Abstract
Extracellular matrix interactions play essential roles in normal physiology and many pathological processes. While the importance of ECM interactions in metastasis is well documented, systematic approaches to identify their roles in distinct stages of tumorigenesis have not been described. Here we report a novel screening platform capable of measuring phenotypic responses to combinations of ECM molecules. Using a genetic mouse model of lung adenocarcinoma, we measure the ECM-dependent adhesion of tumor-derived cells. Hierarchical clustering of the adhesion profiles differentiates metastatic cell lines from primary tumor lines. Furthermore, we uncovered that metastatic cells selectively associate with fibronectin when in combination with galectin-3, galectin-8, or laminin. We show that these molecules correlate with human disease and that their interactions are mediated in part by α3β1 integrin. Thus, our platform allowed us to interrogate interactions between metastatic cells and their microenvironments, and identified ECM and integrin interactions that could serve as therapeutic targets.
INTRODUCTION
Cancer metastasis is a poorly understood multistep process that results in 90% of cancer-related deaths1–2. At the time of initial diagnosis, almost half of lung adenocarcinoma patients have detectable metastases and the majority of the remaining half will relapse with metastatic disease after surgical removal of the primary tumor and adjuvant chemotherapy3. Despite the ominous nature of metastatic disease, the molecular mechanisms that drive each step are poorly characterized and few effective therapies exist4. Recently, it has become apparent that the tumor microenvironment dramatically impacts metastatic progression5. Changes in cancer cell-extracellular matrix (ECM) interactions likely influence each stage of the metastatic cascade, starting with the loss of basement membrane adhesion to colonization of distant sites. Furthermore, alterations in matrix production and crosslinking can promote metastasis6–8. Consequently, inhibiting interactions of tumor cells with their microenvironments by targeting adhesion molecules is an area of active investigation9–10.
While a variety of techniques exist for studying microenvironmental interactions, it has been challenging to date to interrogate the functional implications of specific cell-ECM interactions in a high-throughput manner. Injection of metastatic cells into embryos documented the anti-tumor effects of the embryonic microenvironment11–12, and coculture studies have identified the roles of carcinoma-associated fibroblasts on tumor progression13. ECM-coated transwells have been used to study the effects of small numbers of individual candidate ECM molecules on 2D invasion14, and 3D collagen gels have been useful particularly in the study of matrix metalloproteinase activity15. In vivo studies using gene-targeted mice have documented the importance of several ECM molecules and their receptors in transplant-based models of cancer and metastasis16–17. Each of these techniques has documented key microenvironmental regulators of metastasis, but they have not allowed an unbiased systematic evaluation of the role that ECM components play.
Cell-ECM interactions are particularly difficult to study due to their complexity of synergistic and antagonistic interactions in vivo18. Experiments targeting integrins, a central family of cell surface receptors that mediate ECM interactions, have implicated integrin-ECM interactions as important regulators of cancer progression9,19–20. However, in addition to adhesion, integrins regulate stress transmission and bidirectional signaling, and typically bind multiple ECM molecules21. Furthermore, transmembrane collagens, syndecans, lectins, carbohydrates, gangliosides, glycolipids, CD44, and dystroglycans are among a host of non-integrin ECM receptors. Thus, techniques that allow the specific unbiased interrogation of cell-ECM adhesion are required to directly query the diversity of potential interactions.
In this study, we describe a high-throughput platform capable of systematically uncovering cell-ECM interactions, and use this method to characterize the global changes in ECM adhesion in a model of cancer progression. We previously described a first-generation platform that utilized robotic spotting technology to generate arrays with combinations of five ECM molecules found in normal basement membrane and connective tissue22. Since then, others have utilized similar platforms to investigate ECM responses23–26. While these platforms have demonstrated feasibility of such approaches in physiologic processes such as differentiation of stem cells, they have not yet been applied to increase our understanding of disease states. Furthermore, their limited size (typically five different ECM molecules) has prevented them from querying the diversity of ECM interactions present in the human body. Here, we present an expanded ECM microarray platform containing 768 unique pairwise ECM molecule combinations expressed differentially in development, regeneration, and disease including an expanded representation of proteoglycans and glycosaminoglycans which are difficult to study through integrin manipulation alone, and apply them to investigate changes in adhesion throughout metastatic progression. We have established a highthroughput pipeline to generate these microarrays that utilizes liquid handlers for mixing of source ECM, optimized cell-seeding devices, and automated image capture and analysis. We studied the adhesion profiles of lung adenocarcinoma cell lines generated from a genetically engineered mouse model where discrete stages of metastatic progression have been defined, and correlated the findings with in vivo ECM distributions in mice and humans with metastatic lung cancer27–29. This approach is easily extensible to other disease states, ECM combinations, and phenotypic readouts.
RESULTS
Extracellular matrix microarrays to probe cell-ECM adhesion
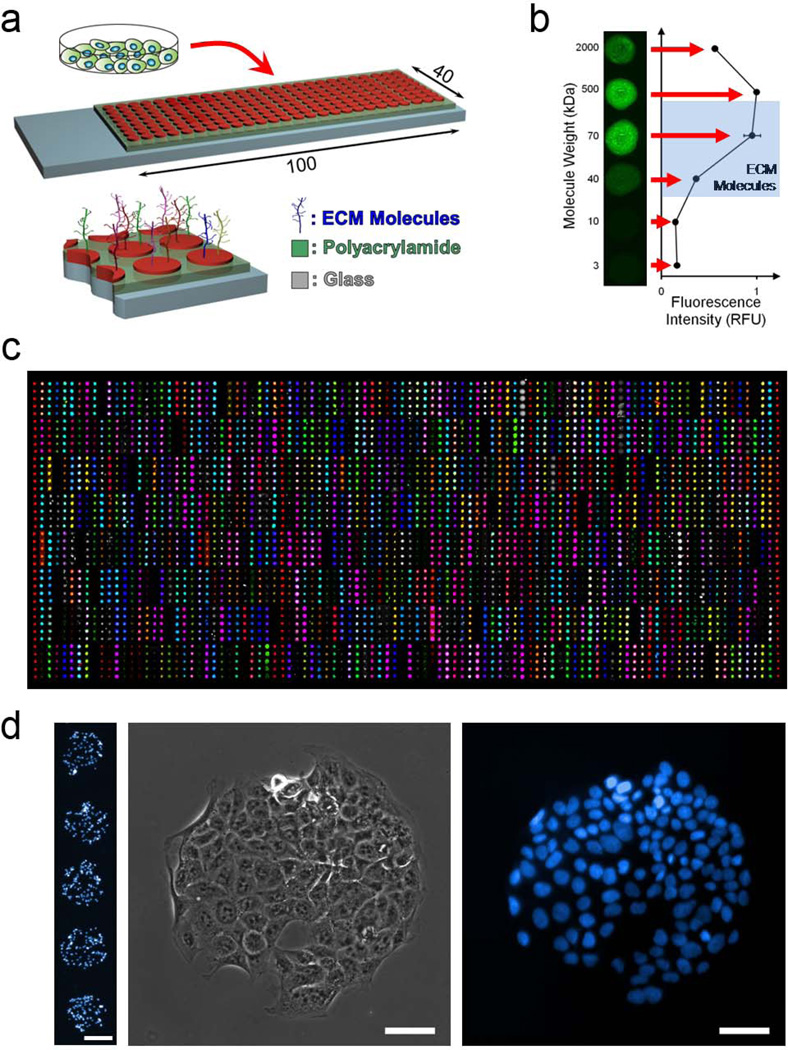
To allow the unbiased study of the ECM adhesion characteristics of any cells-of- interest, we developed a novel high-throughput platform. We expanded, automated, and optimized our adhesion platform22–23 to include every single and pairwise combination of 38 unique ECM molecules (Supplementary Table S1). Thus, these arrays contain 768 different combinations in quintuplicate and 160 control spots, for a total of 4000 arrayed features. To fabricate the arrays, the 38 ECM molecules and controls are transferred from a 96-well source plate to two low-volume 384-well plates and mixed thoroughly using a robotic liquid handler. These 384-well plates are then used as source plates for deposition of the matrix combinations onto the slides by a DNA microarray spotter. Prior to deposition of the molecules, slides are coated with a polyacrylamide hydrogel that is allowed to dry after soaking to remove any unpolymerized monomer. The dehydrated hydrogel acts to entrap molecules without requiring their chemical modification (Fig. 1a). Our data indicate that molecules larger than ∼10kDa can be robustly entrapped in the hydrogel (Fig. 1b), and we verified their entrapment using NHS-Fluorescein labeling or antibody-mediated detection after entrapment (Fig. 1c). Of the 38 molecules that we tested by these methods, all showed excellent reproducibility and uniformity within the expected region of printing (Fig. 1c).
Figure 1. Extracellular Matrix Microarray Platform Presents Combinations of ECM Molecules for Cell Attachment.
