Abstract
Previous analyses of complexes of 40S ribosomal subunits with the hepatitis C virus (HCV) internal ribosome entry site (IRES) have revealed contacts made by the IRES with ribosomal proteins. Here, using chemical probing, we show that the HCV IRES also contacts the backbone and bases of the CCC triplet in the 18S ribosomal RNA (rRNA) expansion segment 7. These contacts presumably provide interplay between IRES domain II and the AUG codon close to ribosomal protein S5, which causes a rearrangement of 18S rRNA structure in the vicinity of the universally conserved nucleotide G1639. As a result, G1639 becomes exposed and the corresponding site of the 40S subunit implicated in transfer RNA discrimination can select  . These data are the first demonstration at nucleotide resolution of direct IRES–rRNA interactions and how they induce conformational transition in the 40S subunit allowing the HCV IRES to function without AUG recognition initiation factors.
. These data are the first demonstration at nucleotide resolution of direct IRES–rRNA interactions and how they induce conformational transition in the 40S subunit allowing the HCV IRES to function without AUG recognition initiation factors.
INTRODUCTION
The mechanism of translation initiation in eukaryotes requires the presence of a 7-methyl guanosine cap at the 5′-ends of messenger RNA (mRNA) [reviewed in (1)]. The cap initiates a cascade of events involving protein initiation factors that allows the 40S ribosomal subunits loaded with the ternary complex eIF2• •GTP to scan mRNA and to recognize the initiator AUG codon [reviewed in (2,3)]. The uncapped genomic RNAs of a number of viruses are enabled to bypass the requirement for a 5′-cap in AUG codon recognition by highly structured elements in the 5′ untranslated regions (UTR) upstream of the initiator AUG codon, named Internal Ribosome Entry Sites (IRESs) (4). One such RNA is the genomic RNA of hepatitis C virus (HCV), whose IRES is able to initiate translation in the absence of the majority of initiation factors and to form the correct 48S translation initiation complex in the presence of just initiation factor eIF3 and the ternary complex eIF2•
•GTP to scan mRNA and to recognize the initiator AUG codon [reviewed in (2,3)]. The uncapped genomic RNAs of a number of viruses are enabled to bypass the requirement for a 5′-cap in AUG codon recognition by highly structured elements in the 5′ untranslated regions (UTR) upstream of the initiator AUG codon, named Internal Ribosome Entry Sites (IRESs) (4). One such RNA is the genomic RNA of hepatitis C virus (HCV), whose IRES is able to initiate translation in the absence of the majority of initiation factors and to form the correct 48S translation initiation complex in the presence of just initiation factor eIF3 and the ternary complex eIF2• •GTP (4,5).
•GTP (4,5).
The HCV IRES has a length of ∼300 nt. It forms a complex secondary structure with three distinct domains (6). The molecular mechanism of HCV IRES-driven translation initiation is only partially understood. The low resolution cryo-electron microscopy (cryo-EM) images of the HCV IRES in binary complexes with 40S subunits (7) and 80S ribosomes (8) provides only a general idea of the location of the IRES and its domains on the 40S subunit. Cross-linking (5,9–12) and protein fluorescent labelling (13) studies have revealed ribosomal proteins neighbouring the IRES RNA and its subdomains in the binary complex with the 40S ribosomal subunit. This information helped to localize the HCV IRES binding site on the 40S subunit and to identify the ribosomal proteins that could be responsible for IRES binding and function. Another result of the cryo-EM studies of the HCV IRES•40S subunit complex was the observation that the IRES causes conformational changes in the structure of the 40S subunit and that the IRES domain II is responsible for them (7). It has been hypothesized that these conformational changes involve the 18S ribosomal RNA (rRNA) and lead to opening of the mRNA-binding channel latch formed by non-covalent junction of helices (h) 18 and 34 in vacant 40S subunits (7). As a result, a new connection stabilizing the open latch emerges between h16 and ribosomal protein (rp) S3 that is accompanied by subunit head rotation (7); this could facilitate location of the coding part of HCV RNA in the mRNA-binding channel. Similar conformational changes were observed upon joint binding of initiation factors eIF1 and eIF1A to the 40S subunit (14) hinting at common features in canonical and HCV IRES-driven translation initiation, although a rotation of the subunit head was observed only with the IRES structure (7). However, no contacts between the HCV IRES and 18S rRNA in the 40S subunit have been found, and probing of 18S rRNA structure with nucleotide base-specific probes has revealed no IRES-inducible changes (12,15). Thus, we know neither whether 18S rRNA is involved in the molecular mechanism of the HCV IRES-driven translation initiation nor what might be the structural basis for IRES-specific conformational changes of the 40S subunit.
Here, we use direct hydroxyl radical probing to demonstrate alterations in the 18S rRNA structure caused by binding of the HCV IRES RNA to the 40S subunit of a human ribosome. This method was chosen because the backbone of 18S rRNA on the 40S subunit surface looked more accessible than nucleotide bases to chemical probes. Hydroxyl radical footprinting of free 40S subunits and their complexes with the intact HCV IRES or its derivatives reveals that the IRES protects the apical loop of h26 in the 18S rRNA from hydroxyl radicals. Additionally, the triplet CCC in this loop is protected from dimethylsulphate (DMS) modification. Furthermore, we show that IRES-binding causes a rearrangement of the 18S rRNA backbone in the region of nucleotide G1639, and that this rearrangement is provided by interplay between the IRES domain II and the initiator AUG codon. These findings shed light on the molecular mechanism of the initial step of HCV IRES-driven translation initiation.
