Abstract
Although the DNA methyltransferase 2 family is highly conserved during evolution and recent reports suggested a dual specificity with stronger activity on transfer RNA (tRNA) than DNA substrates, the biological function is still obscure. We show that the Dictyostelium discoideum Dnmt2-homologue DnmA is an active tRNA methyltransferase that modifies C38 in tRNAAsp(GUC) in vitro and in vivo. By an ultraviolet-crosslinking and immunoprecipitation approach, we identified further DnmA targets. This revealed specific tRNA fragments bound by the enzyme and identified tRNAGlu(CUC/UUC) and tRNAGly(GCC) as new but weaker substrates for both human Dnmt2 and DnmA in vitro but apparently not in vivo. Dnmt2 enzymes form transient covalent complexes with their substrates. The dynamics of complex formation and complex resolution reflect methylation efficiency in vitro. Quantitative PCR analyses revealed alterations in dnmA expression during development, cell cycle and in response to temperature stress. However, dnmA expression only partially correlated with tRNA methylation in vivo. Strikingly, dnmA expression in the laboratory strain AX2 was significantly lower than in the NC4 parent strain. As expression levels and binding of DnmA to a target in vivo are apparently not necessarily accompanied by methylation, we propose an additional biological function of DnmA apart from methylation.
INTRODUCTION
Dnmt2 is a member of the eukaryotic DNA methyltransferase family. A few model organisms, especially Drosophila melanogaster, Schizosaccharomyces pombe, Entamoeba histolytica and Dictyostelium discoideum contain only one Dnmt2 homologue but lack the more active homologues Dnmt1 and Dnmt3. Though highly conserved during evolution, loss of Dnmt2 homologues has no obvious phenotypic effects in Mus musculus (1), D. melanogaster (2), S. pombe (3) and D. discoideum (4,5). In Danio rerio, disruption of dnmt2 has been reported to cause pleiotropic effects (6) and in E. histolytica a gene disruption could not be obtained, suggesting that Dnmt2 was required for viability (7). On closer inspection, more subtle long-term effects of Dnmt2 loss were observed: in D. melanogaster, for example, H4K20me3 was strongly reduced, and telomeric sequences were lost (2). Durdevic et al. (8) recently showed that in D. melanogaster, Dnmt2 was also involved in virus control. At least in D. melanogaster, E. histolytica and D. discoideum, low amounts of DNA methylation, especially on retroelements, were detected and attributed to Dnmt2, although these data are controversially discussed (9–11).
In 2006, Bestor’s group reported that Dnmt2 rather methylates transfer RNA (tRNA) than DNA in M. musculus, D. melanogaster and Arabidopsis thaliana. Specifically, they found C38 in tRNAAsp as the major or only substrate for the enzyme (1). A dual role of Dnmt2 had been discussed (12), and Jurkowski et al. (13) could show that tRNAAsp was methylated by an enzymatic mechanism characteristic for DNA methyltransferases rather than by the reaction pathways of enzymes that methylate RNAs.
tRNAAsp methylation activity was also reported for the Dnmt2-homologues from E. histolytica (14) and D. melanogaster (15). Schaefer et al. (15) showed that D. melanogaster tRNAVal(CAC) and tRNAGly(GCC) were also methylated at position C38 in vivo and by human Dnmt2 (hDnmt2) in vitro. They further found that dnmt2 knockout flies were more sensitive to oxidative stress. A more detailed analysis of tRNAAsp(GUC) and tRNAGly(GCC) suggested that methylation protected tRNAs from stress-induced cleavage (15). Double-knockout mutant mice of Dnmt2 and Nsun2, the second known m5C-tRNA-methyltransferase in higher eukaryotes, showed a phenotype with impaired cellular differentiation, an overall reduction in protein synthesis and early lethality (16). Recently, we identified tRNAGlu(UUC) as an additional novel substrate of Pmt1, the Dnmt2-homologue in S. pombe (17). Pmt1-dependent tRNA methylation seemed to be regulated by nutrient conditions. Nutritional control was also reported for Ehmeth, the Dnmt2 homologue from E. histolytica that is inhibited by the glycolytic enzyme enolase (14,18).
Here, we demonstrate that recombinant D. discoideum DnmA and hDnmt2 can methylate tRNAAsp(GUC), tRNAGlu(UUC) and tRNAGly(GCC) from D. discoideum in vitro with different efficiencies. Both enzymes formed covalent complexes [22] with specific tRNAs with similar kinetics, but they were significantly slower for the minor substrate tRNAGlu than for the major substrate tRNAAsp. Ultraviolet (UV)-crosslinking and immunoprecipitation (CLIP) experiments showed that specific fragments of the three target tRNAs associated with DnmA in vivo. However, in vivo, only methylation of tRNAAsp(GUC) by DnmA was confirmed, whereas methylation of the target nucleotide C38 in other substrates was not detectable by bisulfite sequencing.
As revealed by quantitative reverse transcription (RT)-PCR, dnmA is differentially expressed in development, cell cycle and in the recovery phase after temperature stress. The increase in dnmA expression levels correlated with elevated tRNAAsp methylation in development but not after temperature stress. The other targets identified by in vitro methylation and by CLIP were apparently also not methylated in vivo in development.
Our data document that additional RNA molecules can serve as substrates for Dnmt2 binding and that the full range of targets is probably not yet recognized. The results also suggest that binding of the methyltransferase to an RNA molecule not necessarily results in methylation but may have different biological functions.
MATERIALS AND METHODS
Dictyostelium discoideum cultures and nomenclature
Dictyostelium discoideum AX2-214 was grown in HL5+ medium (ForMedium) containing 50 µg/ml Ampicillin, 0.25 µg/ml Amphotericin-B, 100 µg/ml Penicillin/Streptomycin (PAA) at 22°C, constant light under selective conditions as required. NC4 cells were grown in a suspension of Klebsiella aerogenes in phosphate buffer. When indicated, AX2 cells were also grown in bacterial suspension to allow for comparison with NC4.
For cold treatment cells (1 × 106/ml) were shaken at 4°C for 2–14 h. Cells were allowed to recover at 22°C for 2½ h before RNA isolation. For synchronization, cells at a density of 1–3 × 105/ml were incubated at 4°C over night. Before cold treatment, cells were briefly cooled down in a water bath with ice. For synchronization, cells were then warmed up in a 25°C water bath before cultivating at 22°C. Synchronization was measured by counting cells every 30 min during a period of at least 7 h. Samples for RNA isolation were taken at the times indicated.
Gene names and accession numbers refer to DictyBase (http://dictybase.org/, Jan. 3rd, 2013). Complete sequences for tRNAs relevant for the experiments are given in Supplementary Table S2. In silico folded clover leaf structures for tRNAAsp(GUC), tRNAGlu(UUC) and tRNAGly(GCC) are given in Supplementary Figure S1.
Nucleotides in tRNAs have been numbered according to Motorin et al. (19).
Expression and purification of recombinant DnmA and human hDnmt2
DnmA and hDnmt2 were both cloned with N-terminal His6-tags in pET28a(+) (Novagen) into the NdeI and XhoI sites. Recombinant proteins were expressed in Escherichia coli (DE3) Rosetta pLysS cells. Induction and protein purification were done as described previously (13). Briefly, the cells were induced at an OD600 of 0.6 with 1 mM IPTG and incubated for 3 h at 37°C for hDnmt2 and at 22°C for DnmA. After harvesting, cells were resuspended in sonication buffer [30 mM KPi (pH 7.0), 300 mM KCl, 10% (v/v) glycerol, 10 mM imidazole, 0.1 mM Dithiotreitol (DTT)] containing protease inhibitors (complete mini, Roche) and lysed by sonication with a Dr Hielscher up200s sonifier (5–10 times, cycle 0.5/amplitude 50%). The cell lysate was cleared by centrifugation (10.000 RCF), and the supernatant was incubated on NiNTA-Sepharose for 15 min at 4°C. After washing with sonication buffer, His6-tagged proteins were eluted with a buffer containing 30 mM KPi (pH 7.0), 300 mM KCl, 10% glycerol, 200 mM imidazole and 0.1 mM DTT. The eluate was dialysed in two steps [30 mM KPi (pH 7.0), 200 mM KCl, 20% glycerol, 1 mM DTT, 0.1 mM EDTA and 30 mM KPi (pH 7.0), 100 mM KCl, 50% glycerol, 1 mM DTT, 0.1 mM EDTA].
