Abstract
Synthetic acid tolerance, especially during active cell growth, is a desirable phenotype for many biotechnological applications. Natively, acid resistance in Escherichia coli is largely a stationary-phase phenotype attributable to mechanisms mostly under the control of the stationary-phase sigma factor RpoS. We show that simultaneous overexpression of noncoding small RNAs (sRNAs), DsrA, RprA and ArcZ, which are translational RpoS activators, increased acid tolerance (based on a low-pH survival assay) supra-additively up to 8500-fold during active cell growth, and provided protection against carboxylic acid and oxidative stress. Overexpression of rpoS without its regulatory 5′-UTR resulted in inferior acid tolerance. The supra-additive effect of overexpressing the three sRNAs results from the impact their expression has on RpoS-protein levels, and the beneficial perturbation of the interconnected RpoS and H-NS networks, thus leading to superior tolerance during active growth. Unlike the overexpression of proteins, overexpression of sRNAs imposes hardly any metabolic burden on cells, and constitutes a more effective strain engineering strategy.
INTRODUCTION
Strains tolerant to acid, oxidative and general stress from toxic chemicals are important in bioprocessing and bioremediation applications, where the robustness and prolonged productivity of the cells under stressful bioprocessing conditions are critically important (1–4). Acid resistance (AR) in Escherichia coli and other pathogens are also of physiological importance to human health. Orally ingested microbes pass through the acidic stomach environment, which typically kills most of them (5), but acid-resistant pathogens can survive this natural defense barrier and cause serious infections (6). Toxicity of carboxylic acids is not only due to high H+ concentrations in the medium, but also to anion-specific inhibition in a pKa- and pH-dependent fashion (4,7–9).
The AR systems of E. coli have been extensively studied and can be classified as amino acid dependent or independent (10). They include the glutamate-dependent AR2 (11), the arginine-dependent AR3 (11) and the lysine-dependent AR4 system (12). Their general mechanism is based on consumption of protons during the decarboxylation of the respective amino acid, followed by an antiport of the decarboxylation product in exchange for an extracellular amino acid (11,12). The amino acid independent AR1, also known as the glucose-repressed or oxidative AR system (6,10), depends on the stationary-phase sigma factor RpoS (10,13), engages the FoF1 ATPase to extrude protons out of the cell under ATP consumption (14), but remains less characterized than AR2-AR4. RpoS is involved in the regulation of AR2 by regulating GadX (10), which also acts as a global regulator of the acid fitness island (AFI), a chromosomal locus coding for several AR-associated genes in E. coli (15). The complex regulation of AR2 is orchestrated by GadE, which is under the control of three regulatory circuits, the signal transduction system EvgA/EvgS, the TrmE circuit as well as the GadXW circuit (10).
Another global regulator, the nucleoid associated protein H-NS, was first shown to regulate AR2 by inhibiting gadA and gadX transcription (16), but more recently, its global influence on AR has been firmly demonstrated (17). H-NS is now seen as a top-level regulator of AR: it enhances degradation of rpoS mRNA (18), and represses the expression of specific regulators of the amino acid–dependent AR systems [GadX, AdiY and CadC (16,17)], elements of the GadE regulatory circuits [EvgA, YdeO, GadX, RcsB (17,19,20)] and genes found on AFI [hdeABD (21)].
Acid tolerance response (ATR) (14,22) describes an inducible AR providing lower protection than the stationary-phase AR systems, but which is also active in exponentially growing cells (14,22). ATR is induced by mild acid treatment (pH between 4.7 and 5.5) (22), which induces acid-shock proteins (23) as well as upregulates AR and AFI genes (24–26).
Besides its role in AR1 and AR2, RpoS is the general stress sigma factor in E. coli regulating the response to several stresses, including acid stress, starvation, hyperosmolarity and suboptimal temperature (27,28). RpoS expression is also induced by carboxylic acids (29). RpoS is heavily regulated at all levels, but predominantly at the translational and posttranslational levels (27), and is reported to directly or indirectly influence the expression of >500 genes (13). Thus, for engineering tolerant phenotypes, RpoS is an ideal target, but the complex regulation of RpoS (27) poses many challenges, and thus little has been so far reported in terms of synthetic strategies involving RpoS for developing desirable strains.
By screening coexisting genomic libraries, we have recently (30) identified the noncoding small RNA (sRNA) ArcZ as imparting acid tolerance in actively growing E. coli cells. ArcZ activates RpoS translation by binding to the 5′-untranslated region (5′-UTR) region of the rpoS mRNA to free up the ribosomal binding site, a well-established mechanism by which two other sRNAs, DsrA and RprA, also activate RpoS translation (31–33). DsrA has been reported to be also involved in regulating H-NS by increasing hns mRNA turnover (34). Thus, we wanted to examine if the overexpression of these three RpoS translational activators could be used to alter RpoS expression aiming to engineer acid-tolerant, actively growing E. coli cells. Acid tolerance during active growth is most desirable as most acids are produced during active growth in industrial fermentations (4), when RpoS protein levels are reported to be low (27). Our hypothesis is that overexpression of these three sRNAs will lead to increased RpoS protein synthesis during active growth, so that these cells can mount an effective protective response to acid stress. We show that simultaneous overexpression of all three sRNAs leads to dramatically enhanced acid tolerance during active cell growth and in a supra-additive fashion, and we explore the potential mechanisms that underlie this strong protective phenotype.
MATERIALS AND METHODS
Strains and media
Strains and plasmid used in this study are listed in Supplementary Table S1. Cultures were grown in lysogeny broth (LB) broth or M9 media at 37°C under shaking at 250 rpm with ampicillin (100 µg/ml) or spectinomycin (100 µg/ml). M9 media were supplemented with 1.5 mM glutamate as indicated.
Construction of sRNA overexpression and related plasmids
Noncoding sRNAs, DsrA, RprA and ArcZ were directly amplified from wild-type E. coli MG1655 via colony polymerase chain reaction (PCR) and cloned via TOPO-TA cloning into pCR®8/GW/TOPO vector (Invitrogen, Carlsbad, CA, USA). Small RNAs were amplified as complete transcription units (TUs) including their native promoter and terminator. Combinations of two or three sRNA TUs were constructed via overlap extension PCR (35). The DNA coding for two sRNAs to be combined was first amplified with primers containing a homologous sequence of the other sRNA as an overhang (bold part of the primers listed in Supplementary Table S2). This homologous sequence was used to assemble the DNA for the two individual sRNA TUs, as one PCR product in a second PCR. For example, to assemble the DNA for the DsrA and ArcZ TUs into a single PCR product, the individual sRNA TUs were amplified with primers dsrA-for and DA-dsrA-rev as well as DA-arcZ-for and arcZ-rev in the first PCR, and then both products were added as template (10 ng each) and amplified with the dsrA-for and arcZ-rev primer pair in a second PCR. Supplementary Table S3 lists the primer combinations used in each PCR to construct all sRNA overexpression vectors. For the ternary combination, the DsrA-RprA PCR product was combined with the ArcZ PCR product as described above. The assembled PCR products containing multiple sRNA TUs were cloned into pCR®8/GW/TOPO. These entry vectors were then shuffled via in vitro recombination using the Gateway® system (Invitrogen, Carlsbad, CA, USA) into the commercial destination vector pDEST™14 yielding the final sRNA overexpression plasmids listed in Supplementary Table S1. As a control plasmid, the control entry plasmid pENTR™-gus was recombined with pDEST™14 to generate pCTRL.
