1) that was composed of functional (positive data set) and non-functional (negative data set) interactions. We then calculated the correlation of MRE effectiveness to previously described features such as duplex free energy (ΔGduplex), duplex loop position and class(13,32), target site accessibility (ΔGaccess) (10,12,14,15), seed-match accessibility (ΔGseed) (16,17,35), free energy gain (ΔΔG) (10) and local AU content (13). To reduce possible bias, we built the training data set to include human and mouse validated interactions, with both perfect and imperfect seed-matches. To unambiguously compute the features that depend on the surrounding context of MRE, we picked sites that reside at least 100 nt away from the 3′-UTR boundaries.
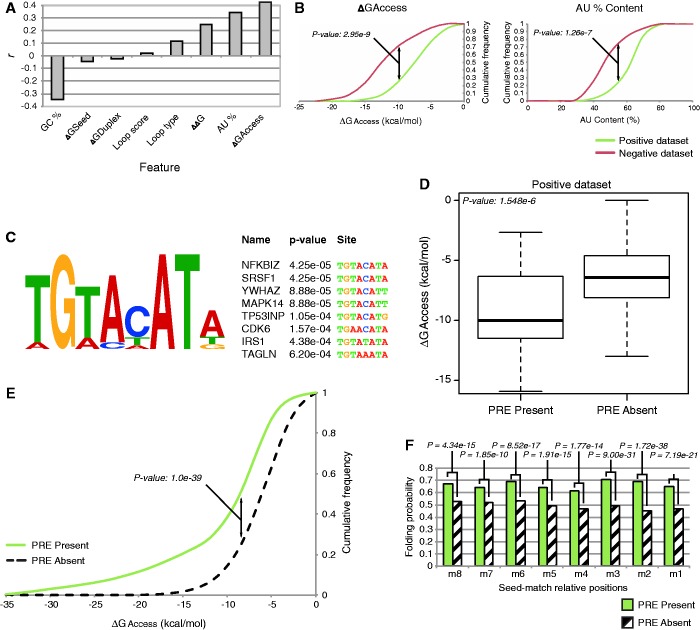
In agreement with previous studies (10,12,13), target site accessibility and local AU content exhibited a higher correlation between the examined features (Figure 1A, r = 0.34 for AU content, r = 0.43 for ΔGaccess, Matthews correlation coefficient) because they stratified well the data set between the functional and non-functional sites (Figure 1B, P-value: 2.95e-9 for ΔGaccess, P-value: 1.26e-7 for AU content). However, ∼20% of the interactions that were functional in our data set were located within poorly accessible contexts. Although a small fraction of these sites could be FP, the majority of these sites were validated by western blot analysis or by using the whole 3′-UTR for the luciferase assay, ruling out the possibility of experimental bias, thus suggesting that other determinants are involved in the functionality of these sites. As RNA-binding proteins binding to their cognate target can influence the miRNA targeting to MRE (14,15,18), we extracted a window of ∼100 nt around the seed-match of the functional MRE that was classified as not accessible and performed discriminative motif-discovery using the MEME algorithm (36). This analysis showed enrichment for the motif TGTANATA surrounding the MRE (Figure 1C, e-value: 1.2e-22), which belongs to the recognition element of the family of Pumilio RNA-binding proteins. On the basis of these results, we classified the functional MRE for its presence within the surrounding sequences of the identified motif. This classification stratified well the positive data set in both the accessible and inaccessible sites (Figure 1D, P-value: 1.55e-6). Notably, the coupling of the target site accessibility with the presence of a nearby PRE showed a marked increase in the correlation to MRE effectiveness that was achieved (r = 0.78, Matthews correlation coefficient).
Figure 1.
PRE is enriched in validated and predicted thermodinamically inaccessible regions. (A) Correlation of different features to miRNA binding effectiveness measured on the training dataset. Local target accessibility (ΔGaccess) and AU content exhibit the greatest correlation. (B) Cumulative frequency plots show that functional targets (positive data set) are more likely to reside within 3′-UTR regions with a higher AU content, and that there is higher target accessibility with respect to non-functional targets (negative data set). P-values are given by Welch’s t-test. (C) Motif discovery analysis performed using MEME shows an enrichment for the PRE motif belonging to the Pumilio family proteins (e-value: 1.2e-22). Top enriched mRNA bearing PRE motifs are shown. (D) Presence of the PRE motif well stratifies the functional interactions between highly accessible and poorly accessible targets. The P-value is given by Welch’s t-test. (E) Cumulative frequency plot of top 20 PRE-associated human miRNA families. Seed-matches with nearby PRE motifs are enriched within poorly accessible regions with respect to those lacking a close PRE. The P-value is given by the Kolmogorov–Smirnov test. (F) The base probability of the seed-match positions for the top 20 PRE-associated miRNA families in human. Sites with nearby PRE motifs exhibit a higher probability of being already engaged in a bond within the local 3′-UTR secondary structure. Positions m8 to m1 are paired, respectively, to miRNA’s seed positions 1–8. P-values per base are given by a Chi-Squared test.