(a) ECM microarrays are generated by spotting nearly 800 unique combinations of ECM molecules on glass slides coated with polyacrylamide followed by seeding of cells onto the slides. (b) Polyacrylamide acts to entrap molecules of a large range of molecular weights. (c) Verification of presentation of all molecules by immunolabeling (colored spots) or NHS-fluorescein labeling (grayscale spots) of all molecules subsequent to array generation and rehydration. (d) Representative images of cells adhered to ECM spots demonstrating selective adhesion in the locations of ECM. Scale bar on five-spot image is 200µm. Scale bars on single-spot images are 50µm.
To measure cell-ECM interactions, cells are seeded onto the arrays in serum-free media and allowed to adhere for 1.5 hours at 37°C (Supplementary Fig. S1a). To ensure uniform seeding, the slides are agitated every fifteen minutes. Furthermore, the top surfaces of the slides are held flush with the bottom of the plate through the use of a custom-designed seeding device that employs a vacuum seal (Supplementary Fig. S1b and 1c). This device minimizes seeding variability between experiments and avoids cell loss by preventing cells from settling below the slide surface or on the backs of the slides. Uniformity of seeding across individual arrays and between replicate arrays was confirmed using test slides composed of only one matrix molecule. To quantify cells bound to each spot, nuclei are stained according to conventional fluorescence staining protocols, and the slides are imaged using an automated inverted epifluorescent microscope with NIS Elements software (Fig. 1d and 2a). Large images are cropped to individual spots and indexed using MATLAB (Mathworks), and adhesion is quantified using CellProfiler software to detect and count nuclei (Fig. 2b)30. Subsequent image and data analysis is performed in MATLAB.
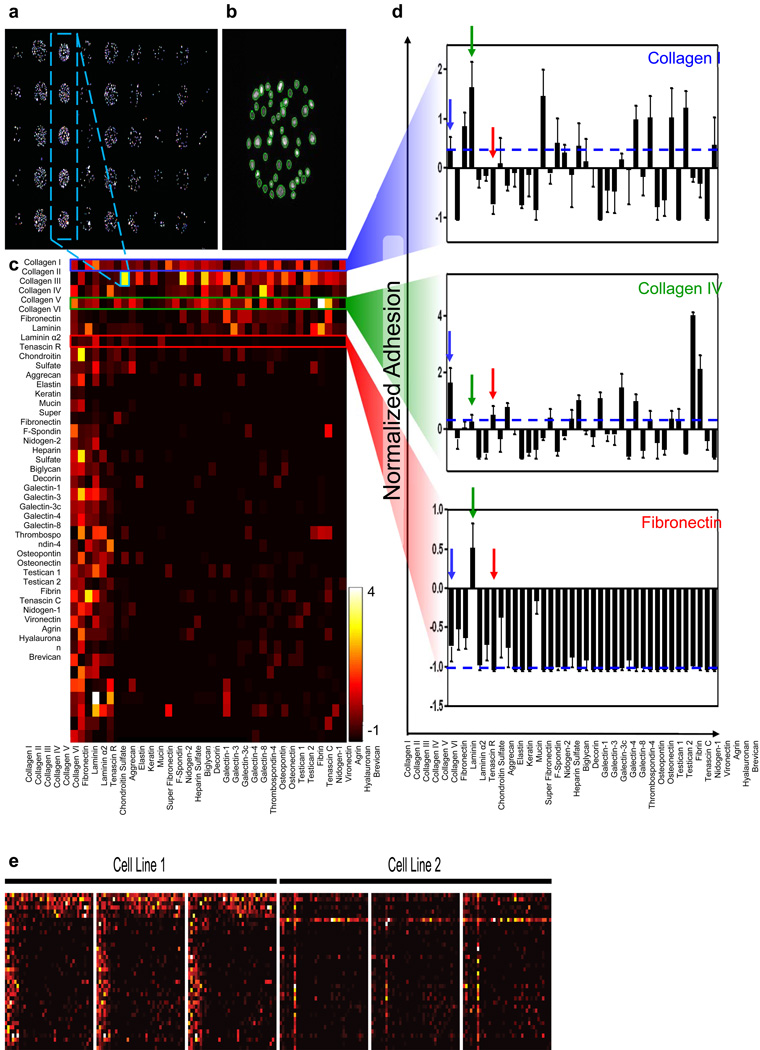
Figure 2. Combinatorial Adhesion Profiles are generated using ECM microarrays.
(a) Nuclear stain of cells seeded on the ECM microarrays. (b) Identification of individual nuclei on one spot using CellProfiler30. (c) Quantification of adhesion to all molecule combinations for one cell line. (d) Selected adhesion profiles for three molecules: Collagen I (blue), Collagen IV (green), and Fibronectin (red) in combination with all other molecules. Dashed blue lines represent adhesion to that molecule alone. Arrows denote combinations with the other two molecules or alone. Error bars are s.e.m. of three replicate slides. (e) Comparisons of three replicate slides for two representative cell lines. Scale bars in (a) and (b) are 450µm and 100µm, respectively.
ECM microarrays identify distinct adhesion profiles
To uncover changes in the global adhesion profile of cancer cells during cancer progression and metastatic spread, we analyzed a panel of murine lung adenocarcinoma cell lines derived from non-metastatic primary tumors (TnonMet), primary tumors that metastasized (TMet), or lymph node (N) and liver (M) metastases (Supplementary Table S2)29. These cell lines were derived from a genetically-engineered mouse model of metastatic lung adenocarcinoma in which tumors were initiated in KrasLSL-G12D/+;p53flox/flox mice with lentiviral-Cre vectors. The stable and random integration of the lentiviral vector allowed the clonal relationship between the multifocal primary tumors and metastases to be established29. Analysis of the adhesion profiles of these cell lines highlighted the diverse adhesion of each line to different ECM combinations (Fig. 2c). Our analysis of these cell lines revealed highly reproducible adhesion between replicate spots and arrays, confirming the quantitative nature of the assay (Fig. 2d). We examined the profiles to interrogate whether various populations exhibit enhanced adhesion to combinations of ECM molecules, relative to the same molecules spotted in isolation. This analysis revealed that different pairwisecombinations of ECM molecules result in additive, synergistic, and antagonistic effects on adhesion. For example, for the TnonMet cell line shown in Figures 2c and 2e, many molecules improve adhesion to collagen I, while others reduce cell binding in comparison to the molecule in isolation (blue line, Fig. 2e). A similar range of responses was observed for other molecules, including collagen IV and fibronectin (Figure 2e). These types of combinatorial effects were present for many molecules and, while the specific patterns varied, all cell lines tested exhibited examples of increased and reduced binding to various ECM combinations. For each of the eleven murine lung adenocarcinoma cell lines tested, distinct profiles that were highly reproducible across replicate slides were obtained (Fig. 2d).
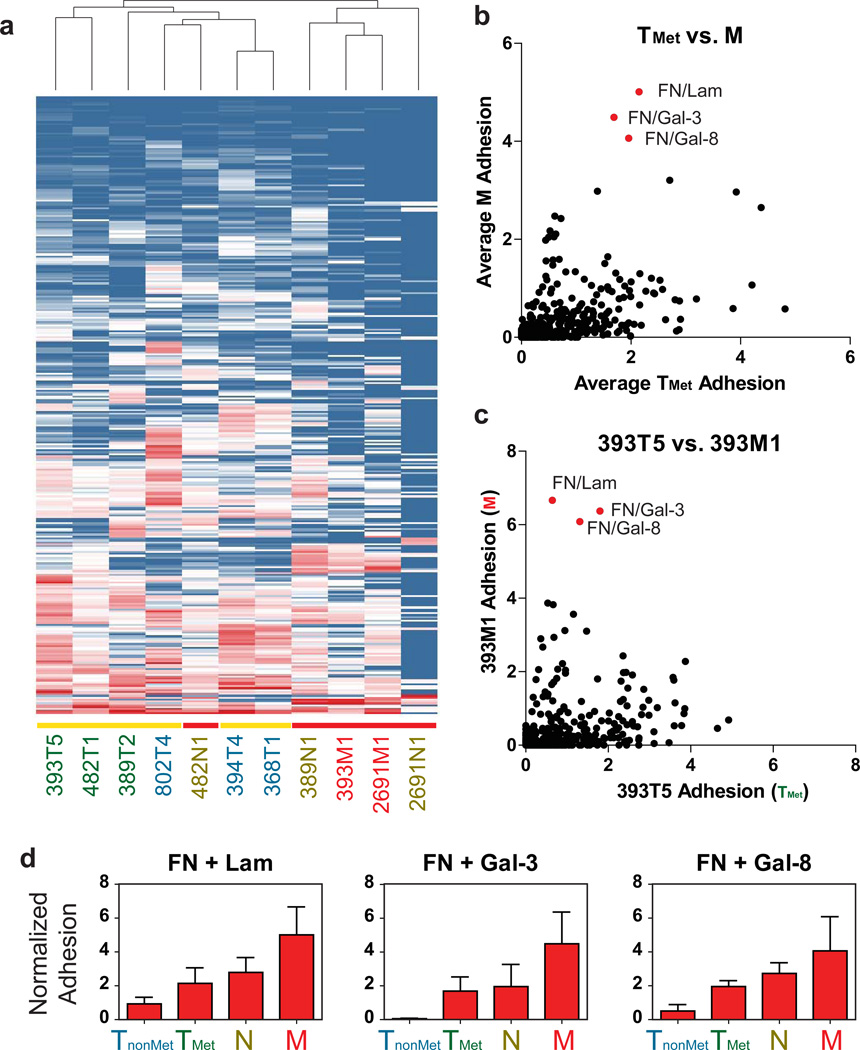
We used the ECM microarrays to compare the adhesion profiles of populations from each of the TnonMet, TMet, N, and M classes of cell lines. We applied unsupervised hierarchical clustering analysis of the adhesion values in a manner analogous to clustering of gene expression microarray data. Interestingly, all the cell lines derived from metastases (N or M), save for one lymph node line, clustered independently from the cell lines derived from primary lung tumors (TnonMet or TMet) (Fig. 3a). This result is particularly notable, since two of the metastatic lines (393M1 and 389N1) were generated from metastases that originated from two of the primary tumors (393T5 and 389T2, respectively), yet clustered more closely to the other metastases than the lines derived from those primary tumors. Thus, there is a conserved change in the ECM adhesion profile of cancer cells present in a metastatic site versus those that remain in the primary tumor. Furthermore, this differential clustering was not evident from unsupervised hierarchical clustering of the gene expression of these lines (ref29 and Supplementary Fig. S2), suggesting that the metastasis-specific adhesion phenotype provides a complementary, non-overlapping view of the molecular mediators that influence metastatic progression.
