Abstract
Although extensive studies have demonstrated the functional impairment of antigen-specific CD4+ and CD8+ T-cells during chronic hepatitis C virus (HCV) infection, the functional status of global CD4+ and CD8+ T-cells remains unclear. In this report, we recruited 42 long-term (~20 years) treatment-naïve chronic HCV (CHC) patients and 15 healthy donors (HDs) to investigate differences in global CD4+ and CD8+ T-cells function. We show that CD4+ and CD8+ T-cells from CHC patients underwent increased apoptosis after TCR stimulation. Furthermore, IFN-γ, IL-9 and IP-10 were elevated in CHC patients’ plasma and promoted activation-induced T-cells death. Global CD4+ and CD8+ T-cells also showed unique transcriptional profiles in the expression of apoptosis-related genes. We identified BCL2, PMAIP1, and CASP1 in CD4+ T-cells and IER3 and BCL2A1 in CD8+ T-cells from CHC patients as HCV-specific gene signatures. Importantly, the gene expression patterns of CD4+ and CD8+ T-cells from CHC patients differ from those in CD4+ and CD8+ T-cells from human immunodeficiency virus type 1 (HIV-1) or hepatitis B virus (HBV) infected individuals. Our results indicate that chronic HCV infection causes a systemic change in cytokine levels that primes T-cells for activation-induced apoptosis. Furthermore, HCV infection programs unique apoptosis-related gene expression profiles in CD4+ and CD8+ T-cells, leading to their enhanced activation-induced apoptosis. These results provide novel insights to the pathogenesis of chronic HCV infection.
Introduction
Approximately 170 million people are chronically infected with hepatitis C virus (HCV) [1,2]. The functional exhaustion of HCV-specific T-cells contributes to failed viral clearance in chronically infected patients [3–5]. HCV-specific CD8+ T-cells exhibit various degrees of functional impairment, including impaired proliferation, reduced effector cytokine production, and enhanced apoptosis [6–8]; however, the mechanisms underlying these changes are incompletely understood. Deregulated expression of inhibitory receptors on HCV-specific CD8+ T-cells, increased Treg numbers, and enhanced cytokine levels impact the function of antigen-specific T-cells during chronic HCV infection [3,4]. Nevertheless, the impact of chronic HCV infection on the function of global CD4+ and CD8+ T lymphocytes remains poorly understood. Addressing this issue will elucidate mechanisms by which the host environment impacts antigen-specific T-cells during chronic HCV infection.
In this study, we investigated the functional status of global CD4+ and CD8+ T-cells in a group of long-term (~20 years) treatment-naive chronic HCV infected (CHC) patients. We found that long-term chronic HCV infection did not cause a significant change in the numbers of total lymphocytes or major lymphocyte subpopulations or in total T-cells proliferation. In contrast, both CD4+ and CD8+ T-cells from CHC patients exhibited dramatically enhanced activation-induced apoptosis. Furthermore, IFN-γ, IP-10, and IL-9 were significantly elevated in the plasma of CHC patients. These cytokines sensitized primary T-cells from healthy donors (HDs) to activation-induced death. Consistent with these findings, CD4+ and CD8+ T-cells from CHC patients displayed deregulated expression of apoptosis-related genes. B-cell lymphoma 2 (BCL2), phorbol-12-myristate-13-acetate-induced protein 1 (PMAIP1), and caspase 1 (CASP1) in CD4+ T-cells and immediate early response 3 (IER3) and BCL2-related protein A1 (BCL2A1) in CD8+ T-cells from CHC patients are identified as HCV-specific gene signatures. These gene signatures were specific to CHC and differed from those in T-cells from chronic human immunodeficiency virus type 1 (HIV-1) or hepatitis B virus (HBV) patients. Taken together, our results demonstrate that chronic HCV infection induces a cytokine environment that sensitizes T-cells to activation-induced apoptosis and induces unique apoptosis-related gene signatures.