In human, the Pumilio family presents two members, PUM1 and PUM2, which are ubiquitously expressed across all tissues and developmental stages (2,4,37) (Supplementary Figure S1A).
To exclude the possibility that our observations were caused by a bias that was introduced during the data set construction, we investigated, genome-wide, the involvement of Pumilio proteins in the regulation of miRNA accessibility. To this end, we downloaded the 3′-UTR sequences for all human RefSeq genes from UTRdb (8,26) and calculated the fraction of perfect seed-matches for each conserved miRNA family that resided within 50 nt from a PRE motif. Next, we selected the top PRE-associated miRNA families and evaluated the local accessibility for any possible seed-match. This analysis showed a strong preference for the sites that fall in the proximity of a PRE to reside within more thermodynamically inaccessible regions when compared with the sites that lack a nearby PRE (Figure 1E, P-value: 1.0e-39). Then, we computed the ensemble of the possible minimum free energy structures for a 100 nt window centered on the predicted MRE, and we calculated the probability that each base of the seed-match was already engaged in a bond within the local 3′-UTR’s secondary structure. The probability of finding a seed-match base that was in an unpaired state and that was, therefore, competent to achieve miRNA-MRE duplex nucleation was significantly lower for the sites with a nearby PRE motif (Figure 1F). Similar results were obtained when performing the same analysis in mouse and fly (Supplementary Figure S1B and C). Overall, in human, the Pumilio-dependent MREs are predicted to be 4.38% of the inaccessible sites (Supplementary Figure S1D). Taken together, these results suggest that a regulation of miRNA targeting by Pumilio proteins is general and highly conserved.
An algorithm that accounts for both structural and contextual features outperforms state-of-the-art methods
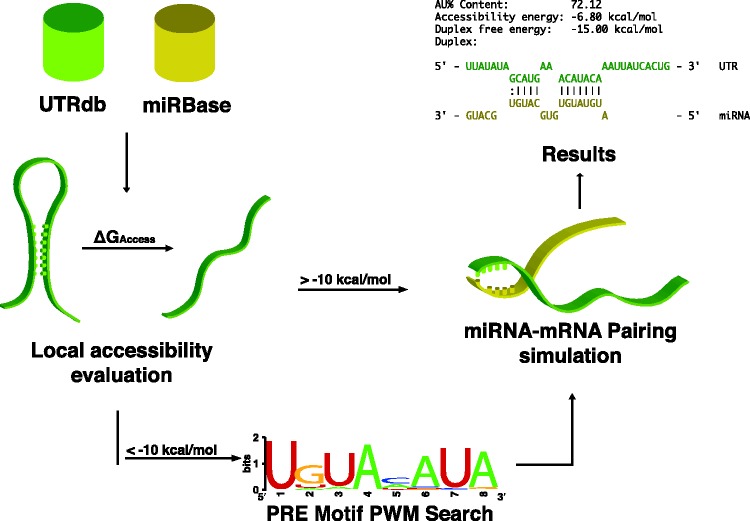
Next, we incorporated the aforementioned findings into a computational method, called MREdictor (Figure 2), to search for MRE for a given miRNA within 3′-UTR sequences. Our algorithm models the miRNA–MRE interaction by considering the presence of Pumilio-binding elements to assess the actual accessibility of the MRE.
Figure 2.
Schematic representation of MREdictor’s pipeline. Given a target to test and a miRNA, MREdictor obtains sequences for the 3′-UTR from UTRdb and for the miRNA from miRBase. For every possible seed-match, the local accessibility is evaluated, and regions exceeding the ΔGaccess energy cost of −10 kcal/mol are subjected to a Positional Weight Matrix scan for possible PRE motifs. If no PRE is discovered, the site is discarded. Sites that pass this first filtering step are then subjected to a simulation of duplex formation and are filtered according to their free energy (ΔGduplex).
For each possible seed-match, the local accessibility is evaluated, as previously described (11,10), over a window of 200 nt centered on the seed-match. For the sites that reside within inaccessible regions (ΔGaccess < −10 kcal/mol), MREdictor scans the window for possible PRE motifs by using a PWM derived from available PUM2 PAR-CLIP (38) data (Supplementary Table S2), to identify PRE sites in the proximity of the MRE.