Figure 3. ECM Microarrays identify key adhesive changes in metastatic progression.
(a) Unsupervised hierarchical clustering of adhesion profiles generated by the ECM microarrays. Vertical axis represents different ECM combinations. Horizontal axis represents different cell lines. Yellow bars indicate primary tumors (TnonMet and TMet lines). Red bars indicate nodal (N) or distant metastases (M). (b) Average adhesion of metastatic cell lines (M) to each combination compared to those of the metastatic primary tumor cell lines (TMet). (c) Comparison of 393M1 adhesion for each combination to its matching primary tumor line, 393T5. Red dots indicate top ECM combinations exhibiting preferential adhesion by metastatic lines over the metastatic primary tumor lines. (d) Top three combinations exhibiting the greatest increase in adhesion across tumor progression as represented by the four classes of cell lines (TnonMet, TMet, N, and M). Error bars in (d) are s.e.m. of the different cell lines of each class (n = 3 cell lines per class) with the exception of the M class where there are two lines, and thus the error bars are the range of the means.
Identification of metastasis-associated ECM molecules
In light of the hierarchical clustering results, we asked whether there were particular combinations of molecules that are favored by metastatic cells rather than by cells from primary tumors. Thus, we compared the average adhesion of the liver metastasis-derived cell lines (M) for each ECM combination to the average adhesion of the TMet lines (Fig. 3b). While many of the M lines exhibit elevated binding to combinations containing fibronectin, pairings that combined fibronectin with any of galectin-3, galectin-8, or laminin had the highest differential adhesion between line classes. To explore changes in adhesion that specifically correlated with changes in metastatic progression, we compared the TMet cell line 393T5 and the clonally-related liver metastasis-derived cell line 393M1. This pair of lines was derived from a primary tumor and a metastasis that disseminated from that tumor, as confirmed by examination of the lentiviral integration site 29. Furthermore, the differential adhesion to the aforementioned ECM combinations was clear in both the group-wise comparison (Fig. 3b) and in the direct comparison of this primary tumor-liver metastasis pair (Fig. 3c).
Collectively, the patterns observed suggest that combinations of molecules may play a more significant role in the adhesion profile of a given population than the tendency to bind to any of the ECM molecules alone. Interestingly, the trend towards increased binding to fibronectin/galectin-3, fibronectin/laminin, and fibronectin/galectin-8 combinations was consistent across tumor progression when we compared the average adhesion of all TnonMet, TMet, N, and M cell lines (Fig. 3d). Binding to these molecules, when presented alone, showed minimal (fibronectin) or no trend (laminin, galectin-3, and galectin-8) across the four groups of cell lines (Supplementary Fig. S3a, red bars). When in combination, however, these pairs demonstrate enhanced effects that exceed the additive values of their individual adhesion. In contrast, other combinations demonstrated a reduced adhesion trend in relatively more metastatic populations, including a variety of collagens and osteopontin (Supplementary Fig. S3a–c). Taken together, these data suggest that adhesion to fibronectin in combination with any of galectin-3, galectin-8, or laminin is highly associated with tumor progression in this model system.
We also noted that some combinations of molecules appear to elicit antagonistic effects on adhesion. We looked more closely at the adhesion profile of the M line 393M1 (Supplementary Fig. S4a) and observed that, while the metastasis-associated molecules laminin, galectin-3, and galectin-8 all increase adhesion to fibronectin, other molecules appeared to decrease adhesion to it (Supplementary Fig. S4b). In vitro adhesion assays using co-adsorbed ECM to multiwell polystyrene plates confirmed that the addition of these molecules does indeed decrease the adhesion of this line to fibronectin (Supplementary Fig. S4c). Collectively, the existence of both synergistic and antagonistic effects highlights the importance of investigating combinations of ECM molecules rather than isolated components.
ECM molecules are present in sites of endogenous tumors
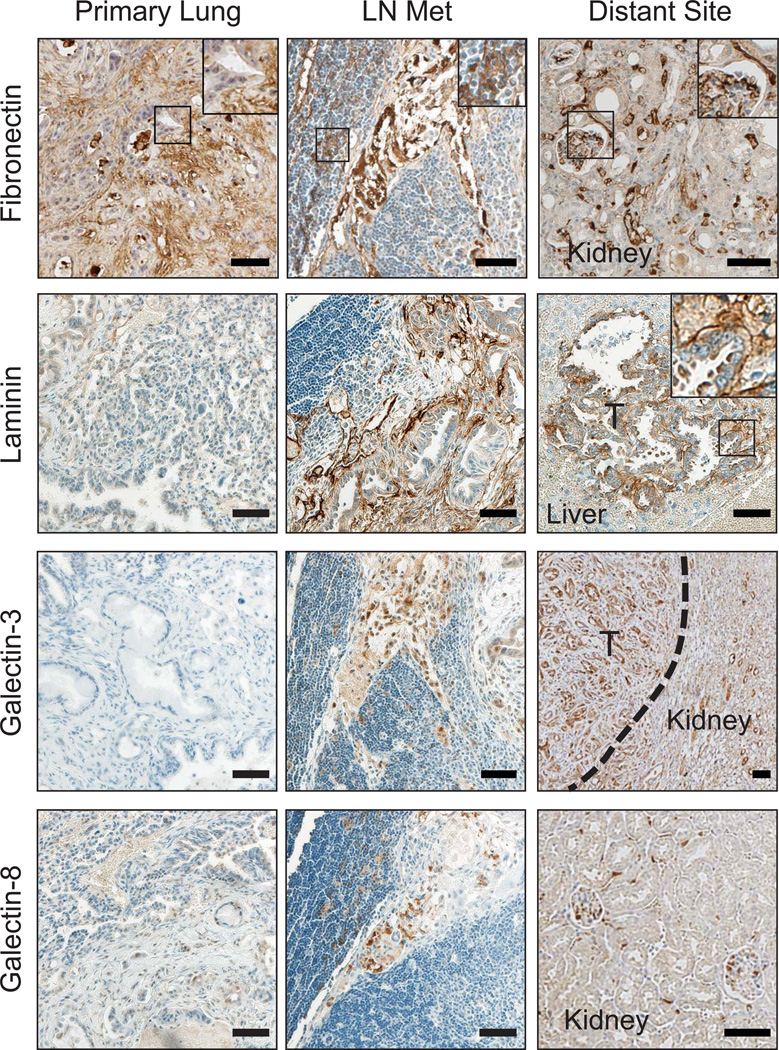
Next we sought to correlate our in vitro adhesion profiles with ECM expression in vivo. To investigate whether the identified ECM molecules may be important in natural tumorigenesis, organs containing primary autochthonous tumors and their metastases were resected from KrasLSL-G12D/+;p53flox/flox mice and stained. Trichrome staining of lungs with extensive tumor burden revealed a significant presence of ECM deposition in the tumor-bearing lung (Supplementary Fig. S5a and ref 31). Previously, we found that primary tumors that have acquired the ability to metastasize (TMet tumors) upregulate the chomatin-associated protein Hmga229. Therefore, we used Hmga2 immunohistochemistry in addition to histological characteristics to identify areas of highly aggressive cancer cells (Supplementary Fig. S5a). As anticipated, primary lung tumors were positive for collagen I (black arrowheads), collagen VI (open black arrowheads), and osteopontin (red arrowheads), with the most intense staining overlapping with the high-grade tumor areas (Supplementary Fig. S5a and S5c). In particular, osteopontin staining strongly co-localized with Hmga2pos regions, suggesting that increased osteopontin production is associated with metastatic primary lung tumors. Furthermore, little to no laminin, galectin-3, or galectin-8 staining was detected in the primary tumors (Fig. 4). Interestingly, fibronectin staining in the tumor was strong, revealing a correlation between increasingly metastatic populations and the presence of fibronectin early in the metastatic cascade (Fig. 4).
