Abstract
The P0 scaffold protein of the ribosomal stalk is mainly incorporated into pre-ribosomes in the cytoplasm where it replaces the assembly factor Mrt4. In analyzing the role of the P0 carboxyl terminal domain (CTD) during ribosomal stalk assembly, we found that its complete removal yields a protein that is functionally similar to Mrt4, whereas a chimeric Mrt4 containing the P0 CTD behaves more like P0. Deleting the P0 binding sites for the P1 and P2 proteins provoked the nuclear accumulation of P0ΔAB induced by either leptomycin B-mediated blockage of nuclear export or Mrt4 deletion. This effect was reversed by removing P1/P2 from the cell, whereas nuclear accumulation was restored on reintroduction of these proteins. Together, these results indicate that the CTD determines the function of the P0 in stalk assembly. Moreover, they indicate that in cells lacking Mrt4, P0 and its stalk base partner, the L12 protein, bind to pre-ribosomes in the nucleus, a complex that is then exported to the cytoplasm by a mechanism assisted by the interaction with P1/P2 proteins. Furthermore, in wild-type cells, the presence of nuclear pre-ribosome complexes containing P0 but not L12 is compatible with the existence of an alternative stalk assembly process.
INTRODUCTION
The ribosomal stalk is a universal domain of the large ribosomal subunit that is essential for the interaction and function of several soluble translation factors (1). In eukaryotes, a protein complex formed by two heterodimers of the acidic proteins P1 and P2 binds to P0 to form the basic stalk structure. The P0-(P1/P2)2 pentamer binds via the N-terminal domain (NTD) of P0 to the highly conserved 25S rRNA GAR region next to the ribosomal protein L12, which forms part of the stalk base (2,3). Archaeal ribosomes contain a simpler eukaryotic-type stalk whose crystal structure was recently elucidated, facilitating the resolution of its eukaryotic counterpart (4). The eukaryotic stalk structure is highly dynamic, and it appears that the acidic P1/P2 heterodimers can be exchanged for free cytoplasmic proteins (5–7), supporting the view that this ribosomal structure undergoes an assembly/disassembly cycles during protein synthesis, fulfilling a regulatory role in ribosome function and hence, in translation (8).
Defining the mechanism of stalk assembly is fundamental to understand this regulatory process. Of the four stalk components, P0, P1, P2 and L12, only the assembly of P0 has been studied in detail. Experimental evidence indicates that in Saccharomyces cerevisiae, P0 is incorporated into the pre-ribosome particle in the late cytoplasmic stage of ribosome assembly (9). This step requires the prior displacement from the pre-60S particle of the ribosome assembly factor Mrt4 (10–12), a process mediated by the Yvh1 protein (10,11) and probably also by Drg1 (9). Mrt4 displays a high degree of amino acid sequence homology with the NTD of P0 (13), which forms the rRNA-binding site of this stalk component (14). Both proteins bind to the same site in the highly conserved GAR region of the 25S rRNA, and thus they cannot coincide in the ribosome structure (13). In agreement with the mechanism proposed for cytoplasmic assembly, fluorescent P0 is detected mainly in the cytoplasm in the presence of the nuclear export inhibitor leptomycin B (LMB) (10–12). However, a fraction of P0 is also found in complexes associated with the assembly factor Nop7 (12), which have been described in the nucleus (15).
Thus, there is convincing experimental data demonstrating that P0 is assembled in the cytoplasm, although it is also present in nuclear particles. In an attempt to reconcile these apparently contradictory findings, it has been proposed that P0 assembly occurs through different pathways, and that nuclear pre-ribosomal particles containing P0 may bypass the LMB-induced blockade of nuclear export (12).
To further investigate this hypothesis, and given the importance of the specific carboxyl terminal domain (CTD) in P0 function (16,17), we analyzed the effects of modifications to the P0 CTD in the ribosome assembly process. Our results clearly demonstrate a key role of this P0 domain in the nuclear export of pre-ribosomal particles, consistent with our previous findings (12).
MATERIALS AND METHODS
Yeast strains
The S. cerevisiae strains used in the present study are listed in Supplementary Table S1. The D45dM, D45Nop7-TAP and D45dMNop7-TAP strains were generated specifically for this study. The former was generated from D45 using a NAT/MRT4 deletion cassette that carried nourseothricin (NAT) as a selection marker, which was obtained from the pYM17 plasmid template (18) by PCR with the 5′MRT4-nat and 3′ MRT4-nat (Supplementary Table S3) oligonucleotide primers. Deletion of Mrt4 was confirmed by immunoblotting using specific antibodies against this protein (13). Saccharomyces cerevisisae W303D7-GFP was generated by inserting at the appropriate position in S. cerevisiae W303 RPP0 gene a PCR fragment encoding yeGFP derived from plasmid pYM44 as described previously (18). D45Nop7-TAP and D45dMNop7-TAP were generated as described previously for W303Nop7-TAP and W303dMNop7-TAP (13).
All strains were grown at 30°C in rich medium (YEP) or synthetic dropout medium containing 2% glucose. For in vivo depletion of P0, the conditional P0 null strains (dGP0) were grown in 2% galactose medium (YPGal) at 30°C until the mid-exponential phase (OD600 = 0.5–0.6) and then transferred to 2% glucose medium (YPD) for 18 h.
Plasmids
The plasmids used are summarized in Supplementary Table S2. pFLhisP0, pFLhisP0-C, pFLhisP0D7 pFL37Mrt4/P0, pFL37P0ΔAB, pUG23-eGFP, YCplac111-Mrt4-eGFP and YCplac111-P0-eGFP have been described previously (see Supplementary Table S2).