MATERIALS AND METHODS
HCV IRES RNA transcripts preparation
HCV IRES RNAs were prepared by T7 transcription of DNAs generated by PCR using plasmid pXL40-372.NS’ (16) containing the 5′-UTR of the HCV genome RNA as a template. Primers for PCR were as follow: T7-IRES-HCV (5′-AAATTAATACGACTCACTATAGGGAGACTCCCCTGTGAGGAAC TAC-3′), T7-IRES–HCVdII (5′-AAATTAATACGACTCACTATAGGGAGACCCTCCCGGGAGAGCC–3′), IRES-HCV(AUG)-R (5′-CATGGTGCACGGTCTACG–3′), IRES-HCV(UUU)-R (5′-AAAGGTGCACGG TCTACG–3′), IRES-HCV(FL)-R (5′-GGTTTTTCTTTGAGGTTTAGG–3′) and IRES HCVdIV-R (5′-TACGAGACCTCCCGGGGCAC–3′). DNA polymerase Pfu (Fermentas) and the following pairs of primers were used to generate DNA templates for synthesis of RNA transcripts: T7-IRES-HCV and IRES-HCV(AUG)-R (for IRESaug), T7-IRES–HCVdII and IRES-HCV(AUG)-R (for IRESaugΔDII), T7-IRES-HCV and IRES-HCV(FL)-R (for IRESfl), T7-IRES–HCVdII and IRES-HCV(FL)-R (for IRESflΔDII), T7-IRES-HCV and IRES-HCV(UUU)-R (for IRESuuu) and T7-IRES-HCV and IRES HCVdIV-R (for IRESΔDIV). RNA transcripts were synthesized by incubation of 2 µg of DNA template in 250 µl of reaction mixture containing 160 mM HEPES-KOH (pH 7.5), 27 mM MgCl2, 2.7 mM spermidine, 13 mM DTT, 5.3 mM NTPs, 500 U of T7 RNA polymerase and 0.2 U of inorganic pyrophosphatase (Sigma) at 37°C for 6 h; the DNA template was then degraded by incubation of the reaction mixture with 1 U of DNase (Ambion) at 37°C for 30 min, and RNA was purified by gel-filtration on a Sephadex G-50 column. RNA transcripts labeled with 32P were synthesized as described (17).
Isolation of human 40S ribosomal subunits and formation of their complexes with the HCV IRES transcripts
The 40S subunits of human ribosomes were isolated as described (18) and stored in liquid nitrogen. Before use, the 40S subunits were re-activated by incubation in buffer A containing 20 mM Tris–HCl (pH 7.5), 100 mM KCl, 2.5 mM MgCl2 and 0.25 mM spermidine at 37°C for 10 min. Binding of the 40S subunits (0.5 µM) with HCV IRES RNA transcripts (0.5 – 5 µM, as specified) was performed in buffer A by incubation at 25°C for 30 min (for IRESaug, IRESaugΔDII, IRESuuu and IRESΔDIV) or at 37°C for 2 h (for IRESfl and IRESflΔDII). The extent of binding of 32P-labelled RNA to 40S subunits was examined by a nitrocellulose filtration assay as described previously (9), and the amounts of 32P-labelled RNA retained on filters were quantified using a Kodak phosphorimager screen and Molecular Imager FX (Bio-Rad).
Hydroxyl radical and DMS footprinting
Hydroxyl radical cleavage of 18S rRNA in 40S ribosomal subunits was performed by addition of 2 µl of freshly made solution containing 12 mM Fe(NH4)2(SO4)2, 62 mM ascorbic acid, 25 mM EDTA-KOH (pH 7.5) and 0.6% H2O2 to 20 µl of 0.5 µM solution of 40S subunits, either complexed with a HCV IRES RNA transcript or free pre-incubated under HCV IRES-binding conditions, in buffer A. After incubation for 3 min at 25°C, the reaction was stopped by adding 20 µl of 0.1 M thiourea, and the RNA was isolated by phenol deproteination. To perform experiments on DMS probing, binding of 40S subunits with HCV IRES RNA transcript was carried out in buffer B [20 mM HEPES-KOH (pH 7.5), 100 mM KCl and 2.5 mM MgCl2]. DMS modification was performed by adding 1 µl of freshly made solution of 0.8 M DMS in ethanol to 20 µl of ribosome solution (see earlier in the text) in buffer B. After incubation for 10 min at 25°C, the reaction was stopped with 1 µl of 2-mercaptoethanol, and the RNA was isolated by phenol deproteination. For reverse transcription, 5′-32P labelled primers complementary to the human 18S rRNA regions 161–180, 331–350, 506–525, 655–674, 821–840, 911–930, 1081–1100, 1228–1247, 1348–1366, 1503–1521, 1681–1699 and 1830–1848 were used. For primer annealing, 2 pmol of 18S rRNA and 5 pmol of primer were incubated in 5 µl of water for 1 min at 90°C and cooled in ice. Reaction mixtures of 10 µl, containing 2 pmol of 18S rRNA, 5 pmol of primer, 0.25 mM dNTPs and 0.5 U of AMV reverse transcriptase (NEB) in 50 mM Tris–HCl (pH 8.3), 75 mM KOAc, 8 mM Mg(OAc)2 and 10 mM DTT, were incubated for 30 min at 37°C. For sequencing reactions, the reaction mixtures contained additionally 50 µM each of four ddNTPs. The products of the reaction were ethanol precipitated, dissolved in deionised formamide (containing 0.1% bromophenol blue and 0.1% xylene cyanol) and separated by denaturing PAGE on 8% gel. The dried gel was exposed on a Kodak phosphorimager screen and quantified using the Quantity One software (BioRad).
Structure modelling
Modelling and fitting of structures of ribosomes and their complexes were done using PyMOL software (19).
