Abstract
We report that the metazoan Wnt protease and signaling inhibitor TIKI shares sequence homology with bacterial TraB/PrgY proteins, inhibitors of pheromone signaling essential for propagation of antibiotic resistance. Our analysis suggests that these proteins represent an ancient metalloprotease clan regulating cellular communications across biological kingdoms.
Signaling by the secreted Wnt family of lipoproteins has essential functions in animal development and homeostasis. Diverse mechanisms of regulating autocrine and paracrine Wnt signaling have evolved, mostly involving actions of proteins that block the assembly of the Wnt-Frizzled-LRP5/6 receptor signaling complex, or inhibit cell-to-cell movement of Wnts (MacDonald et al., 2009). The discovery of the membrane-tethered TIKI metalloprotease reveals a distinct mode of attack by cleaving the amino terminus of mature Wnt proteins, thereby inactivating the Wnt ligand (Zhang et al., 2012) (Figure 1A). Provocatively, TIKI proteins display sequence homology with the recently merged TraB/PrgY/gumN family of bacterial proteins (PFAM families PF01963 and PF07446). The biochemical nature of TraB/PrgY from the gut flora Enterococcusfaecalis, like that of gumN from the plant pathogen Xanthomonas, has not been characterized (Chandler and Dunny, 2004; 2008).
Figure 1.
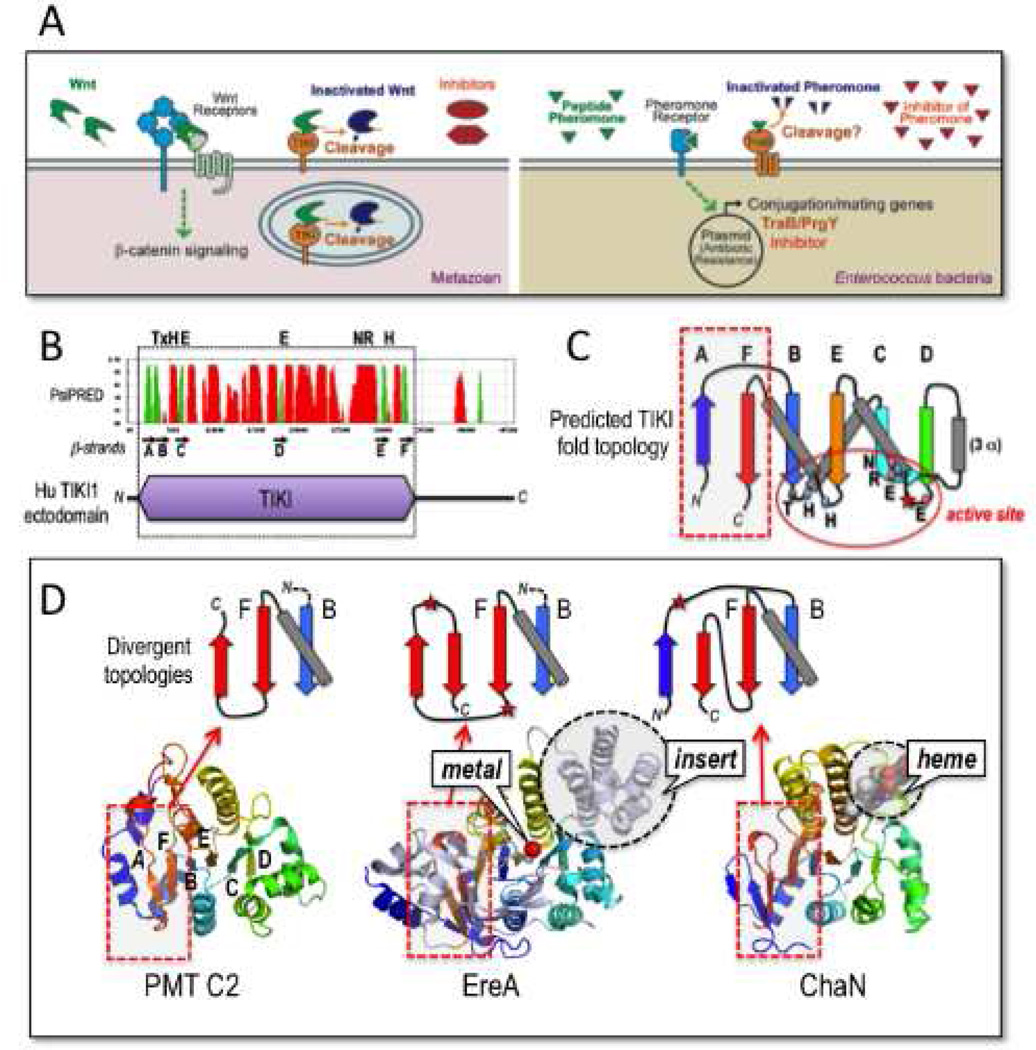
TIKI and TraB/PrgY proteins and links to an ancient metalloprotease fold. (A) Comparisons of the mechanism of action by which TIKI cleaves and inactivates Wnt ligands in animals versus that proposed for TraB/PrgY in cleavage and inactivation of the mating pheromone in Enterococcus bacteria (reviewed by Chandler and Dunny, 2004). TIKI cleaves Wnts in the secretory pathway/vesicle and on the cell surface. The functional parallel extends to Wnt antagonists (sFRP and DKK) and the bacterial pheromone inhibitor. (B) Structural organization of TIKI reveals aglobular ectodomain (purple) rich in secondary structures (predicted by PsiPRED; red peaks are helices, green peaks mark β-strands). A set of β-strands, labeled A to F (black arrows), and conserved putative active site residues (on top) are highlighted. (C) The predicted α+β fold architecture of the TIKI domain. β-strands A to F form a largely parallel β-sheet core that clusters conserved putative active site residues (also see Figure S1A). (D) The predicted TIKI domain fold topology is related to an ancestral core β-sheet fold found in EreA, ChaN and PMT C2 domain proteins (PDB 2QGM, 3B55, 2G5G and 2EBF, respectively). Additional structural embellishments (grey helical insertin EreA), ligands (red metal ion in EreA, and bound heme in ChaN), and chain insertions (red stars) are depicted. Red dashed boxes show predicted regions of fold divergence between TIKI and EreA/ChaN/PMT C2 domains (see also Figure S1B). The divergent β-strand topologies of the latter group, as highlighted by diagrams above the respective structures, contrast with the tight β-hairpin turn between antiparallel βA and βB strands predicted in TIKI/TraB proteins (panel C); notably, the PMT C2 C-terminal βA strand serves as template for the TIKI βA strand. These peripheral zones of topological divergence do not impact the respective active site pockets. An example of a minimal α/β fold of the core TIKI/TraB/PrgY topology, without inserts, embellishments, and metal-coordinating residues, is exhibited by the domain structure of Sua5 (PFAM code PF03481; PDB 2YV4, from hyperthermophilic archaeon P. horikoshii), whose biochemical function is unknown.