Seed-matches passing these preliminary filtering steps are then subjected to a duplex formation simulation and are subsequently filtered according to their duplex free energy (ΔGduplex). This filtering step is crucial and must account for the type of MRE. It is well accepted that a perfect seed-match is sufficient for productive pairing of the miRNA to its target, whereas an imperfect seed-match requires an extended compensatory pairing of the 3′ tail of the miRNA (7,9,13,39). Therefore, the use of a single threshold energy value to filter any MRE is incorrect because imperfect seed-matched sites require an extended 3′ pairing and will have a lower duplex free energy compared with the perfect matches. To account for this consideration, MREdictor uses different threshold values for duplexes that have at least seven consecutive bases paired in the seed or less than seven bases paired in the seed.
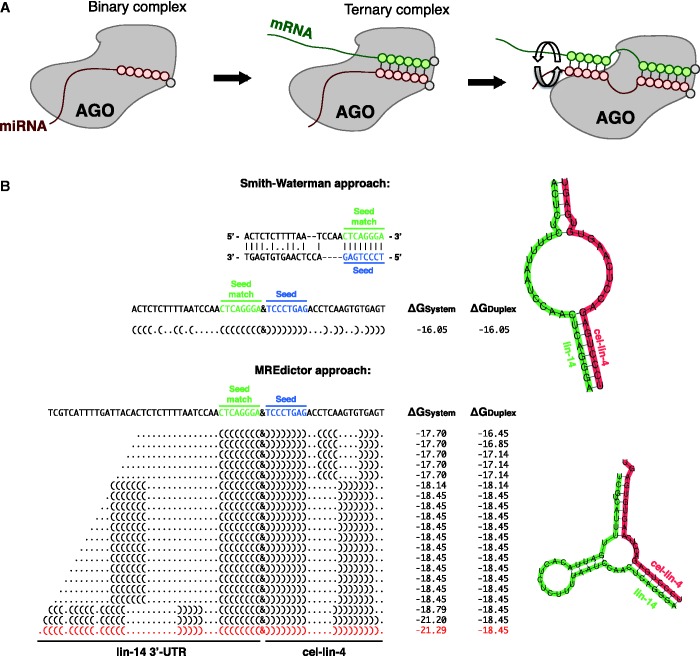
Moreover, the pairing between miRNA and its cognate MRE is a two-step process that starts from the seed-match and extends to the miRNA 3′-end. Structural evidence shows that the miRNA 5′-end is anchored in a solvent-accessible binding pocket within the MID domain of Ago proteins, whereas the 3′-end is solvent inaccessible and locked in a binding pocket of the PAZ domain (1,3,5,6,10,12). Binding of the miRNA 5′-end to its cognate mRNA leads to a conformational change, with the subsequent release of its 3′-end from the PAZ domain leading to a productive pairing.
To account for this mechanism, instead of using the Smith–Waterman approach (13,40,41), which maximizes only the interactions between the miRNA and MRE, MREdictor simulates the duplex formation as a two-step reaction of duplex nucleation at the seed level with progressive pairing under the constraints imposed by the seed (Figure 3). Compared with the Smith–Waterman approach, this method considers the seed-constraint compliant structure, which minimizes both the duplex free energy and the free energy of the MRE rearranged to interact with the miRNA and maximizes the duplex bonds.
Figure 3.
Two step calculation of miRNA-MRE pairing. (A) Two-step model of the miRNA–mRNA interaction. (B) Comparison of a Smith–Waterman-like approach and the MREdictor method for miRNA-MRE duplex modeling, which consist of the nucleation of the miRNA-MRE duplex at the seed level followed by an extended 3′-end pairing (see ‘Materials and Methods’ section), applied to a well-known worm MRE. Previous studies have shown that, in Caenorhabditis elegans, the lin-4 miRNA can target lin-14, forming a bulged structure within lin-14 3′-UTR (25,33,34). A Smith–Waterman-like approach tends to maximize only the number of interactions between the miRNA and the target, minimizing the number of gaps and the extension of the gaps; thus, it fails to correctly predict the duplex secondary structure and underestimates the duplex free energy.