RESULTS
RNA-transcripts used in footprinting experiments and their binding to human 40S ribosomal subunits
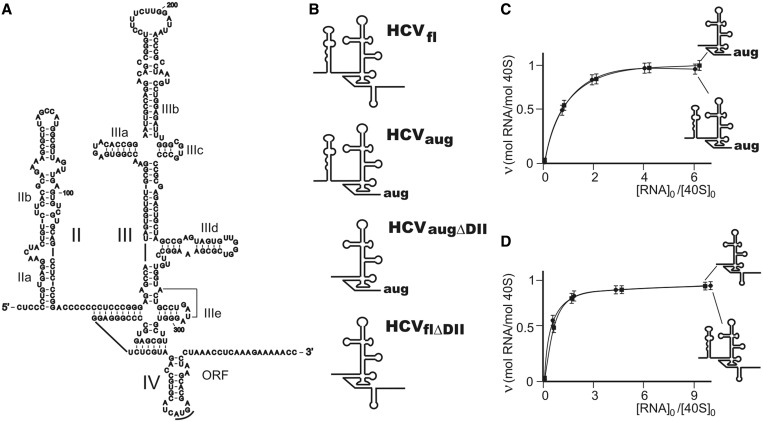
Previous studies revealed that domains II, IV and subdomain IIIb of the HCV IRES are not required for its effective binding to the 40S ribosomal subunit (20). The HCV IRES domain IV forms a hairpin containing the AUG codon in its apical part (Figure 1A). This domain should be unwound upon binding of the HCV IRES to the 40S subunit to place the AUG codon and the initial part of the viral RNA open reading frame (ORF) into the mRNA-binding channel of the 40S subunit. Therefore, in 18S rRNA footprinting experiments, we used both a full-sized HCV IRES transcript containing 40–372 nt of HCV genomic RNA (IRESfl) (16) and a truncated transcript (40–344 nt) lacking the ORF downstream of the initiator AUG codon (IRESaug) to exclude possible footprints caused by ORF binding and to study only effects of the 5′-UTR of HCV genomic RNA (Figure 1A and B). It has been shown that such truncated IRES transcript retains functional activity up to the formation of the 80S initiation complex (23). Additionally, to monitor possible effects of the HCV IRES domain II on the 18S rRNA structure, we also used forms of the IRES, in which domain II was deleted (IRESflΔDII and IRESaugΔDII) (Figure 1A and B). Both IRESaug and IRESaugΔDII transcripts were able to form binary complexes with human 40S ribosomal subunits. Short (30 min) incubation of 40S subunits with these RNA transcripts at 25°C led to saturation of the subunits by RNA; the extent of binding was approximately the same for both IRES forms (∼1 mol subunits per mol RNA) (Figure 1C). Stoichiometric binding of two other RNA transcripts, IRESfl and IRESflΔDII, to the 40S subunits required longer incubations (2 h) of the components at higher temperatures (37°C) as compared with the binding of 40S subunits to IRES derivatives without the ORF (Figure 1D), which may reflect the need for unwinding of the IRES domain IV.
Figure 1.
The HCV IRES constructs. (A) The HCV IRES secondary structure (6,21,22). Domains (large roman numbers) and subdomains (small roman numbers) are indicated, the initiator AUG codon is underlined, and the ORF is marked. (B) The cartoons representing simplified secondary structures of the HCV IRES constructs used in the study; their abbreviations are given in the text. (C) Isotherms of binding of IRESaug (circles) and IRESaugΔDII (squares) to 40S ribosomal subunits at 25°C. (D) Isotherms of binding of IRESfl (circles) and IRESflΔDII (squares) to the 40S ribosomal subunits at 37°C.
Direct hydroxyl radical probing of 18S rRNA in human ribosomal 40S subunits bound to the HCV IRES and its truncated forms
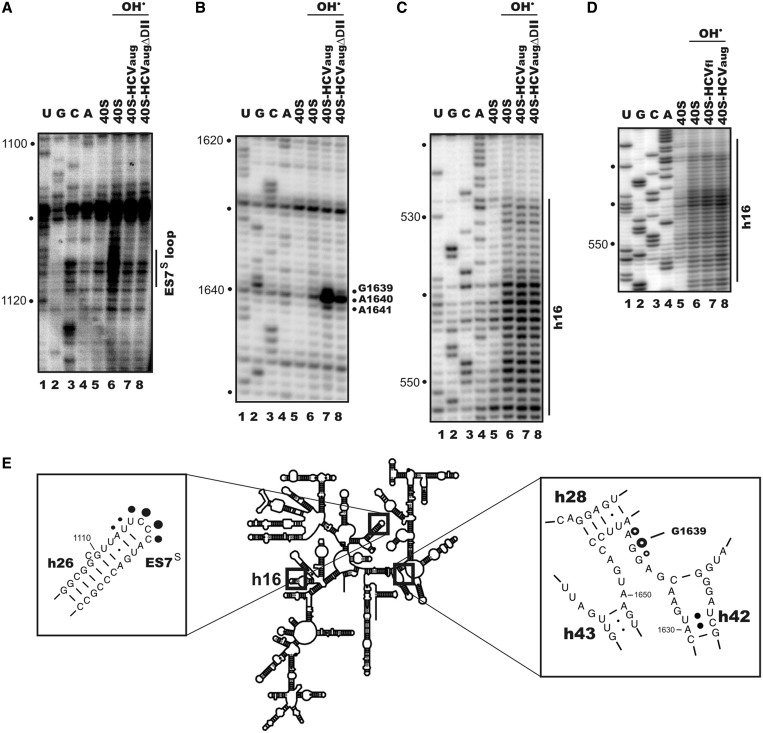
Breaks in the 18S rRNA chain caused by hydroxyl radical treatment of either free 40S subunits or their complexes with the RNA transcripts were detected in a reverse transcription reaction using 32P-labelled primers and assuming that stops of the reverse transcription (signals) occur at nucleotide bases 3′ to the nucleotides whose ribose moieties were subjected to hydroxyl radical attack. The entire 18S rRNA sequence was examined except for the extreme 3′-terminal region 1831–1869 (involved in primer annealing) and region 1221–1247 (involved in primer annealing 5′ to the hypermodified nucleotide m1aspΨ1248, which completely terminates primer extension). With IRESaug and IRESaugΔDII, we found two regions in the 18S rRNA whose accessibilities to hydroxyl radicals in the 40S subunits bound to the RNA were altered as compared with those in free subunits (Figure 2). The first region (A1113–C1118 nt corresponding to stops at positions 1114–1119) was located in the 18S rRNA expansion segment 7 (ES7S) apical loop; this displayed pronounced accessibility to hydroxyl radicals in free 40S subunits and was almost completely inaccessible in the complex with either IRESaug or