Materials and Methods
Subjects
Forty-two anti-HCV Ab+ CHC patients without prior anti-viral treatment from Gansu province, China were recruited. The CHC patients were infected with HCV through blood donation during the 1980s and 1990s (1988-1994). The average time of infection was ~20 years. Fifteen HDs in the same age range were selected as healthy controls. The clinical parameters were measured in Beijing YouAn Hospital (Table 1 ). Written informed consent was obtained from all individuals before their participation in this study, and the study protocol was approved by Beijing YouAn Hospital Ethical Committee.
Table 1. Clinical parameters of CHC patients and HDs.
| Parameter | CHC Patients | Healthy Donors |
|---|---|---|
| Number in each group | 42 | 15 |
| Age (years), median (range) | 51.6 (35-67) | 44.5 (37-56) |
| Sex (M:F) | 25:17 | 1:4 |
| Serum HCV RNA (IU/ml) (range) | 0-6.38×107 | --- |
| Serum HCV RNA gene type | 1b, 1b/2a, 2a, 2a/2b (16:1:19:1) | --- |
| Serum total bilirubin (μmol/L), median (range) | 15.1 (6.9-36.7) | 8.37 (6.1-12.8) |
| Serum direct bilirubin (μmol/L), median (range) | 3.1 (1.7-6.4 ) | 2.02 (0.4-4.3) |
| AST (IU/L), median (range) | 51.2 (18.4-389.0) | 48.45 (26.8-81.9) |
| ALT (IU/L), median (range) | 63.0 (10.6 - 590.8) | 16.2 (8.6-44.3) |
Flow cytometry
Venous blood samples were collected using EDTA as an anticoagulant. Red blood cells were lysed with Lysing Buffer (BD Biosciences, CA, USA), and the cells were stained with APC-anti-CD4 (RPA-T4), PerCP-anti-CD8 (SK1) (BD Biosciences), FITC-anti-CD3 (OKT3), FITC-anti-CD45RO (HI100), and PE-anti-CD45RA (UCHL1) (Biolegend, CA, USA) following the manufacturers’ instructions. Flow cytometry data were collected with a BD FACSAria™ Cell Sorter (BD Biosciences) and analyzed with FlowJo (Tree Star, OR, USA).
PBMC separation and T-cells stimulation
Peripheral blood mononuclear cells (PBMCs) were isolated from fresh blood samples by density gradient centrifugation using Ficoll-Paque PLUS (GE Healthcare, Buckinghamshire, UK). CD4+ and CD8+ T-cells were purified from PBMCs using CD4 and CD8 microbeads with an autoMACS Separator (Miltenyi Biotec, CA, USA) following the manufacturer’s instructions. The purity of separated CD4+ and CD8+ T-cells was >90%. For T-cells stimulation assays, 96-well plates (Costar Corning, MA, USA) were coated overnight at 4°C or for 2-3 hours at 37°C with 1 μg/ml anti-CD3 and anti-CD28 (eBioscience, CA, USA). PBMCs were labeled with 1 μl 5-(and 6-)-carboxyfluorescein diacetate, succinimidyl ester (CFSE, 5mM, Sigma-Aldrich, MO, USA) in a volume of 1 ml PBS and seeded at 1-1.5×106 cells/ml in the coated 96-well plates for 3 days. T-cells proliferation was measured by CFSE dilution, and T-cells apoptosis was determined using Pacific Blue Annexin V (Biolegend, CA, USA). PBMCs without stimulus were cultured for 3 days to determine the spontaneous apoptosis of T-cells.
Cytokine measurement and sensitization culture
Plasma samples were collected by centrifuging fresh blood samples at 400×g for 5 minutes. Twenty-seven cytokines, including Eotaxin, FGF basic, G-CSF, GF-CSF, IFN-γ, IL-1β, IL-1Ra, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12 (p70), IL-13, IL-15, IL-17, IP-10, MCP-1 (MCAF), MIP-1a, MIP-1b, PDGF bb, RANTES, TNF-α and VEGF, were measured with a human Grp I Cytokine 27-Plex Panel (Bio-Rad, CA, USA) using a Luminex 200 (Millipore, MA, USA) according to the manufacturer’s instructions. To test the effects of cytokines on T-cells apoptosis and proliferation, freshly isolated CFSE-labeled PBMCs from HDs were cultured in RPMI 1640 medium (10% FBS and 7.5pg/ml IL-7) containing cytokines at the concentrations identified in the plasma of CHC patients (IFN-γ 300, 500pg/ml; IL-1β 12pg/ml; IL-9 60, 400pg/ml; IP-10 7000pg/ml) (Peprotech, NJ, USA). PBMCs were incubated at 37°C in the presence of 5% CO2 in 96-well plates for 48 hours and then transferred to plates coated with anti-CD3 and anti-CD28 antibodies for stimulation. PBMCs were cultured without cytokines as controls (NC) in the same condition. T-cells apoptosis and proliferation were measured after 3 days of stimulation.
