Abstract
The telomerase enzyme plays a critical role in human aging and cancer biology by maintaining telomere length and extending the proliferative lifespan of most stem cells and cancer cells. Despite the importance of this enzyme, our understanding of the mechanisms that regulate its activity and establish telomere length homeostasis in mammalian cells is incomplete, in part because the perfect repetitive nature of telomeric sequence hampers in situ detection of telomere elongation patterns. Here, we describe a novel assay using a mutant telomerase that adds a well-tolerated variant telomeric repeat sequence to telomere ends. By specifically detecting the addition of these variant repeats, we can directly visualize telomere elongation events in human cells. We validate this approach by in situ mapping of telomere elongation patterns within individual nuclei and across a population of cells.
INTRODUCTION
Human telomeres are composed of tandem arrays of TTAGGG DNA repeats bound by a protective protein complex termed shelterin (1). These nucleoprotein structures cap chromosome ends and prevent them from initiating a DNA damage response. Telomeres shorten with each cell division, and when telomeres become critically short, protection of chromosome ends is compromised and cell proliferation is halted. Proliferative lifespan can be extended by the enzyme telomerase, which elongates telomeres by adding new TTAGGG repeats. The telomerase enzyme minimally consists of the protein reverse transcriptase, TERT, and the template-containing RNA, TER. Through its ability to counteract replicative telomere shortening, the telomerase enzyme plays an important role in maintaining stem cell populations and extending the proliferative lifespan of most cancer cells. Understanding and modulating the function of telomerase is therefore of great interest in the treatment of cancer and in the prevention of other aging-associated diseases (2,3).
Studies in yeast and mammalian cells have identified numerous factors that can influence the activity of telomerase at individual telomeres, including telomere length (4–9), telomeric repeat-containing RNA (10) and shelterin levels (11). To fully understand how such factors guide telomerase function and establish telomere length homeostasis, it is necessary to directly identify which telomeres have been elongated by telomerase. In Saccharomyces cerevisiae, the telomeres consist of degenerate TG1–3 repeats that allow for the detection of specific telomere elongation events by sequencing (4). In contrast, the perfect repetitive nature of mammalian telomeric repeats has hindered such analysis, and as a result, the understanding of mammalian telomerase regulation has lagged that of yeast.
Recently, an elegant method was developed that enables the bulk detection of newly added telomeric sequence at the lagging-strand ends of human telomeres (12). Using this method, the authors showed that most telomeres in HeLa cells are elongated by a single telomerase enzyme during each cell cycle. This result suggests that, unlike yeast telomerase, mammalian telomerase does not preferentially act at short telomeres (4,12). Further experiments in HeLa cells with pre-shortened telomeres showed that short telomeres are sequentially elongated by multiple telomerase enzymes acting in a single cell cycle (13). In combination, these studies support a model for human telomere homeostasis in which telomerase acts at all or most telomeres, but with short telomeres elongating more rapidly owing to extension by multiple telomerase enzymes (13). The assay used in these studies to identify the newly added telomeric sequence requires degradation of the genome and purification of the lagging-strand telomere ends, which precludes the ability to map telomere elongation events to particular chromosome ends or to particular cells of a population (12).
Here, we describe and validate a novel assay that enables direct in situ visualization of telomere elongation patterns within individual nuclei and across cell populations. This assay provides a new approach for dissecting the multiple overlapping regulatory mechanisms that guide telomerase activity and ensure proper telomere length homeostasis.
MATERIALS AND METHODS
Cell culture
Human LOX melanoma cells, MRC-5 fetal lung fibroblasts and HeLa cervical carcinoma cells were grown in Dulbecco’s modified Eagle’s medium (HeLa and MRC-5) or RPMI 1640 (LOX), supplemented with 10% (vol/vol) fetal bovine serum, 1% (vol/vol) GlutaMAX-1 (Gibco) and 1% (vol/vol) penicillin–streptomycin (Gibco). Cells were grown at 37°C in 5% CO2. MRC-5-TERT and HeLa-TERT cells were produced by infecting MRC-5 and HeLa cells with a retrovirus expressing TERT and a neomycin resistance gene. Infected cells were selected with 500 μg/ml G418 for 7 (HeLa-TERT) or 12 (MRC-5-TERT) days, which was sufficient to kill uninfected control cells treated in parallel.
Lentiviral plasmids and production
Lentivirus was produced as previously described (14,15) using a second-generation lentiviral system (16). TER-expressing lentivectors included a wild-type or mutant TER gene driven by the IU1 promoter and a puromycin resistance gene driven by the CMV promoter. The wild-type and mutant TER template sequences were as follows (with predicted telomeric repeat sequences in parentheses): wild-type – 3′-CAAUCCCAAUC-5′ (5′-TTAGGG-3′); TSQ1 – 3′-CCAACGCCAAC-5′ (5′-GTTGCG-3′); and 47A – 3′-CAAACCCAAAC-5′ (5′-TTTGGG-3′). TERT-expressing lentivectors included a wild-type or mutant TERT gene driven by the CMV promoter, followed by an internal ribosome entry site and a puromycin resistance gene. Catalytically dead TERT D868A contains an inactivating mutation in the reverse transcriptase motif (17).
