Abstract
The SIRT6 deacetylase is a key regulator of mammalian genome stability, metabolism and lifespan. Previous studies indicated that SIRT6 exhibits poor deacetylase activity in vitro. Here, we explored the specific conditions that allow SIRT6 to function as a significant deacetylase. We show that SIRT6 associates with the nucleosome and deacetylates histones H3 and H4 when they are packaged as nucleosomes, but not as free histones. In contrast, SIRT1 shows the opposite characteristics. Thus, our results show that SIRT6 activity is nucleosome dependent, and suggest that its binding to the nucleosome might convert it into an active structure.
INTRODUCTION
SIRT6 is one of the seven mammalian sirtuins, SIRT1–7, that possess NAD+-dependent deacetylase activity. To date, SIRT6 was shown to have deacetylase activity on histone H3 lysine 9, lysine 56 (H3K9Ac/56Ac) and CtIP (1–4). In addition, SIRT6 possesses PARP1 mono ADP ribosyl-transferase activity (5).
Recent evidence suggests SIRT6 as a key regulator of metabolism and ageing. SIRT6 levels are elevated in rats fed under dietary restriction (DR) (6). In addition, transgenic mice overexpressing exogenous SIRT6 (MOSES) are protected against the physiological damage of diet-induced obesity, including accumulation of triglycerides and serum LDL cholesterol, increased body fat and reduced glucose tolerance (7). Importantly, in support of these findings, Schwer and his colleagues showed that neuronal SIRT6 is required for normal postnatal somatic growth, and brain-specific SIRT6 knockout mice become obese at adulthood (8). In normal animals, obesity-related metabolic defects become apparent by middle age, and their appearance is delayed in animals fed with DR. Thus, our previous findings suggest that overexpression of SIRT6 might partially mimic DR phenotypes. Indeed, male MOSES mice have extended lifespan compared with their WT littermates, and aged MOSES animals demonstrate improved glucose homeostasis (9).
Additional evidence correlates SIRT6 to ageing, genome stability and metabolism. SIRT6-deficient 129J mice are small, express severe metabolic defects and by 2–3 weeks of age develop abnormalities usually associated with ageing. These mice die at ∼4 weeks of age (10). Recent studies showed that SIRT6 regulates the expression of glycolytic genes, including the glucose transporter GLUT1, in a HIF1α−dependent manner (11). Many cancer cells favor aerobic glycolysis over oxidative phosphorylation to generate energy, a phenomenon known as the Warburg effect. Indeed, a recent study showed that SIRT6 deficiency promotes tumorigenesis by promoting increased glycolysis (12). SIRT6 is tightly bound to chromatin (10), and associates with mononucleosomes (13). It functions at chromatin to attenuate NF-κB signaling via interaction with the NF-κB RELA subunit (p65), and deacetylates H3K9Ac at NF-κB target gene promoters (14).
Several findings suggest a role for SIRT6 in regulating ageing. In primary human fibroblasts, SIRT6 was shown to regulate telomere maintenance and premature cellular senescence (1,15). SIRT6 is also required for the stable association of telomeric chromatin with WRN, a protein mutated in the premature ageing disease, Werner syndrome (1).
Recent studies demonstrated a role for SIRT6 in regulating several DNA repair pathways. First, its absence results in increased sensitivity to H2O2 and methyl methanesulfonate owing to defects in base excision repair (10). Second, it regulates the DNA double-strand break repair pathway by deacetylating histone H3K9Ac and by stabilizing DNA-PKs at chromatin flanking the damage sites (16). Moreover, SIRT6 modifies two key enzymes involved in double-strand break repair, PARP1 (5) and CtIP (4). Thus, SIRT6 might also influence lifespan by regulating telomeres and genome stability.
All together, these findings emphasize the fundamental role of SIRT6 enzymatic activity in regulating survival of the cell and organism.
However, despite our increasing knowledge regarding the physiological relevance of SIRT6, our knowledge regarding the regulation of its enzymatic activity is limited. Interestingly, in comparison to other sirtuins, such as SIRT1, SIRT6 has a poor deacetylase activity in vitro, either on single acetylated lysines or on the acetylated H3K9Ac peptide (1,3,17,18). Yet, SIRT6-deficient cells show a significant increase in the acetylation status of known SIRT6 targets, suggesting that SIRT6 expresses significant deacetylase activity in vivo. These observations become even more puzzling considering the high similarity between the crystal structures of SIRT6 and SIRT2 and SIRT3, which show potent deacetylase activity in vitro (17). Thus, we aimed to explore the discrepancy between SIRT6 enzymatic activity as detected in vitro versus its activity in vivo.