MREdictor identifies the miRNA–mRNA interactions in poorly accessible regions and at non-canonical targets
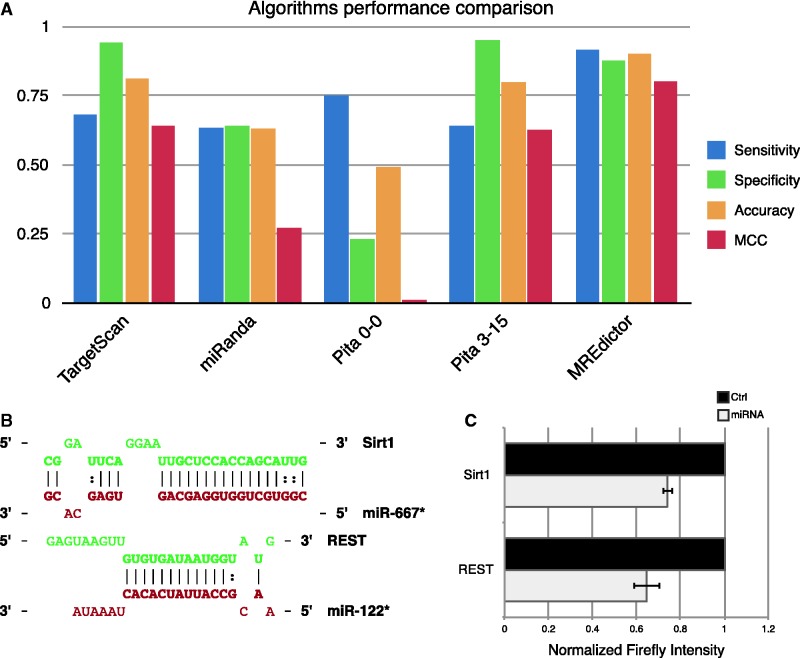
To assess the accuracy of MREdictor, we first created a test data set that was composed of 106 functional and 106 non-functional experimentally validated MREs from individual studies (Supplementary Table S3). Compared with state-of-the-art prediction algorithms (10,13–15,40), MREdictor correctly predicts a larger fraction of the functional sites and has the highest accuracy (ACC = 0.9; Figure 4A). To further test its ability to predict novel putative targets, we performed a whole-genome scan of human and mouse 3′-UTR and randomly picked two non-canonical MREs, which were not predicted by other algorithms. Hsa-miR-122* and mmu-miR-667*, two non-conserved miRNAs, were predicted by MREdictor to target, respectively, REST and Sirt1 on two non-conserved MREs; the interactions lacked perfect seed-pairing, bearing, respectively, a mismatch plus a G:U wobble or two consecutive G:U wobbles at the seed level (Figure 4B).
Figure 4.
Validation of MREdictor method. (A) Comparison of standard performance measures of state-of-the-art algorithms and MREdictor. (B) Schematic representation of two non-canonical MREs predicted by MREdictor but not by other tools, in the absence of a perfect seed-match. Hsa-miR-122* was predicted to target REST (ΔGduplex = −20.9 kcal/mol), whereas mmu-miR-667* was predicted to target Sirt1 (ΔGduplex = −34.7 kcal/mol). (C) Dual luciferase assay validation of the two predicted MREs. Data are averaged over four replicates, and error bars are given for S.Ds.
For each MRE, a window of 200 nt centered on the seed-match was cloned downstream of a firefly luciferase reporter and was cotransfected in HEK293 along with the cognate miRNA or a negative control. For both sites analyzed, we observed a significant repression of the reporter with respect to the controls (Figure 4C). Although MREdictor identified also a perfect seed-matched MRE for mmu-miR-667* on Sirt1 mRNA (located 475 nt downstream of the imperfect site), it was not included in the firefly luciferase construct as we here aimed to measure only the functionality of the predicted imperfect seed-matched site.