Lentiviral infections and growth curves
The day before infection, 1.2 × 105 cells were seeded on a 10-cm plate and allowed to attach overnight. On Day 0, approximately 75 transducing units of lentivirus per cell was added to each plate. For LOX and HeLa cells, 8 μg/ml polybrene was also added to the media at the start of infection. After 8 h, the virus-containing media were replaced with fresh media. Puromycin selection (1.5 μg/ml) was initiated 24–36 h after the introduction of virus and lasted 2–3 days, by which time uninfected control cells treated with puromycin had died. Selected cells were harvested at indicated time points for fluorescence in situ hybridization (FISH) analysis. For growth curves, cells were plated immediately following selection in 6-well dishes at 2 × 104 (LOX), 3 × 104 (HeLa and HeLa-TERT) or 3.5 × 104 (MRC-5-TERT) cells/well. Cells were split as needed to maintain logarithmic growth, and viable cells were counted by hemocytometer. For experiments analysing TSQ1 incorporation after infection with TERT-expressing virus (Figures 1B and 4, and Supplementary Figure S4), MRC-5 cells were first infected with lentivirus expressing TSQ1 TER and subjected to puromycin selection as mentioned above. The TSQ1-expressing MRC-5 cells were next infected with lentivirus expressing wild-type or catalytically dead TERT using the infection protocol outlined above. Cells were harvested for FISH analysis 48 h after infection with TERT-expressing virus.
Figure 1.
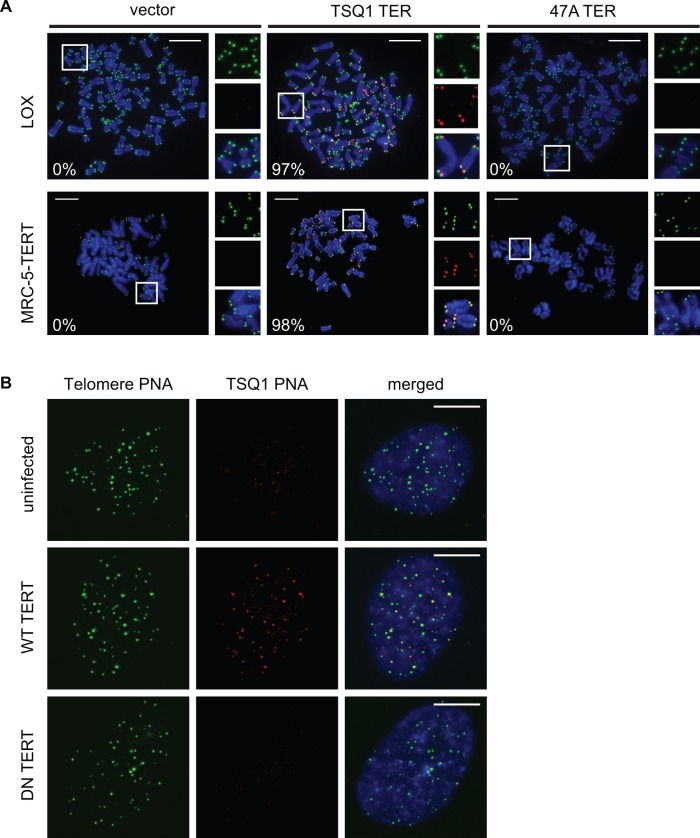
Robust telomeric incorporation of TSQ1 variant repeats. (A) Metaphase spreads of LOX (top) and MRC-5-TERT (bottom) cells 5 days after infection with control lentivirus (vector) or lentivirus expressing TSQ1 or 47A mutant TER. Wild-type telomeric repeats (green) and TSQ1 variant repeats (red) were detected by FISH. Regions in white boxes are enlarged to the right of the corresponding image. For each condition, between 24 and 35 nuclei were manually scored in a blinded manner and the percentage of TSQ1-positive metaphases is indicated in the bottom left corner. (B) FISH analysis of TSQ1-expressing MRC-5 cells infected with lentivirus expressing wild-type (WT) or catalytically dead dominant-negative (DN) TERT. Cells were fixed 2 days after infection and analysed for colocalization of TSQ1 (red) and wild-type (green) telomeric repeats. At least 30 nuclei from each group were blindly scored, and TSQ1 addition was exclusively observed in cells receiving WT TERT. Scale bars, 10 μm.
Figure 4.
