Abstract
We have developed a novel DNA assay based on exonuclease III (ExoIII)-induced target recycling and the fluorescence quenching ability of graphene oxide (GO). This assay consists of a linear DNA probe labeled with a fluorophore in the middle. Introduction of target sequence induces the exonuclease III catalyzed probe digestion and generation of single nucleotides. After each cycle of digestion, the target is recycled to realize the amplification. Finally, graphene oxide is added to quench the remaining probes and the signal from the resulting fluorophore labeled single nucleotides is detected. With this approach, a sub-picomolar detection limit can be achieved within 40 minutes at 37 °C. The method was successfully applied to multicolor DNA detection and the analysis of telomerase activity in extracts from cancer cells.
Keywords: Graphene oxide, Exonuclease III, DNA Detection
1. Introduction
Detection of nucleic acids has great importance for a wide range of applications, including gene therapy, forensic investigations and clinical diagnosis. Development of simple and sensitive DNA assays to detect unique target sequences has been a major goal of biosensor technology. Thus far, the polymerase chain reaction (PCR) has been the method of choice. Although it is extremely powerful in amplifying oligonucleotide targets, making even single-molecule detection possible, PCR is limited by its own complexity, as well as contamination and false-positive signals.(Lie and Petropoulos, 1998, Lang et al., 1997, Heid et al., 1996)
Recently, molecular beacons, which undergo a conformational change from a closed fluorescence-off state to an open fluorescence-on state in the presence of target nucleic acid, have emerged as an alternative by their simplicity and specificity.(Xiao et al., 2009, Zhang et al., 2001, Tyagi et al., 2000, Tyagi et al., 1998, Tyagi and Kramer, 1996) However, the sensitivity of detection is strictly limited because of the 1:1 hybridization ratio. In response, several signal amplification strategies have been developed to increase fluorescence. Particularly, enzymatic amplification involving DNA enzymes, such as nicking endonucleases (Xu et al., 2009), DNase I (Lu et al., 2010) and exonuclease III, (Zuo et al., 2010), (Zhang et al., 2011) is of great interest. Meanwhile, novel materials, such as cationic polymers (Gaylord et al., 2003), quantum dots (Zhang et al., 2005) and gold nanoparticles (Taton et al., 2000) are employed as detection platforms. Among all of these materials, GO has shown great promise in improving the detection sensitivity of nucleic acids. (Balapanuru et al., 2010, He et al., 2010, Li et al., 2010, Lu et al., 2009) However, most of the GO based detection systems rely on the competition between the formation of target-ssDNA complex and the formation of GO-ssDNA complex. Few studies have focused on GO’s different affinity to long ssDNA (10 to 30 bases) and to single nucleotides.
In this paper, we present a rapid, sensitive and selective method to detect DNA using exonuclease III (ExoIII) as a signal amplifier and GO as a fluorescence quencher. This method takes advantage of the much higher binding affinity of GO to the ssDNA than to the single nucleotides. Exonuclease III catalyzes the digestion of duplex DNAs from blunt 3’-hydroxyl termini, but it has only limited activity on single-stranded DNA. (Wang et al., 2005, CC and IR, 1964) GO is proven to strongly bind single-stranded DNA due to the hydrophobic and - stacking interactions with nucleobases. And due to the multivalent interaction between the GO surface and each nucleotide of a ssDNA, a ssDNA generally exhibits higher binding affinity with GO than a single nucleotide. GO is also recognized as an effective quencher for a variety of fluorophores and greatly decreases the background noise and thus increases the signal-to-noise ratio in fluorescence measurements, in addition to functioning as a convenient and versatile platform for multicolor fluorescence analysis of DNAs. (He et al., 2010, Lu et al., 2009)
2. Experimental
2.1. Chemicals and reagents
The oligonucleotide sequences are listed in Table S1. DNA synthesis reagents were purchased from Glen Research (Sterling, VA). DNA probes were synthesized using standard phosphoramidite chemistry and purified using reversed phase HPLC. Fluorescein-dT and TAMRA-dT phosphoramidites (Glen Research, Sterling, VA) were used in the synthesis of signaling probes (S1 and S2). Exonuclease III and dNTPs were purchased from New England Biolabs (Ipswich, MA, USA). Graphene oxide (1mg/mL) was received as a gift from Professor Hongjie Dai (Department of Chemistry, Stanford University), and 4-(1-pyrenyl) butyric acid (PBA) (Fisher Scientific) was used as a surface-blocking agent.