Here, we show that SIRT6 histone deacetylase enzymatic activity is significantly enhanced in the context of the nucleosome. Thus, our results suggest that SIRT6 activity is nucleosome dependent, and its binding to the nucleosome converts SIRT6 into an active structure.
MATERIALS AND METHODS
Cell culture
HeLa and 293T cell lines were obtained from American Type Culture Collection. Cells were grown in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% FCS, 100 U/ml penicillin and 100 mg/ml streptomycin (all purchased from Biological Industries, Israel) and maintained in a 37°C incubator with 5% CO2 humidified air.
si-RNA delivery
si-RNA duplexes targeting SIRT6 and MeCP2 were obtained from Sigma, si-RNA against GFP was used as a control. Cells were transfected with Dharmafect 1 (Dharmacon) according to the manufacturer’s instructions. Cells were harvested 96 h after transfection with X2 Laemmli buffer, boiled for 10 min and subjected to sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS PAGE).
Antibodies
Antibodies against H4K8Ac and H4K12Ac were purchased from Millipore. Antibodies against H4K5Ac and anti-histones H3, H4, H2A and H2B were all from Abcam. Antibodies against Flag, GST, MeCP2 and SIRT6 were from Sigma. Antibodies against HP1 alpha and H4K16Ac were from Cell Signaling Technology. Antibodies against H3K9Ac and H3K56Ac were from Epitomics. Antibodies against RELA, beta Actin, SIRT1 Ku70 and Ku80 were obtained from Santa Cruz Biotechnology.
Plasmids, transfection and immunoprecipitation
Human SIRT6-Flag and SIRT1-Flag sequences were cloned into pcDNA 3.1+ and pET28a for bacterial expression. RELA N-terminal and C-terminal sequences were cloned into pGEX4t3. The 6XHis SIRT1 bacterial expression vector was previously described (19). Metafectene (Biontex) was used to transfect human embryonic kidney (HEK) 293T cells according to the manufacturer's protocol. Vector encoding the Flag tag was used as a control. Flag-tagged proteins were immunoprecipitated using anti-Flag M2 affinity gel (Sigma), washed four times and eluted with a FlagX3 peptide (Sigma) according to the manufacturer's instructions, with minor adjustments. Each IP experiment was conducted with 4 µg of SIRT6-Flag or SIRT1-Flag and 500 ng of histone octamers or reconstituted nucleosomes. Recombinant proteins were incubated with nucleosomes or histones between 3 h and overnight.
Mass spectrometry
Immunoprecipitated proteins were resolved on SDS PAGE and stained with the standard silver stain protocol. Selected bands were identified by mass spectrometry at the Smoler Proteomic Center, Technion, Israel.
Purification of recombinant proteins
BL21 (DE3) competent cells transformed with pGEX4t3, pET28a and pHis were grown in liquid media to OD600 = 0.8 and induced with 1 mM IPTG for 5 h at 37°C. Bacterial cells were disrupted with a microfluidizer (Microfluidics) at 4°C in the presence of complete mini EDTA-free protease inhibitor cocktail (Roche). 6XHis tagged proteins were immobilized to the Talon metal affinity resin (Clontech), and GST-conjugated proteins, RELA N-term and C-term were immobilized to the GST-binding resin (Novagen). His tagged proteins were eluted with 250 mM imidazole, and 50 mM reduced glutathione (Sigma) was used to elute all GST conjugated proteins. PD10 desalting column (Amersham) was used for buffer exchange. Recombinant proteins were stored at −70°C in freezing buffer (30 mM Tris–HCl, pH 7.4, 50 mM NaCl, 1 mM DTT, 10% Glycerol).