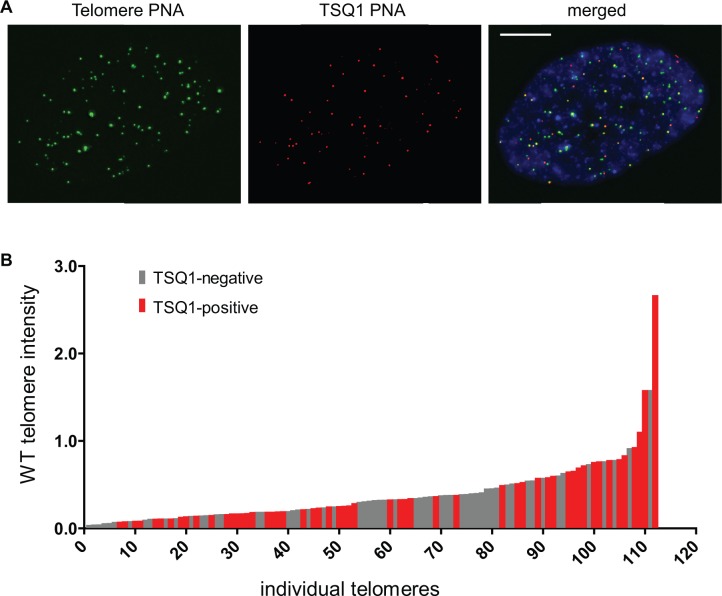
Visualization of telomere elongation patterns within individual nuclei. (A) MRC-5 cells engineered to overexpress TSQ1 were infected with lentivirus expressing wild-type TERT and analysed 48 h later. A representative nucleus stained with wild-type (green) and TSQ1 (red) FISH probes is shown. Scale bar, 10 μm. (B) Plot of TSQ1 incorporation in nucleus from (A). Telomere identification and intensity analysis were performed with CellProfiler image analysis software, and each telomere was manually scored for the presence or absence of a co-localizing TSQ1 focus. WT telomere intensities are plotted in ascending order with TSQ1-negative telomeres in gray and TSQ1-positive telomeres in red.
Metaphase spread preparation and FISH
Cells were treated with 0.1 μg/ml Colcemid for 2–4 h before the harvest. Collected cells underwent hypotonic treatment with 0.075 M KCl for 10 min at 37°C and were subsequently fixed by 4–6 rounds of resuspension in 3:1 methanol/acetic acid. Cells were dropped on microscope slides to prepare metaphase spreads, and the slides were dried for at least 16 h before FISH. For FISH, the cells were rehydrated in 1× phosphate buffered saline (PBS) for 15 min, followed by fixation with 4% paraformaldehyde (in 1× PBS) for 2 min. After a wash in 1× PBS, cells were treated with RNase A (Qiagen catalogue no. 158922, diluted 1:100 in PBS) for approximately 2 h at room temperature. After three 1× PBS washes, cells were treated with 0.1 mg/mL pepsin (Sigma-Aldrich catalogue no. P7012, prepared fresh in 0.01 M HCl) at 37°C for 10 min. The cells were washed twice with 1× PBS, fixed in 4% formaldehyde (in 1× PBS) for 2 min, and rinsed three times in 1× PBS. The cells were dehydrated in ethanol (2 min each in 70% ethanol, 95% ethanol and 100% ethanol) and air dried. The FISH hybridization mix included 70% formamide, 10 mM Tris HCl pH 7.2, 1% (weight/vol) blocking reagent (Roche catalogue no. 11096176001) and 0.5 μg/ml of each peptide nucleic acid (PNA) probe. The following PNA probes (Panagene) were used: wild-type telomere probe (FAM-OO-ccctaaccctaaccctaa), TSQ1 telomere probe (Cy3-OO-ccgcaaccgcaaccgcaa), pan-centromere probe (Cy5-OO-cttcgttggaaacggga) and chromosome 9 centromere probe (Cy5-OO-aatcaacccgagtgcaat) (18). The hybridization mix was added to the cells under a coverslip and the cells were denatured on an 80°C heatblock for 3 min. After a 2 h hybridization at room temperature, the cells were washed twice for 15 min in 70% formamide/Tris HCl pH 7.4 and three times for 5 min in 0.05 M Tris HCl pH 7.4/0.15 M NaCl/0.05% Tween-20. After ethanol dehydration as mentioned above, slides were air dried and mounted in Prolong Gold with DAPI (Invitrogen catalogue no. P-36931).
Microscopy and image analysis
All images were obtained using a Deltavision deconvolution microscope (Applied Precision) with a 60× 1.42 NA Plan Apo objective (Olympus). Deltavision images with Z-stacks were converted to projected Tagged Image File Format using the Fiji image processing package (www.fiji.sc), and TSQ1-positive telomeres were scored by manually counting TSQ1 foci that co-localize with wild-type telomere foci. Enumeration and intensity analysis of wild-type telomere foci in intact nuclei were performed using CellProfiler image analysis software (www.cellprofiler.org; analysis pipelines available on request). Chromosomal fusions on metaphase spreads were manually counted in a blinded manner.
Real-time polymerase chain reaction (PCR)
TERT expression levels were determined using a one-step Brilliant II QRT-PCR kit (Agilent Technologies) and the following TERT and control primer sets: TERT (forward: cctgcactggctgatgagtgtg; reverse: gatgctgcctgacctctgctt); beta-2-microglobulin (forward: tcacgtcatccagcagagaatgga; reverse: cacacggcaggcatactcatcttt). TER expression levels were determined using a two-step protocol. First, cDNA was synthesized using SuperScript III reverse transcriptase with random hexamer primers (Invitrogen). Subsequent Q-PCR was performed using the Brilliant II Q-PCR kit (Agilent Technologies) and the following TER and control primer sets: TER (forward: ttgcggagggtgggcct; reverse: cgggccagcagctgacatt); GAPDH (forward: catgttcgtcatgggtgtgaacca; reverse: atggcatggactgtggtcatgagt). All PCR reactions were performed with the StepOnePlus real-time PCR system (Applied Biosystems). Values were normalized to beta-2-microglobulin or GAPDH and are reported as fold change over vector.