2.2. Apparatus and measurements
An ABI 3400 DNA/RNA synthesizer (Applied Biosystems) was used for DNA synthesis. Probe purification was performed with a ProStar HPLC (Varian) equipped with a C18 column (Econosil 5U, 250 × 4.6 mm) from Alltech Associates using acetonitrile and 0.1 M TEAA as mobile phases. A Cary Bio 100 UV/vis spectrometer (Varian) was used for probe quantitation. Steady-state fluorescence measurements were performed on a FluoroMax-4 spectrofluorometer (Jobin Yvon, Edison, NJ), using a 100µL quartz fluorescence cuvette.
Exonuclease III-aided target recycling was carried out in 120 µL of ExoIII reaction buffer (20mM Tris-HCl, 5mM MgCl2, 50mM NaCl, pH 8.0mM) in a test tube which contained 100 nM signaling probe, 10 µM PBA, 30 units of exonuclease III and varying concentrations of target at 37 °C for 30 minutes. Then, 6 µL graphene oxide (1mg/mL) was added to the reaction solution, followed by a 10 min incubation period for complete quenching of undigested signaling probes. Fluorescence was measured using 488 nm as the excitation wavelength for the FITC-labeled probe and 543 nm as the excitation wavelength for the TAMRA–labeled probe. Each experiment was repeated at least three times.
2.3. Detection of telomerase in HeLa cell extracts
The telomerase was extracted by the CHAPS method. One million HeLa cell pellets were first suspended in 200 µL 1×CHAPS lysis buffer (0.5% CHAPS, 10 mM Tris−HC1, pH 7.5, 1 mM MgC12, 1 mM EGTA, 5 mM β-mercaptoethanol, 0.1 mM PMSF, 10% glycerol) and incubated on ice for 30 min. The mixture was centrifuged at 16000 rpm for 20 min at 4 °C, and the supernatant was collected. The resulting extract was stored at −80 °C.
In the detection, telomerase extract diluted in lysis buffer with the respective number of cells and TS primer (100 nM) were incubated in 20 µL of extension solution (50 mM Tris• HC1, pH 7.5, 1 mM MgC12, 1 mM EGTA, 50 mM KCl) at 30 °C for 0.5 h. Next, a mixture of dNTPs was added, and the final concentration was 0.2 mM. The mixture was incubated at 30 °C for 1 h, after which 20 µL of 10×ExoIII buffer and 160 µL of water were added. Then, signaling probe for telomere extension and ExoIII were added for signal generation. Finally, after incubation at 37 °C for 30 min, the fluorescence signal was measured. For control experiments, telomerase extracts were heat treated (90 °C for 3 min).