IRESaugΔDII (Figure 2A, lanes 6, 7 and 8, and Figure 2E). The second region (G1638–A1640 nt corresponding to stops at positions 1639–1641) was weakly accessible to hydroxyl radicals in free 40S subunits but revealed a strong enhancement of its accessibility in the complex of 40S subunits with IRESaug (the effect was particularly prominent with G1639, whereas effects with A1640 and especially with G1638 were less pronounced) and only weak enhancement in the complex with IRESaugΔDII (Figure 2B, lanes 6, 7 and 8, and Figure 2E). Binding of IRESaug to the 40S subunits did not affect the accessibilities of nucleotides in the 18S rRNA h16 (Figure 2C, lanes 6 and 7, and Figure 2E), although changes in their accessibilities were expected based on cryo-EM data, suggesting that structural rearrangements in this 40S subunit region were induced by the HCV IRES domain II (7). As the 18S rRNA h16 is located close to the mRNA entry site, the absence of respective footprints could relate to the lack of the ORF in IRESaug. To check this, we performed a footprinting assay of 40S subunits in the complex with the IRESfl and again did not find any footprints at h16 (Figure 2D, lanes 6, 7 and 8), indicating that the 18S rRNA h16 backbone is not involved in any interactions between the HCV IRES and 40S ribosome. We note, though, that this does not rule out the possibility that the bases of nucleotides in h16 might be involved.
Figure 2.
Direct hydroxyl radical probing of 18S rRNA from human 40S ribosomal subunits in the complexes with IRESaug or IRESaugΔDII. Reverse transcription analyses of the 18S rRNA regions 1070–1130 (A), 1600–1660 (B), 495–554 (C) and 522–558 (D) with the use of respective 32P-labelled primers are presented. Sequencing lanes are indicated as A, C, G and U. Other lanes correspond to 18S rRNA isolated from free 40S subunits either untreated (40S) or treated with hydroxyl radicals (marked with OH•) and from complexes of 40S subunits with IRESaug (40S-IRESaug) or IRESaugΔDII (40S-IRESaugΔDII) treated with hydroxyl radicals. Positions of reverse transcription stops on the 18S rRNA nucleotides 3′ to the nucleotides, whose ribose moieties were subjected to hydroxyl radical attack, are indicated on the right. (E) Cartoon of the human 18S rRNA secondary structure [adapted from the structures reported in (24,25)]. Positions of IRES-dependent protections and enhancements are shown by filled and open circles, respectively.
Contact of 18S rRNA expansion segment 7 loop with the HCV IRES subdomain IIId
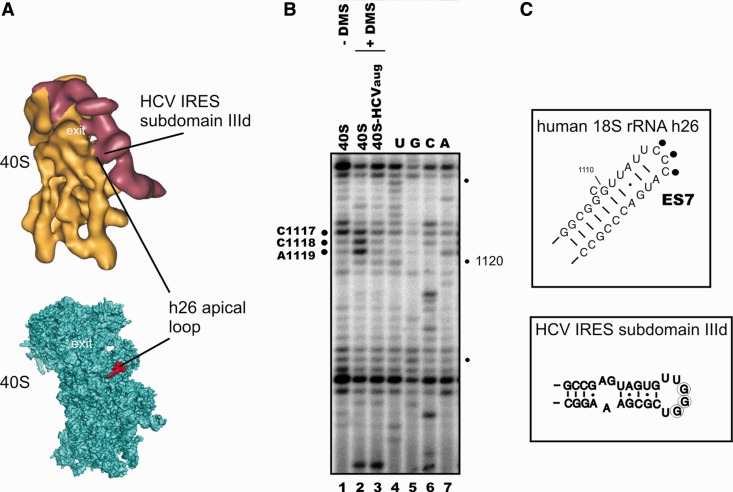
The sequence of 1116–1118 nt in 18S rRNA ES7S is CCC, whereas the HCV IRES contains the triplet GGG in subdomain IIId. To check whether these triplets contact each other upon HCV IRES binding to 40S subunits, we probed by DMS the accessibilities of the N3 positions in cytosines C1116–C1118 of 18S rRNA in the 40S subunits bound to the IRESaug (Figure 3B). The site modified was taken to be the nucleotide 5′ to the site where reverse transcriptase stopped or paused. Two cytosines (C1118 and C1117) were strongly modified in free 40S subunits (corresponding signals were observed at positions 1119 and 1118, respectively), but the extents of their modifications were negligible in 40S subunits bound to IRESaug (Figure 3B, lanes 2 and 3, and Figure 3C). The modification of C1116 (stop at position 1117) in 40S subunits bound to IRESaug was also reduced as compared with that in free 40S subunits, but to a lesser extent than those of C1117 and C1118. The stop at position 1117 was observed with free untreated 40S subunits as well (Figure 3B, lane 1), and this stop could partially mask the protective effect of the IRESaug on the C1116 accessibility to DMS (Figure 3B, lane 3). Thus, the HCV IRES binding to the 40S subunit protects both the backbone and cytidine bases in the apical loop of ES7S from attack by chemical probes, indicating the involvement of these bases in the base pairing.