RESULTS
TSQ1 variant repeat incorporation is well tolerated in primary and cancer cell lines
Our assay builds on work using mutant forms of TER that contain sequence changes in the template region. When overexpressed in cells, mutant TER joins with wild-type TERT to form mutant telomerase, which adds variant repeats to telomere ends (19). Although detection of incorporated variant repeat sequences could in principle be used to map where telomerase has acted, incorporation of these sequences typically induces rapid telomere dysfunction. Because variant repeats cannot properly recruit the shelterin protein complexes that bind and protect wild-type telomeres (19), cells overexpressing mutant TER usually undergo rapid senescence or apoptosis, often accompanied by telomere fusions (2,15,20). This cytotoxicity presents a significant barrier for using mutant TERs to map telomerase activity patterns.
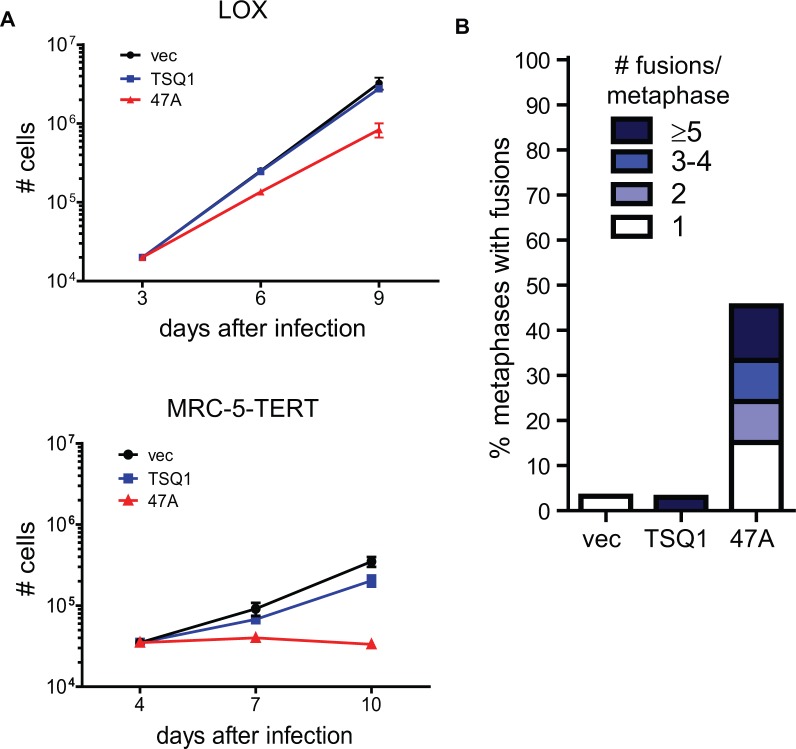
During a recent screen of mutant TER sequences, we serendipitously identified a mutant TER that is well tolerated in human cells despite robust incorporation of variant telomeric repeats. This mutant TER, hereafter referred to as TSQ1 for Tolerated Sequence 1, is designed to add GTTGCG variant repeats. Using PNA probes to detect wild-type (TTAGGG) and TSQ1 (GTTGCG) repeats, robust telomeric incorporation of TSQ1 repeats was specifically detected in human LOX melanoma cells overexpressing TSQ1 (Figure 1A and Supplementary Figure S1). Telomeric incorporation of TSQ1 variant repeats was similarly observed in MRC-5 human diploid fibroblasts that overexpress TERT (MRC-5-TERT, Figure 1A and Supplementary Figure S1). Incorporation of TSQ1 repeats at MRC-5 telomeres required the presence of catalytically active TERT (Figure 1B), confirming that TSQ1 variant repeat incorporation reflects mutant telomerase activity. Although overexpression of a mutant TER that adds TTTGGG variant repeats (47A) displayed the typical acute cytotoxicity and abundant telomere fusions (Figure 2A and B) (2,15,21), TSQ1-treated LOX cells proliferated nearly as well as vector-treated controls during short-term culture (Figure 2A) and displayed very few telomere fusions (Figure 2B). Relatively modest growth effects were also observed in TSQ1-treated MRC-5-TERT cells (Figure 2A) despite the fact that these cells have intact DNA damage checkpoints. We conclude that TSQ1 variant repeats are well-tolerated in human cells during short-term culture, in contrast with other well-characterized mutant TERs.
Figure 2.
TSQ1 incorporation is well tolerated in human cells. (A) Proliferation curves for LOX and MRC-5-TERT cells infected with control lentivirus (vec) or lentivirus expressing TSQ1 or 47A mutant TER. Equivalent cell numbers were plated at Day 3 (LOX) or Day 4 (MRC-5-TERT) after infection and passaged to maintain logarithmic growth. Each point represents the mean ± SD for three independent wells seeded and passaged in parallel. (B) Fusion analysis in LOX cells infected with control lentivirus or lentivirus expressing TSQ1 or 47A mutant TER. LOX metaphase spreads were blinded and manually scored for the presence of chromosome-type fusions. Between 37 and 44 metaphases were scored for each condition. For both (A) and (B), results are representative of at least two independent experiments.