3. Results and discussion
3.1. Principle of amplified DNA detection using exonuclease III and graphene oxide
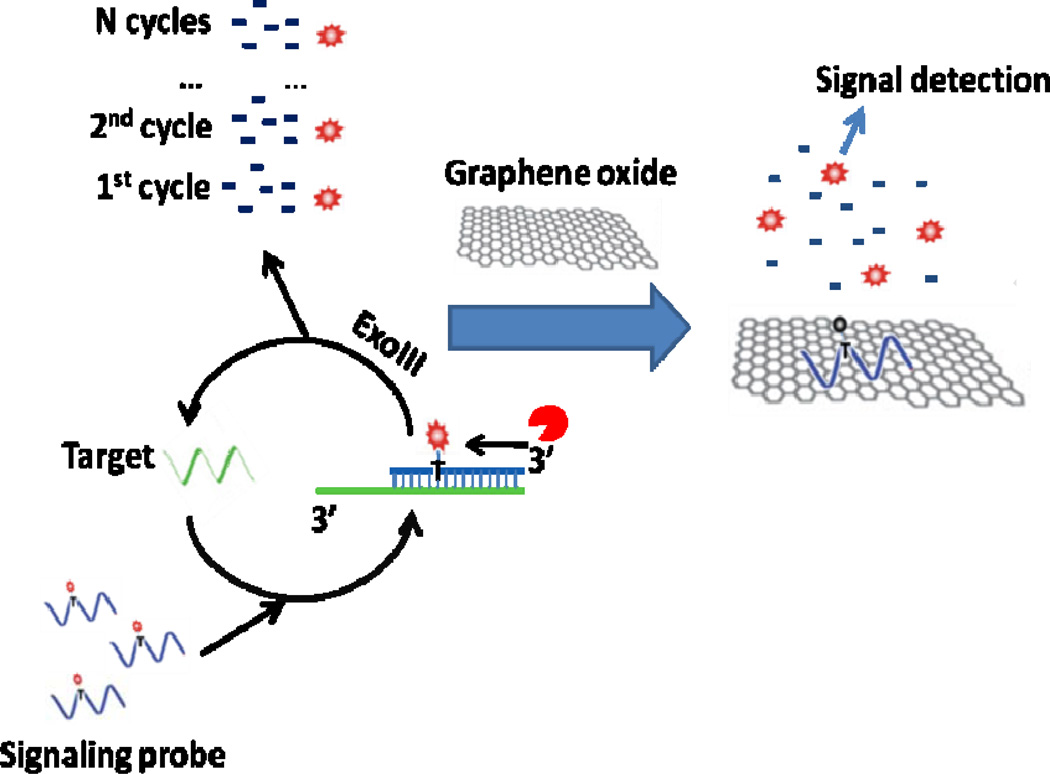
As shown in scheme 1, a single-stranded linear DNA with a fluorophore (red dot in scheme 1)-modified dT base (fluo-dT) at an internal position is employed as the signaling probe. It is designed to hybridize with the perfectly matched target DNA to form a duplex structure with a blunt 3’ terminus on the signaling probe, but with a protruding 3’ terminus on the target strand. After the formation of the duplex, exonuclease III digests the signal probe in a stepwise manner to generate single nucleotides, releasing fluo-dT and the target. In this manner, the released target can then bind to another signaling probe to initiate another round of digestion by exonuclease III and release another fluorophore from a signaling probe. This recycling of target can be repeated multiple times to accumulate free fluo-dTs. Finally, graphene oxide is added and strongly adsorbs single-stranded DNAs, but not the single nucleotides. As a result, undigested signaling probes bind to GO and are quenched, but fluo-dTs stay in solution to give a fluorescence signal.
Scheme 1.
Scheme showing amplified DNA detection using ExoIII-induced target recycling and fluorescence quenching of undigested signaling probes by graphene oxide.
3.2. Verification of amplified detection scheme
In this assay, exonuclease III-induced target recycling is conducted at 37°C for 30 min, followed by addition of graphene oxide. The amplification scheme is verified by comparison with the conventional “premixing” method (Scheme S1), using GO for DNA detection. (He et al., 2010) Upon addition of 5 nM target (T), the conventional “premixing” method shows a small signal difference from the background signal (Figure S1). However, the signal is greatly enhanced by exonuclease III and GO aided amplification, which leads to a huge increase in fluorescence signal (Figure S1). Meanwhile, the background signal resulting from nonspecific digestion of signaling probes (S) by exonuclease III is much lower compared with that of previously reported methods using exonuclease III. (Cui et al., 2010, Zuo et al., 2010) Such low background is achieved by the design of a signaling probe in which the internal fluo-dT is more than 10 bases from the 3’ end. In addition, graphene oxide shows excellent quenching ability for fluorescence dyes. (He et al., 2010, Lu et al., 2009) When considering both factors, this assay allows a high signal-to-noise ratio, leading to a very low limit of detection. In addition to dramatic signal enhancement, rapid detection of DNA target is also achieved, with an assay time of about 40 minutes.