Plasmids encoding P0-C-GFP, P0ΔAB-GFP, Mrt4/P0-GFP. The same cloning strategy was used to obtain plasmids encoding these proteins and carrying either HIS3 or LEU2 as selection markers. Using the plasmid carrying the untagged protein as a template and the oligonucleotides described in Supplementary Table S3, the ORF encoding the modified protein and its 5′ flanking region were amplified by PCR. The PCR fragment flanked by BamHI and XbaI restriction sites was subcloned into the corresponding sites of the eGFP-harbouring vector pUG23 to generate pUG23P0-C-GFP, pUG23P0ΔAB-GFP and pUG23Mrt4/P0-GFP. Using these plasmids as templates and the appropriate oligonucleotide primers (Supplementary Table S3), a PCR fragment was then obtained encoding the eGFP-tagged protein flanked by the XbaI and SacI restriction sites. This fragment was subcloned into the corresponding sites in the pFL36 vector (19), which carries a LEU2 selection marker, yielding pFL36P0-C-GFP, pFL36P0ΔAB-GFP, pFL36Mrt4/P0-GFP.
Plasmid pFL36P0D7-GFP. DNA from S. cerevisiae W303D7-GFP was used as a template to generate a DNA fragment encoding the GFP-tagged P0D7 by PCR using the oligonucleotide primers indicated in Supplementary Table S3. Following digestion with XbaI and SmaI, this fragment was subcloned into the XbaI/EcoRVsites of pFL36.
The integrity of all the plasmids was confirmed by DNA sequencing.
The mobility of the cloned tagged P0 proteins in SDS–PAGE was not as expected in some cases, which is due to the different linkers used to join the proteins to GFP in the distinct plasmids used (see Supplementary Figure S1).
Cell fractionation
Yeasts were fractionated as preciously described (20). In summary, cells were broken with glass beads in the presence of protease inhibitors. The extract was centrifuged in a Sorvall SS-34 rotor at 12 000 rpm for 15 min, yielding the supernatant S30 fraction. The S100 supernatant fraction and the ribosome pellet were obtained by high-speed centrifugation of S30 at 90 000 rpm for 30 min in a Beckman TL100.3 rotor.
Fluorescence microscopy
To study the subcellular localization of proteins the S. cerevisiae strains indicated, transformed with a plasmid encoding the appropriate eGFP-tagged derivative were grown at 30°C in restrictive media to an OD600 = 0.2–04. When required, LMB (0.1 µg/ml) was added 1 h before collecting the cells. The cells were visualized on an Axiovert 200 Zeiss microscope coupled to a Coolsnap FX CCD.
Sucrose gradient analyses
Polysome preparations were obtained from exponentially growing cells and analyzed by 7–50% sucrose gradient centrifugation, as described previously (21). Ten A260 units of extract were loaded in each gradient, and 0.5 ml fractions were collected from gradients, and the proteins recovered were analyzed in western blots.
Affinity purification of TAP-tagged proteins
TAP purifications from W303Nop7-TAP and W303DMNop7-TAP S. cerevisiae strains were performed following a standard procedure described previously (12,15,22). Purified complexes were analyzed by electrophoresis in 12.5% Tris–glycine SDS–PAGE. To normalize the amount of purified complex loaded for comparative studies, a sample was first resolved in the same conditions and silver stained.
Antibodies and western blotting
Total yeast protein extracts were analyzed in western blots using standard procedures. P0 was identified with 3BH5, a monoclonal antibody specific for the CTD. The rabbit antiserum against Mrt4 has been described previously (13), and the monoclonal anti-GFP antibodies were purchased from Roche. Monoclonal antibodies directed against ribosomal protein L3 (a gift of Prof. J. R. Warner) and the L12 protein, obtained in our laboratory, were also used.
RESULTS
Expression of GFP-tagged P0 derivatives
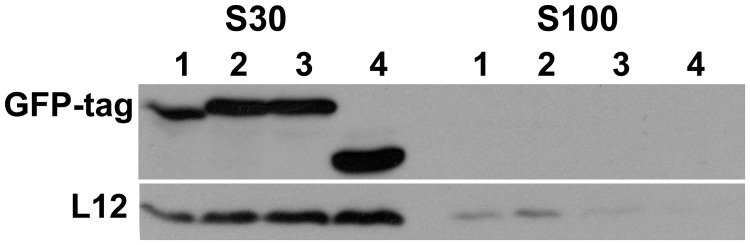
The P0 protein and the three P0 derivatives, P0-C, P0ΔAB and P0D7, all tagged with GFP, were expressed S. cerevisiae AJY1539, a strain that is sensitive to the nuclear transport inhibitor LMB. P0-C lacks the last highly conserved 21 amino acids, which are directly involved in the protein synthesis activity of P0 (16). When absent, native P0 can be substituted by this truncated protein but only in the presence of proteins P1 and P2 (16). P0ΔAB lacks the region from amino acids 198–258, which contains the binding site for proteins P1 and P2 (23). Protein P0ΔAB is functional and can substitute for the loss of parental P0, both in the presence and the absence of P1 and P2 (23,24). In P0D7, the last 132 amino acids of the protein are missing (i.e. the complete CTD). This truncated protein is not found in mature ribosomes, and it is unable to substitute native P0, irrespective of the presence of P1/P2 (16). The expression of the tagged proteins had no significant negative effect on cell growth (Supplementary Figure S2). The expression of the tagged proteins was assessed by SDS–PAGE in the cell extracts from the transformed strains, using the ribosomal L12 protein as a standard. All the proteins were present in similar amounts in total cell extracts (S30 fraction), although they were not detected after removal of the ribosomal particles (S100 fraction: Figure 1). These results demonstrate the absence of a cellular pool of free P0 proteins.
Figure 1.
Expression of tagged P0 derivatives. Equivalent amounts of the cell extracts from the AJY1539 strains expressing the four GFP-tagged proteins (1, P0wt; 2, P0-C; 3, P0ΔAB; 4, P0D7) were obtained before (S30 fraction) and after ribosome removal (S100 fraction), and they were resolved by SDS–PAGE. The proteins were detected using monoclonal antibodies against GFP and the ribosomal protein L12.