TSQ1 variant repeat incorporation is heterogeneous across a population of HeLa cells
TSQ1 displays two characteristics that make it a promising in situ marker of telomerase-directed telomere elongation. First, incorporation of TSQ1 variant repeats is readily detected at telomeres by simultaneous use of PNA probes targeting wild-type and TSQ1 telomeric repeats. Second, TSQ1 is well-tolerated in human cells during short-term culture, allowing detection of telomere elongation patterns before the onset of overt cytotoxicity. We therefore sought to validate TSQ1 for in situ visualization of telomere elongation patterns.
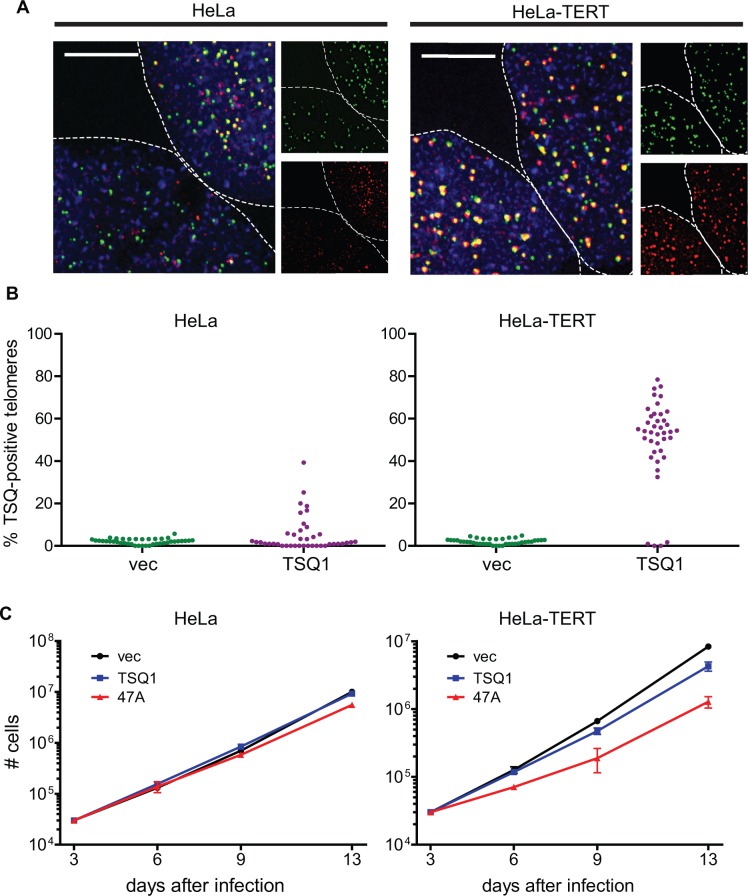
Prior analysis of telomere length and telomerase activity in HeLa cells revealed remarkable cell-to-cell heterogeneity with respect to both metrics (22). Based on these results, we predicted that TSQ1 variant repeat incorporation would vary across a population of HeLa cells. Indeed, our analysis identified significant heterogeneity, with ∼30% of HeLa cells showing TSQ1 repeat incorporation (Figure 3A and B). Consistent with mutant repeat incorporation in a minority of cells, proliferation of HeLa cells was not markedly impacted by overexpression of TSQ1 or 47A mutant TER, with TSQ1 treatment being better tolerated than 47A treatment at later timepoints (Figure 3C and Supplementary Figure S2). We also note that overexpression of wild-type TER for 6 days failed to result in a significant increase in the bulk telomere length of HeLa cells as measured by quantitative FISH (Supplementary Figure S3). Thus, under conditions where a population-based assay to assess telomere repeat addition does not detect significant telomere lengthening, TSQ1 incorporation enables clear identification of the particular cells within a population that are undergoing telomere elongation.
Figure 3.
Variable TSQ1 incorporation across a population. (A) FISH analysis of HeLa and HeLa-TERT cells 6 days after infection with TSQ1 lentivirus. Each image shows portions of two adjacent nuclei (outlined). Individual wild-type telomeric repeat (green) and TSQ1 variant repeat (red) channels are broken out to the right of each merged image. Both HeLa-TERT nuclei display abundant TSQ1-positive telomeres, whereas only one of the two HeLa cell nuclei displays abundant TSQ1-positive telomeres. Scale bars, 10 μm. (B) Plot showing the percentage of TSQ1-positive telomeres for TSQ1-treated (purple) and vector-treated (green) HeLa and HeLa-TERT nuclei. Forty nuclei were scored for each condition, with each point on the plot representing a single nucleus. The few telomeres scored as TSQ1-positive in vector-treated cells reflect the low-level background seen with the TSQ1-specific FISH probe. (C) Proliferation curves for HeLa and HeLa-TERT cells infected with control lentivirus (vec) or lentivirus expressing TSQ1 or 47A mutant TER. Equivalent cell numbers were plated 3 days after infection and passaged to maintain logarithmic growth. Each point represents the mean ± SD for three independent wells seeded and passaged in parallel. For (A–C), results are representative of at least two independent experiments.