UV-crosslinking and RNA immunoprecipitation
For UV CLIP, 2 × 108 D. discoideum cells expressing DnmA-GFP in the dnmAKO-background and cells expressing GFP only were harvested (390 RCF, 5 min), washed once and resuspended in 20 ml of phosphate buffer. Ten microlitre aliquots of the suspension were spread in 10 cm petri dishes and UV irradiated (254 nm) with 250 mJ/cm2. Cells were spun down and collected in 4 ml of modified RIPA-Buffer [50 mM Tris/HCl (pH 7.5), 150 mM NaCl, 0.5% NP40, 0.5% sodium deoxycholate, 1 mM EDTA] (20). Lysis was supported by three times sonication (cycle 0.5 and amplitude 50%) with a Dr Hielscher up200s sonifier. Samples were centrifuged (20 000 RCF, 15 min, 4°C), and the supernatant was transferred to a fresh tube; it was again centrifuged, and the supernatant collected for a third centrifugation step. Hereafter, the supernatant was pre-incubated with 250 µl Sephadex™ G-50 Medium (GE Healthcare) and then transferred to 50 µl GFP-trap beads (ChromoTek, Martinsried) (21). Incubation was carried out for 15 min at 4°C on a rotating wheel, followed by two washing steps with 1.5 ml stringency RIPA buffer A [50 mM Tris/HCl (pH 7.5), 1 M NaCl, 1% NP40, 1% sodium deoxycholate, 1 mM EDTA, 0.1% SDS, 2 M Urea] and two washing steps with stringency RIPA buffer B [50 mM Tris/HCl (pH 7.5), 1 M NaCl, 1% NP40, 1% sodium deoxycholate, 1 mM EDTA, 0.1% SDS, 1 M Urea].
For further analysis, aliquots were taken from input (supernatant after preincubation with Sephadex G50) and the last wash. Beads were equilibrated [50 mM Tris/HCl (pH 7.5), 300 mM NaCl, 0.5% NP40, 0.5% sodium deoxycholate, 1 mM EDTA, 0.1% SDS] and resuspended in RIPA elution buffer [50 mM Tris/HCl (pH 7.5), 300 mM NaCl, 0.5% NP40, 0.5% sodium deoxycholate, 5 mM EDTA, 0.5% SDS]. To each sample, proteinase K was added at a final concentration of 1 mg/ml and incubated for 1 h at 42°C and 5 h at 56°C. Samples were extracted with phenol/chloroform/isoamyl alcohol solution (Roth), and RNA was precipitated with isopropanol and 20 µg of glycogen.
Deep sequencing of co-immunoprecipitated RNAs
cDNA synthesis and Illumnia sequencing was carried out according to previously published protocols (22). Specifically, the two RNA samples from the CLIP experiment were poly(A)-tailed using poly(A) polymerase. After that, the 5′PPP was converted into 5′P using tobacco acid pyrophosphatase. Then, an RNA adapter was ligated to the 5′-phosphate of the RNA. First-strand cDNA synthesis was performed using an oligo(dT)-adapter primer and the M-MLV reverse transcriptase. The resulting cDNAs were PCR-amplified to ∼20–30 ng/μl using a high-fidelity DNA polymerase. Adaptors that were used for ligation are listed in Supplementary Table S1.
The cDNA libraries were sequenced using an Illumina GAIIx machine with 100 cycles resulting in 5 674 433 (DnmA-GFP pull down) and 5 540 257 (GFP control) reads. After removal of the barcodes and adaptor sequences, the reads were aligned to the genome of D. discoideum (sequence accession numbers: NC_000895, NC_001889, NC_007087, NC_007088, NC_007089, NC_007090, NC_007091, NC_007092) using the short read mapper segemehl (23). For unmapped reads, polyA-tails were computationally searched in the 3′-end and clipped if found. The resulting sequences were mapped again to the reference sequences. In all, 1 249 579 reads of the DnmA-GFP pulldown and 514 809 reads of the control sample could be successfully aligned. Enrichment of RNAs was calculated by comparison of normalized read numbers from the DnmA-GFP pull down and the control. In case of multiple genes resulting in the same transcript, the reads are mapped to the first copy in the genome starting from chromosome 1.
Cloning and in vitro transcription of RNAs
The genes for tRNAAsp(GUC-1) (DDB_G0294707), tRNAPhe(GAA-2) (DDB_G0294663), tRNALeu(UAG-1) (DDB_0234827), tRNALeu(AAG-8) (DDB_0235066), tRNAVal(CAC-1) (DDB_0234789) and tRNAGly(GCC-1) (DDB_0234894) were cloned into pJET1/blunt vector (Fermentas) according to the suppliers suggestions. In the case of multiple identical genes for one tRNA, only one gene is listed here. The annotation of tRNAs, sequences, potential isoacceptor tRNAs and in silico folding are shown in Supplementary Table S2 and Supplementary Figure S1. Genes were produced by recursive PCR using overlapping oligos (Supplementary Table S1). For tRNAAsp(GUC), tRNALeu(UAG), tRNALeu(AAG), tRNAVal(CAC) and tRNAGly(GCC), the respective oligos 2 and 3 served as template and oligo 1 and 4 as primers. A restriction site was added at the 3′-end and a T7-promotor at the 5′-end. For tRNAPhe(GAA), oligo 1, 2 and 3 are used as template. Amplification was done with oligo 4 and 5. The tRNAGlu(UUC) (DDB_G0294709) and suppressor tRNAGlu(CUA) (kindly provided by T. Winckler, Jena) were recloned into pJET1/blunt vector from pGEM T vector. The mutated versions tRNAAsp(GUC)C38A and tRNAGlu(UUC)C38A were constructed by using the respective oligonucleotides (Supplementary Table S1).
For in vitro transcription, 500 ng of linearized plasmid was incubated at 37°C for 2 h in 50 µl with 1 µl T7-RNA polymerase, 1 mM NTPs, 1× transcription buffer (Fermentas) and RiboLock™ RNase Inhibitor (Fermentas). After transcription, samples were treated with DNaseI for 15 min, extracted with an equal volume of phenol/chloroform/isoamyl alcohol (Roth) and purified over a Sephadex G-50 spin column.
For radio-labelled in vitro transcription, 2 µg of linearized plasmid were incubated at 37°C for 2 h in 100 µl volume with 2 µl T7-RNA polymerase, 2 mM ATP, GTP and CTP, 1 mM UTP and 2 µl [α32P] UTP (110 TBq/mmol, Hartmann), 1× transcription buffer and RiboLock™ RNase Inhibitor (Fermentas). After transcription, samples were treated as aforementioned.
Preparation of small RNA fraction
RNA isolation was done using the Trizol®-method (24). For enrichment of the small RNA fraction, 0.5 M NaCl/5% PEG 8000 was added to the RNA solution (in DEPC treated H2O) and incubated at −20°C for 30 min. After centrifugation (10.000 RCF, 4°C, 30 min), the supernatant was collected in a fresh tube and precipitated with three volumes of 100% EtOH.
SDS-EMSA
Detection of denaturant-resistant DnmA-tRNA was carried out by SDS-electrophoretic mobility shift assay as described elsewhere (25) with some modifications. Briefly, DnmA (1–9 µg) was incubated in 20 µl of 10 mM KPi (pH 7.0), 22.5 % glycerol, 25 mM KCl, 10 mM DTT, 2 mM MgCl2 and 100–500 µM S-adenosyl-methionine (SAM) with the respective 32P labelled tRNA (100–665 ng) at 22°C. Reactions were terminated at the indicated times by addition of 10 µl of SDS-stop mixture [150 mM Tris–HCl (pH 8.0), 6% SDS (w/v), 30 mM EDTA, 0.025% Bromophenol blue] and heated for 10 min at 65°C. Samples were cooled down to room temperature and subjected to electrophoresis on 6 or 12% SDS–polyacrylamide gels, sealed in Saran Wrap and exposed to an imager plate for 12–40 h at RT. Plates were read in a phosphoimager (Fujifilm FLA-7000). In some cases, samples were transferred from the gel to a nitrocellulose membrane for immunodetection of DnmA using an anti-His antibody and a secondary mouse antibody coupled to alkaline phosphatase.