Another plasmid was constructed by cloning the rpoS gene without its 5′-UTR under the control of the lac promoter. Escherichia coli MG1655 gDNA was used to amplify rpoS (with rpoS-for and rpoS-rev), digested with SphI and SbfI and cloned into the MCS of pUC19 to generate pUC-RpoS. From this plasmid, rpoS together with the lac promoter was amplified (pUC19for and pUC19rev) and cloned into pCR®8/GW/TOPO. Finally, the rpoS insert was shuffled into pDEST™14 yielding pRpoS. Transcription of rpoS was induced with isopropyl β-d-1-thiogalactopyranoside (IPTG). Because the 5′-UTR was not amplified, the generated rpoS mRNA from this plasmid cannot form a stem loop, and thus translation can occur from the unobstructed ribosomal binding site.
Control plasmids carrying multiple copies of DsrA were constructed by cloning additional TUs of DsrA into plasmid pDsrA. First, a DsrA TU (as used before to construct pDsrA) was inserted via blunt-end ligation in the BsaAI site of pDsrA. The resulting plasmid was designated as pDD and contained two copies of the DsrA TU. Next, the pDD plasmid was digested with PvuII and another DsrA TU was inserted via blunt end ligation. The resulting plasmid, now carrying three DsrA TUs, was designated as pDDD. Phusion or Taq Polymerase (NEB, Ipswich, MA, USA) was used for the PCRs.
Isolation of total and low molecular mass RNA
Total RNA was isolated from cell pellets frozen at −85°C via the RNeasy Mini Kit (Qiagen, Hilden, Germany). For sRNA isolation, total RNA was first isolated via the miRNeasy Mini Kit (Qiagen). From this isolation, low molecular mass (LMM) RNA was isolated by sequential RNA precipitation. One hundred fifty micrograms of total RNA was mixed with 0.5 M sodium chloride (NaCl) and 50% PEG 8000 in a final volume of 250 µl. After 30 min incubation on ice, the high molecular mass RNA was collected by centrifugation at >13 000 rpm for 15 min at 4°C. The supernatant, containing the LMM RNA, was mixed with 750 µl 100% ethanol, incubated overnight at −20°C and collected by centrifugation at >13 000 rpm for 30 min. The pellet of the LMM RNA was washed with ice-cold 75% ethanol and dissolved in 30 µl RNase-free H2O, from which 10 µl (50 µg equivalent of total RNA) or 5 µl (25 µg equivalent of total RNA) were separated via gel electrophoresis for northern blots.
Northern-blot analysis
LMM RNA was separated via gel electrophoresis on a 5% Ready Gel® TBE-Urea gel (BioRad, Hercules, CA, USA). Gels were prerun at 170–180 V for 20 min. An equivalent volume of LMM RNA of 50 or 25 µg total RNA was loaded into each well and separated at 170–180 V. As a marker, 2 µg of the low Range ssRNA ladder (NEB) was used. After electrophoresis, gels were washed with DEPC water, stained with ethidium bromide and visualized to ensure separation (separation of the ssRNA ladder) and equal loading (5s RNA band). After destaining the gels in 0.6× Tris/Borate/EDTA buffer (TBE) for 10 min, the gels were electro-blotted (400 mA) onto a BrightStar®-Plus positively charged nylon membrane (Ambion, Foster City, CA, USA) in cold 0.6× TBE at 4°C for 90 min. After blotting, membranes were baked at 80°C for 2 h before prehybridization with ULTRAhyb®-Oligo hybridization buffer (Ambion) at 42°C for 2 h.
Probes for sRNA detection were generated as single-stranded γ-32P end-labeled oligo (ssOligo) probes. ssOligo (10 pmol) was end-labeled with γ-32P (7000 Ci/mmol; MP Biomedicals, Solon, OH, USA) using the OptiKinase™ Kit (USB, Cleveland, OH, USA). Nonincorporated radionucleotides were removed using Micro Bio-Spin 30 columns (BioRad) and cleaned up probes were used to hybridize the membranes for 16–20 h at 42°C. Membranes were washed twice in 2× SSC, 0.1% sodium dodecyl sulphate and 0.1× saline-sodium citrate buffer (SSC), 0.1% sodium dodecyl sulphate for 15 min before exposing the membranes to a phosphor screen. Screens were visualized on a phosphorimager (GE Healthcare, Piscataway, NJ, USA).
Western-blot analysis
Crude cell extracts were prepared and western blots performed as described (36). Briefly, 10–50 µg of total protein was separated via gel electrophoresis on a Ready Gel® 12% Tris–HCl gel (BioRad). Separation was monitored with the Kaleidoscope™ standard (BioRad). After blotting, the nitrocellulose membranes were first blocked with 2% milk in TBST (20 mM TrisߝHCl pH 7.4, 137 mM NaCl and 0.2% Tween-20) buffer overnight. Primary antibody against RpoS (Neoclone, Madison, WI, USA) was diluted 1:5000 to 1:10 000 and hybridized for 1 h at room temperature. Secondary anti-mouse antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) was diluted 1:3000 and incubated for 1 h at room temperature.
Quantitative reverse transcription PCR
Two micrograms of total RNA was reverse transcribed via the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). Quantitative reverse transcription PCR was performed with SYBR® Green PCR Master Mix (Applied Biosystems) as described (37). Briefly, a 25-µl reaction mix containing 20 ng of cDNA, 1 µM of each primer (see Supplementary Table S2), 12.5 µl of SYBER® Green Master Mix and sterile water was analyzed on a iCycler iQ5 Multicolor Real-Time PCR Detection System (BioRad). The E. coli hcaT gene was used as a housekeeping gene to normalize cycle threshold (CT) values. Differences in relative expression levels were calculated with the 2^-(ΔΔCT) method (38).
Low-pH tolerance assays
Tolerance to low-pH was tested as described (30). Briefly, 50 ml Falcon tubes filled with 10 ml LB (or M9, when specified) medium with antibiotics were inoculated with 2% overnight cultures and grown with open caps to ensure adequate oxygen transfer. Growth was monitored via optical density measurements at 600 nm (OD600). Cultures were sampled at different growth phases (based on OD and time), diluted 1:10 in medium containing the stressor (high H+, carboxylic acids or H2O2; see below) and exposed for 1 h. Serial dilutions were plated before and after exposure to the stressful medium and survival rates calculated as survival (%) = colony forming units (CFU) (post stress)/CFU(prior stress) × 100. The following stress conditions were tested: LB containing 5 g/l acetate (∼pH 4.0), 5 g/l butyrate (∼pH 4.2), 4 g/l lactate (∼pH 3.8) or 4 mM H2O2 or M9 media adjusted to pH 2.5 with HCl. Glutamate was added (where indicated) to a concentration of 1.5 mM.
Statistical analysis
After testing for equal variances of the data, either a homo- (equal variance) or heteroscedastic (nonequal variance) one-sided two-sample t-test was performed to test for significance. Analyses were performed with Minitab16 and MS Excel.