Figure 4. Metastasis-associated ECM molecules are present in the sites of metastases but not primary tumors.
Immunostaining of the metastasis-associated ECM molecules in the lungs, lymph nodes, and distant metastases of mice bearing endogenous lung adenocarcinomas (KrasLSL-G12D/+;p53flox/flox mice). Insets are magnified views of boxed areas showing ECM molecule fibrils. Number of tissues examined for each organ: Lungs: 10; lymph nodes: 5; livers/kidneys: 22. ‘T’: tumor. Dotted line depicts edge of tumor and normal kidney. Scale bars are 50µm.
We next asked whether the lymph node and distant organ metastases contained the metastasis-associated ECM molecules. Again, trichrome staining revealed the presence of significant matrix deposition within the lymph nodes (Supplementary Fig. S5b). As expected, the entirety of the lymph node tumors were histologically high-grade and were Hmga2pos (Supplementary Fig. S5b). There was also clear expression of all four of the metastasis-associated molecules (fibronectin, laminin, galectin-3, and galectin-8) within the lymph node metastases (Fig. 4). Furthermore, there was essentially no collagen I or collagen VI (Supplementary Fig. S5c). Interestingly, osteopontin was present in the metastases (Supplementary Fig. S5c) and had its highest expression along the invasive front (Supplementary Fig. S5a,d).
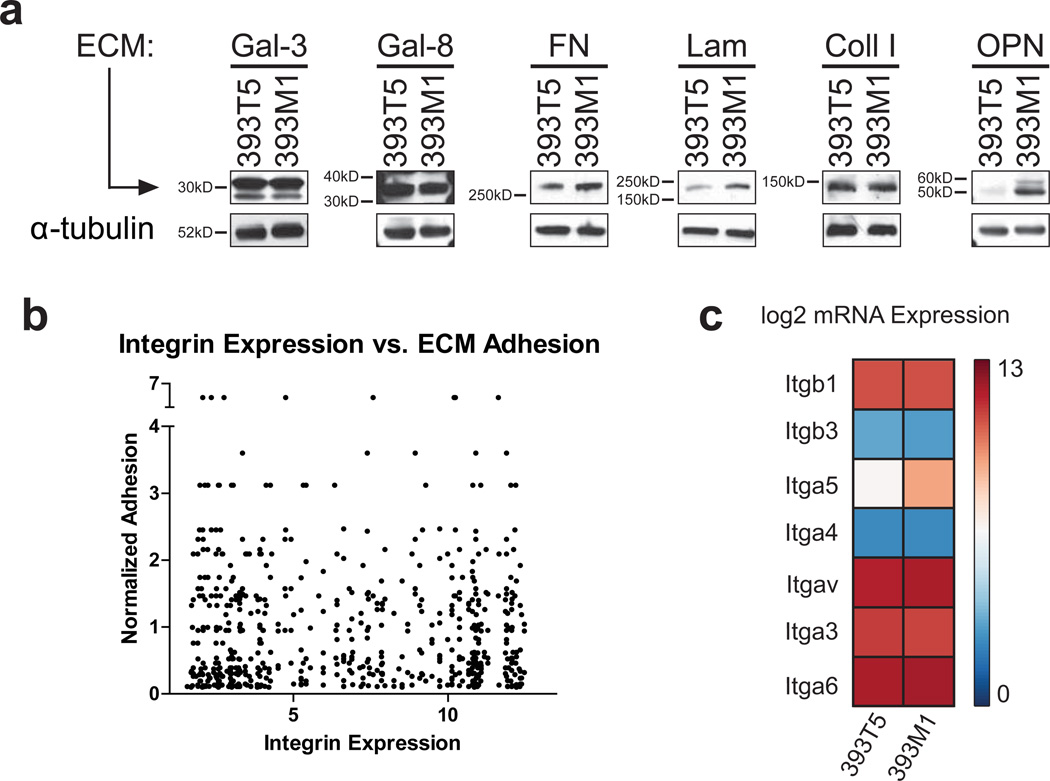
We also examined common metastatic sites for the presence of the metastasis-associated molecules (Fig. 4). Both galectin-3 and galectin-8 were distinctly visible in these sites. Laminin and fibronectin both appeared to line the sinusoids of the livers of the mice and were also present in the metastases formed there. To determine whether these differences between the primary and metastatic sites were due to altered matrix production by the tumor cells, we performed immunoblots on the 393T5 and 393M1 TMet and M cell lines. While the M line showed slight increases in fibronectin and laminin production compared to the TMet line, production of both galectins was constant (Fig. 5a). Furthermore, collagen I production was constant, and osteopontin production was actually increased in the M line. Taken together, these data suggest that the ECM microarrays identified molecules that were found within the physiologically relevant sites of mice bearing autochthonous tumors, and that production of these molecules is not solely performed by the tumor cells present in those sites.
Figure 5. ECM production and integrin mRNA expression by cell lines have minimal correlation with adhesion.
(a) Western blot analysis of the metastasis- and primary tumor-associated ECM molecules produced by the 393T5 (TMet) and 393M1 (M) cell lines. (b) Comparison of ECM adhesion for all cell lines to gene expression of the cognate integrins from gene expression microarray data. (c) Integrin subunit mRNA expression from Affymetrix microarray analysis in 393T5 and 393M1 cell lines.
Integrin surface expression correlates with ECM binding profiles
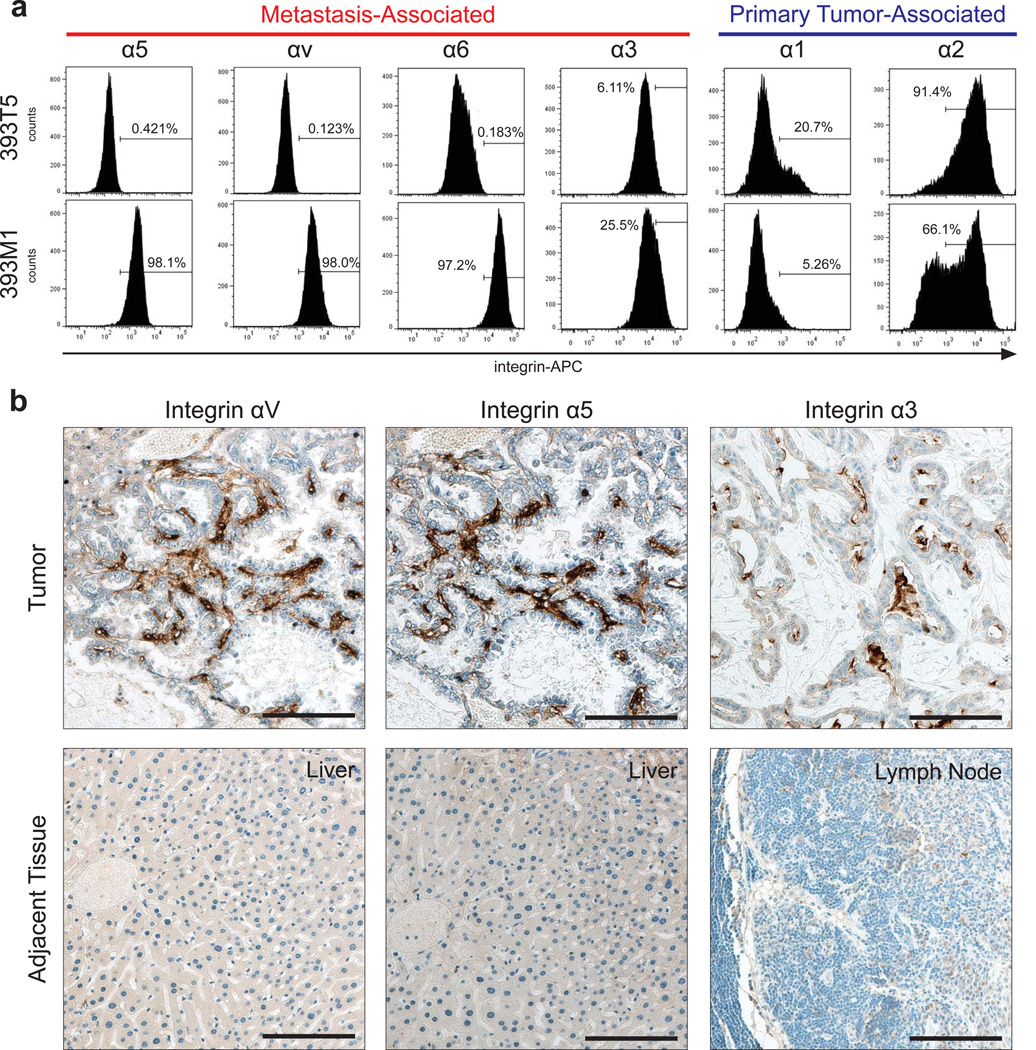
We noted that comparisons of adhesion trends on our ECM arrays did not necessarily correlate with transcriptional profiles of the cognate integrins (Fig. 5b). Thus, to correlate our findings with the presence of receptors for these metastasis-associated ECM molecules, we examined the clonally-related pair of representative TMet and M cell lines for surface expression of their cognate integrins. While the mRNA expression patterns did not show significant upregulation of the metastasis-associated integrins in the M line by gene expression microarray (Fig. 5c), flow cytometry analysis of the integrin subunits corresponding with either the primary tumor-associated molecules or metastasis-associated molecules revealed that the receptor expression trends were consistent with the observed binding patterns. Specifically, integrin subunits known to bind fibronectin (α5 and αv), laminin (α6 and α3), and galectins (α3) were all more prevalent on the metastasis-derived line, while those associated with collagens (α1 and α2) were relatively higher on the primary tumor-derived line (Fig. 6a). Nonetheless, the surface expression trends were consistent for the other TMet and M lines as well (Supplementary Fig. S6a). Furthermore, within a given cell line, we observed relatively homogeneous surface expression of the metastasis-associated integrins (Supplementary Fig. S6b) suggesting that variations in adhesion between lines are due to global increases in surface receptor expression, rather than binding patterns of select subpopulations.