Reconstitution of poly-nucleosomes core particles
Pure chicken histone octamers (Abcam cat-ab45275) were subjected to the standard salt dialysis protocol, as described (20). Briefly, 12 µg of pure chicken octamers and 15 µg of sonicated phage lambda DNA (NEB) were incubated for 30 min in a 2 M dialysis buffer (2 M NaCl, 20 mM Tris–HCl, pH = 7.2) in 700 µl of reaction volume. The mixture was dialyzed against descending concentrations of NaCl (1.2 M, 1 M, 0.8 M and 0.6 M) for 2 h each at 4 C. Finally, the complexes were dialyzed overnight in 10 mM Tris, pH = 7.2, without NaCl. Histone octamers that were used as a control in the deacetylation assays were dialyzed overnight in maintenance buffer (10 mM Tris, pH = 7.2, no NaCl or DNA). To verify the integrity of the reconstituted nucleosomes, micrococcal nuclease (MicN) treatment was performed, and the protected DNA was examined for its length (Supplementary Figure S1). To validate the integrity of the histone octamers, 4 µg of histone octamers were separated on 15% SDS PAGE gel using reduced or non-reduced sample buffer (Supplementary Figure S2).
Ethidium bromide and MicN treatment
Total cell extracts or reconstituted nucleosomes were treated with 300 µg/ml ethidium bromide (EtBr) for 30 min on ice, followed by 10 min centrifugation at 17 000 g. The supernatant was subjected to various immunoprecipitation (IP) procedures. For MicN treatment, reconstituted nucleosomes were incubated for 3 h at 37°C in the presence of protease inhibitor cocktail (Calbiochem) and 1000 Kunitz units of MicN (NEB). The nuclease-treated nucleosomes were subjected to various IP procedures.
In vitro histone and nucleosome deacetylation assays
In vitro deacetylation reactions were performed with 3 µg of recombinant SIRT6-Flag and 1.5 µg of SIRT1, both expressed in bacteria, in deacetylation buffer (5 mM NAD+, 30 mM Tris–HCl, pH = 8, 4 mM MgCl2, 1 mM DTT) in a total volume of 50 µl. Histone octamers or histones bound in nucleosomes were used at ∼300 ng/reaction according to antibody sensitivity and acetylation abundance. After incubation at 37°C, Laemmli buffer was added, and samples were boiled for 10 min.
For total cell extract deacetylation reactions, 30–50 µg of HEK 293T cell extract was added at the same conditions as above with addition of 5 mM sodium butyrate. For protein extraction, cells were lysed in lysis buffer [50 mM Tris (pH 8), 1% NP-40, 150 mM NaCl, 1 mM MgCl2, 10% glycerol, 1 mM DTT and complete mini EDTA free protease inhibitor cocktail (Roche)]. Cell extract was sonicated and cleared by centrifugation.
RESULTS
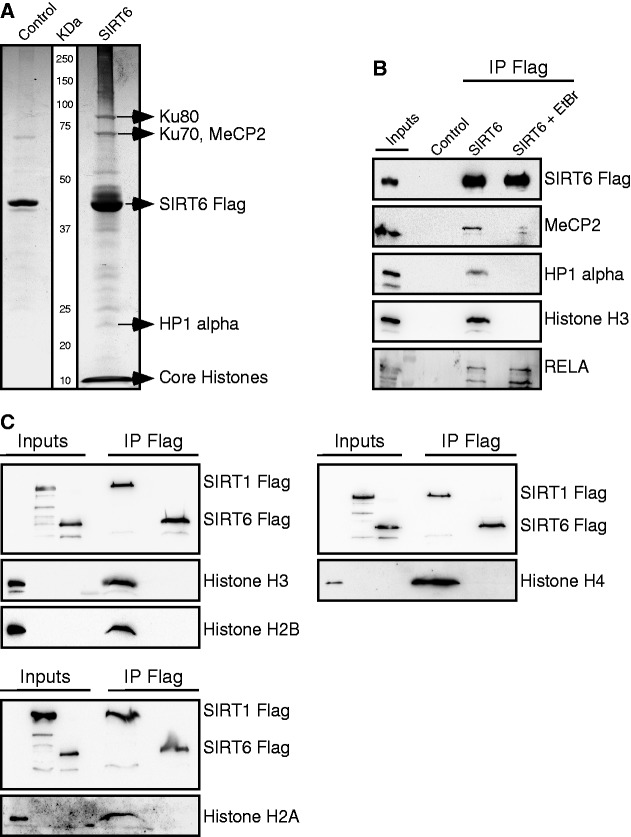
To identify SIRT6-interacting proteins, Flag-tagged SIRT6 was expressed in HEK 293T cells, and SIRT6-associated complexes were isolated on an anti-Flag antibody column. SIRT6-associated proteins were separated by electrophoresis, and selected bands were identified by mass spectrometry (Figure 1A). Co-immunoprecipitation was performed to verify these associations (Figure 1B). Similarly, the association of endogenous SIRT6 with MeCP2, histone H3 and RELA was verified (Supplementary Figure S3). However, considering