Enterococcus faecalis is a multidrug-resistant pathogen and common source for hospital infections. These bacteria harbor plasmids that carry antibiotic resistance genes, and transfer these plasmids and thus antibiotic resistance through mating conjugation (Chandler and Dunny, 2004). Mating is initiated by a pheromone (usually a hepta- or octa-peptide) that functions to induce the expression of plasmid-encoded conjugation factors, which mediate mating (Figure 1A). Self-induction (mating between bacteria that already harbor the plasmid) is unproductive and energy-consuming, and is suppressed by two plasmid-encoded factors (Figure 1A): an inhibitor peptide that antagonizes the pheromone, and the transmembrane protein TraB/PrgY, which reduces the pheromone level or activity via an unknown mechanism (Chandler and Dunny, 2004; 2008). Multiple mating pheromones exist and multiple versions of the inhibitor peptide and TraB/PrgY are encoded by these plasmids, conferring the bacterium with exquisite regulation of its specificity and sensitivity to a given pheromone (Chandler and Dunny 2004). Based on homology with TIKI proteins, it appears likely that TraB/PrgY acts as a protease to inactivate the mating pheromone (Figure 1A). The existence of TraB/PrgY and the inhibitor in the bacterial pheromone response pathway bears striking functional parallels to that of TIKI and Wnt antagonists in morphogen signaling in animal development (Figure 1A).
The human TIKI ectodomain features an amino-terminal region of 330 residues, which is predicted to be globular and rich in secondary structure (Figure 1B) and is homologous to predicted TraB/PrgY products, as these proteins are each marked by a pair of widely spaced GX2H motifs and a conserved glutamate (PFAM PF01963) (Figure S1A). To uncover the corresponding catalytic site signposts in TIKI, we employed a structure-aided method (Pei et al., 2008) to accurately align representative TIKI and TraB/PrgY proteins, and also sought a three-dimensional template for the family by sensitive fold recognition and structure prediction methods (Söding, 2005). These efforts reliably identified a distant fold match for TIKI and TraB/PrgY proteins with a small group of enigmatic proteins, referred to as “EreA/ChaN-like” by the SCOP domain fold database. EreA is an erythromycin esterase (PDB 2QGM and 3B55) (Morar et al., 2012), and ChaN (PDB 2G5G) is a putative heme-transport protein (Chan et al., 2006). The most compact version of the EreA/ChaN-likeα+β fold is present in the C2 domain of a bacterial (Pasteurella multocida) toxin (PMT; PDB 2EBF); this domain of unknown function is fused to a carboxyl terminal Cys-protease module (Kitadokoro et al., 2007). Comparison of these available structures shows a conserved parallel β-sheet core (with helices packed on both sides) with a variable N-terminal end and distinct helical inserts into a central loop, with the differences manifesting as divergent embellishments to the core (Figure 1C and 1D). Intriguingly, a series of conserved amino acids (His39, Glu66, Glu140, Arg283, Asn284, and His311 in human TIKI1) map to the putative active site pocket in the core (Figures 1C and S1A), implying a catalytic assembly for the TIKI metalloprotease and TraB/PrgY proteins. This core has degenerated in PMT and other C2-related domains in MARTX-type bacterial toxins (Figure S1B).