Figure 6. Integrin Surface Expression Correlates with ECM binding profiles.
(a) Flow cytometry of integrin surface expression in 393T5 (TMet) and 393M1 (M) cell lines. Integrin subunits that bind to metastasis-associated molecules show increased surface presentation in the metastatic line (α5, αv, α6, α3), while those that bind to primary tumor-associated molecules show decreased presentation (α1 and α2). (b) IHC for metastasis-associated integrins in mice bearing autochthonous tumors with spontaneous metastases to the liver and lymph nodes. Scale bars are 100µm.
Immunohistochemistry revealed that these integrins were also present in the metastases of mice bearing autochthonous tumors, but not the adjacent tissue (Fig. 6b). The finding that the transcriptional levels of the integrins do not agree with the adhesion trends suggests that post-transcriptional regulation, post-translational modifications such as altered glycosylation, or alterations in activation state of the integrins are likely responsible for the changes in adhesion. Thus, by utilizing our platform that investigates specific ECM binding rather than receptor gene or protein expression, we are able to identify candidate ECM interactions that might otherwise have been overlooked.
Integrin α3β1 mediates adhesion and seeding in vitro and in vivo
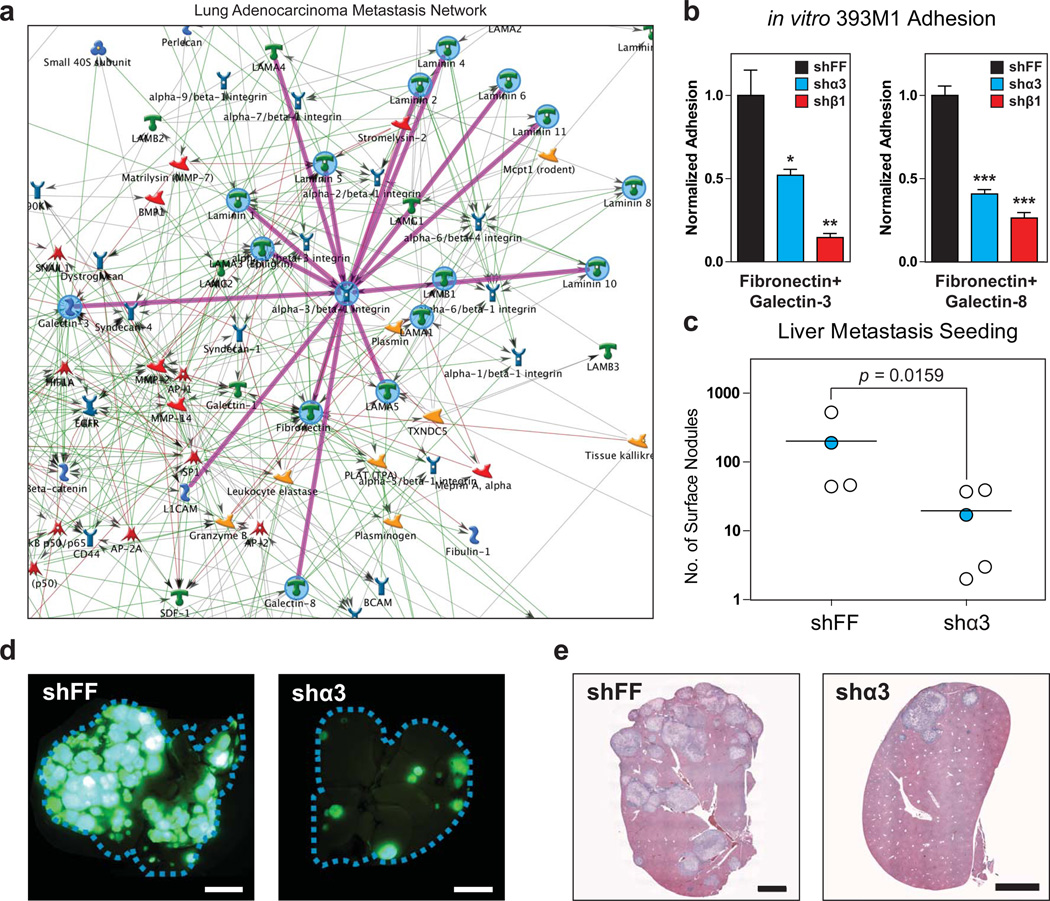
To examine which candidate receptor/ECM interactions may participate in the observed binding patterns, we performed in silico network mapping of the metastasis-associated ECM molecules using GeneGO software (Metacore) of manually curated molecular interactions. We generated a network map that we termed the Lung Adenocarcinoma Metastasis (LAM) network that has a greatest disease association with ‘Neoplasm Metastasis’ (P = 1.094 × 10−45, hypergeometric test, Fig. 7a, Supplementary Fig. S7). A network generated using the same parameters but with the primary-tumor-associated molecules did not exhibit any disease association with metastasis (Supplementary Fig. S8). Analysis of the LAM network identified integrin α3β1 as the surface receptor with the greatest number of edges (Fig. 7a). Based on this finding, we performed a knockdown of both the α3 and β1 subunits (ITGA3 and ITGB1, respectively) using short-hairpin-mediated RNA-interference (Supplementary Fig. S9). Knockdown of these genes in the metastatic line, 393M1, resulted in reduced adhesion to the metastasis-associated molecules in vitro when compared to the control hairpin targeting the firefly luciferase gene (Fig. 7c).
Figure 7. Integrin α3β1 mediates adhesion and seeding in vitro and in vivo.
(a) in silico network mapping using GeneGO (MetaCore) generates the Lung Adenocarcinoma Metastasis Network. Analysis of the network reveals that integrin a3b1 is the surface receptor with the most edges (a). Knockdown of both α3 and β1 integrin subunits by shRNA reduces adhesion to metastasis-associated molecules in vitro (b) and prevents metastatic seeding in vivo (c-e). shFF is the control hairpin targeting firefly luciferase. One-way ANOVA with Tukey’s Multiple Comparison Test was used to analyze the data in figure (b). Error bars in (b) represent standard error (n = 3). (c) Number of liver tumor nodules of the surface of livers 2.5 weeks after intrasplenic injection. Mann-Whitney (non-parametric) test was used to analyze significance. (d) Fluorescence imaging of whole livers after resection. Cell lines express nuclear-excluded ZSGreen. Scale bars are 0.5cm. (f) Hematoxylin and eosin stain of liver slices. Scale bars are 2mm. Blue data points in (d) correspond to images in (e) and (f). All results shown are representative of multiple independent experiments.
We next assessed whether this integrin dimer plays a role in metastatic seeding in vivo. Thus, we conducted experimental metastasis assays by intrasplenic injection of 393M1-shα3 or 393M1-shFF cells into wild type mice, and monitoring for liver tumor formation. We found that mice injected with the 393M1-shα3 cells formed fewer tumor nodules than the controls (Fig. 7d–f). Taken together, these findings suggest that the α3β1 integrin dimer plays a role in adhesion of metastatic cells to the metastasis-associated ECM molecules and in metastatic seeding.