In vitro methylation assay
Methylation of RNA substrates was carried out as described previously (13): 3 µM of recombinant hDnmt2 and 3–12 µM of DnmA were incubated with 500 ng of tRNA-transcript (or 10 µg of small enriched RNA), 1× methylation buffer [50 mM Tris/HCl (pH 7.0), 50 mM NaCl, 5 mM MgCl2, 1 mM DTT] and 1.7 µM [3H]-SAM (Hartmann), 1 u/µl RiboLock™ RNase Inhibitor (Fermentas) at 22°C for 1 min to 1.5 h as indicated in the text. The reaction was stopped by adding 1 mM non-radioactive SAM and 2 mg/ml proteinase K for 30 min. Loading buffer [formamide, 5 mM EDTA (pH 7.4), bromophenol blue] was added, and samples were run on a 12% denaturing (7 M Urea) polyacrylamide gel (1 mm) for 2–5 h at 400 V. The gel was stained with ethidiumbromide, fixed (10% methanol, 10% acetic acid) and immersed for 1 h in amplify solution (Amersham), dried and exposed to X-ray film (Contatyp CX-BL+ Medical X-ray film) for 16 h up to 2 weeks at −80°C.
In vitro blocked methylation assay
The in vitro blocked methylation assay is based on the assumption that RNA methylation is inhibited if the target site is covered by hybridization with a complementary DNA oligo. RNA (500 ng of transcript or 10 µg of small enriched RNA) was mixed with 2 µM of the respective oligonucleotide in 1 × STE-buffer [10 mM Tris/HCl (pH 8), 50 mM NaCl, 1 mM EDTA], heated up to 80°C in a thermocycler and then cooled down stepwise to room temperature. The blocked RNA was subsequently used in a 3H-methylation assay as described earlier in the text. Blocking oligos are shown in Supplementary Table S1 (oligo 2 for tRNAAsp and tRNAGlu, respectively).
dnmA expression analysis by real-time quantitative PCR
For analysing dnmA expression levels, RNA was extracted by the Trizol®-method (24). qPCR was performed in an Eppendorf realplex light cycler with the SensiFAST SYBR No-ROX One-Step Kit (Bioline) according to the suppliers instructions. Relative quantification was done with the ΔΔCt-method using gapdh (DDB_G0275153) as a normalization control (housekeeping gene).
Microscopy
For fluorescence analysis, cells were fixed in 4% (w/v) paraformaldehyde dissolved in 20 mM phosphate buffer (pH 6.7) for 10 min at 22°C and stained with DAPI.
Images were acquired on a Leica DMIRB inverted microscope equipped with a DC350 camera and IM50 Acquisition software (Leica Microsystems; Wetzlar, Germany).
For the movie, DnmA-GFP cells (in DnmAKO background) were analysed with a Zeiss Cell Observer SD with Rapp UGA-40-2 L Scanner. Images of living cells were taken in 15 s intervals at 7 layers with a 0.8 µM distance between each layer during a period of 120 min. Further image analysis was done with ImageJ 1.46. The movie is displayed with eight frames per second.
RNA bisulfite sequencing
RNA bisulfite sequencing was carried out as described elsewhere (26). Primer sequences are listed in the Supplementary Material (Supplementary Table S1).
RESULTS
DnmA is an active tRNAAsp methyltransferase in vitro and in vivo
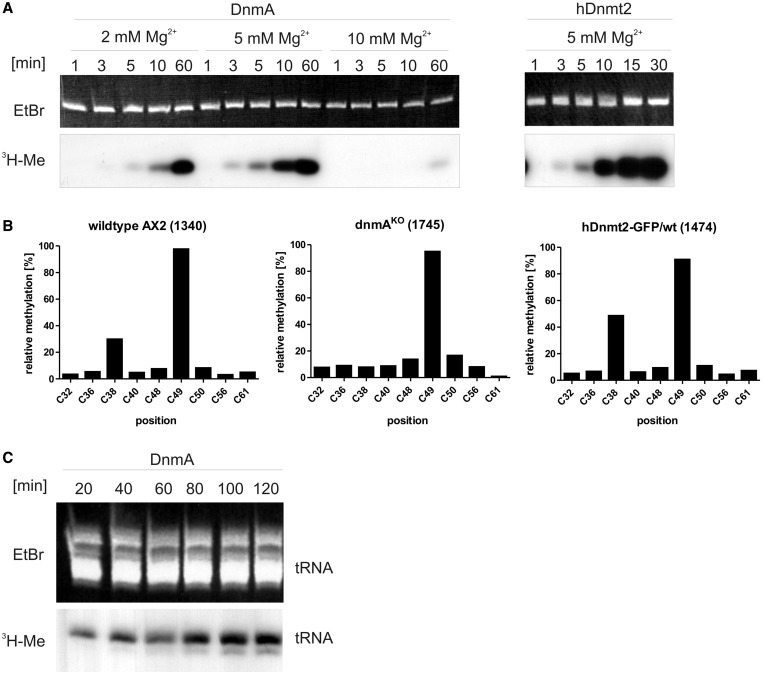
Previous experiments have shown that hDnmt2 can methylate a D. discoideum RNA in the size range of tRNAs in vitro (13). Here, we investigated whether the D. discoideum Dnmt2-homologue DnmA is also an active tRNAAsp methyltransferase. In vitro transcribed D. discoideum tRNAAsp(GUC) was used as a substrate for methylation by DnmA and hDnmt2 in vitro. The assays showed that the tRNA was efficiently methylated by both enzymes (Figure 1A). The reaction was time dependent, the activity was sensitive to Mg2+ concentrations and almost lost at 10 mM Mg2+. In contrast to the human enzyme, DnmA was inactive at 37°C (data not shown). Therefore, all reactions were carried out at 22°C where hDnmt2 still showed good activity.
Figure 1.
In vitro methylation of tRNAAsp(GUC). (A) In vitro methylation of tRNAAsp(GUC) by DnmA at 2 mM, 5 mM and 10 mM MgCl2 and by hDnmt2 at 5 mM Mg2+. The upper panel shows ethidium bromide staining of the in vitro transcripts separated in a denaturing polyacrylamide gel, the lower panel shows incorporated 3H-Me in the tRNAs. Reactions were run for the times indicated. (B) In vivo methylation of cytosines in tRNAAsp(GUC) from different D. discoideum strains. Results of the RNA bisulfite sequencing (454 pyrosequencing) are given in percentage of reads. Numbers of sequence reads are shown in brackets. C49 is methylated by a different methyltransferase and thus serves as an internal standard. All 22 tRNAAsp(GUC) genes result in the same transcript, and no isoacceptors are encoded in the D. discoideum genome. (C) In vitro methylation of small enriched RNA of a dnmAKO strain (ex vivo methylation). The methylation reaction was done for the times indicated.
To investigate methylation in vivo, RNA bisulfite analysis of tRNAAsp(GUC) from D. discoideum was performed. Figure 1B shows that in wild-type cells, tRNAAsp(GUC) was methylated at position C38 to ∼30%, whereas methylation was at background levels in DnmA knockout strains. Ectopic overexpression of hDnmt2 in wild-type cells increased tRNAAsp(GUC) C38 methylation to ∼50% indicating cross-species activity of Dnmt2-homologues. C49 was methylated at ∼90%, independently of DnmA and served as an internal control for the bisulfite reaction.
As tRNAAsp appeared to be not the only target for Dnmt2 in other organisms (e.g. D. melanogaster), we examined if more cellular-derived RNAs could be methylated in vitro (ex vivo methylation experiments). Total RNA from dnmAKO AX2 cells was subjected to in vitro methylation by recombinant DnmA for various times. Methylated RNAs were separated by gelelectrophoresis and visualized by autoradiography. Figure 1C shows that a substantial RNA fraction in the tRNA size range could be methylated. At longer incubation times, a second minor methylated RNA species appeared, indicating that at least one further substrate could be methylated in vitro.
Identification of further substrates—in vitro binding and methylation of tRNAGlu
The group of Helm (personal communication) had proposed a sequence pattern (C32, C34, A37, C40) that may serve as a target recognition motif for C38 methylation by hDnmt2. We therefore examined all D. discoideum tRNAs in silico to identify candidates for the additional methylated band in the ex vivo experiment. In addition to tRNAAsp(GUC), we found tRNAGlu(UUC) and tRNAGlu(CUC) to match the pattern (Figure 2 and Supplementary Table S2).
Figure 2.