RESULTS
Supra-additive effects of DsrA, ArcZ and RprA overexpression on acid tolerance during active cell growth
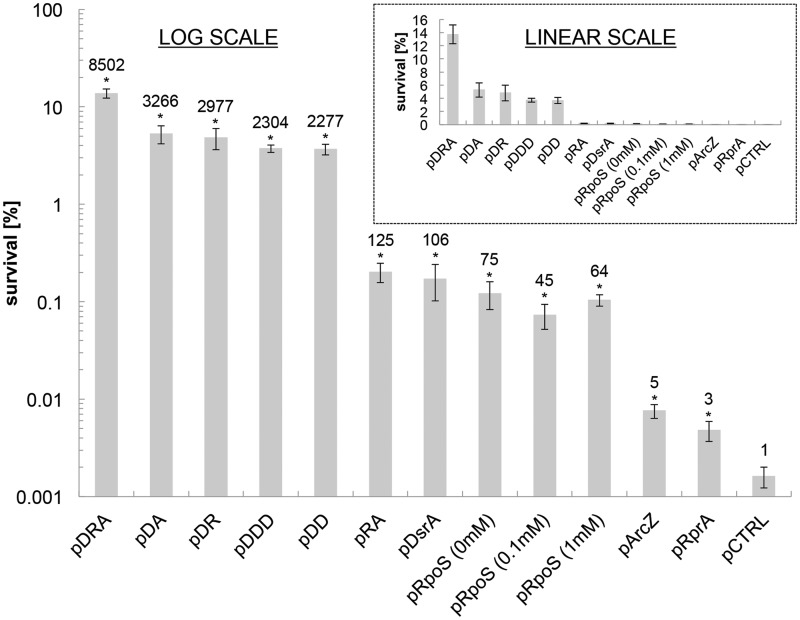
Actively growing cells (OD600 = 1 in LB medium) of E. coli strains expressing ArcZ, RprA and DsrA individually from plasmids off their native promoters were tested for survival in acidified (pH 2.5) minimal M9 media with no amino acid supplementation. AR based on this survival assay in actively growing cells was increased for all three strains compared with control (Figure 1). The highest survival was observed for DsrA overexpression (from plasmid pDsrA), followed by ArcZ overexpression (from pArcZ) and RprA overexpression (from pRprA). In view of our previous finding that combined overexpression of ArcZ and recA has a supra-additive effect on acid tolerance (30), we investigated if the combinatorial overexpression of the three sRNAs would further increase acid tolerance. We constructed the following overexpression strains: E. coli MG1655(pDR) (combined expression of DsrA and RprA), MG1655(pDA) (combined expression of DsrA and ArcZ), MG1655(pRA) (combined expression of RprA and ArcZ) as well as MG1655(pDRA) (combined expression of the triple combination of DsrA, RprA and ArcZ). Both the binary and triple combination (Figure 1) increased acid tolerance during active cell growth in a supra-additive fashion. The highest AR was observed for the triple combination (Figure 1), which displayed an 8500-fold higher survival to low pH compared with the plasmid-control strain. Overexpression of a single sRNA, namely of the most effective sRNA, DsrA, from plasmids carrying two copies [strain MG1655(pDD)] or three copies [strain MG1655(pDDD)] of the DsrA, did not elicit as strong a pH-tolerant phenotype as the triple sRNA combination, namely strain MG1655(pDRA) (Figure 1). Thus, the phenotypic behavior observed from MG1655(pDRA) is not the result of further saturation of the individual sRNA regulatory function, but is rather due to the supra-additive interaction of the three sRNAs.
Figure 1.
Survival of sRNA overexpression strains in exponential growth phase (OD600 = 1) after 1 h exposure to acidified (pH 2.5) M9 media. The fold improvement of each strain compared with the plasmid control strain [MG1655(pCTRL)] is shown above each survival bar. Plasmid inserts are as follows: pDRA = DsrA, RprA and ArcZ, pDA = DsrA and ArcZ, pDR = DsrA and RprA, pDDD = DsrA, DsrA and DsrA, pDD = DsrA and DsrA, pRA = RprA and ArcZ, pDsrA = DsrA, pRpoS = RpoS, pArcZ = ArcZ and pRprA = RprA). pRpoS was investigated at three different induction levels (0, 0.1 and 1 mM IPTG). Error bars indicate the standard error of at least three biological replicates, and statistical significance was tested with a two-sample t-test (*P < 0.05). The insert shows survival in a linear scale.
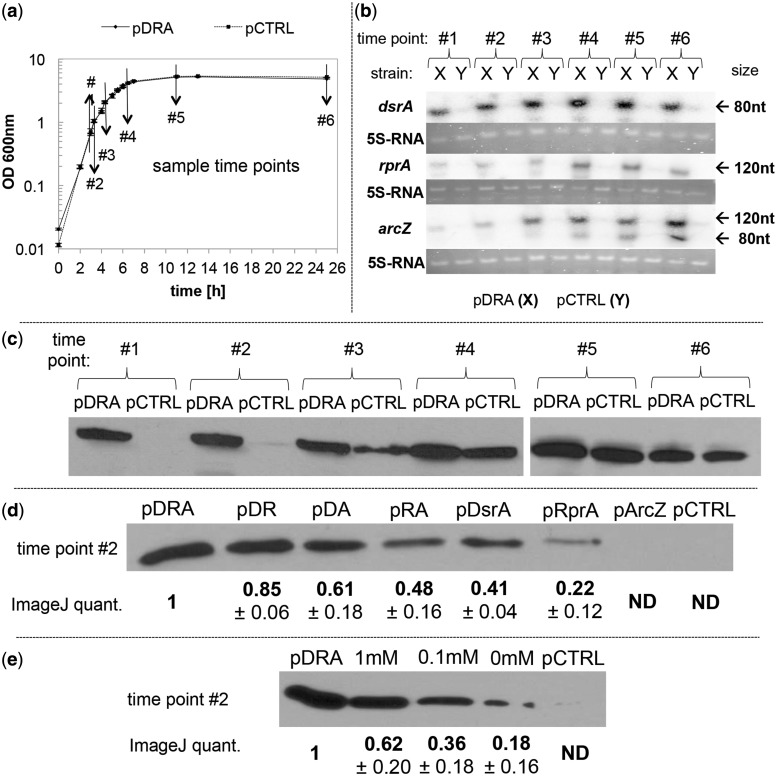
The three sRNAs on all expression plasmids were cloned as individual TUs under their native regulation. To ascertain their transcription as well as investigate their temporal transcription patterns, we used northern blot analysis (Figure 2a and b). Transcription of all three sRNAs was significantly higher in strain MG1655(pDRA) throughout the batch culture compared with the plasmid control strain MG1655(pCTRL). For both DsrA and RprA, maximal transcript levels were observed at the transition between the exponential and early stationary phase (time points 4 and 5) of culture, which is consistent with previously described native transcription (39,40). ArcZ displayed a continuously increasing expression with apparent maxima in stationary phase (time points 5 and 6) as in native transcription (33). The northern blot for ArcZ showed a second smaller-size transcript, which, as previously described, corresponds to a 55-bp truncated form (33). These data show the expected much higher (compared with plasmid control) levels of expression for all three sRNAs and that overexpression did not alter their native temporal patterns.
Figure 2.
Transcriptional analysis of the three sRNAs using northern blots, and western analysis of RpoS protein levels during batch growth. (a) Growth profile of MG1655(pDRA) and MG1655(pCTRL) indicating the sampling points for western and northern analyses. (b) Transcription profile of DsrA (87 nt transcript), RprA (106 nt transcript) and ArcZ (121- and ∼80-nt transcript) of MG1655(pDRA) (X) and MG1655(pCTRL) (Y). LMM RNA of 50 µg (DsrA and ArcZ) or 25 µg (RprA) equivalent of total RNA (see ‘Materials and Methods’ section) was loaded for each sample and probed with an oligonucleotide probe (see ‘Materials and Methods’ section). 5S-RNA and ssRNA ladder (NEB), visualized via ethidium bromide staining before blotting, were used as loading control and size marker. (c) Time course analysis of RpoS protein levels in MG1655(pDRA) and MG1655(pCTRL). In (c–e), strains are indicated by the plasmid they carry. Fifty micrograms of total cell protein were loaded for each sample and detected with monoclonal anti-RpoS antibody. (d) RpoS protein levels in sRNA overexpression strains MG1655(pDRA), MG1655(pDR), MG1655(pDA), MG1655(pRA), MG1655(pDsrA), MG1655(pRprA), MG1655(pArcZ) and the control strain MG1655(pCTRL). Ten micrograms of total protein (extracted from samples taken at time point #2) were loaded for each strain and detected with monoclonal anti-RpoS antibody. Three biological replicates were analyzed; one representative blot is shown. RpoS protein amount was quantified via ImageJ and the relative fold change ± standard error compared with RpoS levels of MG1655(pDRA) is shown below the band for each strain. (e) RpoS expression of MG1655(pRpoS) compared with MG1655(pDRA) and MG1655(pCTRL). RpoS expression was induced with IPTG at various concentrations. Samples were taken at time point #2, and 10 µg of total protein were loaded and detected with monoclonal anti-RpoS antibody. Quantitation as in (d).