To exclude the possibility that the observed heterogeneity resulted from variable TSQ1 expression, we performed the same experiment in HeLa cells overexpressing TERT (HeLa–TERT, Supplementary Figure S2). Strong TSQ1 incorporation was observed in ∼90% of the population (Figure 3A and B), indicating that the vast majority of cells express TSQ1 and that variable TERT levels likely underlie at least some of the heterogeneity observed in HeLa cells. Overexpression of either TSQ1 or 47A mutant TERs had a more pronounced growth effect in HeLa–TERT cells than in HeLa cells (Figure 3C), consistent with higher levels of variant repeat incorporation when TERT is abundant. Comparing the degree of growth inhibition, TSQ1 mutant TER is much better tolerated than 47A mutant TER. The lack of a strong growth effect in TSQ1-treated HeLa–TERT cells despite robust TSQ1 incorporation in almost all cells suggests that the heterogeneity observed in HeLa cells is not the result of variable TSQ1 tolerance across the population. Rather, these results argue that TSQ1 successfully identifies the individual cells within a heterogeneous population with active telomere elongation.
TSQ1 addition within individual nuclei is largely independent of wild-type telomere length
We next used the TSQ1 assay to examine telomere elongation patterns within individual nuclei. As outlined in the Introduction, recent work has indicated that human telomerase elongates most or all telomeres during each cell cycle, in contrast with the preferential elongation of short telomeres observed in yeast (4,12,13). Based on these studies, we predicted that TSQ1 incorporation would not occur preferentially at short telomeres. MRC-5 cells expressing TSQ1 were used for these experiments because they show clear TSQ1 variant repeat addition within 48 h of infection with TERT-expressing lentivirus (Figure 1B). In addition, MRC-5 cells have a doubling time of ∼40 h (Figure 2A), ensuring that analysis performed 48 h after infection would limit our analysis to cells that have undergone at most two cell divisions. In nuclei with detectable TSQ1 variant repeat incorporation, quantitative FISH was used to determine the relative length of each telomere, and telomeres were plotted in ascending order. Each telomere was then manually scored as positive or negative for TSQ1 incorporation, and this information was incorporated into the telomere length plot so that the distribution could be analysed with respect to wild-type telomere length. Although several nuclei showed preferential elongation of longer telomeres, the majority of nuclei analysed did not show strong preferential elongation of either long or short telomeres (Figure 4A and B and Supplementary Figure S4). The lack of a strong and consistent elongation bias across nuclei indicates that telomere length is not a major determinant of telomerase activity patterns. Furthermore, the aggregate pattern argues that, under the conditions tested, telomerase does not preferentially elongate short telomeres, a result that is consistent with prior studies (12,13). Finally, to demonstrate that the TSQ1 approach can also map chromosome-specific telomere elongation patterns, we performed TSQ1 incorporation analysis on MCR-5-TERT metaphase spreads using a chromosome 9-specific centromeric probe (Supplementary Figure S5). This analysis revealed diverse chromosome 9 elongation patterns, confirming that analysis of TSQ1 incorporation can be applied to single chromosomes.
DISCUSSION
Despite the important role of telomerase in cancer and age-related pathologies, our understanding of the regulatory mechanisms that shape telomerase activity patterns remains incomplete. Progress in this area has been hindered by an inability to distinguish newly added telomeric repeats from pre-existing repeats. Here, we identify a variant telomeric repeat sequence—TSQ1—that is unexpectedly well tolerated in human cells during short-term culture (Figure 2). We show that incorporation of variant repeats at telomeres can be detected by simultaneous use of FISH probes for wild-type and TSQ1 sequences, and that incorporation of TSQ1 variant repeats at telomeres requires the presence of both TSQ1 mutant TER and wild-type TERT (Figure 1). These properties of TSQ1 make it a useful tool to directly visualize telomere elongation patterns in situ. In this report, we describe and validate a novel assay that uses TSQ1 to map telomerase activity patterns.
To validate the utility of TSQ1 as an in situ marker of telomere elongation, we examined the pattern of TSQ1 addition in individual nuclei. Although S. cerevisiae telomerase preferentially elongates the shortest telomeres (4), recent work in multiple cell lines indicates that human telomerase acts at all or most telomeres during each cell cycle (12,13). In agreement with the recent studies, we find that TSQ1 addition occurred without a strong, consistent bias for long or short telomeres in MRC-5 human fibroblasts expressing exogenous TERT (Figure 4). A similar pattern was observed in HeLa cells treated with TSQ1 (data not shown). Interestingly, we did not detect TSQ1 addition at every telomere, even with simultaneous overexpression of TSQ1 TER and TERT (Figure 4). There are several possible explanations for this observation. First, TSQ1-treated cells still express wild-type telomerase, and our assay will not detect telomere elongation events that add wild-type telomeric repeats. Second, a portion of TSQ1 elongation events may fall below the threshold required for FISH detection. Finally, owing to its template mutations, TSQ1 mutant telomerase does not align perfectly with the wild-type telomeric overhang, and the resulting mismatches may serve as an initial barrier to elongation. Nevertheless, even if TSQ1 enables visualization of only a subset of elongation events, the aggregate analysis of TSQ1 addition provides a snapshot of telomerase activity patterns. For example, if telomerase acted exclusively at short telomeres, we would expect detection of TSQ1 addition events only at short telomeres. The fact that we do not observe this pattern provides strong evidence that, under the conditions tested, telomerase does not preferentially elongate short telomeres. In the future, it will be interesting to determine whether human telomerase preferentially elongates short telomeres in the context of very low telomerase levels.