3.3. Calibration curve and sensitivity of this assay
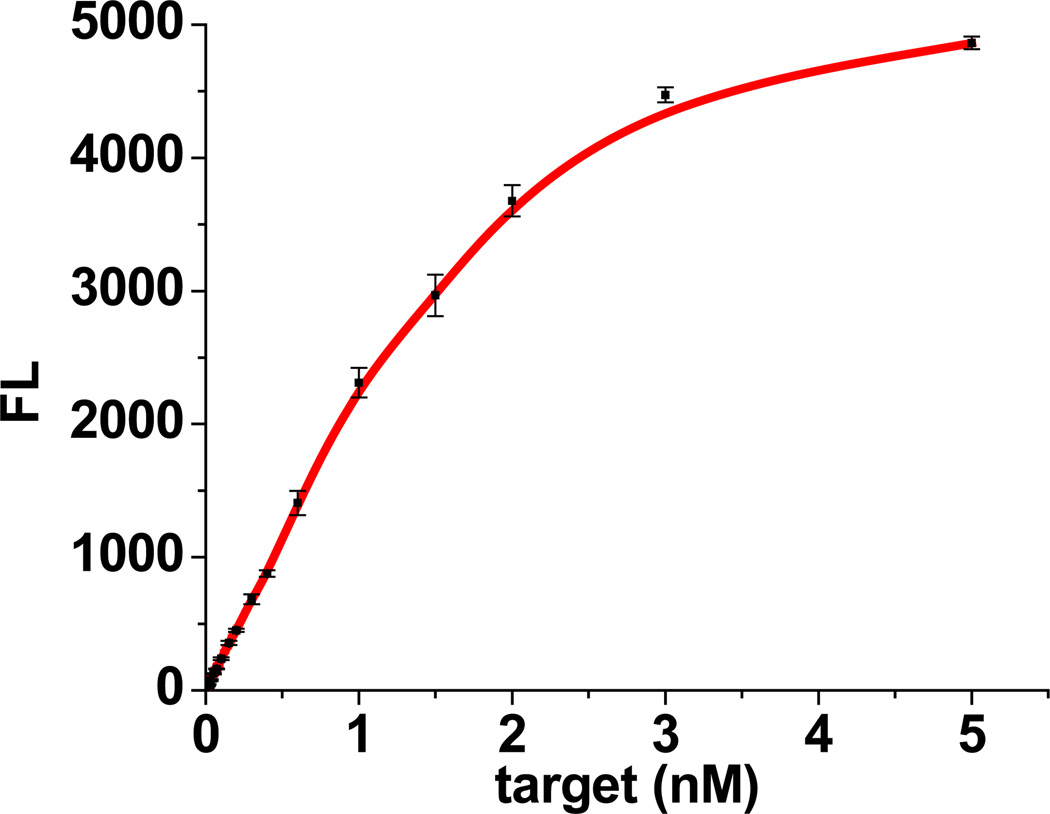
Based on the experimental results shown in Figure S2–S4, the optimized condition for detection is determined to be 100 nM of signaling probe, 30 units of exonuclease III and 30 min of reaction time at 37 °C. Figure S5 shows fluorescence spectra generated by this assay for target DNA of different concentrations. In the absence of target (T), a very low fluorescence signal is obtained as a consequence of the resistance of single-stranded DNA to exonuclease III digestion. However, upon the addition of target (T), an increase in the fluorescence signal is observed in response to target (T) of increasing concentrations from 2 pM to 5 nM (Figure 1). Within the range from 2 pM to 1 nM, the peak intensity increases linearly with target (T) concentration (Figure S6). Based on the 3 rule, the limit of detection is estimated to be 0.5 pM, which is about 20-fold lower than that of previous exonuclease III-aided strategies.(Cui et al., 2010, Zuo et al., 2010) Exhibiting good selectivity, our assay can distinguish single-base mismatched target. As shown in Figure S7, a single-base mismatched target produces a signal less than half that of perfectly matched target of equal concentration.
Figure 1.
Plot of fluorescence intensity at 516 nm vs. target concentration.