Target site sequences of tRNAs used for in vitro methylation. The position of the putative signature nucleotides are indicated and printed in bold if present. The anticodon is underlined. tRNAGlu(CUC) (not shown) also harbours the nucleotide pattern. Except for the anticodon, both tRNAGlu are identical in the anticodon stem-loop region (Supplementary Figure S2). Only a few differences appear in the acceptor stem.
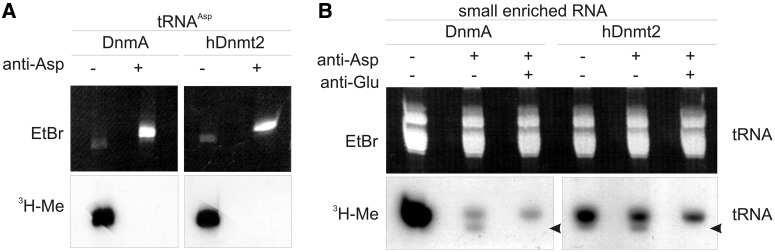
To test for tRNAGlu as a potential substrate, we modified the methylation assay by addition of a 50 nt DNA antisense oligonucleotide to tRNAAsp(GUC) covering the methylation target site. Figure 3A shows, as a control, that methylation of in vitro transcribed tRNAAsp(GUC) was completely blocked. We then performed ex vivo methylation assays with enriched small RNAs isolated from D. discoideum after blocking tRNAAsp(GUC) with the antisense oligo. Figure 3B shows that the majority of 3H-incorporation disappeared. However, with both DnmA and hDnmt2 a significant amount of methylation was still detectable in the size range of tRNAs. Assuming that this signal contained tRNAGlu, we used a second antisense oligo directed against tRNAGlu(UUC) for blocking methylation. The oligo also covered the isoacceptor tRNAGlu(CUC) with minor mismatches (Supplementary Figure S2). A further reduction of ex vivo methylation was observed. Notably, the lower band in the tRNA size range disappeared (arrow in Figure 3B), suggesting that this represented a tRNAGlu population that could be methylated at least in vitro. The residual methylation after blocking with both oligos may represent additional tRNAs or other unknown targets. As we only concentrated on small RNAs, larger methylated molecules and less abundant targets may have escaped detection.
Figure 3.
Ex vivo methylation and blocking assay. (A) Methylation of in vitro transcribed tRNAAsp(GUC) was completely blocked when a complementary antisense oligo was hybridized. (B) Using enriched small RNA from dnmAKO cells, ex vivo methylation in the tRNA size class was differentially lost when antisense oligos to tRNAAsp(GUC) and tRNAGlu(UUC) were hybridized before the methylation reaction. The oligo against tRNAGlu(UUC) also covers tRNAGlu(CUC) with minor mismatches (see Supplementary Figure S2). Even with both oligos, a significant amount of 3H incorporation still remained. The upper panel shows the ethidiumbromide stained gel to demonstrate equal loading, the lower panel shows the fluorogram. The arrow marks a band that was specifically lost when the anti tRNAGlu(UUC) oligo was used.
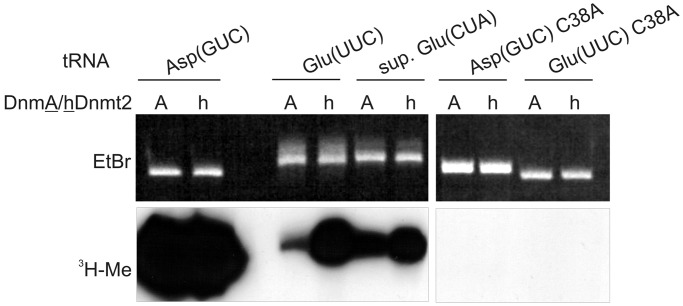
To further analyse methylation on tRNAAsp and tRNAGlu (exemplified by tRNAGlu(UUC)), we carried out assays with wild-type sequences and C38A mutated in vitro transcripts (Figure 4). We also included a suppressor tRNAGlu(CUA) that differed in only 2 nt from tRNAGlu(UUC) (Figure 2). Assays were done in parallel with the recombinant hDnmt2 and D. discoideum DnmA. The experiment showed that tRNAAsp(GUC) was the best substrate for both DnmA and hDnmt2 (Figure 4). Substantial methylation was also observed for tRNAGlu(UUC) and suppressor tRNAGlu(CUA). Moreover, the complete loss of methylation in the C38A mutations of tRNAAsp(GUC) and tRNAGlu(UUC) confirmed that methylation occurred at position C38 and largely ruled out that methyl group incorporation was due to another contaminating methyltransferase in the recombinant protein preparation (17).
Figure 4.
C38 is the target for in vitro methylation of tRNAAsp(GUC) and tRNAGlu(UUC). In vitro methylation of tRNAAsp(GUC), tRNAGlu(UUC) and a suppressor tRNAGlu(CUA). C38A mutations confirmed C38 as the target nucleotide. tRNAAsp(GUC) methylation was most efficient.
The methylation experiments using in vitro transcribed tRNAs showed that no other modifications were necessary for C38 methylation by DnmA and hDnmt2 (13) and that the processed ends of a mature tRNA were not required for target recognition, as the tRNA transcripts contain no 3′-CCA. Even tRNAGlu embedded in a long dsRNA molecule (∼1000 nt) with small stem-loop structures at the ends (27) was a specific substrate of DnmA and hDnmt2 (Supplementary Figure S3).
By EMSA, no difference in in vitro binding could be seen for the specific target tRNAGlu(UUC) and non-target RNAs, indicating that DnmA associated non-specifically with any nucleic acid (data not shown). However, specific covalent enzyme-substrate intermediates can be detected by denaturing SDS–PAGE (25). Figure 5A and Supplementary Figure S4 show that radioactively labelled tRNA transcripts that can be methylated in vitro by DnmA formed denaturation resistant complexes while non-target RNAs did not. To confirm that the denaturation resistant complexes were really due to binding of the enzyme to the substrate, SDS gels were blotted, and His-tagged DnmA was detected by a specific anti-His antibody. A small amount of the tagged protein was shifted to the same position as the labelled tRNA (Supplementary Figure S5).
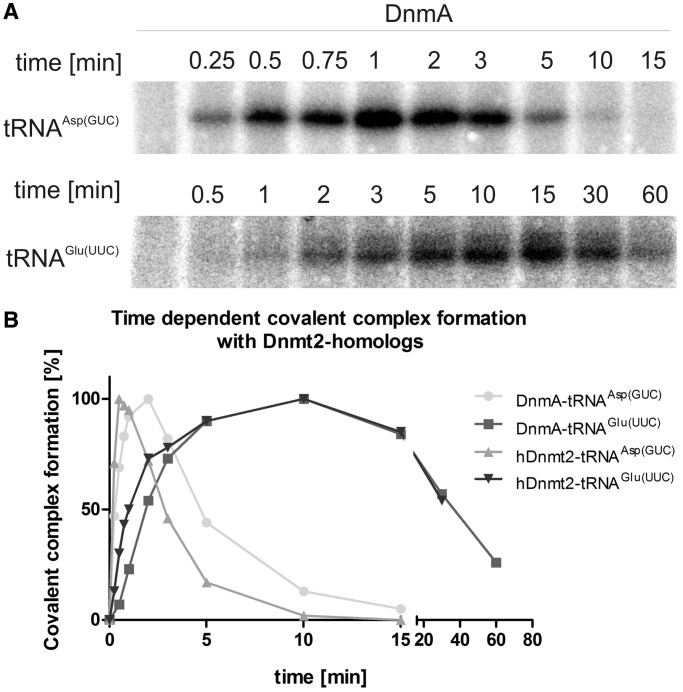
Figure 5.
Formation and turnover of tRNA-methyltransferase complexes. Turnover of covalent complexes of tRNA substrates with DnmA and hDnmt2. (A) Examples of time courses on covalent complex formation with tRNAAsp(GUC) and tRNAGlu(UUC). (B) Complex bands in A were quantified and presented on a time scale.