In addition to analysing TSQ1 addition within individual cells, we also examined TSQ1 incorporation across a population of cells. To enable direct comparison between the TSQ1-derived results and those obtained using other approaches, we focused our analyses on HeLa cells because the telomerase and telomere dynamics of this cell line have been relatively well characterized. A prior study of HeLa cell clones showed that only five of eight single-cell-derived clones were positive for telomerase activity when measured by the Telomere Repeat Amplification Protocol (TRAP) (22). Consistent with this high degree of heterogeneity, we only detected TSQ1 addition in approximately one-third of the HeLa cells (Figure 3). The lower percentage of telomerase-positive cells identified in our study may reflect differences in assay sensitivity. In particular, FISH detection of incorporated TSQ1 sequence may miss cells with low levels of TSQ1 incorporation. Alternatively, the percentage of telomerase-positive clones identified by TRAP in the earlier study may be inflated. Stochastic switching of cells from a telomerase-negative to a telomerase-positive state was reported in the prior study (22), and it is possible some clones that were initially telomerase-negative became positive during the 14 population doublings required to get sufficient outgrowth for TRAP. In either case, TSQ1 incorporation allows for rapid and direct in situ identification of the individual cells within a population undergoing robust telomere elongation.
Heterogeneous telomerase activity and telomere elongation patterns may have significant implications for clonal evolution, genome instability, and the success of telomerase-targeted therapeutics, yet relatively little work has been done to characterize this heterogeneity and define its causes. In our HeLa cell experiments, heterogeneous TERT expression may be the major driver of variable telomere elongation. TERT overexpression indeed resulted in a more homogenous telomere elongation pattern across the HeLa cell population (Figure 3), consistent with prior work showing that simultaneous overexpression of TER and TERT induces massive telomere elongation (23). Alternatively, the absence of detectable TSQ1 incorporation in many TSQ1-treated HeLa cells may stem from negative regulation of telomere elongation. Numerous factors have been shown to inhibit telomerase activity at telomeres, including the shelterin protein POT1 (11) and telomeric repeat-containing RNA (24). We are now in a position to ask whether the expression or localization of such negative regulatory elements correlates with TSQ1 incorporation, and whether experimental manipulation of these factors alters the TSQ1 incorporation patterns both within single cells and across populations.
There are several limitations to the TSQ1 approach that bear mention. First, the TSQ1 assay requires overexpression of TSQ1 mutant TER and therefore cannot be used to assess the role of endogenous TER in establishing telomerase activity patterns. Furthermore, because TER overexpression has been shown to increase telomerase activity in many cell types including HeLa (23,25), this approach does not examine telomere elongation dynamics under homeostatic conditions. Despite the requirement for TER overexpression, the TSQ1 assay highlights significant telomere elongation heterogeneity in the HeLa cell population and therefore enables analysis of the myriad other factors that establish that heterogeneity.
The addition of TSQ1 repeats at telomeres may also interfere with the sequence-specific binding of telomeric proteins—including the POT1-TPP1 (11,26) and CST (27) complexes—that regulate telomerase function. As a result, it is likely that TSQ1 incorporation disrupts the normal feedback mechanisms that regulate telomerase repeat addition processivity and thus the extent of telomere elongation. For this reason, we have focused our analysis on the overall pattern of TSQ1 addition rather than the extent of TSQ1 addition at any given telomere. We also note that although TSQ1 variant repeats are unusually well tolerated in human cells, the incorporation of variant repeats nevertheless induces low-level telomere dysfunction that inhibits cell proliferation over long-term culture (Figures 2 and 3). Because such telomere dysfunction may ultimately induce telomere fusions and alternative lengthening of telomere-like telomeric phenotypes (2,20,28,29), we have limited our analysis of TSQ1 addition in individual nuclei to very short-term culture (48 h in the case of the MRC-5 experiments described in Figure 4), and we have monitored for the appearance of telomere fusions that might skew our results. Finally, although TSQ1 is better tolerated than other mutant TERs in all of the cell types we have tested, we find that some cell types tolerate TSQ1 better than others (Figures 2 and 3 and data not shown). It will therefore be necessary to evaluate the degree to which TSQ1 induces proliferative inhibition and genome instability in other cell types before using this strategy in new contexts.
In conclusion, TSQ1 provides a technically simple method for in situ mapping of telomerase activity patterns in many cell types. To our knowledge, it is the only assay that can be used to track in situ elongation of individual telomeres. By providing a snapshot of recent telomerase activity, TSQ1 will enable the exploration of telomere elongation patterns and the dissection of the molecular mechanisms that shape those patterns.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health grant [K08 CA134552]; the Program for Breakthrough Biomedical Research, which is funded in part by the Sandler Foundation; the University of California Cancer Research Coordinating Committee (CRCC). Funding for open access charge: University of California San Francisco, Department of Pathology.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank I. Listerman for critical review of the manuscript.