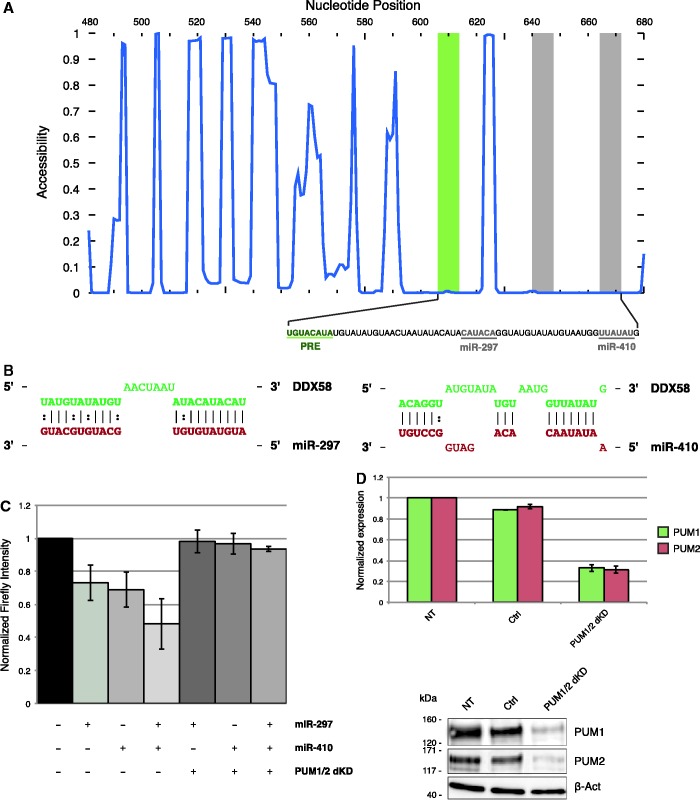
Next, we tested the potentiality of MREdictor to predict sites that were dependent on Pumilio proteins binding. To this end, we recomputed the association of each miRNA family to the PRE motif, accounting for the local accessibility (Supplementary Table S4). We then selected an extremely poorly accessible region within the DDX58 3′-UTR (Figure 5A, ΔGaccess = −20.75 kcal/mol), which was predicted to be bound by both hsa-miR-297, one of the top PRE-associated miRNA families (P-value: 4.46e-06) and hsa-miR-410 (P-value: 6.46e-04; Figure 5B). HEK293 were cotransfected with a luciferase reporter bearing the DDX58 3′-UTR window together with the cognate miRNA or a negative control. For both sites, we observed a significant downregulation of the reporter’s activity with respect to the control (Figure 5C). The co-transfection of the two miRNAs together showed increased repression, which suggested a cooperative action of the two MREs. Next, we verified the dependency of the two sites on Pumilio protein binding. In HEK293, PUM1 is the most abundant Pumilio family member; however, we observed an increase in the PUM2 levels when performing the single knockdown of PUM1 (Supplementary Figure S2A and B). This mutual regulation, together with the previously reported observation that a large fraction of PUM1-bound mRNAs are also PUM2 targets (16,17,24), suggests that these two proteins have redundant functions. Therefore, we performed the knockdown of both members (Figure 5D). PUM1/2 double knockdown almost completely abolished the effect of hsa-miR-410 and hsa-miR-297 on DDX58 3′-UTR (Figure 5C).
Figure 5.
Validation of PUM-dependent sites predicted by MREdictor. (A) Accessibility plot of a DDX58 3′-UTR window containing two MREs for miR-297 and miR-410, which are predicted to be dependent on Pumilio proteins binding to their cognate PRE motif. (B) Schematic representation of the two MREs, as predicted by MREdictor. (C) Dual luciferase assay validation of the two predicted MREs within DDX58 3′-UTR. Data are averaged over four replicates, and error bars are given for S.Ds. (D) RT-qPCR analysis and western blot of HEK293 FT cells transfected either with a negative control or two siRNAs targeting PUM1 and PUM2.
Pumilio proteins are necessary for miRNA-mediated repression at thermodynamically inaccessible sites
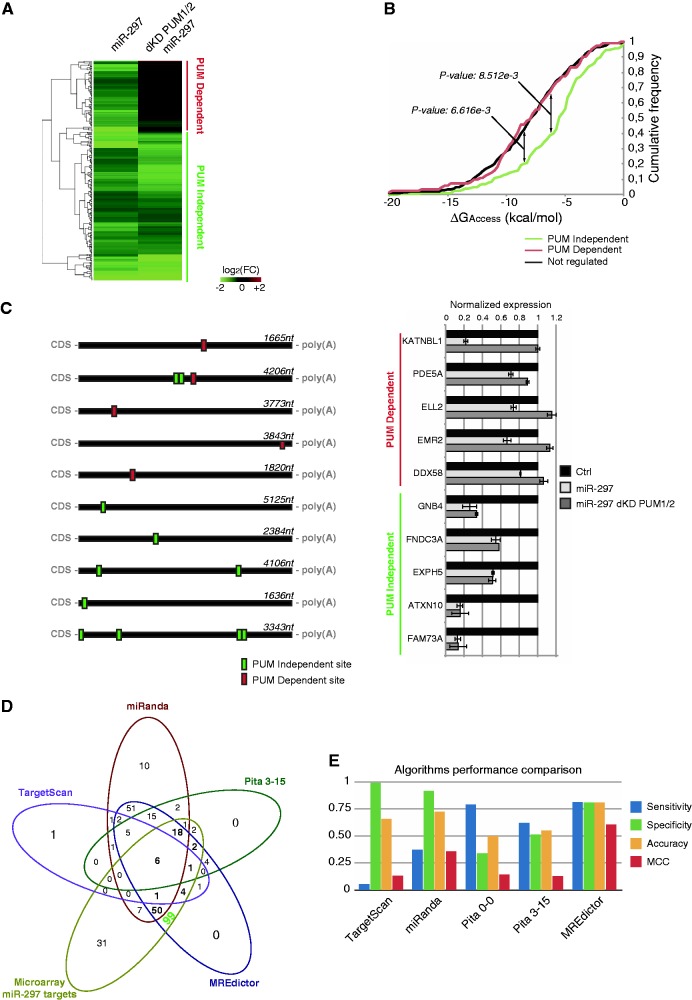
It is a widely accepted assumption that miRNAs can determine downregulation of their targets by triggering mRNA degradation (11,18,42). To verify that we observed a general phenomenon, we analyzed the role of Pumilio proteins in the positive regulation of miRNA-mediated repression by microarray analysis on HEK293 transfected either with a negative control, hsa-miR-297 or hsa-miR-297 along with PUM1/2 double knockdown (Supplementary Table S5). Although this method underestimates the actual number of true targets, as miRNA that affect translational inhibition without affecting RNA stability are not considered by this assay, it provides a good approximation to estimate the Pumilio function on miRNA targeting. The results were then clustered according to the gene expression relative to the control. This analysis revealed two main clusters of transcripts regulated by miR-297 (Figure 6A). A first cluster was composed of genes whose downregulation was not significantly affected by PUM1/2 knockdown (PUM-independent cluster). A second cluster was composed of genes whose downregulation was significantly reduced or completely abolished by PUM1/2 knockdown (PUM-dependent cluster).