We then performed time courses for the reaction and found that covalent complex formation was only transient under the conditions used. For tRNAAsp(GUC), adducts reached a maximum after 1 min and were undetectable after 15 min, whereas the tRNAGlu(UUC) complex only began to accumulate after 1 min but persisted for almost 60 min (Figure 5A and B). This coincided with the high methylation activity on tRNAAsp(GUC-1) and a lower one on tRNAGlu(UUC-2). The human enzyme appeared to be slightly faster in the reaction but otherwise showed a similar behaviour compared with D. discoideum DnmA.
tRNAAsp(GUC), tRNAGlu(UUC/CUC) and tRNAGly(GCC) bind specifically to DnmA in vivo
The blocking assay (Figure 3) suggested that further substrates could be targeted by DnmA. To identify such targets and to further investigate the interaction between DnmA and RNAs in vivo, we established a UV-CLIP procedure for D. discoideum. DnmA-GFP expressing cells and control cells expressing GFP only, were UV cross-linked, and complexes were precipitated under denaturing conditions by a GFP nanobody (21). Then, co-precipitated RNAs were isolated, converted into strand-specific cDNA libraries and analyzed by Illumina sequencing (Figure 6 and Supplmentary Figure S6).
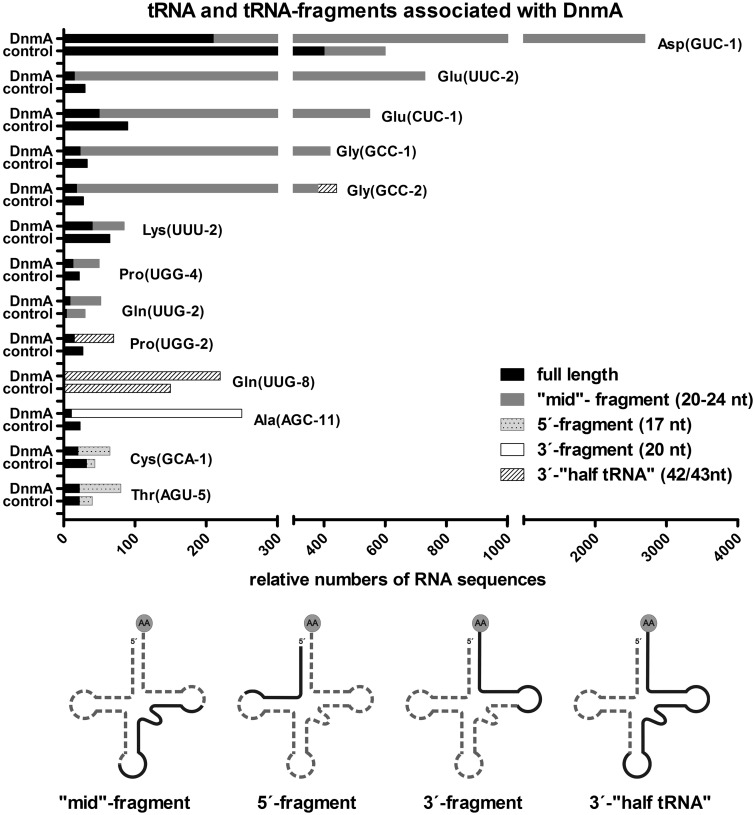
Figure 6.
tRNA and tRNA fragments associated with DnmA-GFP. After UV crosslinking, RNA bound to DnmA was co-immunoprecipitated under denaturing conditions (CLIP). The number of normalised reads for tRNAs detected by Illumina sequencing is shown for DnmA CLIP and for the control CLIP with GFP. In addition to full-length tRNAs, four classes of tRNA fragments were found, and these are indicated by different shading of the bars. Size and localization of fragments is shown schematically on the simplified clover leaf structure. tRNAs that are not significantly enriched with DnmA are shown in Supplementary Figure S6. In case of multiple gene copys resulting in the same RNA transcript, the sequencing reads were mapped to the first copy in the genome starting from chromsome 1. Only this copy is listed. Sequences and detailed information on gene copies and potential isoacceptors are listed in Supplementary Table S2.
Various tRNA fragments were enriched in the DnmA CLIP sample compared with the control. By their length and position, fragments appeared not to be random but specific. We found mid-fragments of 20–24 nt (position 32–36 to 53–56) that covered the C38 methylation site in the respective tRNAs, 5′-fragments of 17 nt (nucleotide position 1–17), 3′-fragments of 20/21 nt (nucleotide position 54 to the end) and 3′-half tRNAs of 41–43 nt (position 30–32 to the end). Mid-fragments of tRNAAsp(GUC), tRNAGlu(UUC/CUC) and tRNAGly(GCC) were significantly enriched in the DnmA-GFP fraction in comparison with the control CLIP assay. tRNAAsp(GUC), tRNAGlu(UUC) and tRNAGlu(CUC) harbour the sequence pattern (C32, C34, A37 C40), which has been suggested as a signature for C38 methylation (Figure 2, Helm, personal communication), tRNAGly(GCC) contains the pattern except for C40. For tRNAAla(AGC), which does not contain C38 and the signature pattern, a different fragment from the 3′-end was enriched in the DnmA-CLIP sample. For other tRNAs, no significant accumulation was observed in the experiment (Figure 6 and Supplementary Figure S6). For several tRNAs, varying amounts of full-length molecules were found, but these were never enriched in the DnmA sample in comparison with the control.
tRNAGly(GCC) is an additional methylation substrate in vitro
The CLIP data suggested that tRNAGly(GCC) was another target for DnmA. We therefore cloned this and several other tRNA genes that contained the pattern only partially (Figure 2). We generated in vitro transcripts and examined methylation by DnmA and hDnmt2 in vitro. Indeed, tRNAGly(GCC) was methylated by both enzymes (Figure 7), while none of the other tRNAs (tRNALeu(UAG-1), tRNALeu(AAG-8), tRNAVal(CAC-1) and tRNAPhe(GAA-2)) resulted in detectable methylation signals in the assay (Supplementary Figure S7). Interestingly, the in vitro methylation efficiency of the three tRNA substrates (tRNAAsp(GUC), tRNAGlu(UUC), tRNAGly(GCC)) correlated roughly with the number of tRNA fragments found in the RNA-CLIP experiment.
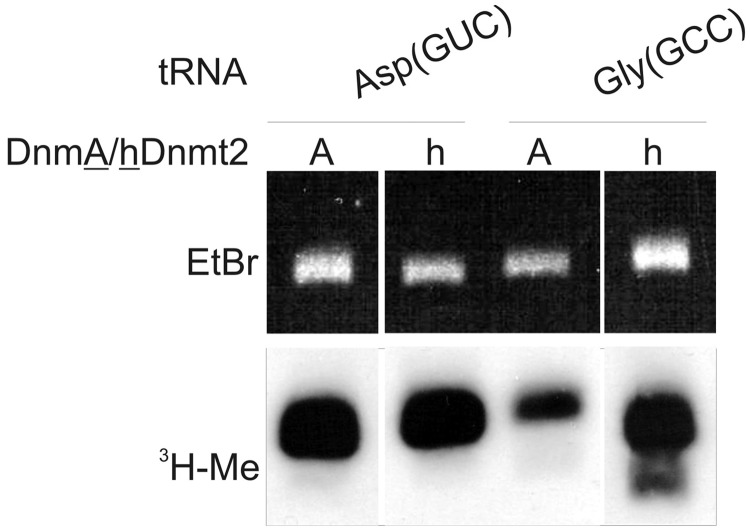
Figure 7.
In vitro methylation of tRNAGly(GCC) by recombinant DnmA and hDnmt2. In vitro transcribed tRNAs were methylated in vitro as described before. The top panel shows the ethidium-bromide stained gel, the bottom panel the fluorogram of 3H-labelled methylated tRNAs. As a control tRNAAsp(GUC) methylation is shown.
The data supported the conclusion from the ex vivo blocking assay (Figure 3B) that in addition to D. discoideum tRNAAsp and tRNAGlu, other RNAs were substrates for DnmA and hDnmt2 in vitro.
Expression of dnmA is regulated during development, cell cycle and on temperature stress
DnmA expression in growing vegetative D. discoideum cells is low. To examine tRNA methylation in vivo, it was therefore advantageous to determine conditions under which dnmA was most highly expressed. Prompted by previous studies (14,15,17), we assumed that the biological function of DnmA was connected to developmental and/or environmental conditions. We therefore examined expression levels of dnmA under different regimes. dnmA mRNA was quantified by qPCR over the 24 h developmental cycle of D. discoideum (data not shown) and confirmed the data deposited in Dictyexpress (http://dictyexpress.biolab.si/).
Interestingly, an ∼5-fold increase in expression was observed at 16 h in development. This time coincides with a presumed synchronous mitotic division (28) where a multitude of cell-cycle regulated genes are expressed [(29) and McWilliams, personal Communication].