Figure 3.
Interaction between the HCV IRES subdomain IIId apical loop and the loop of 18S rRNA ES7. (A) Comparison of the cryo-EM derived structure of the 40S ribosomal subunit bound with the HCV IRES (8) (on the top) and the crystal structure of the 40S ribosomal subunit (24) (on the bottom) taken in the same orientations. Positions of the 18S rRNA h26 apical loop and the IRES subdomain IIId are indicated. (B) Reverse transcription analysis of the 18S rRNA h26 from free human 40S ribosomal subunits either untreated (lanes 40S, − DMS) or treated with DMS (40S, + DMS) and from complex of 40S subunits with IRESaug treated with DMS (40S-IRESaug, + DMS). A, C, G and U are sequencing lanes. Positions of reverse transcription stops on 18S rRNA nucleotides 3′ to the nucleotides protected from modification by IRESaug are marked on the left. (C) Secondary structures of the human 18S rRNA h26 upper part (on the top) where nucleotides protected by IRESaug from DMS modification (circles) are indicated, and of the HCV IRES subdomain IIId (on bottom) with marks corresponding to the conserved GGG triplet (encircled) protected by 40S subunits from RNase digestion (20,26).
The joint effect of the HCV IRES domain II and initiator AUG codon on the accessibility of the nucleotide G1639
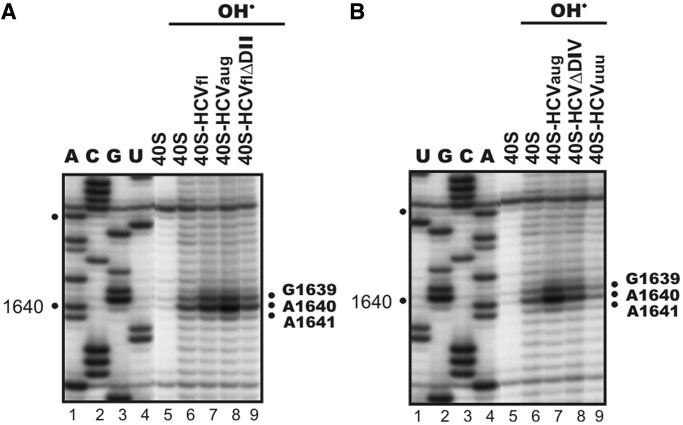
To clarify the reasons for the strong enhancement of accessibility of G1639 to hydroxyl radicals on binding of the IRESaug to the 40S subunit, we performed additional footprinting of this region using IRESfl and IRESflΔDII, as well as derivatives of IRESaug, in which the AUG codon was substituted for UUU (IRESuuu) or domain IV was completely deleted (IRESΔDIV). First, we checked whether the initial part of the ORF of the viral genomic RNA affects the domain II-dependent alteration of the G1639 accessibility to hydroxyl radicals. Comparison of the signals intensities at A1640 (corresponding to the hydroxyl radical attack at G1639) observed for complexes of 40S subunits with IRESaug (Figure 4A, lane 8) or IRESfl (Figure 4A, lane 7) with those detected for the complexes with the respective IRES forms lacking domain II, IRESaugΔDII (Figure 2B, lane 8) or IRESflΔDII (Figure 4A, lane 9), revealed that the strong signal enhancement at A1640 appeared only when HCV IRES contained domain II (compare Figure 4A, lanes 7, 8 and 9 and Figure 2B, lanes 7 and 8). In addition, the results showed that the ORF downstream of the AUG codon did not contribute to the change of accessibility of G1639 induced by HCV IRES binding (Figure 4A, lanes 7 and 8). However, deletion of the IRES domain IV (IRESΔDIV) resulted in a strong reduction of the signal at A1640 (Figure 4B, lane 8) as compared with that observed for IRESaug (Figure 4B, lanes 7). This implies that the 5′ part of domain IV, including the AUG codon, is necessary for the domain II-dependent enhancement of accessibility of G1639. We mutated the initiator AUG codon in IRESaug to UUU, an elongation codon (IRESuuu). Strikingly, this mutation did not cause any significant enhancement of the accessibility of G1639 to hydroxyl radicals in the complex with the 40S subunit as compared with that in free 40S subunits (Figure 4B, lanes 6 and 9). Thus, one can conclude that the enhancement of accessibility of 18S rRNA nucleotide G1639 occurs due to the joint action of two distant HCV IRES regions, namely, domain II and the initiator AUG codon.
Figure 4.
Direct hydroxyl radical probing of 18S rRNA region G1639 in human 40S ribosomal subunits complexed with HCV IRES constructs. (A) Reverse transcription analysis of the 18S rRNA isolated from free 40S subunits either untreated (40S) or treated with hydroxyl radicals (marked with OH•) and from complexes of 40S subunits with IRESfl (40S-IRESfl), IRESaug (40S-IRESaug) or IRESflΔDII (40S-IRESflΔDII) treated with hydroxyl radicals. (B) Reverse transcription analysis of the 18S rRNA isolated from free 40S subunits either untreated (40S) or treated with hydroxyl radicals (marked with OH•) and from complexes of 40S subunits with IRESaug (40S-IRESaug), IRESΔDIV (40S-IRESΔDIV) or IRESuuu (40S-IRESuuu) treated with hydroxyl radicals. Positions of reverse transcription stops on 18S rRNA nucleotides 3′ to the nucleotides, whose ribose moieties were subjected to hydroxyl radical attack, are indicated on the right in panels (A) and (B).
DISCUSSION
We have found that the HCV IRES bound to the 40S ribosomal subunit protects the backbone and bases of the triplet CCC in the 18S rRNA ES7S from attack by chemical probes, and nucleotide G1639 displays enhanced accessibility to hydroxyl radicals when the 40S subunits are complexed with the IRES. Our findings are the first evidence that the HCV IRES directly contacts the 18S rRNA ES7S upon binding to the 40S subunit and that together the IRES domain II and initiator AUG codon activate the 40S subunit in the region of nucleotide G1639.