If one assumes that the impact of these three sRNAs on acid tolerance is mediated by RpoS, would direct rpoS overexpression from an inducible promoter generate a comparably strong phenotype? To address this question, rpoS was cloned without its regulatory 5′-UTR portion (27) under control of the lac promoter (plac) to generate plasmid pRpoS, which was transformed also into MG1655, and rpoS expression was induced by varying the IPTG concentration. Survival to low pH of this strain was similar to what was observed by overexpressing DsrA alone, and, surprisingly, independent of induction level (Figure 1). Apparently, leaky expression from plac without any IPTG addition was sufficient to saturate the effect of direct rpoS expression from this construct. It has been reported that ‘overexpression of RpoS in exponential phase cells is not sufficient for full induction of many RpoS-dependent genes’ (27), and this would suggest that if the observed acid-tolerant phenotype is at least in part related to RpoS expression, then this plasmid-borne rpoS overexpression is not sufficient to engage the genes necessary for strong pH tolerance as in the MG1655(pDRA) strain. Western-blot data presented below corroborate this hypothesis.
Unlike the phenotype of known AR mechanisms, increased survival to low pH is observed during active growth, without amino acids and without acid stimulation
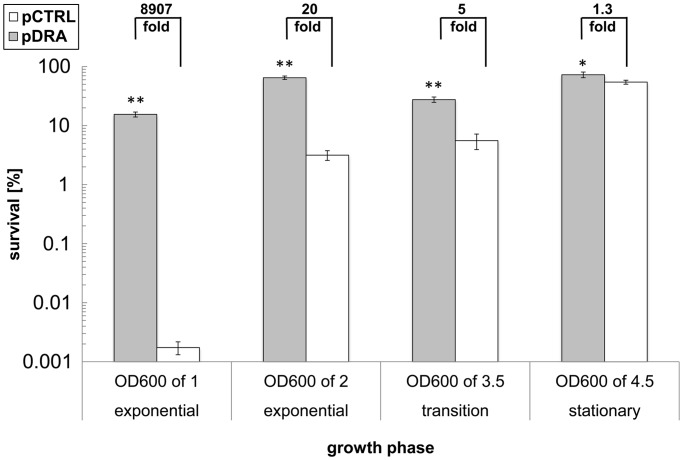
We have shown in Figure 1 that the three sRNAs impart acid tolerance during early to mid-exponential growth and without acid induction. We also wanted to investigate if their overexpression would also increase AR in post-exponential phases of culture. Thus, we carried out assays for survival to low pH on cells grown to an OD600 of 2 (late exponential to early transition phase), 7 h (early stationary phase) as well as 12 h (stationary phase) (Figure 3). Overexpressing the triple sRNA combination increased AR for all phases of culture, but the highest effect was observed for cells in exponential growth phase.
Figure 3.
Survival of MG1655(pDRA) (gray bars) and MG1655(pCTRL) (open bars) cells from different phases of culture in acidified pH-2.5 M9 media. Cells were sampled from the exponential growth phase (OD600 of 1 and 2), transition phase (OD600 of 3.5; 7 h after inoculation) and mid-stationary phase (OD600 of 4.5; 12 h after inoculation) of culture. Increased survival is shown as fold improvement above each set of comparisons. Error bars indicate the standard error of at least three biological replicates, and statistical significance was tested with a two-sample t-test (**P < 0.05, *P < 0.10).
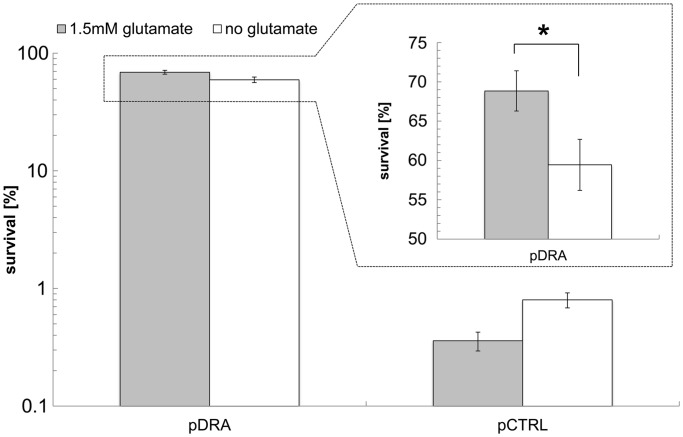
As discussed, to resist acid stress, E. coli engages four distinct mechanisms (AR1–AR4), which are largely associated with the stationary phase of culture (AR1–AR3) and/or are acid induced (AR1, AR4), and three of which (AR2–AR4) require the presence of amino acids, glutamate, arginine and lysine, respectively (10). In the experiments of Figure 1, cells were grown in LB media but acid-tolerance assays were carried in the defined medium M9 without the presence of any amino acids. Could there be a carryover of amino acids from the LB medium into the M9 medium, thus enhancing acid tolerance? Is it possible that the observed effects are glucose repressible like the AR1 system? To examine these questions, strains MG1655(pDRA) and MG1655(pCTRL) were grown in M9 media (22.2 mM glucose) for 7 h (exponential growth phase, OD600 between 0.3 and 1.5; different range than cultures in LB media) and we examined the acid tolerance of MG1655(pDRA) and MG1655(pCTRL) in the presence as well as absence of 1.5 mM glutamate, which is essential for the AR2 system with the highest observed protection against low pH (10). Survival of MG1655(pDRA) increased several orders of magnitude compared with the control-strain MG1655(pCTRL) and was largely independent of glutamate supplementation (Figure 4). For the control strain MG1655(pCTRL), glutamate addition offered no protection, indicating that, as would be expected, the glutamate-dependent AR2 mechanism is not engaged in these exponential phase cells. For the sRNA-overexpression strain MG1655(pDRA), 1.5 mM glutamate addition does provide a statistically significant increase in acid tolerance (Figure 4). However, the difference in survival observed in glutamate supplemented versus unsupplemented media is low compared with the overall increased survival of MG1655(pDRA) versus the MG1655(pCTRL) strain, which shows that the contribution of the glutamate-dependent mechanism is small. These data suggest that the observed AR is largely independent of glutamate and any amino acid supplementation because the used M9 media did not have any other amino acid supplementations.
Figure 4.
Survival of exponential-growth E. coli MG1655(pDRA) and MG1655 (pCTRL) cells grown on M9 media after 1 h exposure in acidified pH-2.5 M9 media. Cells were tested in M9 media supplemented either with 1.5 mM glutamate (gray bars) or without any glutamate (open bars). The difference in survival of E. coli MG1655(pDRA) in glutamate-supplemented versus nonsupplemented media is shown in the insert in a linear scale. Error bars indicate the standard error of at least three biological replicates, and statistical significance was tested with a two-sample t-test (*P < 0.05).