Therefore, we synchronized vegetative D. discoideum cells to examine dnmA expression during the cell cycle. For these experiments, the D. discoideum wild-type strain NC4, the parent of the axenic laboratory strain AX2 was used (30). This was prompted by the observation that D. melanogaster and E. histolytica laboratory strains may lose dnmt2 expression (Reuter, Ankri, personal communication).
The data confirmed our assumption of a cell-cycle-dependent transcriptional regulation of dnmA (Figure 8A). The peak of expression coincided with late mitosis or early S-phase, as D. discoideum does not display a G1 phase. Using AX2 cells cotransformed with PCNA-RFP and DnmA-GFP, we observed GFP accumulation in the nucleus during S phase (indicated by PCNA expression) and a loss of DnmA from the nucleus during mitosis (Figure 8B). This is further exemplified by the movie in the Supplementary Material (Supplementary Movie S1).
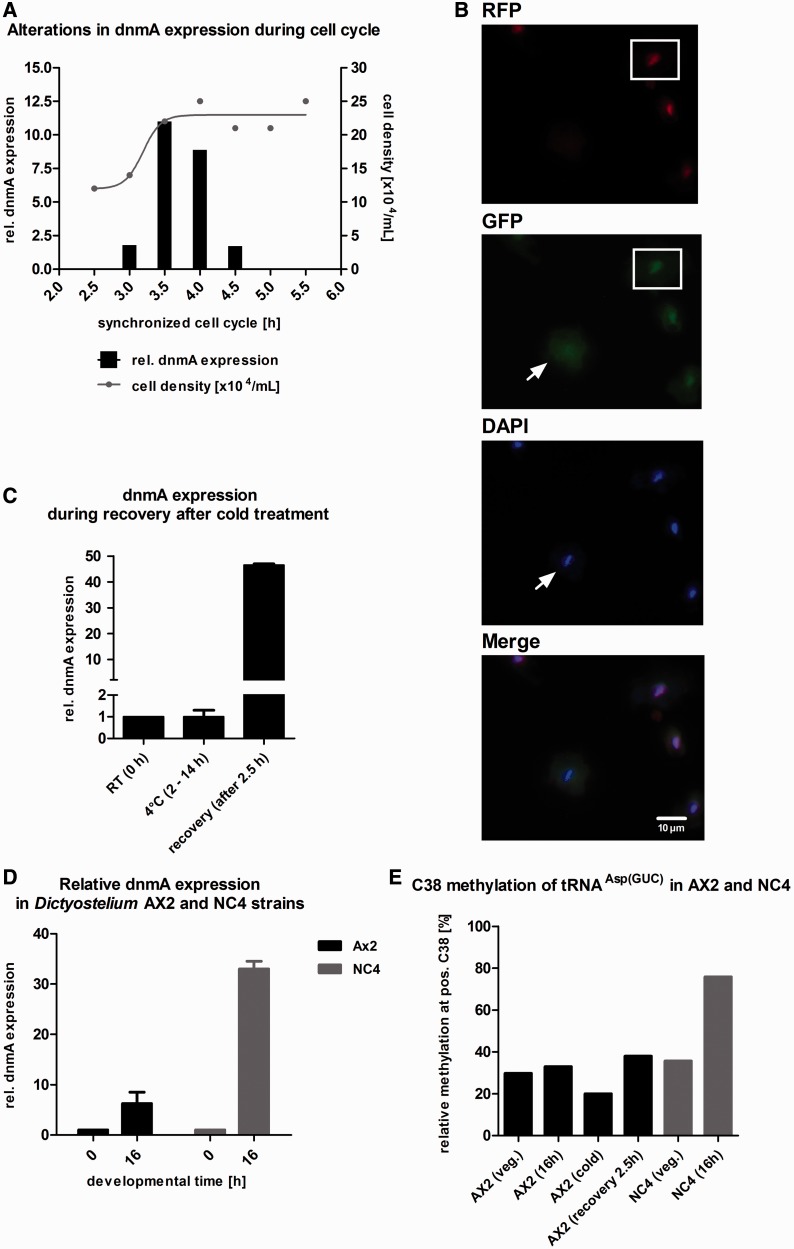
Figure 8.
Expression, localization and tRNAAsp(GUC) methylation activity of DnmA under various conditions. (A) dnmA expression is regulated during the cell cycle. Cells were arrested in the cell cycle by cold treatment and then released by transfer to 22°C. Cells were counted every 30 min (grey dots and grey line), and samples were taken for qPCR (black bars). After cell division, dnmA expression increased ∼5-fold and then rapidly declined to basal levels. Normalization was done on vegetative growing cells. dnmA expression is only shown from 3 to 4.5 h during recovery. (B) DnmA is lost from the nucleus during mitosis (arrow). The three cells on the right are in S-phase as indicated by the RFP-PCNA marker and accumulate DnmA in the nucleus (for further details see movie in Supplementary Material). (C) Relative quantification of dnmA expression levels in AX2 cells after 2.5 h recovery from cold shock at 4°C. Expression of dnmA increased >40-fold and returned within 30 min to basal levels (n = 3). (D) At 16 h of development, dnmA expression increased ∼46-fold in the NC4 strain. In AX2 cells, expression increased only ∼5-fold. (E) At 16 h of development, in vivo methylation of C38 in tRNAAsp(GUC) increased up to 75% in the D. discoideum strain NC4, whereas no significant increase was observed in AX2 cells.
In other organisms, dnmt2 was observed to be upregulated during stress response (3,15). We therefore examined expression after anoxia, heat shock and cold shock but did not find any significant changes (data not shown). Only during recovery from cold shock, we detected a strong transient increase in dnmA mRNA by ∼47-fold (Figure 8C).
Thus, dnmA expression was differentially regulated during the cell cycle, development and on temperature stress. Furthermore, DnmA localization changed specifically during the cell cycle.
Though the basic expression in vegetative cells was indistinguishable, dnmA expression in NC4 was ∼30-fold higher at 16 h of development, whereas developmental upregulation was only 5-fold in AX2. The data are summarized in Figure 8D.
NC4, the parent strain of AX2, displays increased tRNAAsp methylation during development
Based on these observations, we asked the question whether expression levels correlated with tRNA methylation activity in vivo. Bisulfite sequencing was done with primers for tRNAAsp, tRNAGlu and tRNAGly on RNA isolated from vegetative AX2 cells, AX2 cells developed for 16 h, AX2 cells subjected to cold shock, AX2 recovered for 2.5 h after cold shock, vegetative NC4 cells and NC4 cells developed for 16 h. The results on C38 methylation of tRNAAsp(GUC) are summarized in Figure 8E. The methylation status of all C residues from all tested tRNAs in this study is shown in Supplementary Figure S8. Indeed, tRNAAsp(GUC) methylation at C38 increased to 76% in development in NC4 cells. No increase was observed in AX2 cells during development, possibly due to insufficient sensitivity of the assay. Interestingly, no increase in tRNAAsp(GUC) methylation was observed at the highest dnmA expression levels during stress recovery. In Figure 8E, the 30–35% methylation level in all tRNAAsp(GUC) samples represents significant methylation as compared with other C residues (see Supplementary Figure S8).
We did, however, not detect any significant methylation of tRNAGlu(UUC/CUC) and tRNAGly(GCC) under any conditions (Supplementary Figure S8). In contrast, C49/C50 methylation, which does not depend on Dnmt2 enzymes, was found in all samples and thus served as an internal control. In all, 1.175 bisulfite-sequence reads were examined for tRNAGlu(UUC/CUC) and 4.040 for tRNAGly(GCC). No methylation above background was found. Though it cannot be excluded that the failure to detect C38 methylation of tRNAGlu and tRNAGly was due to inefficient reverse transcription or low levels of methylation (see ‘Discussion’ section), we conclude that tRNAAsp is the major or possibly the only in vivo target for methylation by DnmA in D. discoideum.
DISCUSSION
The biological function of Dnmt2 methyltransferases is largely unknown, and its role in DNA methylation is controversially discussed (9,10). In contrast, tRNA methylation by Dnmt2 is robust in vitro and in vivo and evolutionary conserved. In addition to the originally described tRNAAsp(GUC) (1), two further tRNAs targets (tRNAVal(CAC) and tRNAGly(GCC)) have been defined in D. melanogaster (15) and tRNAGlu(UUC) in S. pombe (17).