Cellular distribution of truncated P0 derivatives in wild-type cells
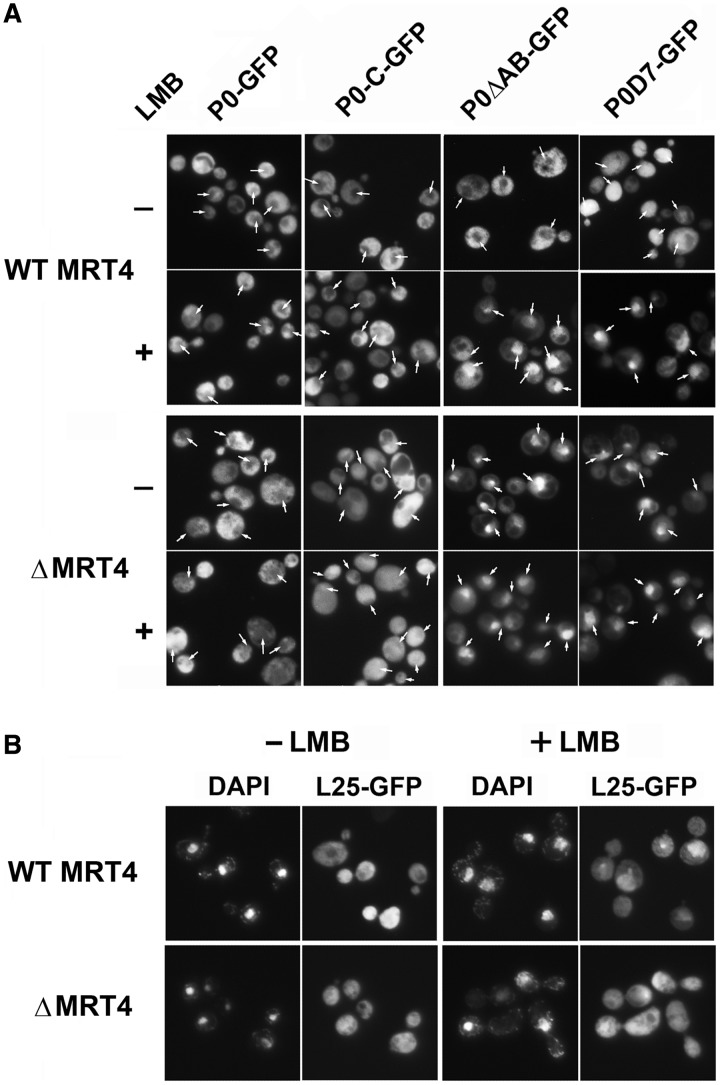
We next studied how the truncated P0 proteins affected the assembly of the ribosomal stalk in strain AJY1539. We analyzed fluorescence in cells grown in the absence or presence of LMB (Figure 2A upper panel). In agreement with previous reports, wild-type P0-GFP was found mainly in the cytoplasm, whereas the nucleus, identified by DAPI staining (Supplementary Figure S3), displayed no fluorescence either in the presence or absence of LMB. Removal of the last 21 amino acids in P0-C did not appreciably alter the distribution of this protein. P0ΔAB exhibited a cytoplasmic localization in the absence of the inhibitor, but unlike native P0, it was detected predominantly in the nucleus in the presence of LMB. The absence of the entire CTD also altered the distribution of P0D7. This protein was more homogeneously distributed throughout the cell and fluorescence was also detected in the nucleus in the absence of LMB, whereas in the presence of LMB, it concentrated in the nucleus, as observed for P0ΔAB.
Figure 2.
(A) Cellular distribution of pre-ribosomal particles carrying different GFP-tagged ribosomal proteins. P0-C-GFP, P0ΔAB-GFP and P0D7-GFP, as well as wild-type P0-GFP, were expressed in the AJY1539 (WT MRT4) and AJY1539dM (ΔMRT4) strains in the presence (+) or absence (−) of LMB. Cells were also stained with DAPI to identify the nucleus (indicated with an arrow in GFP pictures, the original DAPI-stained cells are shown in Supplementary Figure S3). (B) Ribosomal protein L25-GFP was expressed in the same strains as described in panel (A), and the nuclear location was determined by DAPI staining.
Cellular distribution of truncated P0 derivatives in the absence of Mrt4
Given the close structural and functional relationship between P0 and Mrt4, we investigated how the elimination of the assembly factor affected the cellular distribution of truncated P0 derivatives by expressing the fluorescent proteins in the MRT4-deleted AJY1539dM strain (Figure 2A, lower panel). The absence of Mrt4 did not alter the cytoplasmic distribution of P0 and P0-C, although it promoted the accumulation of P0ΔAB and P0D7 in the nucleus in both the presence and absence of LMB.
The nuclear accumulation of some truncated P0 proteins in the AJY1539dM strain suggests a possible role of Mrt4 in nuclear export. To investigate this possibility, we analyzed the cellular distribution of pre-ribosomal particles carrying the L25-GFP ribosomal protein in MRT4-deficient cells (Figure 2B). The fluorescence level increased in the nucleus of cells lacking Mrt4 in the absence of LMB. Moreover, the inhibitor promoted the nuclear accumulation of L25-GFP in the parental AJY1539 strain but not in the MRT4-deficient cells.
Incorporation of fluorescent proteins into ribosomal particles
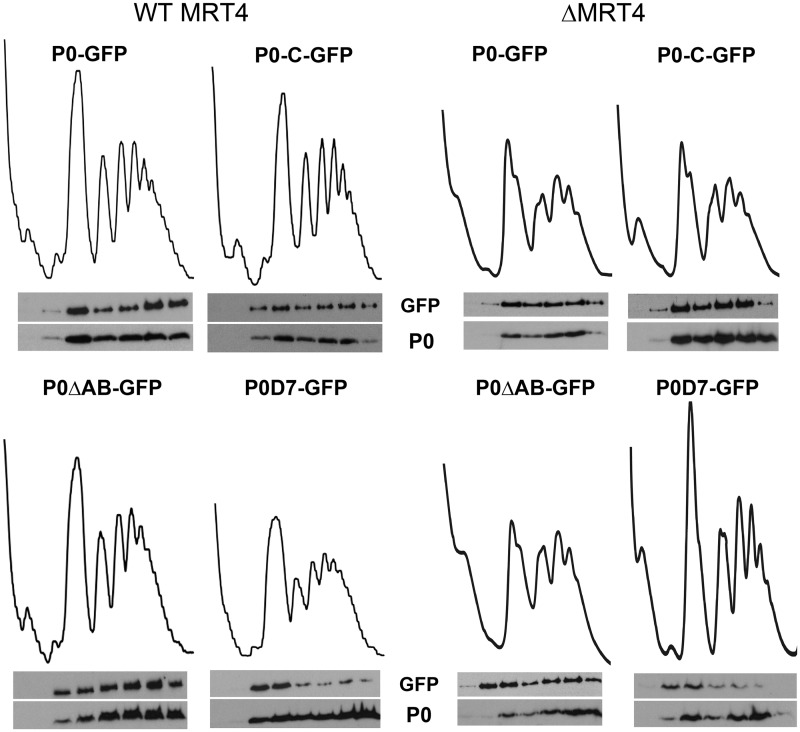
Total extracts were also resolved in sucrose gradients, and the presence of the tagged protein and the native untagged P0 in the gradient fractions was determined (Figure 3A). The top fractions of the gradient have been omitted, as they did not contain P0 proteins, consistent with the analysis of the S100 extract (Figure 1). The tagged P0, P0-C and P0ΔAB were detected in the 60S, monosome and polysome fractions along with untagged wild-type P0, indicating that despite carrying a GFP-blocked carboxyl, these proteins are assembled into the ribosomes, and they are functional. By contrast, P0D7-GFP was detected mainly with the subunits and 80S peaks, and only traces were detected in polysomes.