3.4. Multiplexed detection using this assay
It has been recognized that simultaneous detection of multiple targets holds new promise in molecular diagnostics.(Song et al., 2009, Tyagi et al., 1998, Sidransky, 1997) The use of GO as a fluorescence quencher makes multicolor DNA analysis feasible.(He et al., 2010) With this assay, sensitive multicolor DNA analysis can be realized easily and rapidly as shown in Scheme S2. Two probes (S1 and S2) labeled with FITC and TAMRA, respectively, are employed to demonstrate amplified multicolor DNA analysis of targets T1 and T2. In the presence of 100 pM T1 only, a strong fluorescence signal is detected in the channel of S1 (FITC, excitation =488 nm), while no signal is observed in the channel of S2 (TAMRA, excitation =543 nm) (Figure S8a, red). Similarly, 100 pM of T2 leads only to a fluorescence signal for TAMRA (Figure S8a, black). However, the presence of both targets T1 and T2 leads to a fluorescence signal in both channels (Figure S8b, black).
Furthermore, to verify the general applicability of this assay for real-sample analysis, we investigated multiplexed DNA detection in 50% serum. Following the same procedure for multiplexed DNA detection in buffer, we achieved the signal for targets spiked in the 50% serum. As shown in Figure S9, both background and signal increase compared with the values in buffer due to the complexity of serum. However, multiplexed DNA detection is successfully performed with results similar to that in buffer. Therefore, the graphene-ExoIII based DNA assay shows good potential for clinical applications.
3.5. Application of this assay for telomerase detection
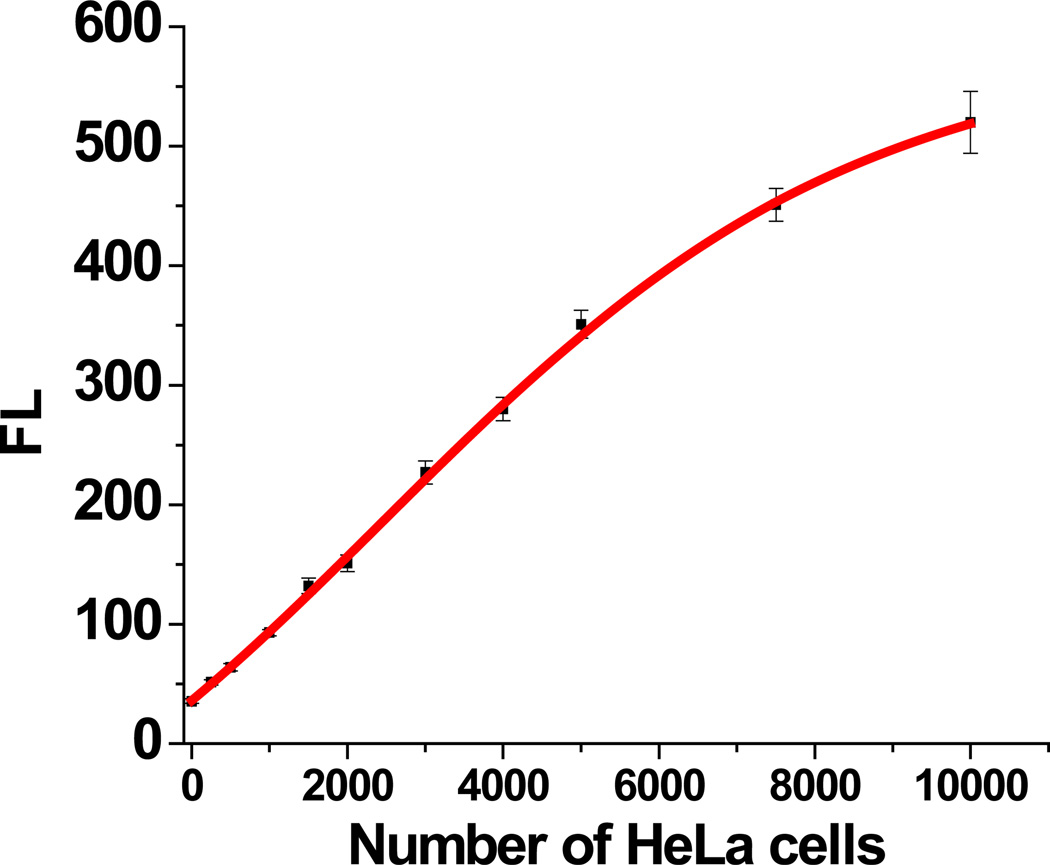
In order to demonstrate the feasibility of our assay in practical applications, experiments were designed to analyze telomerase activity in cell extracts from HeLa cells. Telomerase extension reaction of TS primer was carried out using a previously reported method with modifications (Ding et al., 2010, Herbert et al., 2006). Afterwards, our ExoIII/GO-based assay was performed to detect telomerase extension products with signaling probe (S-tel) complementary to telomere repeats. As shown in Figure S10, cell extract from 5,000 HeLa cells (red curve) exhibits a much higher fluorescence signal than that of heat-treated cell extract from 20,000 HeLa cells (black curve). The calibration curve of fluorescence signal vs number of HeLa cells (Figure 2) shows that the telomerase activity in the extract from as few as 250 cells can be detected.