Galectin-3/8 are present in human lung cancer metastases
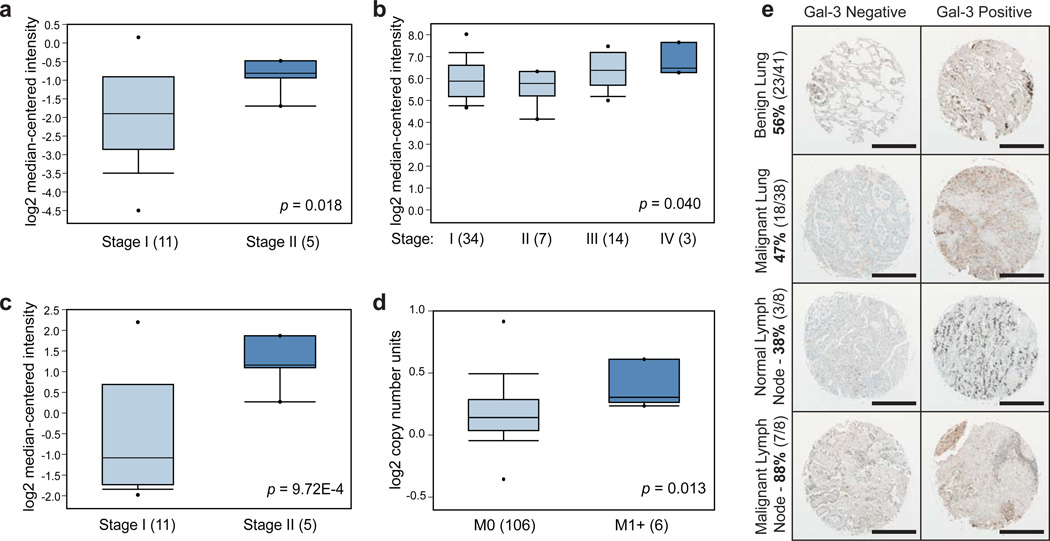
Based on the in vitro adhesion data and in vivo mouse findings, we sought to explore the role of the metastasis-associated ECM molecules in human samples. Using Oncomine32, a human genetic dataset analysis tool, we examined the correlation of ECM gene expression and disease severity (e.g. clinical stage or the presence of metastases). Results of these queries demonstrate that increased expression of LGALS3 or LGALS8 (galectin-3 and galectin-8, respectively) correlate with increased clinical stage or the presence of metastases (Fig. 8a). We next investigated whether galectin-3 protein is present at higher levels in malignant human lung tumors compared to benign non-neoplastic human lung tissue using samples taken from lungs and lymph nodes of patients. Staining for galectin-3 in human tissue microarrays revealed a higher presence of the molecule in lymph nodes of patients with malignant disease (88%) compared to those without cancer (38%) (Fig. 8b). Furthermore, there was a higher fraction of galectin-3-positive lymph nodes (88%) than positive primary lung tumor samples (47%), confirming its association with the metastatic site over the primary tumor (P < 0.05, Fisher’s Exact Test). Thus, the ECM microarrays were capable of identifying interactions associated with metastasis in human lung cancer.
Figure 8. Metastasis-associated molecules are present in the metastases of human lung cancers.
(a-d) Oncomine32 results for human lung cancer expression of LGALS3 and LGALS8. (a) LGALS3 Expression in Hou Lung: Large Cell Lung Carcinoma – Advanced Stage. (b) LGALS3 Expression in Bild Lung: Lung Adenocarcinoma – Advanced Stage. (c) LGALS8 Expression in Hou Lung: Large Cell Lung Carcinoma – Advanced Stage. (d) LGALS8 Copy Number in TCGA Lung 2: Lung Adenocarcinoma – Advanced M Stage. LGALS3 and LGALS8 are overexpressed in Stage II lung cancer compared to stage I (P = 0.018 and 9.72E-4, respectively)(a,c). Microarray data source GSE1918846. (b) LGALS3 is overexpressed in Stage IV lung cancer compared to other stages (P = 0.040). Microarray data source GSE314147. (d) LGALS8 has increased copy number in advanced M stage lung cancer (P = 0.013) in the “Lung Carcinoma DNA Copy Number Data” dataset available from The Cancer Genome Atlas website (https://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp). (e) Representative images of human tissue microarray staining results for galectin-3 presence or absence in the primary sites and lymph nodes. Scale bars are 500µm. Box and whisker plots in (a-d): dots represent maximum and minimum values, whiskers show 90th and 10th percentiles, boxes show 75th and 25 percentiles, and line shows median. P-values in (a-d) were computed by Oncomine software using Student’s t-test (a, c-d) or Pearson’s correlation analysis (b).
DISCUSSION
Our ECM microarrays provide a high-throughput multiplexed platform capable of measuring a variety of cellular responses to ECM. Here, we show they are capable of identifying adhesion patterns that differentiate metastatic populations from primary tumors. We found that metastatic lung cancer cells preferentially bind to fibronectin in combination with laminin, galectin-3, or galectin-8 compared to cells derived from primary tumors. These changes in adhesion correlate with changes in surface presentation of various integrins. In particular, α3β1 mediates adhesion to these molecules in vitro and permits metastatic seeding in vivo. Furthermore, metastases derived from both a genetically-engineered mouse lung cancer model and from human lung cancers express the metastasis-associated ECM molecules. It is worth noting that the combinations of these ECM components elicited the strongest effects, highlighting the importance of using a platform that is capable of measuring responses to more than individual molecules.
Galectins are a class of lectins that bind β-galactosides and can associate with other ECM molecules such as fibronectin33. Galectin-3 is associated with metastasis in a variety of cancers34–35 and can bind to the oncofetal Thomsen-Friedenreich antigen, a carbohydrate antigen overexpressed by many carcinomas36. Our platform confirmed its importance in lung adenocarcinoma, and also identified galectin-8 as having similar importance. While galectin-8 is known to affect adhesion of cells to other matrix molecules, its role in cancer and metastasis has been less clear as it has been found to have both a positive and negative association with adhesion and tumorigenesis37–38. Using the ECM microarrays, we showed that binding to galectin-8 in combination with fibronectin is strongly associated with metastatic progression in lung adenocarcinoma.
Furthermore, in addition to many collagens, we found that loss of adhesion to osteopontin accompanied metastatic progression (Supplementary Fig. S3a–c). Osteopontin levels correlate with prognosis in patients with metastatic disease39, and secretion of osteopontin by primary tumors results in mobilization of bone marrow-derived stromal precursors that help establish the metastatic niche40. In addition to confirming the presence of the metastatic molecules at the sites of metastases, we found that the invasive portions of primary tumors and the invasive front of the metastases secrete osteopontin (Supplementary Fig. S5b). A metastatic tumor line also produces more osteopontin than its corresponding primary (Fig. 5a). These findings suggest that while some primary tumors may activate bone marrow cells by secreting osteopontin, in our model metastatic cells may contribute to this recruitment at a comparable or higher level than the instigating primaries, despite their own loss of adhesion to the immobilized molecule.
The use of gene expression signatures for patient stratification in the clinic has become more widespread41–45, but while genomic approaches have been beneficial for identifying candidate genes, the diversity of findings makes the development of broad therapeutic options seem nearly impossible. By assaying for conserved mechanisms at the phenotypic level, however, relevant targets can be identified and therapeutics can be developed for a broad spectrum of patients. Our results highlight the utility of phenotypic screening approaches for identifying clinical biomarkers. While we identify α3β1 integrin as a therapeutic target, we also demonstrate that the adhesion signatures generated by the ECM microarrays are capable of differentiating between genetically similar populations with varying metastatic potential. Furthermore, no increase in the mRNA levels of the galectins or their receptors was observed by gene expression microarrays in the M lines (Fig. 5 and ref29), despite the association of these molecules with metastasis. The presence of galectin-3 and galectin-8 in human samples (Fig. 8) demonstrates the relevance of this platform to human disease, and thus, we envision that these arrays may be a useful clinical tool for stratification of cancer patients beyond traditional TNM staging.
The value of the ECM microarray platform extends beyond the specific application of cancer metastasis. While this study documents the ability to profile adhesion patterns, cells bound to the arrays can be kept in culture for multiple days to monitor long-term responses to ECM such as cell death, proliferation, and alterations in gene or protein expression. Toward that end, one could use multiplexed antibody staining to probe the effects of ECM on stem cell differentiation or activation. Orthogonal screens can be performed to look at the effects of growth factors, small molecules, or RNA-interference agents in the context of ECM. Reduction of requisite cell numbers can be achieved using miniaturized arrays to screen rare cell populations such as circulating tumor cells or cancer stem cells and to help expand those populations in vitro for further biological studies. Overall, the ECM microarrays will enhance our ability to study a host of questions as they pertain to both basic biological and clinical settings.
METHODS
Murine Lung Adenocarcinoma cell lines
Cell lines have been described29. Briefly, tumor initiation was achieved using intratracheal injection of lentiviral Cre recombinase. Tumors were resected, digested, and plated onto tissue culture treated plastic to generate cell lines29. Cell lines were subsequently cultured in DMEM, 10% FBS, penicillin/streptomycin, and glutamine. These lines were derived from both primary lung tumors and their metastases. See Supplementary Table S2 for nomenclature regarding cell line origins.