Figure 3.
Total extracts from parental AJY1539 (WT MRT4) and MRT4-deleted AJY1539dM (ΔMRT4) cells expressing the indicated fluorescent proteins were resolved by sucrose gradient centrifugation. The presence of the tagged proteins in the gradient fractions was tested using an anti-GFP antibody (GFP), and a P0-specific antibody was used to detect the native protein P0.
The absence of Mrt4 diminished all the peaks in the gradient, although it had no effect on the overall distribution of the tagged proteins, except for the notable increase of P0ΔAB in the 60S subunit fraction (Figure 3B). Moreover, the expression of P0D7 protein to some extent complemented the absence of Mrt4, and it increased the size and sharpness of the peaks in the polysome profile, especially that of the 80S particles. Indeed, P0D7 was able to partially rescue the cell growth deficiency caused by the MRT4 deletion (Supplementary Figure S4).
Presence of truncated proteins in nuclear pre-ribosomal particles
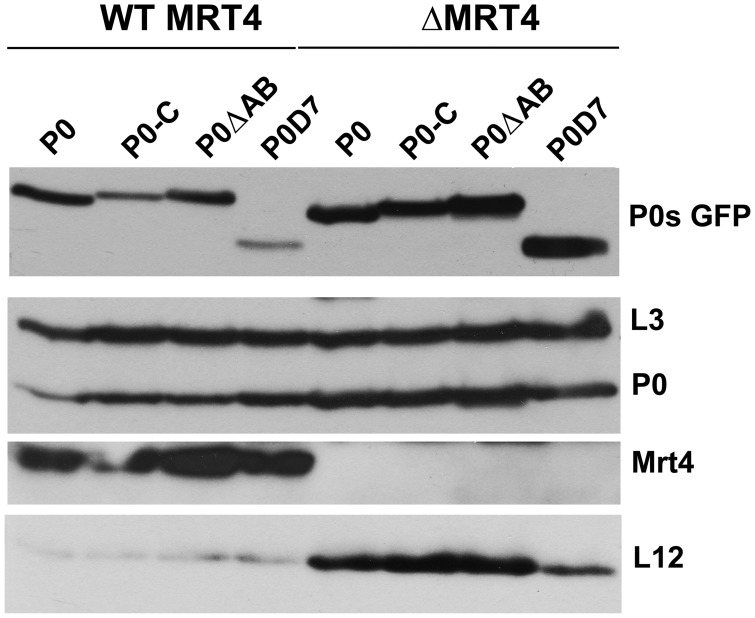
The fluorescent truncated proteins were expressed in S. cerevisiae W303Nop7-TAP and in the MRT4-deleted W303dMNop7-TAP strain. Nop7 pre-ribosomal protein complexes from all strains were TAP-purified, resolved by SDS–PAGE, and the proteins analyzed in western blots (Figure 4). In complexes from the parental W303 strain, the four truncated proteins were detected to varying extents, with the weakest band observed for P0D7. In the W303dM particles, all proteins markedly increased, particularly P0D7, which was the most abundant tagged protein in the Mrt4-deficient complexes. The presence of wild-type P0 protein in the Nop7-protein complexes has been reported previously, confirming that this is not due to contamination introduced during the purification (12). These results indicate, therefore, that the tagged P0 proteins are incorporated into pre-ribosomes in the nucleus. Protein L12, the partner of P0 in the ribosomal stalk base, was only detected in the nuclear complexes formed in cells lacking Mrt4.
Figure 4.
Detection of truncated proteins in nuclear pre-ribosomal particles. Extracts from W303 (WT MRT4) and W303dM (ΔMRT4) S. cerevisiae strains expressing Nop7-TAP and transformed with the appropriate plasmids to express different eGFP-tagged P0 derivatives were used to purify nuclear complexes using the TAP method. The presence of the indicated proteins in the complex was determined by SDS–PAGE and western blots with specific antibodies.
Expression of truncated P0 proteins in the absence of P1 and P2
We analyzed the distribution of the truncated fluorescent proteins in the absence of P1and P2 proteins, expressing the proteins in S. cerevisiae D45, a yeast strain that lacks P1 and P2 proteins (25), and in D45dM, an MRT4-deleted strain derived from D45. In D45, the P0-GFP and P0-C-GFP proteins were mainly cytoplasmic, and, as previously found in wild-type cells (Figure 2A upper panels), P0D7-GFP was distributed homogenously throughout the cell (Supplementary Figure S5, upper panel). By contrast, P0D7-GFP was found exclusively in the nucleus in the D45dM strain, whereas both P0-GFP and P0-C-GFP were also found in the nucleus (Supplementary Figure S5, lower panel), even though they were mostly cytoplasmic in the equivalent AJY1539dM strain that contains P1 and P2 (Figure 2A, lower panel).