Figure 6.
Microarray validation of MREdictor predictions. (A) Heatmap showing the two clusters of genes that emerge after microarray analysis of HEK293 cells transfected with miR-297 in the presence or absence of PUM1/2. In the PUM-dependent cluster, the miR-297-mediated repression is abolished (or significantly reduced) following PUM1/2 double knockdown. (B) Cumulative frequency plot of the PUM-dependent, PUM-independent and non-regulated gene clusters. Genes within the PUM-dependent cluster are more likely to reside within poorly accessible regions compared with the PUM-independent cluster, which is similar to the non-regulated genes. The P-value is given by the Welch t-test. (C) Schematic representation of the 3′-UTR of five genes from each cluster, and RT-PCR validation of microarray analysis. Data are averaged over triplicate experiments, and error bars are given for the S.Ds. (D) Venn diagram showing the overlap between the predictions for miR-297 of different tools and MREdictor compared with microarray downregulated genes. MREdictor successfully predicts 99 unique targets, which are not predicted by other programs. (E) Comparison of standard performance measures of state-of-the-art algorithms and MREdictor predictions for miR-297, calculated over microarray data.
The unbiased de novo motif discovery performed on the 3′-UTR sequences of the PUM-independent cluster’s genes showed enrichment for the miR-297 seed-match motif CATACA (e-value: 5.6e-31). The genes of the PUM-dependent cluster, in addition to the seed-match motif, also showed enrichment for the PRE motif (e-value: 6.1e-47). We then focused our attention on a subset of 222 genes from both clusters that bear at least one perfect 7–8mer match for miR-297 within their 3′-UTRs. Computation of local accessibility for each MRE showed that the predicted sites for the PUM-dependent data set exhibited a strong preference for residing within poorly accessible regions compared with the PUM-independent data set (Figure 6B, P-value: 8.512e-3). Notably, a third cluster of 402 genes that did not exhibit any significant downregulation following miR-297 (although they had a perfect 7–8mer seed-match) showed a similar preference (Non-regulated cluster, P-value: 6.616e-3). This finding suggests that Pumilio proteins are necessary to disrupt local stable secondary structures within the mRNA. We validated our findings by RT-qPCR on a selected group of genes that were correctly predicted by MREdictor as being either PUM-dependent or PUM-independent (Figure 6C). We observed that PUM1/2 double knockdown totally abolished the miR-297 mediated repression of the KATNBL1 (C15ORF29), ELL2, EMR2 and DDX58 genes, which belong to the PUM-dependent cluster, whereas the expression of PDE5A was rescued only partially, which is consistent with the presence of two PUM-independent sites within its 3′-UTR in addition to one PUM-dependent site. RNA immunoprecipitation assay confirmed that all of the PUM-dependent transcripts are actually bound by PUM1 and PUM2 (Supplementary Figure S3). Interestingly, PUM proteins also bind to some of the PUM-independent transcripts. In these cases, however, PUM silencing did not affect the miR-297-mediated repression, which demonstrated that the effect observed on the PUM-dependent transcripts is not due to other known RNA decay functions of Pumilio proteins (19,43).