the weak endogenous interaction between SIRT6 and MeCP2, its physiological relevance should be further investigated. Given the known localization of SIRT6 on chromatin (10), EtBr was used to examine which of the SIRT6 interactions is DNA dependent (Figure 1B). EtBr abolished the association of SIRT6 with MeCP2, HP1-α and histone H3, but not its interaction with RELA (Figure 1B). Similarly, MicN also blocked the association between SIRT6, HP1-α and histone H3, but not its interaction with RELA (Supplementary Figure S4). Interestingly, in contrast to SIRT6, knockdown of MeCP2 by siRNA had no effect on RELA-regulated genes (Supplementary Figure S5). These findings support the above conclusion that the association between SIRT6 and MeCP2 must be further examined. Together, these findings show that SIRT6 association with Histone H3 and other chromatin-associated proteins is DNA dependent.
Figure 1.
SIRT6 interacts with multiple chromatin-related proteins in a DNA-dependent manner. (A) SIRT6-Flag overexpressed in HEK 293T cells and co-immunoprecipitated (IP) with anti-Flag beads. Associated proteins were separated on SDS PAGE, silver stained and identified by mass spectrometry. HEK 293T cells overexpressing empty Flag plasmid were used as control. (B) Disruption of DNA-dependent interactions by EtBr. (C) In contrast to recombinant bacterial expressed SIRT1-Flag, recombinant bacterial expressed SIRT6-Flag did not co-immunoprecipitate with any of the histone octamers, H2A, H2B, H3 and H4, in vitro.
To verify that the association between SIRT6 and the core histones is DNA dependent, in vitro interaction assays were carried out (the various recombinant proteins are shown in Supplementary Figure S6). As shown in Figure 1C, SIRT6 did not associate with any of the core histones. In contrast, SIRT1, a known histone deacetylase in vitro (21), associated with all of the core histones.
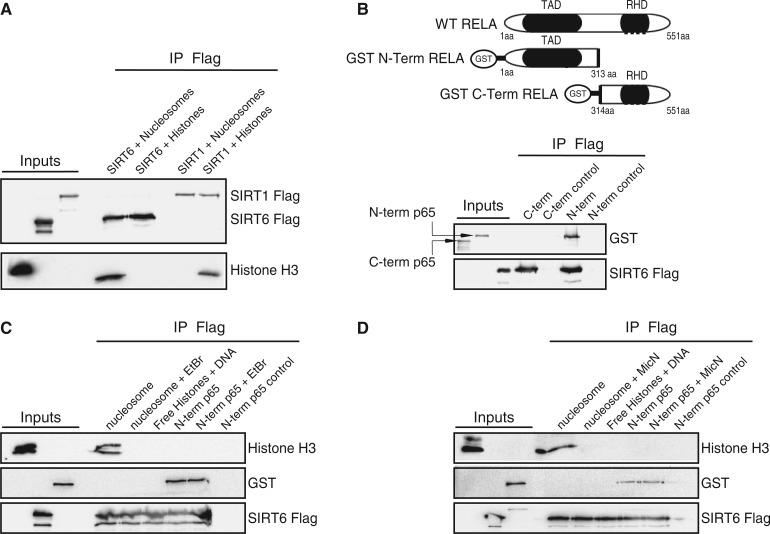
The requirement for DNA to enable the association between SIRT6 and its known substrate, histone H3, suggests that in vivo the histone is arranged in a structure facilitating this association. To further explore the structural requirements of the histone substrate, histone octamers were packaged into poly-nucleosomes. Interestingly, once packed as part of the nucleosome complex, histone H3 and SIRT6 were found to associate (Figure 2A). However, this association disappeared in the presence of EtBr or MicN (Figures 2C and D). MicN was used at high concentration to ensure the DNA degradation inside the nucleosome and its dissociation into free histones. N-terminus RELA (a.a 1–313) was used as a positive control. As seen in Figure 2B, only the N-terminal portion of RELA was associated with SIRT6, and this association was DNA independent (Figure 2C and D). To confirm that DNA does not cause SIRT6 to interact with free histones, free DNA was added to isolated histones. Under these conditions, SIRT6 failed to bind histone H3 (Figure 2C and D). Thus, these results show that the nucleosome complex is essential for the association between SIRT6 and histones, whereas SIRT1 associates with free histones.