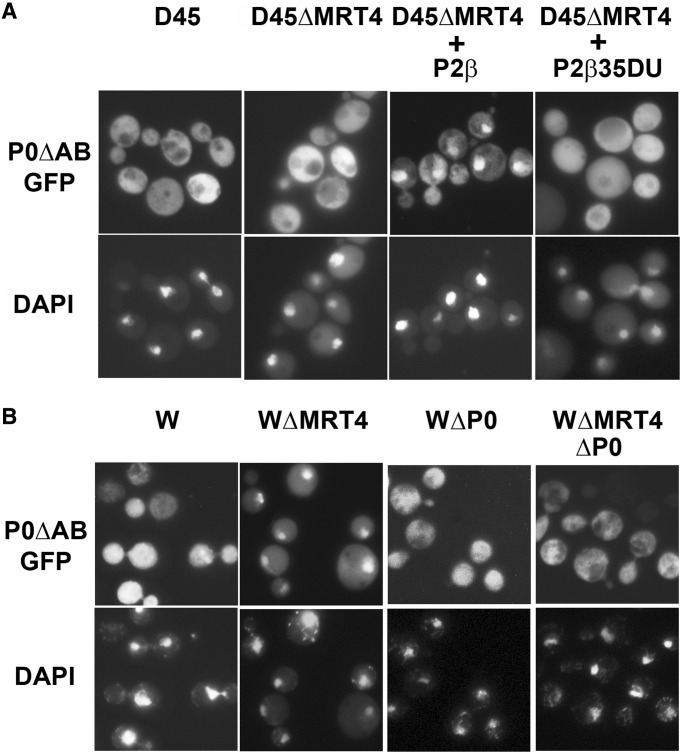
The distribution of P0ΔAB-GFP in the absence of the stalk proteins was particularly interesting. This protein exhibited a cytoplasmic location in parental AJY1539 and a nuclear distribution in the MRT4-deleted AJY1539dM (Figure 2A); yet, it was detected only in the cytoplasm in both D45 strains (Figure 5A, panels 1 and 2), indicating that the absence of Mrt4 induced nuclear accumulation of the truncated protein only if proteins P1/P2 are present. To determine whether this difference is dependent on the P1/P2 proteins, the D45dM strain was transformed using a plasmid encoding the yeast protein P2α, which also resulted in the accumulation of the corresponding P1 partner (26). In this transformed strain, P0ΔAB once again accumulated in the nucleus (Figure 5, panel 3). Moreover, this redistribution was not observed when D45dM was transformed using a plasmid encoding P2β35DU, a P2α derivative carrying the initial NTD amino acids from protein P2β and that is unable to bind to the ribosome (27) (Figure 5, panel 4).
Figure 5.
Effect of the P1 and P2 proteins on the cellular distribution of P0ΔAB. (A) -P0ΔAB-GFP was expressed in the P1- and P2-deprived strain D45 (D45) and in the MRT4-deleted D45dM (D45ΔMRT4), as well as in D45dM after transformation with a plasmid encoding either the functional stalk protein P2α (D45ΔMRT4 + P2α) or the inactive stalk protein derivative P2β35DU (D45ΔMRT4+P2β35DU). GFP fluorescence (GFP) and the DAPI-stained nuclei are shown in the upper and lower panels, respectively. (B)- P0ΔAB-GFP was expressed in S. cerevisiae strains W303 (W), W303dM (WΔMRT4) deprived of Mrt4, W303dGP0 (WΔP0) deprived of P0 and W303dGP0dM (WΔP0ΔMRT4) deprived of P0 and Mrt4. The cells were grown in glucose for 18 h to deplete cells of P0. The samples were stained with DAPI (lower panel) to identify the position of the nucleus.
To test whether the presence of a wild-type P0 affects the distribution of P0ΔAB in the absence of the Mrt4 and P1/P2 proteins, the truncated protein was expressed in S. cerevisiae W303dGP0dM, an Mrt4-deficient and conditional P0 null mutant strain that does not express P0 in glucose media (28). In these conditions, this protein was found in the cytoplasm (Figure 5B).
When Nop7 pre-ribosomal particles from the corresponding D45Nop7-TAP strains were analyzed, all the truncated proteins were detected (Supplementary Figure S6). L12 protein was present in the Nop7-TAP purifications from ΔMRT4 cells only.
The influence on cellular distribution of replacing the Mrt4 CTD with the equivalent domain from P0
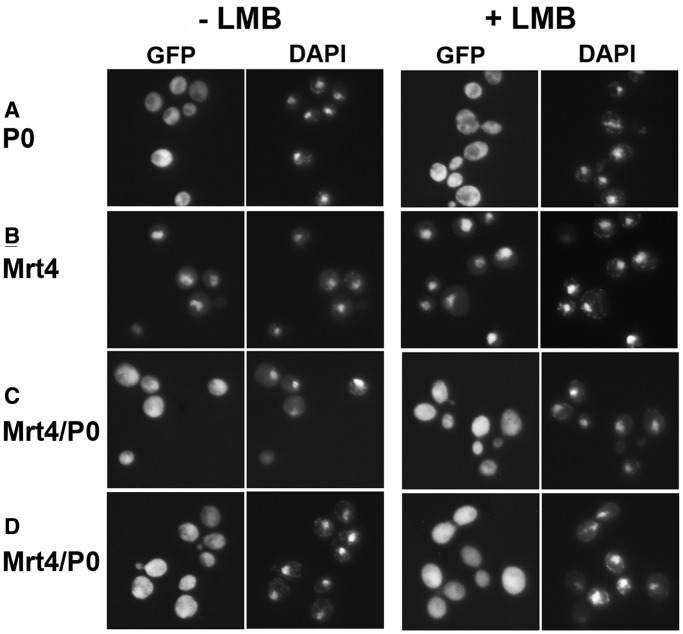
We extended our analysis of the stalk assembly process testing the effect of this domain when present in a nuclear protein. To this end, we used Mrt4/P0, a previously described protein chimera containing the CTD of P0 and the NTD of Mrt4 (13). When we compared the cellular localisation of tagged Mrt4/P0, Mrt4 and P0 (Figure 6), P0 was detected in the cytoplasm but was almost totally absent from the nucleus (Figure 6A), as reported previously (10–12). Mrt4 accumulated in the nucleus (Figure 6B) when the proteins were expressed in AJY1539, either in the presence or absence of LMB, whereas Mrt4/P0 fluorescence showed comparable levels in the nucleus and cytoplasm (Figure 6C). The distribution of Mrt4/P0 was not affected by the absence of Mrt4 when the chimera was expressed in the AJY1539dM strain under the same conditions (Figure 6D).