The absence of mild acid-stress pretreatment and the presence of glucose suggest that the engineered system does not display the requisite characteristics of AR1, AR4 or ATR. If one or more of these latter mechanisms may still be engaged in the strong acid-tolerant phenotype, then overexpression of the three sRNAs would appear to override the expected requirement for acid pretreatment and the suppressive effect of glucose.
RpoS protein expression levels are largely commensurate with observed acid tolerance levels but cannot alone explain the supra-additive effect of the triple sRNA combination on acid tolerance
Temporal western analysis of RpoS expression of E. coli MG1655(pDRA) and MG1655(pCTRL) was carried out to investigate if combinatorial overexpression of the three sRNAs alters the protein-expression profile of RpoS. In unmodified E. coli and in the absence of stress, RpoS protein expression is initiated during the transition phase and continues to high levels in stationary phase (27,28). This RpoS profile was observed for the control strain MG1655(pCTRL): low protein levels were detected in late exponential phase (time point #3) and continued to increase into the stationary phase (Figure 2c). In contrast, high levels of RpoS protein were observed for MG1655(pDRA) in early exponential phase (time point #1, OD600 ∼0.75) and maintained high through the stationary phase (Figure 2c). Thus, combinatorial overexpression of the three sRNAs leads not only to increased RpoS expression, but, significantly, it alters the temporal expression profile by activating RpoS expression during exponential growth.
Transcription of rpoS has not been reported to be influenced by the expression levels of these three sRNAs, and because all three of these sRNAs bind to the same part of the 5′-UTR of the rpoS mRNA, one would expect that simultaneous overexpression of the three sRNAs would not dramatically affect RpoS protein levels. Yet, it does and in a profound way (Figure 2d). This suggests that the combinatorial overexpression of the three sRNAs leads to elevated translation of rpoS mRNA. The highest RpoS amount was detected by overexpressing all three sRNAs [MG1655(pDRA)]. We quantitated the relative levels of RpoS protein by setting this observed maximum to 1 (Figure 2d). The two strains with the second and third highest tolerance (Figure 1), namely MG1655(pDR) and MG1655(pDA), also show the second and third highest RpoS levels. Strains MG1655(pRA) and MG1655(pDsrA), which have similar survival rates (Figure 1), exhibit similar RpoS protein levels (Figure 2d), which are about half of what was observed for MG1655(pDRA). Surprisingly for two strains displaying similar levels of acid tolerance (Figure 1), RpoS protein was detected for MG1655(pRprA) but not for MG1655(pArcZ) (Figure 2d). Moreover RpoS levels of MG1655(pRpoS) for different induction levels (Figure 2e) do not correlate with the observed phenotypic behavior (Figure 1). This could be an indication that the amount of RpoS in exponential phase cells must reach a certain threshold to effectively compete with the major sigma factor RpoD to activate the part of the RpoS regulon that is involved in acid tolerance. Despite these exceptions, these data suggest that there is an overall good qualitative correlation between high RpoS protein levels and increased acid tolerance. Yet, these data cannot explain the observed supra-additive effect of the three sRNAs on acid tolerance (Figure 1).
Combinatorial sRNA overexpression impacts rpoS and hns mRNA level as well as mRNA levels of core members of their respective regulons
Degradation of rpoS mRNA by endonuclease RNase E is blocked by DsrA and RprA binding, which leads to mRNA stabilization and increased rpoS mRNA levels (41). Thus, we wanted to investigate if the observed high RpoS protein levels are the result of increased mRNA levels in the cell and if so, whether the combinatorial overexpression of the three sRNAs enhances this effect. As expected, overexpression of the sRNAs increases the mRNA levels of rpoS (Figure 5a). Relative to MG1655(pCTRL),the highest mRNA levels were found in strain MG1655(pDsrA), and the lowest levels in the strain expressing the triple combination, but there was no statistically significant difference in rpoS mRNA levels between the three strains relative to MG1655(pRpoS) or among themselves (Figure 5a). Thus, overexpression of these sRNAs stabilizes rpoS mRNA, but the double or triple combinations do not improve rpoS mRNA stability if not the opposite. Therefore, the hypothesis that the increased RpoS protein levels are the result of increased rpoS mRNA levels due to combinatorial overexpression of the three sRNA is not valid. This suggests that other mechanisms must account for the observed increased RpoS-protein levels in the strains overexpressing the double and triple combinations of the three sRNAs. A possible mechanism and hypothesis is that the three sRNAs provide a protective effect on RpoS protein stability. Thus, chloramphenicol chase experiments were performed to investigate the degradation rates of RpoS in the presence of pDRA, pDR, pDsrA, pRpoS and pCTRL (Supplementary Data). Overexpression of the three sRNAs increased RpoS stability with the highest half-life being observed for the triple combination (pDRA) (Supplementary Figure S1). The tested hypothesis appears to be correct and thus explains the lack of correlation between mRNA and proteins levels for RpoS.
Figure 5.
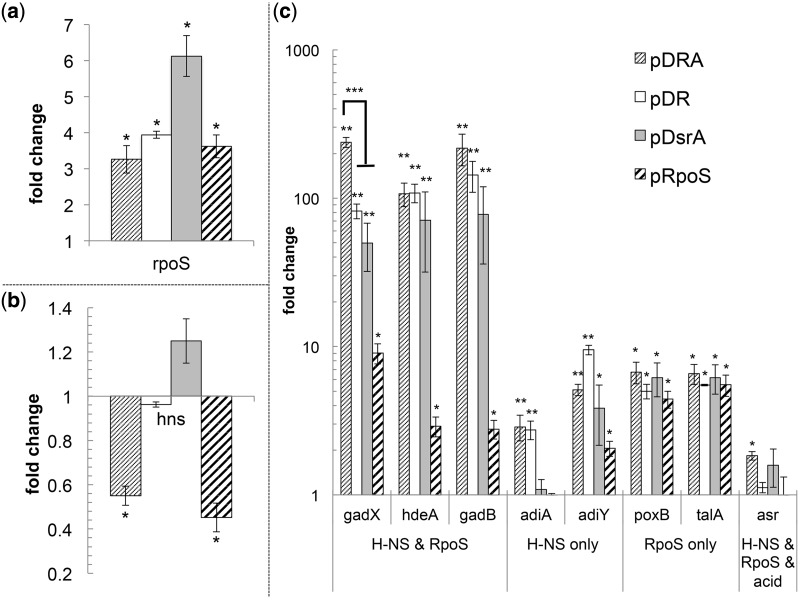
Transcriptional changes due to combinatorial sRNA overexpression. Relative gene expression (fold change) of rpoS (a); hns (b); and of genes regulated by RpoS and H-NS (gadX, hdeA and gadB), by H-NS (adiA and adiY), by RpoS (poxB and talA) as well as asr (its transcription is acid induced and regulated by RpoS and H-NS) (c) in various overexpression strains (carrying the plasmid indicated) in comparison with the control strain MG1655(pCTRL). Error bars indicate the standard error of at least three biological replicates, and statistical significance (P < 0.05) was tested with a two-sample t-test. Single asterisk indicates significant change in comparison with MG1655(pCTRL); double asterisks indicate significant change compared with both MG1655(pCTRL) and MG1655(pRpoS); triple asterisks indicate significant change of MG1655(pDRA) compared with all other strains.