Microarray analysis
Purified CD4+ and CD8+ T-cells were lysed in Trizol (Invitrogen, CA, USA), and RNA extraction and microarray assays were performed by CapitalBio Corporation (Beijing, China). The quality and integrity of total RNA were determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA). T-cells gene expression patterns were detected with an Affymetrix GeneChip® Human Genome U133 Plus 2.0 Array (Affymetrix, CA, USA). Five patients and 5 HDs randomly selected from sample groups (HCV-h group, HCV-l group, and HD group) were analyzed for each cell type.
Statistical analysis
CEL files were imported to Partek Genomics Suite software (Partek Inc., MO, USA) as raw data using default settings. After background correction, normalization, and summarization using RMA (Robust Multichip Average) analysis, expression data were log2 transformed for further analysis. Differential gene expression between CHC patients and HDs was determined by one way ANOVA. False discovery rate (FDR) correction was applied using the step-up method with an FDR of 5% as the cut-off. Significantly modulated genes were defined as those with P <0.05 and ≥1.5-fold change in mean differential expression. Significantly modulated apoptosis genes were hierarchically clustered using Partek Genomics Suite software with default settings. Additionally, principal components analysis (PCA) was used to cluster patients in 3 dimensions. The significantly changed genes were used as input for the gene function and pathway enrichment analysis. GeneGo Metacore software (GeneGO Inc., CA, USA) was used to identify the most significant biological pathways associated with the modulated genes using default parameters. Different HCV patients groups (HCV-h or HCV-l) were performed separately and compared to each other for common pathways. In addition, common significantly modulated genes shared by both HCV patient groups were imported for pathway analysis. DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov/) was used to identify functional categories based on the annotation sources GOTERM-BP (biological process). Different HCV patient groups were performed separately and the FDR for enrichment analysis was set as 5%. Significantly modulated genes were classified into different functional categories according to the enrichment results, as listed in Table S1 and S2. Microarray data for gene expression in global CD4+ and CD8+ T-cells during chronic HBV [9] and HIV-1 infection [10,11] were obtained from public databases Gene Expression Omnibus (GEO). The same statistical analysis for HBV and HIV-1 data was done as described above. The common significantly modulated genes between chronic HCV infection and chronic HIV-1 or HBV infection were identified by gene symbols, which were also analyzed for pathway and function enrichment. For comparison of special T-cells gene expression signature, log2 values were used and statistic strategy for quantitative real-time PCR was applied. For correlation analysis of apoptosis gene expression level with clinical parameters or plasma levels of cytokines, fluorescent values of all samples from microarray analysis instead of log2 values were used.
GraghPad Prism 5.00 software was applied for data analysis except in microarray analysis, and p-values<0.05 (t-test) were regarded as statistically significant. Microarray raw data are available in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) under accession no.GSE49954.