Figure 2.
Analysis of telomerase activity in cell extracts from HeLa cells: relationship between fluorescence intensities and different amounts of cell extracts from different numbers of cells.
Moreover, different types of cancer cells have different degrees of telomerase activity. It is important to differentiate the telomerase activity among different cancer cells for diagnostic purposes. Therefore, we also applied our assay for analysis of telomerase activity in different cancer cells. The fluorescence signals generated by cell extracts from 5,000 cells were acquired for three different cell lines, including HeLa, MCF-7 and K562 cells. It was found that MCF-7 cells show a lower telomerase activity than HeLa cells, while K562 cells exhibit a higher telomerase activity (Figure S11). This is consistent with previous reports (Hirose et al., 1997, Zhou et al., 2009) and proves that our ExoIII/GO-based platform is able to discriminate between telomerase activities of different cancer cells.
4. Conclusion
In conclusion, we have demonstrated an amplified method for rapid detection of DNA based on ExoIII-induced target recycling and graphene oxide quenching. Binding of target DNA with signaling probe generates a DNA duplex having an overhang on the 3’-end of the target DNA strand to trigger the digestion of signaling probe by ExoIII. This releases the target and allows the the entire reaction to be recycled. With this approach, a sub-picomolar detection limit can be achieved within 40 minutes at 37 °C. The method was successfully applied to multicolor DNA detection and the analysis of telomerase activity in extracts from different cancer cells. The method proposed here avoids the complex temperature cycling protocol used in PCR and does not require a specific recognition site for ExoIII. Furthermore, sensitive detection of DNA in real samples is also achieved. Therefore, this ExoIII-GO based strategy for DNA detection may find wide application in cancer and disease diagnostics, detection of biological warfare agents and forensic analysis.
Supplementary Material
Highlights.
Novel DNA assay based on exonuclease III-induced target recycling
Fluorescence detection and selective quenching of ssDNA by graphene oxide
Sub-pM detection limit within 40 min at 37 °C
Application to multicolor DNA detection and analysis of telomerase activity
ACKNOWLEDGMENT
This work is supported by grants awarded by the National Institutes of Health (GM066137, GM079359 and CA133086).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Heid CA, Stevens J, Livak KJ, Williams PM. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Lang R, Pfeffer K, Wagner H, Heeg K. J. Immunol. Methods. 1997;203:181–192. doi: 10.1016/s0022-1759(97)00028-8. [DOI] [PubMed] [Google Scholar]
- Lie YS, Petropoulos CJ. Curr. Opin. Biotechnol. 1998;9:43–48. doi: 10.1016/s0958-1669(98)80082-7. [DOI] [PubMed] [Google Scholar]