Comparison of the cryo-EM density maps of the 40S subunit (7) and the 80S ribosome (8) complexed with the HCV IRES with the crystal structures of 40S ribosomal subunits from lower eukaryotes (24,27) suggests that ES7S is located in the vicinity of the HCV IRES subdomain IIId loop (Figure 3A). This loop contains a GGG triplet, which is phylogenetically conserved in hepacivirus/pestivirus IRESs (28) and is essential for HCV IRES folding and translation (29). This triplet is protected by the 40S ribosomal subunit from RNase T1 cleavage in the binary complex of the subunit with the HCV IRES (20,26). Point mutations at this triplet [GGG(266–268)CCC] (20,30) or its masking by antisense oligonucleotides (17,31) drastically reduce the IRES affinity for 40S subunits and its translational activity. Therefore, it is reasonable to assume that this triplet contacts the triplet CCC (positions 1116–1118) in the apical loop of ES7S of 18S rRNA by complementary base pairing. Protections of N3 atoms in the CCC triplet by the IRES bound to the 40S subunit (this study) and of the bases of the triplet GGG in the IRES by the 40S subunit (20,26,32), together with the data on neighbourhood of apical loops of 18S rRNA ES7S and the IRES subdomain IIId from cryo-EM studies of the binary complexes (7,8), strongly support the existence of base-specific contacts between these loops. Taking into account the conserved nature of the triplet CCC of 18S rRNA ES7S in vertebrates (25) and the aforementioned conserved triplet GGG of the subdomain IIId apical loop in hepacivirus/pestivirus IRESs, as well as in IRESs of several picornaviruses (28), we suggest that this interaction is universal for the binding of genomic RNAs from many viruses bearing HCV-like IRESs to 40S subunits. Earlier attempts to determine the contacts of the HCV IRES with 18S rRNA in 40S subunits using cross-linking assays with IRES derivatives bearing either thiouridine residues randomly distributed over the RNA-transcript (12) or aryl azide groups introduced at the specific sites in subdomain IIId (9) have failed. However, the absence of the cross-link does not mean lack of the contact: thiouridine residues neighbouring the triplet GGG might have conformations unfavourable for cross-linking to RNA, and the aryl azide groups attached to the subdomain IIId nucleotides through a spacer might be directed away from the 18S rRNA. The reasons why earlier experiments on DMS probing of the 18S rRNA in the 40S•HCV IRES complex have not revealed the protection of C-residues in ES7S (12,15) are not clear.
Nucleotide G1639 in human 18S rRNA is universally conserved (25) and corresponds to G1338 in 16S rRNA of Escherichia coli, thereby suggesting its functional importance. G1338 and its neighbour A1339 have been shown to participate in IF3-dependent discrimination of transfer RNA (tRNA) at the ribosomal P site through their interactions with the characteristic G-C pairs in the anticodon stem of the initiator tRNA (33). X-ray studies of functional complexes of bacterial ribosomes suggest that G1338 and A1339 act as a gate that, when closed, prevents translocation of tRNA from the P- to E-site and opens when the subunit head turns, i.e. that G1338 and A1339 act as a switch during both translation initiation and the translocation (34). Experimental data that could prove the movement of these nucleotides in bacterial ribosomes have not been, to our knowledge, reported. However, a turn of the region corresponding to G1639 region in human 18S rRNA was recently observed in vacant 80S ribosomes from yeast by means of X-ray analysis (35), and this state of the ribosome was attributed to the ratcheted state. The data obtained in the present study allow us to conclude that simultaneous action of two structural elements of the HCV IRES, domain II and the AUG initiation codon, causes substantial rearrangement of 18S rRNA backbone in the region of nucleotide G1639, which is manifested as enhancement of accessibility of this region to hydroxyl radicals. This conformational transition is apparently required for positioning of initiator tRNA on the HCV IRES AUG codon at the P-site during IRES-mediated translation initiation in the absence of the initiation factors participating in the AUG selection and recognition.
Remarkably, hydroxyl radical-generating probes tethered to amino acids in several positions of the N- and C-terminal tails of initiation factor eIF1A produced the strongest cleavages of the 18S rRNA in rabbit 40S subunits in the area overlapping the G1639 region (36). Thus, eIF1A binds to the region of 40S subunit where the IRES-induced conformational change of the 18S rRNA backbone occurs. In addition, both the N- and C-terminal tails eIF1A strongly stimulate binding of the ternary complex eIF2• •GTP to the 40S subunits (36). This is consistent with the data (14) that of two factors, eIF1A and eIF1, only eIF1A stabilizes interaction of the ternary complex with the 40S subunit, although both factors together stabilize a conformational change that opens the mRNA-binding channel latch. Based on these considerations, we propose that the rearrangement of 18S rRNA in the G1639 region induced by joint action of the HCV IRES domain II and AUG codon functionally replaces eIF1A and eIF1 in the mechanism of the HCV IRES-mediated initiation of translation. This conformational change might relate to the rotation of the 40S subunit head (7).
•GTP to the 40S subunits (36). This is consistent with the data (14) that of two factors, eIF1A and eIF1, only eIF1A stabilizes interaction of the ternary complex with the 40S subunit, although both factors together stabilize a conformational change that opens the mRNA-binding channel latch. Based on these considerations, we propose that the rearrangement of 18S rRNA in the G1639 region induced by joint action of the HCV IRES domain II and AUG codon functionally replaces eIF1A and eIF1 in the mechanism of the HCV IRES-mediated initiation of translation. This conformational change might relate to the rotation of the 40S subunit head (7).