Cell Transplantation Assays
All animal procedures were performed in accordance with the MIT Institutional Animal Care and Use Committee under protocol 0211-014-14. Cell injection studies were performed in B6129SF1/J mice (Jackson Laboratory, Stock Number 101043). Intrasplenic injections were performed using 5×105 cells resuspended in 100µL of PBS and injected into the tip of the spleen following existing protocols29. Animals were anesthetized with avertin prior to surgery. Fur was removed from the animals and they were sterilized with Betadine and 70% ethanol. The spleen was exteriorized following incisions in the skin and body wall. Cells were injected into the end of the spleen with a 27gauge syringe and allowed to travel into circulation for two minutes. Spleens were then excised from the animals following cauterization of the splenic vessels. The muscle wall was closed using 5-0 dissolvable sutures, and the skin was closed using 7mm wound clips (Roboz). Mice were euthanized 2.5-4 weeks following injection, and their livers were excised. Quantification of surface nodules and imaging of livers was performed using a dissection microscope. Tissues were embedded in paraffin following fixation in 4% paraformaldehyde and stained using hematoxylin and eosin.
Extracellular Matrix Microarrays Preparation
Vantage acrylic slides (CEL Associates VACR-25C) were coated with polyacrylamide by depositing prepolymer containing Irgacure 2959 photoinitiator (Ciba) between the slide and a glass coverslip22. Following polymerization, slides were soaked in ddH2O and the coverslips were removed. Slides were allowed to dry prior to molecule deposition. Slides were spotted using a DNA Microarray spotter (Cartesian Technologies Pixsys Microarray Spotter and ArrayIt 946 Pins). 768 combinations were spotted in replicates of five. Rhodamine dextran (Invitrogen) was spotted as negative controls and for use in image alignment. The following molecules were used: Collagen I (Millipore), Collagen II (Millipore), Collagen III (Millipore), Collagen IV (Millipore), Collagen V (BD Biosciences), Collagen VI (BD Biosciences), Fibronectin (Millipore), Laminin (Millipore), Merosin (Millipore), Tenascin-R (R&D Systems), Chondroitin Sulfate (Millipore), Aggrecan (Sigma), Elastin (Sigma), Keratin (Sigma), Mucin (Sigma), Superfibronectin (Sigma), F-Spondin (R&D Systems), Nidogen-2 (R&D Systems), Heparan Sulfate (Sigma), Biglycan (R&D Systems), Decorin (R&D Systems), Galectin 1 (R&D Systems), Galectin 3 (R&D Systems), Galectin 3c (EMD Biosciences), Galectin 4 (R&D Systems), Galectin 8 (R&D Systems), Thrombospondin-4 (R&D Systems), Osteopontin (R&D Systems), Osteonectin (R&D Systems), Testican 1 (R&D Systems), Testican 2 (R&D Systems), Fibrin (Sigma), Tenascin-C (R&D Systems), Nidogen-1 (R&D Systems), Vitronectin (R&D Systems), Rat Agrin (R&D Systems), Hyaluronan (R&D Systems), Brevican (R&D Systems). The laminin used is Millipore Catalog No. AG56P, and is a mixture of human laminins that contain the beta1 chain. Source plates used in the spotter were prepared using a Tecan liquid handler. Molecules were prepared at a concentration of 200µg/mL using a buffer described previously 22. Slides were stored in a humidity chamber at 4°C prior to use.
Extracellular Matrix Microarray Seeding and Analysis
Slides were washed in PBS and treated with UV prior to seeding cells. They were placed in a seeding device that holds the top surface of the slides flush with bottom of the well. 400,000 cells were seeded on each slide in 6mL of serum-free medium (DMEM and penicillin/streptomycin). Cells were allowed to attach for two hours at 37°C. After attachment, slides were washed three times, transferred to quadriperm plates (NUNC, 167063), and new medium was added (DMEM, 10%FBS, penicillin/streptomycin, and glutamine). Slides were left at 37°C for two additional hours prior to removal for staining. Slides were washed twice with PBS and fixed with 4% paraformaldehyde. Nuclei were stained using Hoechst (Invitrogen) in combination with 0.1% Triton-X and PBS. Slides were mounted with Fluoromount-G (Southern Biotech 0100-01) and stored at 4°C prior to imaging. Slides were imaged using a Nikon Ti-E inverted fluorescence microscope and NIS Elements Software (Nikon). The entire slide was scanned and images stitched using that software. Image manipulation and analysis was performed in MATLAB (Mathworks) and quantification of nuclei was performed using CellProfiler30. Clustering analysis was performed using Spotfire (Tibco). Replicate spots on each slide were averaged and those whose values were greater than one standard deviation above or below the mean of the replicates were excluded. Slides were normalized to the mean of their non-zero adhesion values. Clustering was performed based on Euclidean distances using Spotfire (TIBCO) with the Hierarchical Clustering algorithm (normalized adhesion > 0.01).
In vitro Adhesion Seeding
in vitro ECM adhesion tests were performed using 96-well-plates (Corning 3603). Plates were coated with 20µg/mL of fibronectin alone or 20µg/mL of fibronectin and 20µL/mL of the second molecule in PBS overnight at 4°C. Plates were then blocked with 1wt% BSA at room temperature for 1hr. Plates were allowed to dry prior to adding 2×104 cells/well in warm serum-free DMEM. Cells were allowed to adhere for 1hr at 37°C and shaken every 15minutes to ensure uniform seeding. Cells were washed, fixed with 4% paraformaldehyde, and stained with Hoechst (invitrogen). Wells were imaged using a Nikon Ti-E inverted epifluorescent microscope and analyzed with Nikon Elements software.
Protein Analysis
Western blot analysis of ECM molecules was performed with the following antibodies: galectin-3 (Abcam ab53082, 1:500), galectin-8 (Abcam ab69631, 1:500), osteopontin (Abcam ab8448, 1:2000), fibronectin (Abcam ab2413, 1:1000), laminin (Abcam b11575, 1:1000), collagen I (Abcam ab34710, 1:5000), and α-tubulin (Cell Signaling 2125, 1:1000). Immunohistochemistry of ECM molecules was performed with the following antibodies: galectin-3 (Abcam ab53082, 1:500), galectin-8 (Abcam ab69631, 1:75), osteopontin (Abcam ab8448, 1:200), laminin (Abcam ab11575, 1:100), fibronectin (Millipore AB2033, 1:80), Hmga2 (Biocheck 59170AP, 1:1000), collagen I (Abcam ab34710, 1:500),collagen VI (Abcam ab6588, 1:100). Integrin staining was performed using the following antibodies: integrin αv (Millipore AB1930, 1:200), integrin α5 (Chemicon AB1928, 1:200), integrin α3 antibody was a gift from JML. Tissue microarrays were acquired from LifeSpan Biosciences (LS-SLUCA50), and were stained with the same galectin-3 antibody. Murine tissues were harvested from KrasLSL-G12D, p53flox/flox mice27–29. IHC was performed following resection from mice, fixation in formalin, and embedding in paraffin. Flow cytometry analysis of integrin expression was performed using the following antibodies: integrin α5 (Abcam and BioLegend-clone 5H10-27, 1:100), integrin αv (BD-clone RMV-7, 1:100), integrin α6 (BD and BioLegend-clone GoH3, 1:100), integrin α3 (R&D, 1:100), integrin α1 (BD-clone Ha31/8 and BioLegend-clone HMα1, 1:100) and integrin α2 (BD-clone HMα2, 1:100).
RNA Isolation and Expression Profiling
Cell lysates were harvested using Trizol (Sigma). Chloroform extraction was performed followed by RNA purification using Qiagen RNeasy spin columns. Lysates were analyzed for RNA integrity and prepared with Affymetrix GeneChip WT Sense Target Labelling and Control Reagents kit, followed by hybridization to Affymetrix Mouse 3’ Arrays (Mouse 430A 2.0) Lysates used for gene expression microarrays were harvested at the same time as the ECM microarrays were seeded to ensure minimal variability introduced by cell culture. R/Bioconductor software was used to process array images. Unsupervised hierarchical clustering analysis was performed in Spotfire (Tibco) for all probesets with variance>0.5 and expression>3.0 using Euclidean distances. Datasets are publically available from NCBI under accession number GSE40222.