The best model for TIKI catalytic function comes from EreA enzymes, in which the esterase active site (for macrolide antibiotic hydrolysis) has been crystallized with a bound, though indistinct metal ion (Morar et al., 2012) (Figure S1C). This appears to be consistent with the observed dependence of the TIKI protease activity on metals (Mn2+ or Co2+, but not on Ni2+, Cu2+ or Zn2+ ions) (Zhang et al., 2012). Catalytic core residues of EreA are matched by PDBSiteScan (Ivanisenko et al., 2004) to the Zn2+-coordinating catalytic geometry of carboxypeptidase A4 (Gomis-Ruth, 2008). A corresponding model for TIKI1 suggests that the conserved Glu140 at the end of β–strand C (Figure S1A) acts as the general base/acid in the catalytic reaction that enlists an activated water molecule in the nucleophilic attack of the scissile peptide bond in substrates (i.e., Wnts) (Figure S1C). Like carboxypeptidase A4, TIKI1 features a similar helix-positioned Arg283-Asn284 pair (Arg-Asp in TraB/PrgY proteins) that could act to stabilize the substrate chain (Gomis-Ruth, 2008). However, the EreA-like TIKI active site is embedded in a metalloprotease fold that is unrelated to carboxypeptidase A4 and other metalloenzymes.
We propose that the TIKI/TraB/PrgY superfamily represents an ancient metalloprotease clan with a common protein architecture––cobbled from the folds of the EreA/ChaN/PMT group––that mediates proteolytic activities. These structural insights should facilitate understanding of how TIKI and TraB/PrgY proteins specifically recognize and cleave their respective Wnt and pheromone substrates, and drive therapeutic targeting of these enzymes for pathogenesis associated with abnormal Wnt signaling or bacterial antibiotic resistance.
Supplementary Material
ACKNOWLEDGEMENTS
X.H. acknowledges support of this work by NIH (RO1 GM057603 and GM057603S1), and by Children's Hospital Boston Intellectual and Developmental Disabilities Research Center (P30 HD-18655).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chan AC, Lelj-Garolla B, F IR, Pedersen KA, Mauk AG, Murphy ME. Cofacial heme binding is linked to dimerization by a bacterial heme transport protein. J Mol. Biol. 2006;362:1108–1119. doi: 10.1016/j.jmb.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Chandler JR, Dunny GM. Enterococcal peptide sex pheromones: synthesis and control of biological activity. Peptides. 2004;25:1377–1388. doi: 10.1016/j.peptides.2003.10.020. [DOI] [PubMed] [Google Scholar]
- Chandler JR, Dunny GM. Characterization of the sequence specificity determinants required for processing and control of sex pheromones by the intramembrane protease Eep and plasmid-encoded protein PrgY. J. Bacteriol. 2008;190:1172–1183. doi: 10.1128/JB.01327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis-Ruth FX. Structure and mechanism of metallocarboxypeptidases. Crit Rev Biochem. Mol. Biol. 2008;43:319–345. doi: 10.1080/10409230802376375. [DOI] [PubMed] [Google Scholar]
- Ivanisenko VA, Pintus SS, Grigorovich DA, Kolchanov NA. PDBSiteScan: a program for searching for active, binding and posttranslational modification sites in the 3D structures of proteins. Nucleic Acids Res. 2004;32:W549–W554. doi: 10.1093/nar/gkh439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitadokoro K, Kamitani S, Miyazawa M, Hanajima-Ozawa M, Fukui A, Miyake M, Horiguchi Y. Crystal structures reveal a thiol protease-like catalytic triad in the C-terminal region of Pasteurella multocida toxin. Proc. Natl. Acad. Sci. USA. 2007;104:5139–5144. doi: 10.1073/pnas.0608197104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morar M, Pengelly K, Koteva K, Wright GD. Mechanism and diversity of the erythromycin esterase family of enzymes. Biochemistry. 2012;51:1740–1751. doi: 10.1021/bi201790u. [DOI] [PubMed] [Google Scholar]
- Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- Zhang X, Abreu JG, Yokota C, MacDonald BT, Singh S, Coburn KL, Cheong SM, Zhang MM, Ye QZ, Hang HC, et al. Tiki1 is required for head formation via Wnt cleavage-oxidation and inactivation. Cell. 2012;149:1565–1577. doi: 10.1016/j.cell.2012.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.