Figure 2.
SIRT6 interacts with core histones in a nucleosome-dependent manner. (A) Co-immunoprecipitation of recombinant bacterial expressed SIRT6-Flag or SIRT1-Flag and Histone H3. Histone H3 was detected as part of histone octamers or in the nucleosome particles. (B) Recombinant SIRT6-Flag co-immunoprecipitated with recombinant truncated amino terminus (a.a 1–313) of RELA but not with RELA carboxyl terminus (a.a. 334–551). Schematic representation of each RELA construct is shown. (C) The effect of EtBr on SIRT6-Flag interactions. (D) The effect of MicN on SIRT6-Flag interactions.
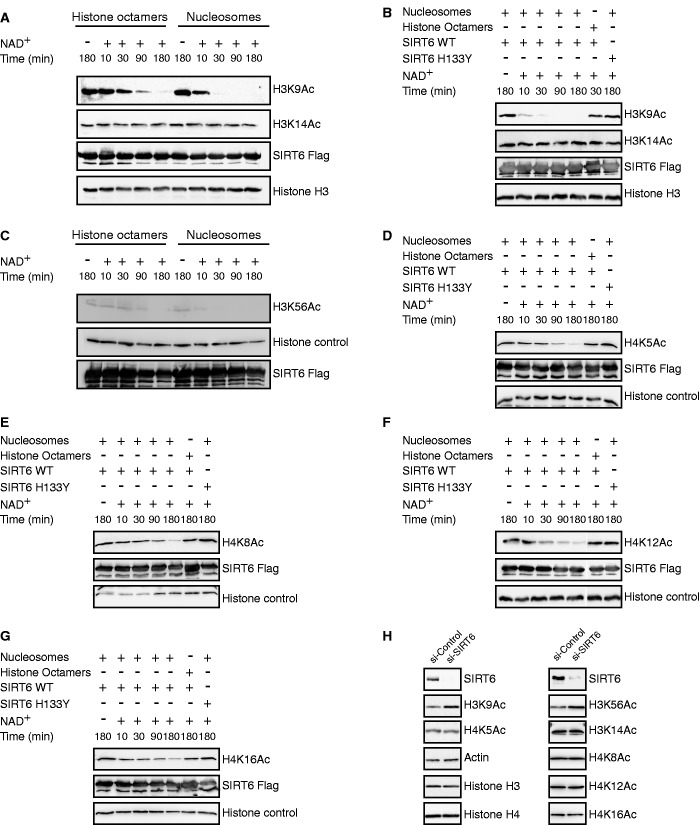
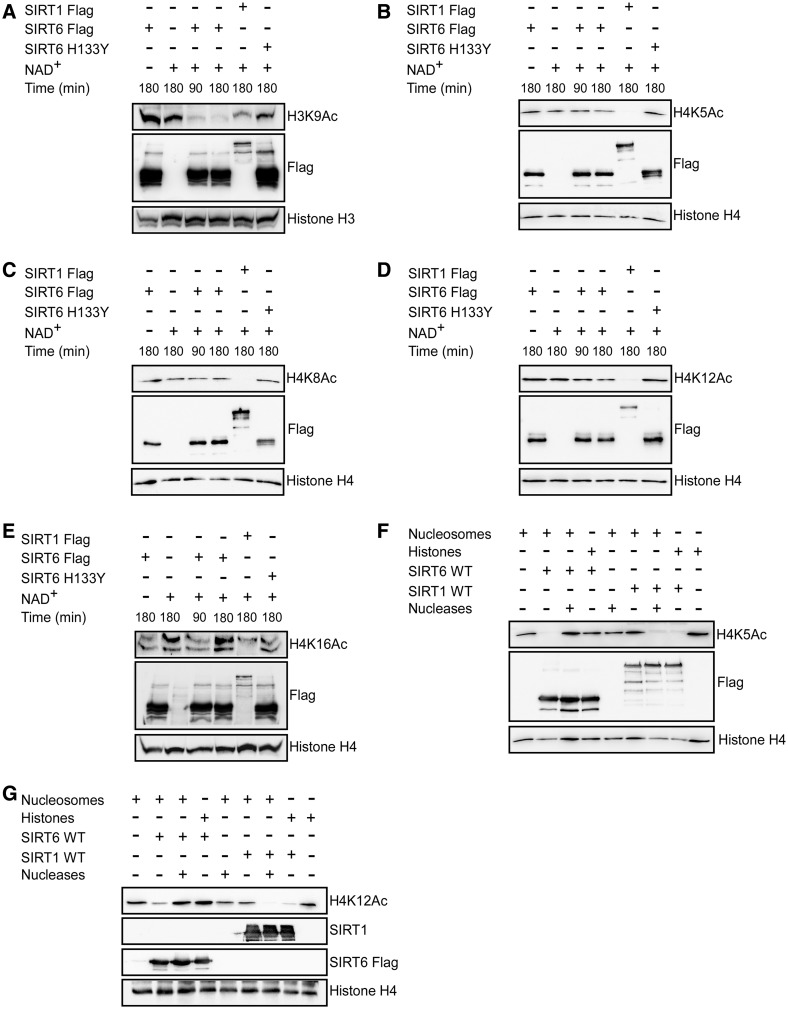
Next, we examined whether the interaction between SIRT6 and the nucleosome affects SIRT6 enzymatic activity. Deacetylation of H3K9Ac by SIRT6 was compared when the acetylated histone was present as free protein, or packaged as nucleosomes. As seen in Figure 3A–C, when bound to the nucleosome, SIRT6 deacetylated H3K9Ac and H3K56Ac at much higher efficiency. Nicotinamide, a known inhibitor of sirtuins, inhibited SIRT6 histone deacetylase activity on the nucleosome, but at much higher concentrations than previously reported for SIRT1 or yeast Sir2 (Supplementary Figure S7). Inactive SIRT6 carrying the H133Y mutation showed no H3K9Ac deacetylase activity (Figure 3B).