To get insight into potential functions of the D. discoideum Dnmt2 homologue DnmA, we examined expression levels in development, in the cell cycle and under different environmental conditions. We confirmed by qPCR an expression peak at 16 h of development. Weijer and co-workers (28) had previously proposed a synchronous cell division at this developmental stage. We therefore investigated dnmA expression in the cell cycle and found a strong transient increase of mRNA in S-phase or shortly thereafter. The expression peak was accompanied by reaccumulation of DnmA in the nucleus. We assume that DnmA is degraded during mitosis and re-synthesised after cell division.
In S. pombe as well as in D. melanogaster, tRNA methylation changes in response to environmental challenges like temperature stress, anoxia or nutrients. We tested heat shock, cold shock, anoxia and various nutrient media but did not observe altered levels. Only during the recovery phase after cold treatment, a transient, almost 50-fold increase in mRNA abundance was seen.
Reuter and Ankri (personal communication) had observed a loss of Dnmt2 expression in laboratory strains of D. melanogaster and E. histolytica. We therefore examined D. discoideum NC4, the parent of the axenic laboratory strain AX2. The developmental regulation in AX2 and NC4 was qualitatively similar, but expression levels were much higher in NC4. The observation supports the assumption that Dnmt2 expression was reduced under optimal growth conditions in the laboratory strain and may actually be selected against.
We show that the D. discoideum Dnmt2 homologue DnmA is an active tRNAAsp(GUC) methyltransferase in vitro and in vivo. To identify further potential targets for DnmA, we established a UV-CLIP method to analyse DnmA–RNA interaction in vivo. In addition to the known target tRNAAsp(GUC), fragments of tRNAGlu(UUC) and tRNAGly(GCC) were most abundant in comparison with the control. Unexpectedly, complete tRNAs were not enriched in the DnmA CLIP fraction but rather specific ‘mid-fragments’ that covered the putative methylation site and the recognition pattern proposed by Helm (personal communication). A minor exception was tRNAGly(GCC), which lacks C40. We do not know whether these fragments were directly or indirectly generated by DnmA in vivo or whether they were protected by DnmA from degradation during the experimental procedure. As no co-crystal of Dnmt2 and a tRNA substrate is available, the exact interface for the interaction is unknown. Jurkowski et al. (31) mapped the binding site on Dnmt2 by mutational analysis. A manual placement of a tRNA into the crystal structure suggests that Dnmt2 mainly contacts the anticodon stem loop. Assuming that mid-fragments are footprints of DnmA, our data support this notion but suggest that the interaction surface extends into the D-loop, which is partially covered by the mid-fragments.
We examined the putative DnmA targets in vitro (by methylation of in vitro transcribed tRNAs), ex vivo (by methylation of RNA isolated from D. discoideum) and in vivo (by bisulfite sequencing). Furthermore, we investigated by SDS–PAGE the kinetics of covalent complex formation between tRNAAsp and tRNAGlu with DnmA and hDnmt2.
All three tRNAs (tRNAAsp, tRNAGlu, tRNAGly) were readily methylated by recombinant DnmA and hDnmt2 in vitro, although the activity was much higher (estimated at least 10-fold) on tRNAAsp compared with tRNAGlu and tRNAGly. For tRNAAsp and tRNAGlu, this correlated well with the formation and decay of covalent RNA—protein complexes where kinetics for tRNAAsp were much faster than for tRNAGlu. Instead of C methylation, the covalent intermediate may also proceed to a hydrogen exchange reaction or to a deamination of the cytosine. In both cases, an intermediate will be detected, but it will not result in a methylated product (32). It may be that slower off rates represent these alternative pathways that would result in unmethylated tRNAs.
Using antisense oligos, we showed that tRNAAsp, tRNAGlu and at least one additional RNA could be methylated ex vivo. A quantification of the results was not possible: the apparently low methylation of tRNAGlu ex vivo could be due to the low activity of the enzyme on this substrate (as suggested by the in vitro experiments, see Figure 5B).
The data for the examination of in vivo methylation by bisulfite sequencing were rather unexpected. Although tRNAAsp(GUC) was consistently found to be methylated at C38, we never detected any methylation at C38 of tRNAGlu(UUC/CUC) or tRNAGly(GCC). There could be two reasons to explain the failure to detect C38 methylation for these in vitro substrates: (i) due to the low activity of DnmA on tRNAGlu(UUC/CUC) and tRNAGly(GCC) as shown in vitro, there was low or no significant methylation in vivo and (ii) methylation escaped detection because other unknown modifications blocked reverse transcription of the methylated molecules. However, if this was the case, one has to assume that C49/C50 methylation occurs independently of these unknown modifications, whereas C38 methylation strictly depends on them. As C38 methylation does not require any other modifications in vitro, we believe that this scenario is less likely. If in vitro methylation activity was comparable with in vivo methylation, 10% methylation of tRNAGlu(UUC/CUC) and tRNAGly(GCC) could be expected relative to tRNAAsp(GUC). As tRNAAsp was methylated to ∼30%, a 3% methylation of tRNAGlu and tRNAGly could be below the detection level of the bisulfite assay.
We find that high expression of dnmA did not necessarily correlate with increased methylation of tRNAs. During recovery from cold shock, D. discoideum cells displayed the highest level of dnmA expression, but this had no effect on tRNAAsp methylation. Only in development and with the high expressing NC4 strain, a significant increase of tRNAAsp methylation to ∼70% was observed. It has to be considered, however, that bisulfite sequencing does not necessarily allow for quantitative evaluation of data: detected C-methylation depends on reverse transcription efficiency, which may be hampered by other modifications that could be different under different conditions. Furthermore, the completeness of bisulfite-mediated conversion could also depend on RNA structure, which may be altered by other modifications. However, incomplete conversion will lead to an overestimation of C-methylation as maybe the case for, for example, C30, C31, C35 in tRNAGly in some samples.
Taken together, the data from CLIP, complex formation and methylation show that all three substrates can be bound and methylated in vitro. The CLIP data show that there is a specific interaction of DnmA with tRNA mid-fragments of tRNAAsp(GUC), tRNAGlu(UUC/CUC) and tRNAGly(GCC) but not with other tRNAs. In contrast, bisulfite sequencing could only detect methylation on tRNAAsp(GUC), even at the highest expression levels of dnmA. Even though low amounts of methylation may have escaped detection in bisulfite sequencing, there is still a discrepancy between dnmA expression levels (e.g. during cold shock recovery), DnmA binding to substrates (as determined by CLIP) and methylation in vivo. We therefore suggest that DnmA binding to specific targets could elicit functions in addition to RNA methylation, e.g. by promoting processing or degradation and/or recruitment of other proteins. At least for some rRNA methyltransferases, such as Bud23 or Nep1 from Saccharomyces cerevisiae, it is known that they execute functions independent from the methylation activity (33–35). As shown by Schaefer and co-workers (15), methylation protects tRNAs from degradation or processing after stress. By a different pathway, binding of DnmA may initiate processing that leads to specific tRNA fragments that could serve for other biological functions (36). Interestingly, tRNAGlu(UUC) is also a substrate for the Dnmt2 homologue Pmt1 in S. pombe (17). However, tRNAGlu(UUC) is found to be methylated to a low but significant extend in vivo in fission yeast when Pmt1 is overexpressed.
The detection of tRNAAla(AGC) fragments in the CLIP assay brings up a new question. tRNAAla(AGC) has a T instead of a C in position 38 and does not contain the signature nucleotides in the anticodon stem-loop. Instead of a mid-fragment, a distinct 3′ fragment was associated with DnmA, suggesting that a recognition motif was located in this part of the molecule.
As methylation also occurred in a large artificial molecule with embedded tRNAGlu (Supplementary Figure S2), it could be that other longer RNAs containing an appropriate recognition motif may also be bound by the enzyme. We thus propose that the biological functions of Dnmt2 proteins could extend far beyond our present knowledge. Investigation of further targets and of the significance of RNA fragments associated with Dnmt2 should shed light on the importance of this enzyme.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [FOR 1082 to W.N. and A.J.] and the Studienstiftung des Deutschen Volkes (to S.M.). Funding for open access charge: [FOR1082].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Thomas Winckler (Friedrich-Schiller-University Jena) for providing the tRNAGlu(UUC) and suppressor tRNAGlu(CUA) plasmids, Susanne Junk for cloning tRNAAspC38A. Matthias Schäfer (DKFZ) and Robin Lorenz (University of Kassel) are acknowledged for RNA bisulfite sequencing and Richard Reinhardt (Max Planck Genome Centre, Cologne) for deep sequencing. Mark Helm (Johannes-Gutenberg-University Mainz) generously provided data before publication. Carsten Seehafer (University of Kassel) helped with the analysis of deep-sequencing data.