REFERENCES
- 1.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 2.Stohr BA, Blackburn EH. ATM mediates cytotoxicity of a mutant telomerase RNA in human cancer cells. Cancer Res. 2008;68:5309–5317. doi: 10.1158/0008-5472.CAN-08-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armanios M, Blackburn EH. The telomere syndromes. Nat. Rev. Genet. 2012;13:693–704. doi: 10.1038/nrg3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 5.Hemann MT, Strong MA, Hao LY, Greider CW. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- 6.Zhu L, Hathcock KS, Hande P, Lansdorp PM, Seldin MF, Hodes RJ. Telomere length regulation in mice is linked to a novel chromosome locus. Proc. Natl Acad. Sci. USA. 1998;95:8648–8653. doi: 10.1073/pnas.95.15.8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouellette MM, Liao M, Herbert BS, Johnson M, Holt SE, Liss HS, Shay JW, Wright WE. Subsenescent telomere lengths in fibroblasts immortalized by limiting amounts of telomerase. J. Biol. Chem. 2000;275:10072–10076. doi: 10.1074/jbc.275.14.10072. [DOI] [PubMed] [Google Scholar]
- 8.Britt-Compton B, Capper R, Rowson J, Baird DM. Short telomeres are preferentially elongated by telomerase in human cells. FEBS Lett. 2009;583:3076–3080. doi: 10.1016/j.febslet.2009.08.029. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Kha H, Ungrin M, Robinson MO, Harrington L. Preferential maintenance of critically short telomeres in mammalian cells heterozygous for mTert. Proc. Natl Acad. Sci. USA. 2002;99:3597–3602. doi: 10.1073/pnas.062549199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feuerhahn S, Iglesias N, Panza A, Porro A, Lingner J. TERRA biogenesis, turnover and implications for function. FEBS Lett. 2010;584:3812–3818. doi: 10.1016/j.febslet.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 11.Loayza D, De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 12.Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell. 2009;138:463–475. doi: 10.1016/j.cell.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y, Abreu E, Kim J, Stadler G, Eskiocak U, Terns MP, Terns RM, Shay JW, Wright WE. Processive and distributive extension of human telomeres by telomerase under homeostatic and nonequilibrium conditions. Mol. Cell. 2011;42:297–307. doi: 10.1016/j.molcel.2011.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu L, Blackburn EH. Human Rif1 protein binds aberrant telomeres and aligns along anaphase midzone microtubules. J. Cell Biol. 2004;167:819–830. doi: 10.1083/jcb.200408181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Rosenberg JE, Donjacour AA, Botchkina IL, Hom YK, Cunha GR, Blackburn EH. Rapid inhibition of cancer cell growth induced by lentiviral delivery and expression of mutant-template telomerase RNA and anti-telomerase short-interfering RNA. Cancer Res. 2004;64:4833–4840. doi: 10.1158/0008-5472.CAN-04-0953. [DOI] [PubMed] [Google Scholar]
- 16.Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 17.Nakayama J, Tahara H, Tahara E, Saito M, Ito K, Nakamura H, Nakanishi T, Ide T, Ishikawa F. Telomerase activation by hTRT in human normal fibroblasts and hepatocellular carcinomas. Nat. Genet. 1998;18:65–68. doi: 10.1038/ng0198-65. [DOI] [PubMed] [Google Scholar]
- 18.Chen C, Wu B, Wei T, Egholm M, Strauss WM. Unique chromosome identification and sequence-specific structural analysis with short PNA oligomers. Mamm. Genome. 2000;11:384–391. doi: 10.1007/s003350010072. [DOI] [PubMed] [Google Scholar]
- 19.Marusic L, Anton M, Tidy A, Wang P, Villeponteau B, Bacchetti S. Reprogramming of telomerase by expression of mutant telomerase RNA template in human cells leads to altered telomeres that correlate with reduced cell viability. Mol. Cell. Biol. 1997;17:6394–6401. doi: 10.1128/mcb.17.11.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guiducci C, Cerone MA, Bacchetti S. Expression of mutant telomerase in immortal telomerase-negative human cells results in cell cycle deregulation, nuclear and chromosomal abnormalities and rapid loss of viability. Oncogene. 2001;20:714–725. doi: 10.1038/sj.onc.1204145. [DOI] [PubMed] [Google Scholar]
- 21.Stohr BA, Xu L, Blackburn EH. The terminal telomeric DNA sequence determines the mechanism of dysfunctional telomere fusion. Mol. Cell. 2010;39:307–314. doi: 10.1016/j.molcel.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bryan TM, Englezou A, Dunham MA, Reddel RR. Telomere length dynamics in telomerase-positive immortal human cell populations. Exp. Cell Res. 1998;239:370–378. doi: 10.1006/excr.1997.3907. [DOI] [PubMed] [Google Scholar]
- 23.Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Redon S, Reichenbach P, Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;38:5797–5806. doi: 10.1093/nar/gkq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pickett HA, Cesare AJ, Johnston RL, Neumann AA, Reddel RR. Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J. 2009;28:799–809. doi: 10.1038/emboj.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 27.Chen LY, Redon S, Lingner J. The human CST complex is a terminator of telomerase activity. Nature. 2012;488:540–544. doi: 10.1038/nature11269. [DOI] [PubMed] [Google Scholar]
- 28.Conomos D, Stutz MD, Hills M, Neumann AA, Bryan TM, Reddel RR, Pickett HA. Variant repeats are interspersed throughout the telomeres and recruit nuclear receptors in ALT cells. J. Cell Biol. 2012;199:893–906. doi: 10.1083/jcb.201207189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brault ME, Autexier C. Telomeric recombination induced by dysfunctional telomeres. Mol. Biol. Cell. 2011;22:179–188. doi: 10.1091/mbc.E10-02-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.