Figure 3.
Nucleosomes enhance SIRT6 deacetylase activity. (A) Time course analysis of a recombinant bacterial–expressed SIRT6-Flag deacetylation of H3K9Ac and H3K14Ac as part of histone octamers (left) or nucleosomes (right). (B) Kinetics of SIRT6-Flag deacetylation of H3K9Ac. (C–G) Kinetics of SIRT6-Flag deacetylation of H3K56Ac, H4K5Ac, H4K8Ac, H4K12Ac and H4K16Ac. (H) Global acetylation of H3K9, H3K14, H3K56, H4K5, H4K8, H4K12 and H4K16 in HeLa cells following SIRT6 knockdown.
The enhanced SIRT6 enzymatic activity on nucleosome targets prompted us to examine whether SIRT6 can deacetylate novel sites on histone H3 when bound to the nucleosome. As seen in Figure 3, SIRT6 failed to deacetylate H3K14Ac (Figure 3A and B) but surprisingly, showed H4K5Ac, H4K8Ac, H4K12Ac and H4K16Ac deacetylase activity (Figure 3D–G). Interestingly, knockdown of SIRT6 by siRNA in HeLa cells, resulted in a small increase in acetylated H3K9Ac and H3K56Ac, while other acetylation sites were unchanged (Figure 3H). This suggests that deacetylation of the novel SIRT6 substrates in vivo need to be verified.