REFERENCES
- 1.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 2.Phalke S, Nickel O, Walluscheck D, Hortig F, Onorati MC, Reuter G. Retrotransposon silencing and telomere integrity in somatic cells of Drosophila depends on the cytosine-5 methyltransferase DNMT2. Nat. Genet. 2009;41:696–702. doi: 10.1038/ng.360. [DOI] [PubMed] [Google Scholar]
- 3.Wilkinson CR, Bartlett R, Nurse P, Bird AP. The fission yeast gene pmt1+ encodes a DNA methyltransferase homologue. Nucleic Acids Res. 1995;23:203–210. doi: 10.1093/nar/23.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katoh M, Curk T, Xu Q, Zupan B, Kuspa A, Shaulsky G. Developmentally regulated DNA methylation in Dictyostelium discoideum. Eukaryot. Cell. 2006;5:18–25. doi: 10.1128/EC.5.1.18-25.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhlmann M, Borisova BE, Kaller M, Larsson P, Stach D, Na J, Eichinger L, Lyko F, Ambros V, Soderbom F, et al. Silencing of retrotransposons in Dictyostelium by DNA methylation and RNAi. Nucleic Acids Res. 2005;33:6405–6417. doi: 10.1093/nar/gki952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rai K, Chidester S, Zavala CV, Manos EJ, James SR, Karpf AR, Jones DA, Cairns BR. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev. 2007;21:261–266. doi: 10.1101/gad.1472907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher O, Siman-Tov R, Ankri S. Pleiotropic phenotype in Entamoeba histolytica overexpressing DNA methyltransferase (Ehmeth) Mol. Biochem. Parasitol. 2006;147:48–54. doi: 10.1016/j.molbiopara.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Durdevic Z, Hanna K, Gold B, Pollex T, Cherry S, Lyko F, Schaefer M. Efficient RNA virus control in Drosophila requires the RNA methyltransferase Dnmt2. EMBO Rep. 2013;14:269–275. doi: 10.1038/embor.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaefer M, Lyko F. Lack of evidence for DNA methylation of Invader4 retroelements in Drosophila and implications for Dnmt2-mediated epigenetic regulation. Nat. Genet. 2010;42:920–921. doi: 10.1038/ng1110-920. author reply 921. [DOI] [PubMed] [Google Scholar]
- 10.Krauss V, Reuter G. DNA methylation in Drosophila - a critical evaluation. Prog. Mol. Biol. Transl. Sci. 2011;101:177–191. doi: 10.1016/B978-0-12-387685-0.00003-2. [DOI] [PubMed] [Google Scholar]
- 11.Raddatz G, Guzzardo PM, Olova N, Fantappie MR, Rampp M, Schaefer M, Reik W, Hannon GJ, Lyko F. Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc. Natl Acad. Sci. USA. 2013;110:8627–8631. doi: 10.1073/pnas.1306723110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeltsch A, Nellen W, Lyko F. Two substrates are better than one: dual specificities for Dnmt2 methyltransferases. Trends Biochem. Sci. 2006;31:306–308. doi: 10.1016/j.tibs.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Jurkowski TP, Meusburger M, Phalke S, Helm M, Nellen W, Reuter G, Jeltsch A. Human DNMT2 methylates tRNA(Asp) molecules using a DNA methyltransferase-like catalytic mechanism. RNA. 2008;14:1663–1670. doi: 10.1261/rna.970408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tovy A, Siman Tov R, Gaentzsch R, Helm M, Ankri S. A new nuclear function of the Entamoeba histolytica glycolytic enzyme enolase: the metabolic regulation of cytosine-5 methyltransferase 2 (Dnmt2) activity. PLoS Pathog. 2010;6:e1000775. doi: 10.1371/journal.ppat.1000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012;19:900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- 17.Becker M, Muller S, Nellen W, Jurkowski TP, Jeltsch A, Ehrenhofer-Murray AE. Pmt1, a Dnmt2 homolog in Schizosaccharomyces pombe, mediates tRNA methylation in response to nutrient signaling. Nucleic Acids Res. 2012;40:11648–11658. doi: 10.1093/nar/gks956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tovy A, Hertz R, Siman-Tov R, Syan S, Faust D, Guillen N, Ankri S. Glucose starvation boosts Entamoeba histolytica virulence. PLoS Negl. Trop. Dis. 2011;5:e1247. doi: 10.1371/journal.pntd.0001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA. 2011;2:611–631. doi: 10.1002/wrna.79. [DOI] [PubMed] [Google Scholar]
- 20.Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 21.Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann S, Otto C, Kurtz S, Sharma CM, Khaitovich P, Vogel J, Stadler PF, Hackermuller J. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput. Biol. 2009;5:e1000502. doi: 10.1371/journal.pcbi.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Connolly MA, Clausen PA, Lazar JG. Purification of RNA from animal cells using trizol. Cold Spring Harbor Protoc. 2006;2006:pii: pdb. doi: 10.1101/pdb.prot4104. prot4104. [DOI] [PubMed] [Google Scholar]
- 25.Dong A, Yoder JA, Zhang X, Zhou L, Bestor TH, Cheng X. Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA. Nucleic Acids Res. 2001;29:439–448. doi: 10.1093/nar/29.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaefer M, Pollex T, Hanna K, Lyko F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 2009;37:e12. doi: 10.1093/nar/gkn954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonin M, Oberstrass J, Vogt U, Wassenegger M, Nellen W. Binding of IRE-BP to its cognate RNA sequence: SFM studies on a universal RNA backbone for the analysis of RNA-protein interaction. Biol. Chem. 2001;382:1157–1162. doi: 10.1515/BC.2001.145. [DOI] [PubMed] [Google Scholar]
- 28.Zimmerman W, Weijer CJ. Analysis of cell cycle progression during the development of Dictyostelium and its relationship to differentiation. Dev. Biol. 1993;160:178–185. doi: 10.1006/dbio.1993.1296. [DOI] [PubMed] [Google Scholar]
- 29.Strasser K, Bloomfield G, MacWilliams A, Ceccarelli A, MacWilliams H, Tsang A. A retinoblastoma orthologue is a major regulator of S-phase, mitotic, and developmental gene expression in Dictyostelium. PLoS One. 2012;7:e39914. doi: 10.1371/journal.pone.0039914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watts DJ, Ashworth JM. Growth of myxameobae of the cellular slime mould Dictyostelium discoideum in axenic culture. Biochem. J. 1970;119:171–174. doi: 10.1042/bj1190171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jurkowski TP, Shanmugam R, Helm M, Jeltsch A. Mapping the tRNA binding site on the surface of human DNMT2 methyltransferase. Biochemistry. 2012;51:4438–4444. doi: 10.1021/bi3002659. [DOI] [PubMed] [Google Scholar]
- 32.Svedruzic ZM, Reich NO. DNA cytosine C5 methyltransferase Dnmt1: catalysis-dependent release of allosteric inhibition. Biochemistry. 2005;44:9472–9485. doi: 10.1021/bi050295z. [DOI] [PubMed] [Google Scholar]
- 33.Thomas SR, Keller CA, Szyk A, Cannon JR, Laronde-Leblanc NA. Structural insight into the functional mechanism of Nep1/Emg1 N1-specific pseudouridine methyltransferase in ribosome biogenesis. Nucleic Acids Res. 2011;39:2445–2457. doi: 10.1093/nar/gkq1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leulliot N, Bohnsack MT, Graille M, Tollervey D, Van Tilbeurgh H. The yeast ribosome synthesis factor Emg1 is a novel member of the superfamily of alpha/beta knot fold methyltransferases. Nucleic Acids Res. 2008;36:629–639. doi: 10.1093/nar/gkm1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White J, Li Z, Sardana R, Bujnicki JM, Marcotte EM, Johnson AW. Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of pre-40S subunits. Mol. Cell Biol. 2008;28:3151–3161. doi: 10.1128/MCB.01674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sobala A, Hutvagner G. Small RNAs derived from the 5′-end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013;10:525–535. doi: 10.4161/rna.24285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.