Figure 6.
Comparative cellular distribution of P0-GFP (A), Mrt4-GFP (B) and Mrt4/P0-GFP (C and D) proteins when expressed in parental S. cerevisiae AJY1539 (A, B, C) and MRT4-deleted AJY1539dM (D) in the presence or absence of LMB. Nuclear location was determined by DAPI staining, and the cells were analyzed as described in Figure 2.
Moreover, like wild-type P0 and all its tested derivatives, Mrt4/P0 was also detected in nuclear Nop7-TAP particles from the parental AJY1539 strain, and its levels were markedly higher in protein complexes purified from the MRT4-deleted AJY1539dM strain (Supplementary Figure S7). Moreover, the Nop7 particles obtained from AJY1539dM also contained a significantly larger amount of ribosomal protein L12. From all these results, we can conclude that P0 CTD converts Mrt4 into a P0-like protein.
DISCUSSION
Mrt4 deletion might affect nuclear export
We have studied the distribution of the P0 protein in cells and, unexpectedly, we found that the nuclear accumulation of pre-60S particles in cells lacking the Mrt4 protein was clearly less sensitive to LMB. Moreover, the presence of LMB and the absence of Mrt4 exerted similar effects on the cellular distribution of some truncated P0 proteins, as discussed later in the text. LMB sensitivity in S. cerevisiae is caused by a single mutation in the CRM1 gene, which prevents its interaction with this inhibitor (29). In sensitive organisms, the drug covalently modifies Crm1 (30), altering its interaction with the Nmd3 receptor and blocking pre-ribosomal nuclear transport (31). It remains unclear how this blockade might be dampened in the absence of Mrt4, although alterations in the Nmd3-60s interaction might be involved. One of the Nmd3 domains in the ribosome-bound protein is located close to the stalk base (32), where the Mrt4 factor associates (13). Nevertheless, alternate explanations for the Mrt4-deletion effect are possible. Thus, alteration of the nuclear turnover of the pre-ribosomes cannot be excluded.
The CTD determines the role and cellular distribution of P0 proteins
We observed notable differences in the cellular distribution of the GFP-tagged truncated P0 proteins. P0-C behaved almost identically to the parental P0 in all tested strains, suggesting that removal of the last 21 amino acids has no effect on the protein assembly process. As the missing amino acids are required for P0 activity during translation (16), including the stimulation of the GTPase associated with soluble factors, our results indicate that this stimulatory capacity seems to be irrelevant for ribosome assembly. As P0 is found in nuclear pre-ribosomal particles, and given the role of different nuclear GTPases in ribosome biogenesis (33), we believe this conclusion might be significant.
The P0D7 protein, in which the CTD is completely removed, failed to function as a P0 protein, and it accumulated in the 60S and 80S peaks. Moreover, in contrast to the other truncated derivatives, which were clearly absent from the nucleus in exponentially growing cells, P0D7-GFP was distributed throughout the cell, including the nucleus. Furthermore, the addition of LMB and the absence of Mrt4 both induced the nuclear accumulation of the GFP-tagged protein. P0D7 was also the least abundant P0 derivative in Nop7-TAP particles when Mrt4 was present and the most abundant in its absence. Additionally, P0D7 partially complemented the absence of Mrt4, enhancing the size and sharpness of the 80S and polysome peaks in the sucrose gradients and partially rescuing cell growth of AJY1539dM strains. Together, these observations suggest that removal of the complete CTD makes the P0 derivative functionally similar to Mrt4.
In contrast to P0D7, the nuclear export and cytoplasmic accumulation of the Mrt4/P0 chimera, which carries the P0 CTD, occurred both in the presence and absence of Mrt4, as seen for wild-type P0. Also, LMB failed to block the export of nuclear complexes carrying the chimera, which like wild-type P0 accumulated in the cytoplasm in the presence of this drug. Moreover, Mrt4/P0 was incorporated into polysomes and partially substituted P0 (12,13). It therefore seems that Mrt4/P0 functions like a protein P0.
The P0ΔAB protein, which has been shown to complement the absence of P0 (23,24,34), was mainly detected in the cytoplasm, like P0 and P0-C. However, in contrast to these two proteins, P0ΔAB accumulates in the nucleus in the presence of LMB and in the absence Mrt4. It was previously suggested that the failure of LMB to induce nuclear accumulation of pre-ribosomes when P0-GFP was used as a reporter might be due to the insensitivity to the drug of particles carrying wild-type P0 (12). The sensitivity of P0ΔAB-GFP pre-ribosomes to LMB supports the previous proposal and, moreover, limits the region of the P0 CTD involved in this process to the P1/P2-binding sites.
Together, the data from all the different P0 derivatives indicate that the CTD is a central element in determining the function and, consequently, the cellular distribution of the proteins.
The stalk proteins P0 and L12 can be assembled in the nucleus
The enhancement of P0 proteins in Nop7 nuclear particles together with the LMB-dependent nuclear accumulation of some truncated P0 derivatives support that in Mrt4-deficient cells, this stalk component can be assembled in the nucleus, but particles carrying a wild-type P0 scape from the drug-induced export blockade. The nuclear assembly of P0 in Mrt4-deficient cells does not contradict the cytoplasmic assembly pathway proposed for the stalk (10–12), which is probably the main mechanism used in exponentially growing wild-type cells. As Mrt4 is not an essential protein, it is reasonable to assume that cells possess an alternative means of assembling functional ribosomes in the absence of this factor. Our data indicate that in this situation P0, and probably ribosomal protein L12, can be directly assembled in the nucleus.