In addition to the positive regulation of rpoS translation, these three sRNAs are reported to interact with other mRNAs in the cell, typically involving negative regulation. Of interest here is the interaction of DsrA with hns mRNA. H-NS is a global regulator involved in the transcriptional regulation of ∼250 genes (19). As discussed, H-NS inhibits RpoS translation and increases RpoS protein degradation (27). H-NS is also involved in AR (17,19): it represses the AR genes hdeAB (21), as well as gadA and gadX, which are part of the AR2 system (16). DsrA negatively regulates H-NS by increasing turnover of hns mRNA, but the observable mRNA levels are not affected as a result of a feedback loop whereby H-NS protein controls hns transcription (42). We found that hns mRNA levels were not significantly altered in strains MG1655(pDR) or MG1655(pDsrA), but were reduced to about half in the triple overexpression strain MG1655(pDRA) as well as in MG1655(pRpoS) in comparison with the control strain MG1655(pCTRL). So, by overexpressing all three sRNAs simultaneously, the hns mRNA turnover is increased significantly compared with the control strain MG1655(pCTRL), as well as the MG1655(pDsrA) and MG1655(pDR) strains. This suggests that in addition to DsrA, ArcZ and RprA are also involved in regulating hns mRNA levels, which, if confirmed, is a novel finding.
Do higher RpoS levels lead to increased transcription of genes in the RpoS regulon? Do the lower hns mRNA levels impact genes under H-NS control? To address these questions, we examined the transcription of a subset of genes associated with AR, which are either under the control of RpoS or H-NS alone or are coregulated by RpoS and H-NS. Specifically, we examined the transcription levels of poxB, talA [under RpoS control (13)], adiA, adiY [under H-NS control (17,43)] as well as hdeA, gadX and gadB [coregulated by RpoS and H-NS (10,17,44)]. As an additional control, we also examined the expression of ars, which, in addition to being controlled by RpoS and H-NS, requires acid induction for its expression (44).
As expected, genes under the control of RpoS only (poxB and talA) increased in all overexpression strains compared with the plasmid-control strain (Figure 5c). Genes under the control of H-NS only (adiA and adiY) showed increased expression in the sRNA overexpression strains MG1655(pDRA) and MG1655(pDR), but not in the RpoS expression strain or MG1655(pDsrA) (Figure 5c). Expression of asr was not altered significantly in any strain [except modestly for MG1655(pDRA)], most likely because we did not use acid induction. The genes under the control of both RpoS and H-NS (gadX, hdeA and gadB) showed dramatically increased transcription in all sRNA overexpression strains compared with the RpoS expression strain. Thus, sRNA overexpression activates transcription of AR genes under the joint RpoS and H-NS control stronger than observed for rpoS overexpression alone.
Transcription levels of gadX were highest in the triple sRNA expression strain, the strain with reduced hns mRNA levels, and the highest RpoS protein levels, in comparison with all other strains. GadX was originally discovered as a major transcriptional regulator of the AR2 system (45), but further studies revealed its global regulatory role in AR, as it controls the majority of genes found in the AFI (15). The triple sRNA overexpression led to the highest RpoS protein level, low hns mRNA levels, the induction of all investigated AR associated genes and most importantly to the highest expression of gadX, resulting in the highest acid survival observed (Figure 1). So, given that the observed increased acid tolerance imparted by the triple sRNA overexpression is largely amino acid independent, does not require acid induction, is growth associated and not glucose repressed, one would ascribe the protection mechanism as associated largely to GadX, HdeA and other genes whose function transcends the AR1-AR4 mechanisms and ATR.
Increased tolerance to carboxylic-acid stress by sRNA overexpression
Acid stress can derive from both inorganic and organic acids (46), the latter being of significance both in food safety (5,47) and bioprocessing (4). Carboxylic acids are high value products as they can be used as precursor in the chemical industry (4). Bioprocess-based productions of carboxylic acids such as butyrate, acetate, lactate and succinate requires strains that can tolerate high concentrations of the carboxylic acid of interest (4,48). Here, we examined if the high AR of strain MG1655(pDRA) would also result in resistance to three important and model toxic carboxylic acids: acetate, butyrate and lactate. Production of carboxylic acids is typically growth dependent (4). Thus, we tested exponential phase cells for survival to 5 g/l acetate (∼pH 4.0), 5 g/l butyrate (∼pH 4.2) and 4 g/l lactate (∼pH 3.8). These concentrations and pH values were determined through screening experiments, whereby conditions that resulted in substantial reduction but did not abolish survival for the control strain were used (Figure 6).
Figure 6.
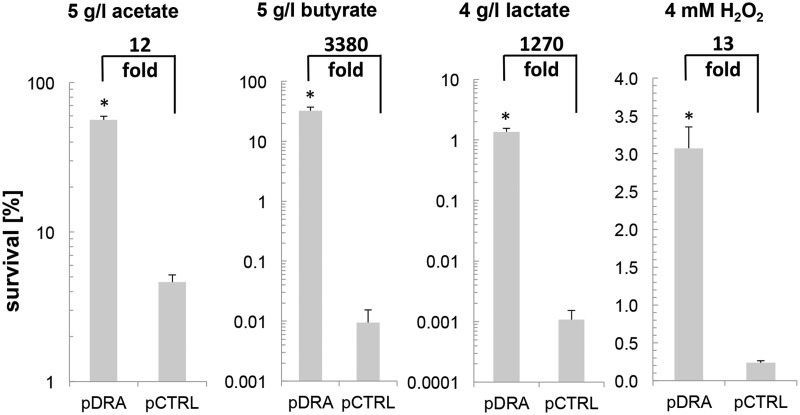
Survival of strain MG1655(pDRA) and MG1655(pCTRL) exposed for 1 h to high concentrations of carboxylic acids or H2O2. From the left: Survival to 5 g/l acetic acid (∼pH 4.0), survival to 5 g/l butyric acid (∼pH 4.2), survival to 4 g/l lactic (∼pH 3.8) acid and survival to 4 mM H2O2. Increased tolerance of MG1655(pDRA) is indicated as fold change compared with the control strain. Error bars indicate the standard error of at least three biological replicates, and statistical significance was tested with a two-sample t-test (*P < 0.05).
Overexpression of the three sRNAs increased E. coli survival to toxic concentrations of the tested carboxylic acids (Figure 6). The highest protection was observed for butyrate, whereby a 3380-fold increase was detected. This was followed by a 1270-fold increase for lactate and 12-fold increase for acetate. The toxicity of carboxylic acids is rather complex: their production by the cells and their transport in and out of the cell does not only result in a pH drop in the medium, but, significantly, it acidifies the cytosol and reduces the transmembrane ΔpH and protonmotive force (4,8,48). Thus, their toxicity is likely more complex than the toxicity imparted from inorganic acids. In this respect, the protection offered by overexpressing the three sRNAs is of both fundamental and practical significance.
Increased tolerance to oxidative stress by sRNA overexpression
Tolerance to oxidative stress is also of major physiological, medical and bioprocessing significance (49,50). Acid stress introduced by carboxylic acids is increasingly viewed as, partially at least, similar to oxidative stress (3,4,48,51). For instance, two mechanisms of acid-mediated generation of reactive-oxygen species were proposed in yeast cells (51). Thus, we wanted to investigate if overexpression of the three sRNAs will also protect the cells from oxidative stress. To do so, we used a standard oxidative-stress assay (52) to test the survival of MG1655(pDRA) in comparison with the plasmid control strain MG1655(pCTRL) to 4 mM H2O2 using active growing cells (Figure 6). Overexpressing the three sRNAs resulted in a 13-fold increase in survival.
DISCUSSION
We have shown that overexpressing the three sRNAs, DsrA, RprA and ArcZ, increased survival of MG1655(pDRA) against acid stress in a supra-additive fashion, during active cell growth, without the presence of amino acids, without acid induction and in the presence of glucose. We also showed that MG1655(pDRA) displays resistance against carboxylic-acid and oxidative stress. Taken together, these data demonstrate that overexpressing the three sRNAs results in protection against a broad range of stresses during active cell growth and without any supplements. While several studies have used growth assays (at a relatively higher pH: 3.9–4.5) to assess tolerance to acid stress (53–55), in this study, we used the widely used survival to low pH assay, which we found to be reproducible and appropriately quantitative.