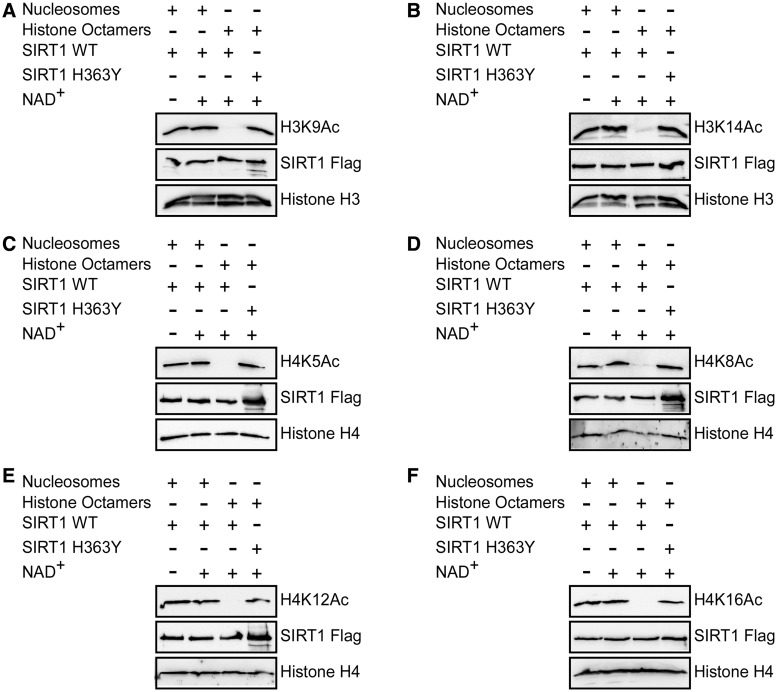
Given that the association between SIRT6 and nucleosome significantly improved its enzymatic activity, we next determined if SIRT1 enzymatic activity improved on its binding to free core histones. Surprisingly, SIRT1 deacetylated histone H3 on K9Ac and K14Ac and histone H4 on K5Ac, K8Ac and K12Ac, when present as free core histones but not as part of the nucleosome (Figure 4A–E). Importantly, these properties of SIRT1 were maintained even for its canonical substrate, H4K16Ac (Figure 4F). However, recombinant SIRT1 deacetylated H3K9Ac, H4K5Ac, H4K8Ac, H4K12Ac and H4K16Ac isolated from HEK 293T cells (Figure 5A–E). This deacetylation activity was specifically mediated by SIRT1, as the acetylation status of these sites was not changed in the presence of cell extract and SIRT6 without NAD+ or cell extract with NAD+ without additional enzymes even after 180 min (Figure 5A–E the first two left lanes of each). Thus, SIRT1 requires an as yet unidentified mediator or modification to deacetylate histones on the nucleosome. SIRT6 efficiently deacetylates H3K9Ac (Figure 5A). However, in comparison to reconstituted nucleosomes (Figure 3D–F), SIRT6 shows only weak deacetylase activity on H4K5Ac, H4K8Ac and H4K12Ac (Figure 5B–E), suggesting that other proteins interfere with its activity.
Figure 4.
SIRT1 shows no catalytic activity on nucleosomes. (A–F) SIRT1-Flag deacetylation assay of H3K9Ac, H3K14Ac, H4K5Ac, H4K8Ac, H4K12Ac and H4K16Ac on histone octamers or nucleosomes.
Figure 5.
SIRT6 and SIRT1 deacetylate histone substrates in total cell extract. (A–E) Recombinant SIRT1-Flag and SIRT6-Flag deacetylation of H3K9Ac, H4K5Ac, H4K8Ac, H4K12Ac and H4K16Ac, when added to total cell proteins extracted from HEK 293T cells. The amount of NAD+ in the cell extract was negligible (3–5 µM). (F–G) Deacetylation of H4K5Ac and H4K12Ac by SIRT1-Flag and SIRT6-Flag after the disruption of the nucleosomes into free histones by nuclease cocktail.
To further demonstrate that SIRT6 activity is nucleosome dependent, a nuclease cocktail was used to dissociate the packaged nucleosomes into free histones. Indeed, in support of our previous observations, after addition of nucleases SIRT6 displayed no deacetylase enzymatic activity on histone H4K5Ac and H4K12Ac (Figure 5F–G). Conversely, supplementation of nucleases into the reaction restored SIRT1 histone H4K5Ac and H4K12Ac deacetylation activity (Figure 5F–G).
DISCUSSION
SIRT6 possesses significant deacetylase activity in vivo. However, the in vitro enzymatic activity of SIRT6 is poor. Here, we show that the association between SIRT6 and free histones was undetectable under the conditions used. Similarly, SIRT6 deacetylates H3K9Ac or other sites on histone octamers at low efficiency. However, by using semi-quantitative methods, we found that SIRT6 significantly binds to and deacetylates histone H3 and H4 when these are packaged as nucleosomes. SIRT6 shows the strongest deacetylase activity on histone H3K9. In contrast to SIRT6, SIRT1 shows highly efficient deacetylase activity on free histones, rather than on the nucleosome. Yet, addition of cell extract significantly promotes the deacetylase activity of SIRT1 on nucleosomes. Thus, our results demonstrate that SIRT6 activity is nucleosome dependent, and suggest that SIRT6 binding to the nucleosome converts it into an active conformation.