Finally, we attempted to determine how many of the sites exhibiting a significant downregulation from microarray analysis were correctly recovered by MREdictor, compared with other miRNA target prediction algorithms (10,13,20,40). To this end, we computed whole-genome predictions for miR-297 (Supplementary Table S6). MREdictor predicted 254 targets for miR-297 in HEK293 cells. Of the 222 genes that resulted to be downregulated by microarray analysis, 181 sites were correctly predicted by MREdictor. Approximately 55% of these genes were not identified by other tools (Figure 6D and Supplementary Figure S3C). Furthermore, we used the cluster of the non-regulated genes as a negative data set and computed standard performance measures for MREdictor (Figure 6E and Supplementary Figure S3D) (30,44–46). Remarkably, our method outperformed existing tools, obtaining the highest overall accuracy (ACC = 0.81) and correctly predicted the dependence on Pumilio binding of ∼3.6% of the identified MREs.
DISCUSSION
It has been recently shown that, beyond the canonical determinants of miRNA pairings to their cognate targets, RNA-binding proteins play a key role in regulating the effectiveness of this regulation. Kedde and colleagues (16,21–23) demonstrated that Pumilio proteins favor the miR-221/222-mediated repression of p27 in mammalian cells by increasing the MRE accessibility. By the analysis of previously validated miRNA targets and by whole-genome analysis of seed-match sequences, we have observed that MREs with nearby Pumilio-binding sites reside preferentially within 3′-UTR less accessible contexts. We further demonstrated, by luciferase and gene expression analysis, that Pumilio proteins are required for miRNA-mediated repression of targets that reside within stable secondary structures.
We have developed a computational tool that accounts for the target’s accessibility and the presence of nearby PRE motifs as well as the actual two-step mechanism of pairing between the miRNA and the MRE.
Our analysis, in agreement with previous studies (10,13,24), showed that target accessibility, which is reflected by the local higher AU content, is a major determinant of miRNA regulation efficacy. However, this result alone cannot explain why many previously validated functional targets reside within poorly accessible contexts. Motif discovery analysis, performed on data sets of previously validated sites, showed that the PRE motif, belonging to the highly conserved family of Pumilio RNA-binding proteins, is highly enriched in the proximity of functional validated targets, which are predicted to be thermodynamically inaccessible. Whole-genome analysis confirmed that PRE is enriched around miRNA predicted seed-match sequences, which reside within locally stable secondary structures (examples are shown in Supplementary Figure S5) not only in humans but also in mouse and drosophila, which suggests a conserved widespread mechanism of action of PUM proteins. Furthermore, we took advantage of high-throughput microarray analysis to demonstrate that, in the absence of Pumilio proteins, miRNA-mediated repression is abolished on MRE residing on low accessibility regions.
A good miRNA targets prediction tool should be able to identify the larger number of bona fide MREs for a given miRNA while keeping the false-discovery rate as low as possible. Our method fulfills these criteria in terms of the overall accuracy because it correctly recovers a higher fraction of the sites when compared with other methods. The strength of this algorithm can be explained by the fact that it finds targets independently from phylogenetic conservation, it models more accurately the actual miRNA–MRE interaction, and it allows us to include in the analysis 3′UTR less-accessible regions. To our knowledge, this is the first time that a computational tool accounts for the contribution of RNA-binding proteins for miRNA targeting prediction.