As discussed, AR is a complex phenotype that engages several mechanisms and notably those of the AR1-AR4 systems (11,12), but several more as already discussed, with more possibly remaining to be explored. Several of these mechanisms are controlled by global regulators like RpoS and H-NS, which can be targeted for synthetic strategies to generate tolerant phenotypes. This is not, however, a simple task, as these regulators are extensively regulated themselves (27,42,56) in complex, interactive and frequently incompletely understood mechanisms even in E. coli. RpoS in particular is an attractive target to engineer tolerant phenotypes (27). We demonstrated that simple overexpression of RpoS is not sufficient to mount the full phenotypic response against acid stress, most likely because of incomplete induction of its regulon deriving from ineffective recruitment of the RNA polymerase (RNAP) owing to limiting RpoS protein levels (57). Our data (Figure 5) would suggest that this could be partially due to regulation by H-NS, which mediates degradation of rpoS mRNA and RpoS protein (27). We demonstrated that this limitation can be overcome by combinatorially overexpressing the three sRNAs (Figure 2), whereby the interaction of all three sRNAs with the rpoS mRNA lead to increased translation resulting in higher RpoS protein amounts in the cell (see Figure 2e). This observed behavior is not a saturation effect of the sRNAs binding to the rpoS mRNA, as we observed, especially for the early time points (#1–#4), similar RpoS protein level (Figure 2c), while the transcription of all three sRNAs is constantly increasing during this period (Figure 2b). Thus, simultaneous interactions between these three sRNAs and the rpoS mRNA must be responsible for this behavior. The detailed elucidation of these interactions is a difficult problem beyond the scope of this work, and will require new tools and strategies to bring to fruition.
We speculate that simultaneous overexpression of the three sRNAs saturates the effect of rpoS mRNA stabilization and translational activation, so that sufficient levels of the three sRNAs are present in the cell to interact with other targets. In this context, we examined the impact of overexpressing the three sRNAs on H-NS, a well-known target of DsrA (42) and an important regulator of AR (17). DsrA binds to hns mRNA and increases mRNA turnover (42), but this does not influence the steady state hns mRNA levels, as H-NS autoregulates itself (42). However, when all three sRNAs were overexpressed, steady state mRNA levels of hns were decreased (Figure 5b), which we speculate is due to the effect of increased mRNA turnover by increased DsrA levels and/or possible additional regulatory effects by ArcZ and RprA. When we examined genes known to be transcriptionally silenced by H-NS (namely, adiA and adiY; Figure 5c), we measured significant upregulation of these genes in strains MG1655(pDRA) and MG1655(pDR), but less so or not at all in MG1655(pDsrA) (Figure 5b). These and the data from strain MG1655(pRpoS), which displays reduced steady state hns mRNA levels but a lesser impact on adiA and adiY expression, demonstrate the complex regulatory network engaged in this acid-tolerance phenotype. Although it is possible that the three sRNAs have additional targets imparting tolerance of acid stress, our data suggest that the supra-additive effect of overexpressing the three sRNAs is largely due to the altered regulatory and heavily interconnected network of RpoS and H-NS, which could not be achieved by overexpressing RpoS directly. Significantly, unlike the overexpression of proteins, overexpression of sRNAs imposes hardly any metabolic burden on cells, and constitutes a more effective strategy of synthetic biology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [58].
FUNDING
National Science Foundation (NSF) (USA) [CBET-1033926]. Funding for open access charge: NSF [CBET-1033926].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
S.M.G. and E.T.P. developed the concept and co-wrote the manuscript; S.M.G. performed most experiments; M.A.H. helped with the western blots; D.I. helped with northern blot experiments; S.A.N. performed the oxidative stress assays; E.T.P. supervised the project and all experiments.
REFERENCES
- 1.Patnaik R. Engineering complex phenotypes in industrial strains. Biotechnol. Prog. 2008;24:38–47. doi: 10.1021/bp0701214. [DOI] [PubMed] [Google Scholar]
- 2.Boyle NR, Gill RT. Tools for genome-wide strain design and construction. Curr. Opin. Biotechnol. 2012;23:666–671. doi: 10.1016/j.copbio.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 3.Nicolaou SA, Gaida SM, Papoutsakis ET. A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: from biofuels and chemicals, to biocatalysis and bioremediation. Metab. Eng. 2010;12:307–331. doi: 10.1016/j.ymben.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Warnecke T, Gill RT. Organic acid toxicity, tolerance, and production in Escherichia coli biorefining applications. Microb. Cell Fact. 2005;4:25. doi: 10.1186/1475-2859-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Biase D, Pennacchietti E. Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: function, distribution and biomedical implications of the gadBC operon. Mol. Microbiol. 2012;86:770–786. doi: 10.1111/mmi.12020. [DOI] [PubMed] [Google Scholar]
- 6.Lin JS, Smith MP, Chapin KC, Baik HS, Bennett GN, Foster JW. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microb. 1996;62:3094–3100. doi: 10.1128/aem.62.9.3094-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beales N. Adaptation of microorganisms to cold temperatures, weak acid preservatives, low pH, and osmotic stress: a review. Compr. Rev. Food Sci. Food Saf. 2004;3:1–20. doi: 10.1111/j.1541-4337.2004.tb00057.x. [DOI] [PubMed] [Google Scholar]
- 8.Husemann MHW, Papoutsakis ET. Solventogenesis in Clostridium-acetobutylicum fermentations related to carboxylic-acid and proton concentrations. Biotechnol. Bioeng. 1988;32:843–852. doi: 10.1002/bit.260320702. [DOI] [PubMed] [Google Scholar]
- 9.Roe AJ, McLaggan D, Davidson I, O'Byrne C, Booth IR. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids. J. Bacteriol. 1998;180:767–772. doi: 10.1128/jb.180.4.767-772.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster JW. Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2004;2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 11.Richard H, Foster JW. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J. Bacteriol. 2004;186:6032–6041. doi: 10.1128/JB.186.18.6032-6041.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meng SY, Bennett GN. Nucleotide-sequence of the Escherichia-coli cad operon - a system for neutralization of low extracellular pH. J. Bacteriol. 1992;174:2659–2669. doi: 10.1128/jb.174.8.2659-2669.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. Genome-wide analysis of the general stress response network in Escherichia coli: sigma(S)-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 2005;187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richard HT, Foster JW. Acid resistance in Escherichia coli. Adv. Appl. Microbiol. 2003;52:167–186. doi: 10.1016/s0065-2164(03)01007-4. [DOI] [PubMed] [Google Scholar]
- 15.Tramonti A, De Canio M, De Biase D. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol. Microbiol. 2008;70:965–982. doi: 10.1111/j.1365-2958.2008.06458.x. [DOI] [PubMed] [Google Scholar]
- 16.Giangrossi M, Zattoni S, Tramonti A, De Biase D, Falconi M. Antagonistic role of H-NS and GadX in the regulation of the glutamate decarboxylase-dependent acid resistance system in Escherichia coli. J. Biol. Chem. 2005;280:21498–21505. doi: 10.1074/jbc.M413255200. [DOI] [PubMed] [Google Scholar]
- 17.Krin E, Danchin A, Soutourina O. Decrypting the H-NS-dependent regulatory cascade of acid stress resistance in Escherichia coli. BMC Microbiol. 2010;10:273. doi: 10.1186/1471-2180-10-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brescia CC, Kaw MK, Sledjeski DD. The DNA binding protein H-NS binds to and alters the stability of RNA in vitro and in vivo. J. Mol. Biol. 2004;339:505–514. doi: 10.1016/j.jmb.2004.03.067. [DOI] [PubMed] [Google Scholar]