Retroviral shRNA Constructs
miR30-based shRNAs targeting integrins β1 (TGCTGTTGACAGTGAGCGCGGCTCTCAAACTATAAAGAAATAGTGAAGCCACAGATGTATTTCTTTATAGTTTGAGAGCCTTGCCTACTGCCTCGGA), α3 (TGCTGTTGACAGTGAGCGCCGGATGGACATTTCAGAGAAATAGTGAAGCCACAGATGTATTTCTCTGAAATGTCCATCCGTTGCCTACTGCCTCGGA), or control firefly luciferase (AAGGTATATTGCTGTTGACAGTGAGCGAGCTCCCGTGAATTGGAATCCTAGTGAAGCCACAGATGTAGGATTCCAATTCAGCGGGAGCCTGCCTACTGCCTCG) were designed using the shRNA retriever software (http://katahdin.cshl.edu/homepage/siRNA/RNAi.cgi?type=shRNA), synthesized (IDT, Coralville, Iowa), and then cloned into the MSCV-ZSG-2A-Puro-miR30 vector48. Packaging of retrovirus and transduction of cells was done as described previously49.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank H. Fleming, S. Katz, L. Ingaharro, and all other members of the S.B. lab for their insightful discussions and general research assistance. They also thank the Swanson Biotechnology Center Core Facilities and the David H. Koch Institute and, in particular, M. Brown, C. Whittaker, G. Paradis, M. Luo, S. Levine, Y. Xue, and C. Kim-Kiselak. M.M.W. is funded by NIH grant K99-CA151968. J.M.L. was supported by a National Research and Service Award (NRSA) from the NIH, and a postdoctoral fellowship from the National Cancer Center. This work was supported by Stand Up to Cancer (SU2C/AACR), the Koch Institute CTC Project, and the Harvard Stem Cell Institute (SG-0046-08-00), the National Cancer Institute (U54CA126515 and U54CA112967), the Howard Hughes Medical Institute (of which R.O.H., T.E.J., and S.N.B. are Investigators), and the Ludwig Center at MIT.
Footnotes
AUTHOR CONTRIBUTIONS
N.E.R.F. and S.N.B. wrote the manuscript. N.E.R.F., M.M.W., and S.N.B. designed the studies. N.E.R.F. and D.F.B.M. designed and produced the ECM microarrays. M.M.W. generated the initial cell lines. N.E.R.F. and M.J.X. performed the ECM microarray seeding. J.M.L. designed and synthesized the hairpins and retroviruses. N.E.R.F. performed all other experiments and all data analysis. All authors contributed conceptual advice and edited the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Gupta GP, Massagué J. Cancer Metastasis: Building a Framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Mehlen P, Puisieux A. Metastasis: a question of life or death. Nat Rev Cancer. 2006;6:449–458. doi: 10.1038/nrc1886. [DOI] [PubMed] [Google Scholar]
- 3.Hoffman PC, Mauer AM, Vokes EE. Lung cancer. The Lancet. 2000;355:479–485. doi: 10.1016/S0140-6736(00)82038-3. [DOI] [PubMed] [Google Scholar]
- 4.Steeg PS, Theodorescu D. Metastasis: a therapeutic target for cancer. Nat Clin Prac Oncol. 2008;5:206–219. doi: 10.1038/ncponc1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naba A, et al. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Molecular & Cellular Proteomics. 2011 doi: 10.1074/mcp.M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levental KR, et al. Matrix Crosslinking Forces Tumor Progression by Enhancing Integrin Signaling. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oskarsson T, et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med. 2011;17:867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weaver VM, et al. Reversion of the Malignant Phenotype of Human Breast Cells in Three-Dimensional Culture In Vivo by Integrin Blocking Antibodies. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dolberg DS, Bissell MJ. Inability of Rous sarcoma virus to cause sarcomas in the avian embryo. Nature. 1984;309:552–556. doi: 10.1038/309552a0. [DOI] [PubMed] [Google Scholar]
- 12.Hendrix MJC, et al. Reprogramming metastatic tumour cells with embryonic microenvironments. Nat Rev Cancer. 2007;7:246–255. doi: 10.1038/nrc2108. [DOI] [PubMed] [Google Scholar]
- 13.Olumi AF, et al. Carcinoma-associated Fibroblasts Direct Tumor Progression of Initiated Human Prostatic Epithelium. Cancer Research. 1999;59:5002–5011. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albini A, et al. A Rapid in Vitro Assay for Quantitating the Invasive Potential of Tumor Cells. Cancer Research. 1987;47:3239–3245. [PubMed] [Google Scholar]
- 15.Hotary KB, et al. Membrane Type I Matrix Metalloproteinase Usurps Tumor Growth Control Imposed by the Three-Dimensional Extracellular Matrix. Cell. 2003;114:33–45. doi: 10.1016/s0092-8674(03)00513-0. [DOI] [PubMed] [Google Scholar]
- 16.Malik G, et al. Plasma Fibronectin Promotes Lung Metastasis by Contributions to Fibrin Clots and Tumor Cell Invasion. Cancer Research. 2010;70:4327–4334. doi: 10.1158/0008-5472.CAN-09-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White DE, et al. Targeted disruption of β1-integrin in a transgenic mouse model of human breast cancer reveals an essential role in mammary tumor induction. Cancer Cell. 2004;6:159–170. doi: 10.1016/j.ccr.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 18.Hynes RO. The Extracellular Matrix: Not Just Pretty Fibrils. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reuter JA, et al. Modeling Inducible Human Tissue Neoplasia Identifies an Extracellular Matrix Interaction Network Involved in Cancer Progression. Cancer Cell. 2009;15:477–488. doi: 10.1016/j.ccr.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avraamides CJ, Garmy-Susini B, Varner JA. Integrins in angiogenesis and lymphangiogenesis. Nat Rev Cancer. 2008;8:604–617. doi: 10.1038/nrc2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hynes RO. Integrins: Bidirectional, Allosteric Signaling Machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 22.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat Meth. 2005;2:119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 23.Flaim CJ, Teng D, Chien S, Bhatia SN. Combinatorial Signaling Microenvironments for Studying Stem Cell Fate. Stem Cells and Development. 2008;17:29–40. doi: 10.1089/scd.2007.0085. [DOI] [PubMed] [Google Scholar]
- 24.LaBarge MA, et al. Human mammary progenitor cell fate decisions are products of interactions with combinatorial microenvironments. Integrative Biology. 2009;1:70–79. doi: 10.1039/b816472j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mei Y, et al. Cell-Compatible, Multicomponent Protein Arrays with Subcellular Feature Resolution. Small. 2008;4:1600–1604. doi: 10.1002/smll. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brafman DA, et al. Investigating the role of the extracellular environment in modulating hepatic stellate cell biology with arrayed combinatorial microenvironments. Integrative Biology. 2009;1:513–524. doi: 10.1039/b912926j. [DOI] [PubMed] [Google Scholar]
- 27.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & Development. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase Nat. Protocols. 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winslow MM, et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473:101–104. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carpenter AEJT, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biology. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson EL, et al. The Differential Effects of Mutant p53 Alleles on Advanced Murine Lung Cancer. Cancer Research. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 32.Rhodes DR, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu F-T, Rabinovich GA. Galectins as modulators of tumour progression. Nat Rev Cancer. 2005;5:29–41. doi: 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- 34.Takenaka Y, Fukumori T, Raz A. Galectin-3 and metastasis. Glycoconjugate Journal. 2002;19:543–549. doi: 10.1023/B:GLYC.0000014084.01324.15. [DOI] [PubMed] [Google Scholar]
- 35.Bresalier RS, et al. Metastasis of human colon cancer is altered by modifying expression of the β-galactoside-binding protein galectin 3. Gastroenterology. 1998;115:287–296. doi: 10.1016/s0016-5085(98)70195-7. [DOI] [PubMed] [Google Scholar]
- 36.Yu L-G. The oncofetal Thomsen-Friedenreich carbohydrate antigen in cancer progression. Glycoconjugate Journal. 2007;24:411–420. doi: 10.1007/s10719-007-9034-3. [DOI] [PubMed] [Google Scholar]
- 37.Nagy N, et al. Galectin-8 expression decreases in cancer compared with normal and dysplastic human colon tissue and acts significantly on human colon cancer cell migration as a suppressor. Gut. 2002;50:392–401. doi: 10.1136/gut.50.3.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zick Y, et al. Role of galectin-8 as a modulator of cell adhesion and cell growth. Glycoconjugate Journal. 2002;19:517–526. doi: 10.1023/B:GLYC.0000014081.55445.af. [DOI] [PubMed] [Google Scholar]
- 39.Singhal H, et al. Elevated plasma osteopontin in metastatic breast cancer associated with increased tumor burden and decreased survival. Clinical Cancer Research. 1997;3:605–611. [PubMed] [Google Scholar]
- 40.McAllister SS, et al. Systemic Endocrine Instigation of Indolent Tumor Growth Requires Osteopontin. Cell. 2008;133:994–1005. doi: 10.1016/j.cell.2008.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van‘t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 42.Beer DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 43.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 44.Paik S, et al. A Multigene Assay to Predict Recurrence of Tamoxifen-Treated, Node-Negative Breast Cancer. New England Journal of Medicine. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 45.van de Vijver MJ, et al. A Gene-Expression Signature as a Predictor of Survival in Breast Cancer. New England Journal of Medicine. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 46.Hou J, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS ONE. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 48.Lamar JM, Stern P, Liu H, Schindler JW, Jiang Z, Hynes RO. The Hippo pathway target, YAP, promotes metastasis through its TEAD interaction domain. Proc. Natl. Acad. Sci. USA. 2012 doi: 10.1073/pnas.1212021109. manuscript in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stern P, et al. A system for Cre-regulated RNA interference in vivo. Proceedings of the National Academy of Sciences. 2008;105:13895–13900. doi: 10.1073/pnas.0806907105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.