In summary, our software outperforms existing state-of-the-art methods in predicting previously validated targets and successfully predicts new canonical as well as non-canonical targets. It is already known that other RNA-binding proteins participate to the regulation of miRNA function (14,15) and many other could be discovered. Once the molecular mechanism of action of these proteins will be characterized and their binding motifs will be defined, they could be introduced into predictive algorithms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Associazione Italiana Ricerca sul Cancro (AIRC); Regione Toscana programma salute; and Ministero della Salute Bando Cellule Staminali. Funding for open access charge: HuGeF.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Lambert NJ, Gu SG, Zahler AM. The conformation of microRNA seed regions in native microRNPs is prearranged for presentation to mRNA targets. Nucleic Acids Res. 2011;39:4827–4835. doi: 10.1093/nar/gkr077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 3.Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 4.Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 2009;10:141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat. Struct. Mol. Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 8.Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin C, Nam J-W, Farh KK-H, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol. Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 11.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 12.Brown KM, Chu CY, Rana TM. Target accessibility dictates the potency of human RISC. Nat. Struct. Mol. Biol. 2005;12:469–470. doi: 10.1038/nsmb931. [DOI] [PubMed] [Google Scholar]
- 13.Grimson A, Farh KH, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 15.Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Ørom UA, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 16.Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JAF, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol. 2010;12:1014–1020. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- 17.Miles WO, Tschop K, Herr A, Ji JY, Dyson NJ. Pumilio facilitates miRNA regulation of the E2F3 oncogene. Genes Dev. 2012;26:356–368. doi: 10.1101/gad.182568.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nolde MJ, Saka N, Reinert KL, Slack FJ. The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3′UTR-mediated repression of the let-7 microRNA target gene, hbl-1. Dev. Biol. 2007;305:551–563. doi: 10.1016/j.ydbio.2007.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 20.Zamore PD, Williamson JR, Lehmann R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA. 1997;3:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 21.Chen G, Li W, Zhang Q-S, Regulski M, Sinha N, Barditch J, Tully T, Krainer AR, Zhang MQ, Dubnau J. Identification of synaptic targets of Drosophila pumilio. PLoS Comput. Biol. 2008;4:e1000026. doi: 10.1371/journal.pcbi.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White EK, Moore-Jarrett T, Ruley HE. PUM2, a novel murine puf protein, and its consensus RNA-binding site. RNA. 2001;7:1855–1866. [PMC free article] [PubMed] [Google Scholar]
- 23.Fox M, Urano J, Reijo Pera RA. Identification and characterization of RNA sequences to which human PUMILIO-2 (PUM2) and deleted in Azoospermia-like (DAZL) bind. Genomics. 2005;85:92–105. doi: 10.1016/j.ygeno.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 24.Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS One. 2008;3:e3164. doi: 10.1371/journal.pone.0003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friend K, Campbell ZT, Cooke A, Kroll-Conner P, Wickens MP, Kimble J. A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nat. Struct. Mol. Biol. 2012;19:176–183. doi: 10.1038/nsmb.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grillo G, Turi A, Licciulli F, Mignone F, Liuni S, Banfi S, Gennarino VA, Horner DS, Pavesi G, Picardi E, et al. UTRdb and UTRsite (RELEASE 2010): a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2010;38:D75–D80. doi: 10.1093/nar/gkp902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vergoulis T, Vlachos IS, Alexiou P, Georgakilas G, Maragkakis M, Reczko M, Gerangelos S, Koziris N, Dalamagas T, Hatzigeorgiou AG. TarBase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res. 2011;40:D222–D229. doi: 10.1093/nar/gkr1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bandyopadhyay S, Mitra R. TargetMiner: microRNA target prediction with systematic identification of tissue-specific negative examples. Bioinformatics. 2009;25:2625–2631. doi: 10.1093/bioinformatics/btp503. [DOI] [PubMed] [Google Scholar]
- 31.Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye W, Lv Q, Wong C-KA, Hu S, Fu C, Hua Z, Cai G, Li G, Yang BB, Zhang Y. The effect of central loops in miRNA:MRE duplexes on the efficiency of miRNA-mediated gene regulation. PLoS One. 2008;3:e1719. doi: 10.1371/journal.pone.0001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 34.Ha I, Wightman B, Ruvkun G. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes Dev. 1996;10:3041–3050. doi: 10.1101/gad.10.23.3041. [DOI] [PubMed] [Google Scholar]
- 35.Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 36.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W773. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castle JC, Armour CD, Löwer M, Haynor D, Biery M, Bouzek H, Chen R, Jackson S, Johnson JM, Rohl CA, et al. Digital genome-wide ncRNA expression, including SnoRNAs, across 11 human tissues using polyA-neutral amplification. PLoS One. 2010;5:e11779. doi: 10.1371/journal.pone.0011779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp A-C, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie F, Zhang B. Target-align: a tool for plant microRNA target identification. Bioinformatics. 2010;26:3002–3003. doi: 10.1093/bioinformatics/btq568. [DOI] [PubMed] [Google Scholar]
- 42.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 43.Van Etten J, Schagat TL, Hrit J, Weidmann CA, Brumbaugh J, Coon JJ, Goldstrohm AC. Human Pumilio Proteins Recruit Multiple Deadenylases to Efficiently Repress Messenger RNAs. J. Biol. Chem. 2012;287:36370–36383. doi: 10.1074/jbc.M112.373522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sturm M, Hackenberg M, Langenberger D, Frishman D. TargetSpy: a supervised machine learning approach for microRNA target prediction. BMC Bioinformatics. 2010;11:292. doi: 10.1186/1471-2105-11-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X, Naqa ElIM. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;24:325–332. doi: 10.1093/bioinformatics/btm595. [DOI] [PubMed] [Google Scholar]
- 46.Mitra R, Bandyopadhyay S. MultiMiTar: a novel multi objective optimization based miRNA-target prediction method. PLoS One. 2011;6:e24583. doi: 10.1371/journal.pone.0024583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.