Recently, it was shown that SIRT6 possesses much higher catalytic activity for deacylation of long-chain fatty acyl groups than deacetylation activity on histone peptides (22). Our findings suggest that the nucleosome association promotes the deacetylase activity of SIRT6. Thus, there is no requirement for additional modifications or the presence of another protein for the nucleosomal effect on SIRT6. We therefore suggest that once bound to the nucleosome, SIRT6 and the nucleosome undergo a conformation change enabling SIRT6 enzymatic activity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank the members of the Cohen lab, in particular Shoshana Naiman, for their helpful comments on the manuscript.
R.G, S.B. and H.Y.C designed the experiments; R.G and S.B. equally conducted the studies; R.G, S.B., Y.K. and H.Y.C. analyzed the data and H.Y.C. wrote the manuscript.
FUNDING
Israel Science Foundation, I-Core foundation, Israeli Ministry of Health and the ERC: European Research Council. Funding for open access charge: ERC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang B, Zwaans BM, Eckersdorff M, Lombard DB. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle. 2009;8:2662–2663. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, Chua KF. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8:2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329:1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanfi Y, Shalman R, Peshti V, Pilosof SN, Gozlan YM, Pearson KJ, Lerrer B, Moazed D, Marine JC, de Cabo R, et al. Regulation of SIRT6 protein levels by nutrient availability. FEBS Lett. 2008;582:543–548. doi: 10.1016/j.febslet.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanfi Y, Peshti V, Gil R, Naiman S, Nahum L, Levin E, Kronfeld-Schor N, Cohen HY. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell. 2009;9:162–732. doi: 10.1111/j.1474-9726.2009.00544.x. [DOI] [PubMed] [Google Scholar]
- 8.Schwer B, Schumacher B, Lombard DB, Xiao C, Kurtev MV, Gao J, Schneider JI, Chai H, Bronson RT, Tsai LH, et al. Neural sirtuin 6 (Sirt6) ablation attenuates somatic growth and causes obesity. Proc. Natl Acad. Sci. USA. 2010;107:21790–21794. doi: 10.1073/pnas.1016306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Bar-Joseph Z, Cohen HY. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 10.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 11.Zhong L, D'Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via HIF1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sebastian C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151:1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tennen RI, Berber E, Chua KF. Functional dissection of SIRT6: identification of domains that regulate histone deacetylase activity and chromatin localization. Mech. Ageing Dev. 2010;131:185–192. doi: 10.1016/j.mad.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tennen RI, Bua DJ, Wright WE, Chua KF. SIRT6 is required for maintenance of telomere position effect in human cells. Nat. Commun. 2011;2:433. doi: 10.1038/ncomms1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCord RA, Michishita E, Hong T, Berber E, Boxer LD, Kusumoto R, Guan S, Shi X, Gozani O, Burlingame AL, et al. SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging (Albany NY) 2009;1:109–121. doi: 10.18632/aging.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan PW, Feldman JL, Devries MK, Dong A, Edwards AM, Denu JM. Structure and biochemical functions of SIRT6. J. Biol. Chem. 2011;286:14575–14587. doi: 10.1074/jbc.M111.218990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimley R, Polyakova O, Vamathevan J, McKenary J, Hayes B, Patel C, Smith J, Bridges A, Fosberr A, Bhardwaja A, et al. Over Expression of Wild Type or a Catalytically Dead Mutant of SIRTUIN 6 Does Not Influence NFkappaB Responses. PLoS One. 2012;7:e39847. doi: 10.1371/journal.pone.0039847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 20.Lee KM, Narlikar G. Assembly of nucleosomal templates by salt dialysis. Curr Protoc Mol Biol. 2001;21:mb2106s54. doi: 10.1002/0471142727.mb2106s54. [DOI] [PubMed] [Google Scholar]
- 21.Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 22.Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, et al. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496:110–113. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.