There are no data pointing to contacts between domains II and IV of the HCV IRES at the 40S subunit decoding groove. However, recent studies have shown that these two distal IRES regions are structurally communicated (37,38); consequently, the joint action of the HCV IRES domain II and AUG codon on the 18S rRNA structure could occur due to an interplay between these two IRES regions. Possibly, this interplay is realized by means of rpS5, as this protein is located in close proximity to domain II (8,23) and can be cross-linked to a thiouridine residue or aryl azide-containing nucleotide incorporated into an mRNA codon bound at the ribosomal E site (39,40). Notably, rpS5 can be directly cross-linked to the HCV IRES by ultraviolet light, and the cross-linking depends on the integrity of the IRES’s domain II and the initial part of the ORF (5,10), which may indicate that rpS5 is involved in the interplay of domain II and the AUG codon. This interplay apparently induces a rearrangement of the 18S rRNA backbone at G1639 region such that nucleotide G1639 appears in a conformation that is favourable for binding to the anticodon stem of initiator tRNA at the P site at the subsequent step of translation initiation.
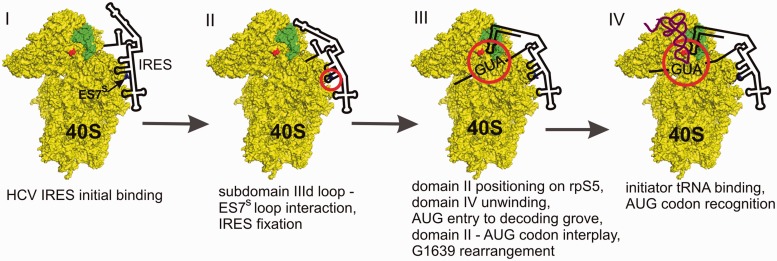
The data obtained in this study provide new insights into the molecular basis of the HCV-like IRES-driven initiation of translation and allow us to hypothesize a model for 48S complex formation (Figure 5). In agreement with our previous data (9,11), the initial binding of the HCV IRES to the 40S subunit occurs through the interaction of the stably structured domain III with ribosomal proteins near the mRNA exit site (Figure 5, state I). The binding is stabilized by the coordinated complementary interactions between HCV IRES subdomain IIId apical loop and the loop of 18S rRNA ES7S (Figure 5, state II). This stabilization causes IRES domain II to be located on the head of the 40S subunit (7) and to come in contact with rpS5 (21), which in turn allows the IRES domain IV to be unwound to place the ORF part into the mRNA-binding channel. The further steps might occur according to the following scenario. As soon as the AUG codon is placed into the decoding grove, an interaction occurs between the start codon and the apical loop of the domain II promoted by rpS5. This induces a conformational transition of the 18S rRNA in the region of nucleotide G1639, which enables initiator tRNA [either as the ternary complex with eIF2 and GTP or alone under stressed conditions (41,42)] to be accepted at the ribosomal P site (Figure 5, state III) for subsequent recognition of the IRES AUG codon and fixation of the IRES ORF at the mRNA-binding channel (Figure 5, state IV).
Figure 5.
A hypothetical model of the process of HCV IRES adaptation to the 40S subunit up to the 48S complex formation. Positions of rpS5 (green), ES7 (indicated by arrow) and G1639 (red) are designated on the simplified view of the eukaryotic 40S ribosomal subunit, in accordance with (27). States of the process are indicated by roman numbers and described in the text. The regions where key events occur are encircled with red.
The results of our study shed light on the intriguing problem of how the HCV subverts the cellular translation machinery, making it capable of synthesizing the viral polyprotein. We have shown for the first time that 18S rRNA makes direct contacts with the HCV IRES, and we have identified the 18S rRNA site involved, which is open in the 40S subunits owing to an IRES-induced conformational change. We have also identified structural elements of the IRES involved in this rearrangement. Thus, we have revealed those18S rRNA nucleotides that are recruited by the HCV IRES to provide the initial stages of translation initiation on the viral RNA without the assistance of initiation factors eIF1A and eIF1. The proposed mechanism could be extended to other HCV-like IRESs from various pestivirus and picornavirus genera, which have Type 3 IRESs capable of direct binding to the 40S subunit. Our findings may promote a deeper understanding of the molecular basis of cap-independent initiation of translation.
ACKNOWLEDGEMENTS
The authors gratefully thank Ian Eperon for critical reading of this manuscript.