- 19.Hommais F, Krin E, Laurent-Winter C, Soutourina O, Malpertuy A, Le Caer JP, Danchin A, Bertin P. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 2001;40:20–36. doi: 10.1046/j.1365-2958.2001.02358.x. [DOI] [PubMed] [Google Scholar]
- 20.Krin E, Danchin A, Soutourina O. RcsB plays a central role in H-NS-dependent regulation of motility and acid stress resistance in Escherichia coli. Res. Microbiol. 2010;161:363–371. doi: 10.1016/j.resmic.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Oshima T, Ishikawa S, Kurokawa K, Aiba H, Ogasawara N. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 2006;13:141–153. doi: 10.1093/dnares/dsl009. [DOI] [PubMed] [Google Scholar]
- 22.Seputiene V, Daugelavicius A, Suziedelis K, Suziedeliene E. Acid response of exponentially growing Escherichia coli K-12. Microbiol. Res. 2006;161:65–74. doi: 10.1016/j.micres.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Stancik LM, Stancik DM, Schmidt B, Barnhart DM, Yoncheva YN, Slonczewski JL. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 2002;184:4246–4258. doi: 10.1128/JB.184.15.4246-4258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson MD, Burton NA, Gutierrez B, Painter K, Lund PA. RcsB is required for inducible acid resistance in Escherichia coli and acts at gadE-dependent and -independent promoters. J. Bacteriol. 2011;193:3653–3656. doi: 10.1128/JB.05040-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tucker DL, Tucker N, Conway T. Gene expression profiling of the pH response in Escherichia coli. J. Bacteriol. 2002;184:6551–6558. doi: 10.1128/JB.184.23.6551-6558.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stincone A, Daudi N, Rahman AS, Antczak P, Henderson I, Cole J, Johnson MD, Lund P, Falciani F. A systems biology approach sheds new light on Escherichia coli acid resistance. Nucleic Acids Res. 2011;39:7512–7528. doi: 10.1093/nar/gkr338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hengge-Aronis R. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 2002;66:373–395. doi: 10.1128/MMBR.66.3.373-395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schellhorn HE, Stones VL. Regulation of Katf and Kate in Escherichia coli K-12 by weak acids. J. Bacteriol. 1992;174:4769–4776. doi: 10.1128/jb.174.14.4769-4776.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicolaou SA, Gaida SM, Papoutsakis ET. Coexisting/Coexpressing Genomic Libraries (CoGeL) identify interactions among distantly located genetic loci for developing complex microbial phenotypes. Nucleic Acids Res. 2011;39:e152. doi: 10.1093/nar/gkr817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl Acad. Sci. USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 2002;46:813–826. doi: 10.1046/j.1365-2958.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- 33.Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 2010;29:3094–3107. doi: 10.1038/emboj.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lease RA, Smith D, McDonough K, Belfort M. The small noncoding DsrA RNA is an acid resistance regulator in Escherichia coli. J. Bacteriol. 2004;186:6179–6185. doi: 10.1128/JB.186.18.6179-6185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2007;2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- 36.Tracy BP, Jones SW, Papoutsakis ET. Inactivation of sigma(E) and sigma(G) in Clostridium acetobutylicum illuminates their roles in clostridial-cell-form biogenesis, granulose synthesis, solventogenesis, and spore morphogenesis. J. Bacteriol. 2011;193:1414–1426. doi: 10.1128/JB.01380-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alsaker KV, Paredes CJ, Papoutsakis ET. Design, optimization and validation of genomic DNA microarrays for examining the Clostridium acetobutylicum transcriptome. Biotechnol. Bioprocess Eng. 2005;10:432–443. [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Repoila F, Gottesman S. Signal transduction cascade for regulation of RpoS: temperature regulation of DsrA. J. Bacteriol. 2001;183:4012–4023. doi: 10.1128/JB.183.13.4012-4023.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Gene Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCullen CA, Benhammou JN, Majdalani N, Gottesman S. Mechanism of positive regulation by DsrA and RprA small Noncoding RNAs: pairing increases translation and protects rpoS mRNA from degradation. J. Bacteriol. 2010;192:5559–5571. doi: 10.1128/JB.00464-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lease RA, Belfort M. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proc. Natl Acad. Sci. USA. 2000;97:9919–9924. doi: 10.1073/pnas.170281497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 44.Seputiene V, Suziedelis K, Normark S, Melefors O, Suziedeliene E. Transcriptional analysis of the acid-inducible asr gene in enterobacteria. Res. Microbiol. 2004;155:535–542. doi: 10.1016/j.resmic.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 45.Tramonti A, Visca P, De Canio M, Falconi M, De Biase D. Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 2002;184:2603–2613. doi: 10.1128/JB.184.10.2603-2613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burton NA, Johnson MD, Antczak P, Robinson A, Lund PA. Novel aspects of the acid response network of E. coli K-12 Are revealed by a study of transcriptional dynamics. J. Mol. Biol. 2010;401:726–742. doi: 10.1016/j.jmb.2010.06.054. [DOI] [PubMed] [Google Scholar]
- 47.Buchanan RL, Doyle MP. Foodborne disease significance of Escherichia coli O157:H7 and other enterohemorrhagic E coli. Food Technol. Mag. 1997;51:69–76. [Google Scholar]
- 48.Alsaker KV, Paredes C, Papoutsakis ET. Metabolite stress and tolerance in the production of biofuels and chemicals: gene-expression-based systems analysis of butanol, butyrate, and acetate stresses in the anaerobe Clostridium acetobutylicum. Biotechnol. Bioeng. 2010;105:1131–1147. doi: 10.1002/bit.22628. [DOI] [PubMed] [Google Scholar]
- 49.Avery SV. Molecular targets of oxidative stress. Biochem. J. 2011;434:201–210. doi: 10.1042/BJ20101695. [DOI] [PubMed] [Google Scholar]
- 50.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abbott DA, Zelle RM, Pronk JT, van Maris AJA. Metabolic engineering of Saccharomyces cerevisiae for production of carboxylic acids: current status and challenges. FEMS Yeast Res. 2009;9:1123–1136. doi: 10.1111/j.1567-1364.2009.00537.x. [DOI] [PubMed] [Google Scholar]
- 52.Macomber L, Rensing C, Imlay JA. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J. Bacteriol. 2007;189:1616–1626. doi: 10.1128/JB.01357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Presser KA, Ratkowsky DA, Ross T. Modelling the growth rate of Escherichia coli as a function of pH and lactic acid concentration. Appl. Environ. Microbiol. 1997;63:2355–2360. doi: 10.1128/aem.63.6.2355-2360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski JL. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 1994;176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Presser KA, Ross T, Ratkowsky DA. Modelling the growth limits (growth/no growth interface) of Escherichia coli as a function of temperature, pH, lactic acid concentration, and water activity. Appl. Environ. Microbiol. 1998;64:1773–1779. doi: 10.1128/aem.64.5.1773-1779.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dorman CJ. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2004;2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 57.Farewell A, Kvint K, Nystrom T. Negative regulation by RpoS: a case of sigma factor competition. Mol. Microbiol. 1998;29:1039–1051. doi: 10.1046/j.1365-2958.1998.00990.x. [DOI] [PubMed] [Google Scholar]
- 58.Mandel MJ, Silhavy TJ. Starvation for different nutrients in Escherichia coli results in differential modulation of RpoS levels and stability. J. Bacteriol. 2005;187:434–442. doi: 10.1128/JB.187.2.434-442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.