Data on the assembly of L12, which together with the P0 NTD forms the ribosomal stalk base, are both scarce and contradictory in contrast to the assembly of bacterial ribosomal protein L11, its prokaryotic counterpart (35). The eukaryotic protein has been detected in some nuclear particles (15,36) but not always (37,38), and no detailed study of L12 assembly has been so far reported. Our analysis revealed that this ribosomal protein was practically absent from Nop7 particles when Mrt4 was present in the cell, and that its levels increased significantly in the absence of Mrt4. The parallel increase in P0 and L12 in particles from Mrt4-deficient cells suggests the joint assembly of both these proteins and a possible role for L12 in putative Mrt4-independent P0 assembly.
The presence of P0 in nuclear Nop7 particles from the parental S. cerevisiae strains confirms a previous report (12) and suggest that an Mrt4-independent pathway may also exist in wild-type conditions, which must be different from the one occurring in the absence of Mrt4, as protein L12 is missing in that case. However, it cannot be fully ruled out that the role of P0 in the wild-type particles is not directly related to stalk assembly.
The ribosomal P1 and P2 stalk proteins assist pre-ribosome nuclear export
The increased P0-GFP and P0-C-GFP fluorescence in the nucleus of the Mrt4-deficient D45dM strain (Supplementary Figure S5), in contrast to the clear cytoplasmic location of these proteins in the Mrt4-deficient AJY1539dM strain (Figure 2A), is consistent with the negative effect on nuclear export produced in the absence of the stalk proteins. Indeed, the cellular distribution of P0ΔAB in the absence of P1 and P2 proteins supports this conclusion. Thus, in the absence of Mrt4, this truncated P0 derivative is located in the cytoplasm when the P proteins are also missing (D45dM), and in the nucleus, when stalk proteins are present (AJY1539dM). However, P0ΔAB was once again detected in the nucleus when D45dM was transformed to express P1 and P2. This was not the case when D45dM was transformed to express P proteins unable to bind to the ribosome. These results show that P1/P2 proteins affect the cellular distribution of particles that contain a P0 protein lacking the P1/P2-binding sites. A simple explanation for these apparently paradoxical results could be the presence of wild-type P0 in the test strain. If P1/P2 proteins help in the nuclear export of particles carrying P0, the wild-type protein should be more efficient than the truncated protein, which would tend to accumulate in the nucleus. In the absence of the stalk P1/P2 proteins, the particles carrying the truncated P0 would not be discriminated, and both type of particles could be exported with a similar efficiency. The mainly cytoplasmic location of protein P0ΔAB in Mrt4-deficient cells that contain P1/P2 proteins and lack wild-type P0 (Figure 5B) is fully consistent with our interpretation.
Although other more complex alternatives cannot be totally excluded, the data reported support a role for the P1 and P2 stalk proteins in assisting the nuclear export of pre-ribosome containing P0. It is likely that these proteins accelerate the process by interacting with the bound P0 at the exit site. This effect is especially evident under conditions affecting the efficiency of export, such as in the presence of LMB or the absence of Mrt4.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [38–41].
FUNDING
Spanish Ministry of Science and Innovation [BFU2009-09738 to J. P. G. B.] and the Fundación Ramón Areces [Institutional Grant to C.B.M.S.O.]. Funding for open access charge: Research grant from Spanish Government.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank M.C. Fernández Moyano for expert technical assistance and Prof. J. de la Cruz and Dr J. J. Garcia-Gomez for reading and commenting the manuscript.
REFERENCES
- 1.Gonzalo P, Reboud JP. The puzzling lateral flexible stalk of the ribosome. Biol. Cell. 2003;95:179–193. doi: 10.1016/s0248-4900(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 3.Lee KM, Yu CW, Chiu TY, Sze KH, Shaw PC, Wong KB. Solution structure of the dimerization domain of the eukaryotic stalk P1/P2 complex reveals the structural organization of eukaryotic stalk complex. Nucleic Acids Res. 2012;40:3172–3182. doi: 10.1093/nar/gkr1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naganuma T, Nomura N, Yao M, Mochizuki M, Uchiumi T, Tanaka I. Structural basis for translation factor recruitment to the eukaryotic/archaeal ribosomes. J. Biol. Chem. 2010;285:4747–4756. doi: 10.1074/jbc.M109.068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zinker S, Warner JR. The ribosomal proteins of Saccharomyces cerevisiae. Phosphorylated and exchangeable proteins. J. Biol. Chem. 1976;251:1799–1807. [PubMed] [Google Scholar]
- 6.Tsurugi K, Ogata K. Evidence for the exchangeability of acidic ribosomal proteins on cytoplasmic ribosomes in regenerating rat liver. J. Biochem. 1985;98:1427–1431. doi: 10.1093/oxfordjournals.jbchem.a135410. [DOI] [PubMed] [Google Scholar]
- 7.Scharf KD, Nover L. Control of ribosome biosynthesis in plant cell cultures under heat shock conditions. II. Ribosomal proteins. Biochim. Biophys. Acta. 1987;909:44–57. doi: 10.1111/j.1432-1033.1986.tb09971.x. [DOI] [PubMed] [Google Scholar]
- 8.Ballesta JPG, Remacha M. The large ribosomal subunit stalk as a regulatory element of the eukaryotic translational machinery. Progr. Nucl. Acid. Res. Mol. Biol. 1996;55:157–193. doi: 10.1016/s0079-6603(08)60193-2. [DOI] [PubMed] [Google Scholar]