- Tyagi S, Bratu DP, Kramer FR. Nat. Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- Tyagi S, Kramer FR. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- Tyagi S, Marras SAE, Kramer FR. Nat. Biotechnol. 2000;18:1191–1196. doi: 10.1038/81192. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Plakos KJI, Lou X, White RJ, Qian J, Plaxco KW, Soh HT. Probe. Angew. Chem., Int. Ed. 2009;48:4354–4358. doi: 10.1002/anie.200900369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Beck T, Tan W. Angew. Chem., Int. Ed. 2001;40:402–405. doi: 10.1002/1521-3773(20010119)40:2<402::AID-ANIE402>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Xu W, Xue X, Li T, Zeng H, Liu X. Angew. Chem., Int. Ed. 2009;48:6849–6852. doi: 10.1002/anie.200901772. [DOI] [PubMed] [Google Scholar]
- Lu CH, Li J, Lin MH, Wang YW, Yang HH, Chen X, Chen GN. Angew. Chem., Int. Ed. 2010;49:8454–8457. doi: 10.1002/anie.201002822. [DOI] [PubMed] [Google Scholar]
- Zuo X, Xia F, Xiao Y, Plaxco KW. J. Am. Chem. Soc. 2010;132:1816–1818. doi: 10.1021/ja909551b. [DOI] [PubMed] [Google Scholar]
- Zhang M, Guan YM, Ye BC. Chem.Commun. 2011;47:3478–3480. doi: 10.1039/c0cc05703g. [DOI] [PubMed] [Google Scholar]
- Gaylord BS, Heeger AJ, Bazan GC. J. Am. Chem. Soc. 2003;125:896–900. doi: 10.1021/ja027152+. [DOI] [PubMed] [Google Scholar]
- Zhang CY, Yeh HC, Kuroki MT, Wang TH. Nat. Mater. 2005;4:826–831. doi: 10.1038/nmat1508. [DOI] [PubMed] [Google Scholar]
- Taton TA, Mirkin CA, Letsinger RL. Science. 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- Balapanuru J, Yang JX, Xiao S, Bao Q, Jahan M, Polavarapu L, Wei J, Xu QH, Loh KP. Angew. Chem., Int. Ed. 2010;49:6549–6553. doi: 10.1002/anie.201001004. [DOI] [PubMed] [Google Scholar]
- Lu CH, Yang HH, Zhu CL, Chen X, Chen GN. Angew. Chem., Int. Ed. 2009;48:4785–4787. doi: 10.1002/anie.200901479. [DOI] [PubMed] [Google Scholar]
- Li F, Huang Y, Yang Q, Zhong Z, Wang L, Song S, Fan C. Nanoscale. 2010;2:1021–1026. doi: 10.1039/b9nr00401g. [DOI] [PubMed] [Google Scholar]
- He S, Song B, Li D, Zhu C, Qi W, Wen Y, Wang L, Song S, Fang H, Fan C. Adv. Funct. Mater. 2010;20:453–459. [Google Scholar]
- CC R, IR L. J. Biol. Chem. 1964;239:251–258. [PubMed] [Google Scholar]
- Wang J, Li T, Guo X, Lu Z. Nucleic Acids Res. 2005;33:23–31. doi: 10.1093/nar/gni021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Ke G, Wang C, Yang CJ. 2010;135:2069–2073. doi: 10.1039/c0an00215a. [DOI] [PubMed] [Google Scholar]
- Song S, Liang Z, Zhang J, Wang L, Li G, Fan C. Angew. Chem., Int. Ed. 2009;48:8670–8674. doi: 10.1002/anie.200901887. [DOI] [PubMed] [Google Scholar]
- Sidransky D. 1997;278:1054–1058. doi: 10.1126/science.278.5340.1054. [DOI] [PubMed] [Google Scholar]
- Herbert BS, Hochreiter AE, Wright WE, Shay JW. Nat. Protoc. 2006;1:1583–1590. doi: 10.1038/nprot.2006.239. [DOI] [PubMed] [Google Scholar]
- Ding C, Li X, Ge Y, Zhang S. Anal. Chem. 2010;82:2850–2855. doi: 10.1021/ac902818w. [DOI] [PubMed] [Google Scholar]
- Hirose M, Abe-Hashimoto J, Ogura K, Tahara H, Ide T, Yoshimura T. J. Cancer Res. Clin. Oncol. 1997;123:337–344. doi: 10.1007/BF01438310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Xing D, Zhu D, Jia L. Anal. Chem. 2008;81:255–261. doi: 10.1021/ac801914b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.