FUNDING
A program of the Presidium of the Russian Academy of Sciences ‘Molecular and cellular biology’ (to G.G.K.) and a grant from the Russian Foundation for Basic Research [12-04-31138 to O.A.K.] Funding for open access charge: Presidium of the Russian Academy of Sciences ‘Molecular and cellular biology’ (to G.G.K.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Merrick WC. Cap-dependent and cap-independent translation in eukaryotic systems. Gene. 2004;332:1–11. doi: 10.1016/j.gene.2004.02.051. [DOI] [PubMed] [Google Scholar]
- 2.Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 3.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell. Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hellen CU. IRES-induced conformational changes in the ribosome and the mechanism of translation initiation by internal ribosomal entry. Biochim. Biophys. Acta. 2009;1789:558–570. doi: 10.1016/j.bbagrm.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Honda M, Beard MR, Ping LH, Lemon SM. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 1999;73:1165–1174. doi: 10.1128/jvi.73.2.1165-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 8.Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure. 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 9.Babaylova E, Graifer D, Malygin A, Stahl J, Shatsky I, Karpova G. Positioning of subdomain IIId and apical loop of domain II of the hepatitis C IRES on the human 40S ribosome. Nucleic Acids Res. 2009;37:1141–1151. doi: 10.1093/nar/gkn1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukushi S, Okada M, Stahl J, Kageyama T, Hoshino FB, Katayama K. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J. Biol. Chem. 2001;276:20824–20826. doi: 10.1074/jbc.C100206200. [DOI] [PubMed] [Google Scholar]
- 11.Laletina E, Graifer D, Malygin A, Ivanov A, Shatsky I, Karpova G. Proteins surrounding hairpin IIIe of the hepatitis C virus internal ribosome entry site on the human 40S ribosomal subunit. Nucleic Acids Res. 2006;34:2027–2036. doi: 10.1093/nar/gkl155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otto GA, Lukavsky PJ, Lancaster AM, Sarnow P, Puglisi JD. Ribosomal proteins mediate the hepatitis C virus IRES-HeLa 40S interaction. RNA. 2002;8:913–923. doi: 10.1017/s1355838202022057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malygin AA, Shatsky IN, Karpova GG. Proteins of the human 40S ribosomal subunit involved in hepatitis C IRES binding as revealed from fluorescent labeling. Biochemistry (Mosk.) 2013;78:53–59. doi: 10.1134/S0006297913010069. [DOI] [PubMed] [Google Scholar]
- 14.Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 15.Shenvi CL, Dong KC, Friedman EM, Hanson JA, Cate JH. Accessibility of 18S rRNA in human 40S subunits and 80S ribosomes at physiological magnesium ion concentrations—implications for the study of ribosome dynamics. RNA. 2005;11:1898–1908. doi: 10.1261/rna.2192805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynolds JE, Kaminski A, Kettinen HJ, Grace K, Clarke BE, Carroll AR, Rowlands DJ, Jackson RJ. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malygin AA, Graifer DM, Laletina ES, Shatskii IN, Karpova GG. Approach to identifying the functionally important segments of RNA, based on complementation-addressed modification. Mol. Biol. (Mosk.) 2003;37:1027–1034. [PubMed] [Google Scholar]
- 18.Matasova NB, Myltseva SV, Zenkova MA, Graifer DM, Vladimirov SN, Karpova GG. Isolation of ribosomal subunits containing intact rRNA from human placenta: estimation of functional activity of 80S ribosomes. Anal. Biochem. 1991;198:219–223. doi: 10.1016/0003-2697(91)90416-q. [DOI] [PubMed] [Google Scholar]
- 19.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 20.Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berry KE, Waghray S, Mortimer SA, Bai Y, Doudna JA. Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning. Structure. 2011;19:1456–1466. doi: 10.1016/j.str.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao WD, Wimmer E. Genetic analysis of a poliovirus/hepatitis C virus chimera: new structure for domain II of the internal ribosomal entry site of hepatitis C virus. J. Virol. 2001;75:3719–3730. doi: 10.1128/JVI.75.8.3719-3730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filbin ME, Vollmar BS, Shi D, Gonen T, Kieft JS. HCV IRES manipulates the ribosome to promote the switch from translation initiation to elongation. Nat. Struct. Mol. Biol. 2013;20:150–158. doi: 10.1038/nsmb.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 25.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolupaeva VG, Pestova TV, Hellen CU. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J. Virol. 2000;74:6242–6250. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 28.Hellen CU, de Breyne S. A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: evidence for modular exchange of functional noncoding RNA elements by recombination. J. Virol. 2007;81:5850–5863. doi: 10.1128/JVI.02403-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jubin R, Vantuno NE, Kieft JS, Murray MG, Doudna JA, Lau JY, Baroudy BM. Hepatitis C virus internal ribosome entry site (IRES) stem loop IIId contains a phylogenetically conserved GGG triplet essential for translation and IRES folding. J. Virol. 2000;74:10430–10437. doi: 10.1128/jvi.74.22.10430-10437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji H, Fraser CS, Yu Y, Leary J, Doudna JA. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc. Natl Acad. Sci. USA. 2004;101:16990–16995. doi: 10.1073/pnas.0407402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tallet-Lopez B, Aldaz-Carroll L, Chabas S, Dausse E, Staedel C, Toulme JJ. Antisense oligonucleotides targeted to the domain IIId of the hepatitis C virus IRES compete with 40S ribosomal subunit binding and prevent in vitro translation. Nucleic Acids Res. 2003;31:734–742. doi: 10.1093/nar/gkg139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 2000;7:1105–1110. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- 33.Lancaster L, Noller HF. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol. Cell. 2005;20:623–632. doi: 10.1016/j.molcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 34.Selmer M, Dunham CM, Murphy FVT, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 35.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 36.Yu Y, Marintchev A, Kolupaeva VG, Unbehaun A, Veryasova T, Lai SC, Hong P, Wagner G, Hellen CU, Pestova TV. Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Res. 2009;37:5167–5182. doi: 10.1093/nar/gkp519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Filbin ME, Kieft JS. HCV IRES domain IIb affects the configuration of coding RNA in the 40S subunit's decoding groove. RNA. 2011;17:1258–1273. doi: 10.1261/rna.2594011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fraser CS, Hershey JW, Doudna JA. The pathway of hepatitis C virus mRNA recruitment to the human ribosome. Nat. Struct. Mol. Biol. 2009;16:397–404. doi: 10.1038/nsmb.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Demeshkina NA, Laletina ES, Meshchaninova MI, Repkova MN, Ven'iaminova AG, Graifer DM, Karpova GG. The mRNA codon environment at the P and E sites of human ribosomes deduced from photo crosslinking with pUUUGUU. Mol. Biol. (Mosk.) 2003;37:147–155. [PubMed] [Google Scholar]
- 40.Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CU, Pestova TV. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008;27:1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim JH, Park SM, Park JH, Keum SJ, Jang SK. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J. 2011;30:2454–2464. doi: 10.1038/emboj.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat. Struct. Mol. Biol. 2008;15:836–841. doi: 10.1038/nsmb.1445. [DOI] [PubMed] [Google Scholar]