- 9.Lo KY, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW. Defining the Pathway of Cytoplasmic Maturation of the 60S Ribosomal Subunit. Mol. Cell. 2010;39:196–208. doi: 10.1016/j.molcel.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lo KY, Li Z, Wang F, Marcotte EM, Johnson AW. Ribosome stalk assembly requires the dual-specificity phosphatase C for the exchange of Mrt4 with P0. J. Cell Biol. 2009;186:849–862. doi: 10.1083/jcb.200904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kemmler S, Occhipinti L, Veisu M, Panse VG. Yvh1 is required for a late maturation step in the 60S biogenesis pathway. J. Cell Biol. 2009;186:863–880. doi: 10.1083/jcb.200904111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez-Mateos M, Garcia-Gomez JJ, Francisco-Velilla R, Remacha M, de la Cruz J, Ballesta JP. Role and dynamics of the ribosomal protein P0 and its related trans-acting factor Mrt4 during ribosome assembly in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:7519–7532. doi: 10.1093/nar/gkp806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez-Mateos M, Abia D, Garcia-Gomez JJ, Morreale A, de la Cruz J, Santos C, Remacha M, Ballesta JP. The amino terminal domain from Mrt4 protein can functionally replace the RNA binding domain of the ribosomal P0 protein. Nucleic Acids Res. 2009;37:3514–3521. doi: 10.1093/nar/gkp209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santos C, Ballesta JP. Characterization of the 26S rRNA-binding domain in Saccharomyces cerevisiae ribosomal stalk phosphoprotein P0. Mol. Microbiol. 2005;58:217–226. doi: 10.1111/j.1365-2958.2005.04816.x. [DOI] [PubMed] [Google Scholar]
- 15.Harnpicharnchai P, Jakovljevic J, Horsey E, Miles T, Roman J, Rout M, Meagher D, Imai B, Guo Y, Brame CJ, et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- 16.Santos C, Ballesta JPG. The highly conserved protein P0 carboxyl end is essential for ribosome activity only in the absence of proteins P1 and P2. J. Biol. Chem. 1995;270:20608–20614. doi: 10.1074/jbc.270.35.20608. [DOI] [PubMed] [Google Scholar]
- 17.Mochizuki M, Kitamyo M, Miyoshi T, Ito K, Uchiumi T. Analysis of chimeric ribosomal stalk complexes from eukaryotic and bacterial sources: structural features responsible for specificity of translation factors. Genes Cells. 2012;17:273–284. doi: 10.1111/j.1365-2443.2012.01586.x. [DOI] [PubMed] [Google Scholar]
- 18.Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 19.Bonneaud N, Ozier-Kalogeropoulos O, Li G, Labouesse M, Minvielle–Sebastia L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez-Gabriel MA, Remacha M, Ballesta JPG. The RNA interacting domain but not the protein interacting domain is highly conserved in ribosomal protein P0. J. Biol. Chem. 2000;275:2130–2136. doi: 10.1074/jbc.275.3.2130. [DOI] [PubMed] [Google Scholar]
- 21.Kressler D, de la Cruz J, Rojo M, Linder P. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:7283–7294. doi: 10.1128/mcb.17.12.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 23.Krokowski D, Boguszewska A, Abramczyk D, Liljas A, Tchorzewski M, Grankowski N. Yeast ribosomal P0 protein has two separate binding sites for P1/P2 proteins. Mol. Microbiol. 2006;60:386–400. doi: 10.1111/j.1365-2958.2006.05117.x. [DOI] [PubMed] [Google Scholar]
- 24.Cárdenas D, Revuelta-Cervantes J, Jiménez-Díaz A, Camargo H, Remacha M, Ballesta JPG. P1 and P2 protein heterodimer binding to the p0 protein of Saccharomyces cerevisiae is relatively non-specific and a source of ribosomal heterogeneity. Nucleic Acids Res. 2012;40:4520–4529. doi: 10.1093/nar/gks036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Remacha M, Santos C, Bermejo B, Naranda T, Ballesta JPG. Stable binding of the eukaryotic acidic phosphoproteins to the ribosome is not an absolute requirement for in vivo protein synthesis. J. Biol. Chem. 1992;267:12061–12067. [PubMed] [Google Scholar]
- 26.Nusspaumer G, Remacha M, Ballesta JP. Phosphorylation and N-terminal region of yeast ribosomal protein P1 mediate its degradation, which is prevented by protein P2. EMBO J. 2000;19:6075–6084. doi: 10.1093/emboj/19.22.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Briceno V, Camargo H, Remacha M, Santos C, Ballesta JP. Structural and functional characterization of the amino terminal domain of the yeast ribosomal stalk P1 and P2 proteins. Int. J. Biochem. Cell Biol. 2008;41:1315–1322. doi: 10.1016/j.biocel.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 28.Santos C, Ballesta JP. Ribosomal protein P0, contrary to phosphoproteins P1 and P2, is required for ribosome activity and Saccharomyces cerevisiae viability. J. Biol. Chem. 1994;269:15689–15696. [PubMed] [Google Scholar]
- 29.Neville M, Rosbash M. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 1999;18:3746–3756. doi: 10.1093/emboj/18.13.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl Acad. Sci. USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho JH, Kallstrom G, Johnson AW. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 2000;151:1057–1066. doi: 10.1083/jcb.151.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sengupta J, Bussiere C, Pallesen J, West M, Johnson AW, Frank J. Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J. Cell Biol. 2010;189:1079–1086. doi: 10.1083/jcb.201001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kressler D, Hurt E, Babetaler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 34.Remacha M, Jimenez-Diaz A, Bermejo B, Rodriguez-Gabriel MA, Guarinos E, Ballesta JPG. Ribosomal acidic phosphoproteins P1 and P2 are not required for cell viability but regulate the pattern of protein expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:4754–4762. doi: 10.1128/mcb.15.9.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shajani Z, Sykes MT, Williamson JR. Assembly of bacterial ribosomes. Annu. Rev. Biochem. 2011;80:501–526. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- 36.Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schafer T, Kuster B, Tschochner H, Tollervey D, et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 38.Dez C, Froment C, Noaillac-Depeyre J, Monsarrat B, Caizergues-Ferrer M, Henry Y. Npa1p, a component of very early pre-60S ribosomal particles, associates with a subset of small nucleolar RNPs required for peptidyl transferase center modification. Mol. Cell. Biol. 2004;24:6324–6337. doi: 10.1128/MCB.24.14.6324-6337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas TJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 40.Hedges J, West M, Johnson AW. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 2005;24:567–579. doi: 10.1038/sj.emboj.7600547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zambrano R, Briones E, Remacha M, Ballesta JPG. Phosphorylation of the acidic ribosomal P proteins in Saccharomyces cerevisiae. A reappraisal. Biochemistry. 1997;36:14439–14446. doi: 10.1021/bi971494o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.