Results
CHC patient characteristics
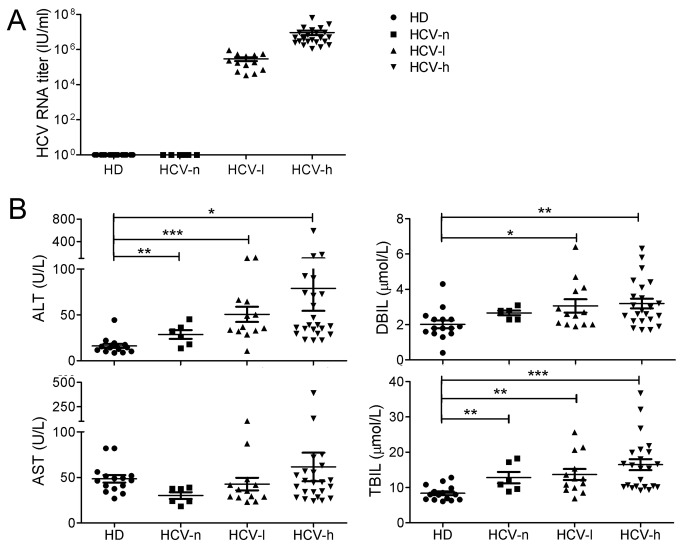
Viral load and CTL function are inversely correlated during HIV and HBV infection [12–15]. To determine the impact of serum HCV titers on the function of global CD4+ and CD8+ T-cells in CHC patients, we divided the CHC patients into 3 groups: HCV-n (negative), HCV-l (low) (104-106 IU/ml RNA), and HCV-h (high) (106-108 IU/ml RNA) (Figure 1A ). Alanine transaminase (ALT) and total bilirubin (TBIL) values were significantly higher in all 3 groups of CHC patients than in HDs (Figure 1B ). Although the direct bilirubin (DBIL) values in HCV-l and HCV-h patients were within the normal range, they were significantly higher than those in HDs (Figure 1B ). Consistent with previous reports, 12% of the CHC patients exhibited abnormal liver function based on TBIL and ALT levels [16]. Six large groups of viral genotypes (1-6) containing over 70 different subtypes are defined based on subgenomic regions [17]. In China, the main subtypes are 1b and 2a (Table 1 ). There was no correlation between HCV genotypes and RNA titers (data not shown). Although the mean ALT, AST, DBIL, and TBIL values in the HCV-h group were higher than those in the other groups (Figure 1B ), we did not find a significant correlation between HCV viral titers and gender, age or ALT, AST, DBIL, and TBIL values in these long-term treatment-naive patients (Figure S1 ).
Figure 1. Clinical parameters of the three groups of CHC patients (HCV-n group, n=6; HCV-l group, n=13; HCV-h group, n=23) and HDs.
(A), HCV RNA titers for the three groups of CHC patients. HCV-n: negative; HCV-l:103-106 IU/ml; HCV-h: ≥106 IU/ml. (B), The concentrations of ALT, AST, DBIL, and TBIL in the plasma of CHC patients compared to HDs. *, p<0.05; **, p<0.01; ***, p<0.001.
Enhanced activation-induced apoptosis of T-cells from CHC patients
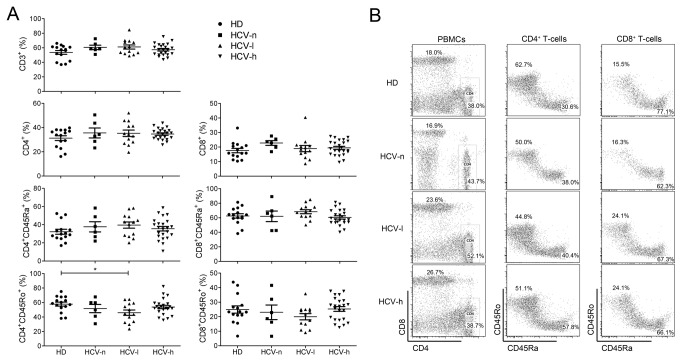
Although HCV-specific T-cells have been thoroughly studied in CHC patients [3], the functional status of total T-cells in CHC patients is not clear. We first assessed the frequencies of lymphocyte subpopulations in CHC patients. Interestingly, the frequencies of most of the T-cells subpopulations in the peripheral blood of CHC patients were similar to those in HDs (Figure 2A and 2B ). The only significant change was a reduction in CD4+CD45RO+ T-cells in HCV-l patients (HD, 57.8% ± 2.8%; HCV-l, 46.0% ± 3.8%) (Figure 2A ), suggesting that long-term chronic HCV infection had a minimal impact on overall lymphocyte composition.
Figure 2. Percentages of T-cells subpopulations in the peripheral blood of CHC patients and HDs.
(A), The percentages of total T-cells (CD3+), CD8+, and CD4+ T-cells in the peripheral blood of patients and HDs. The frequency of memory/activated (CD45RO+) T-cells and naïve (CD45RA+) T-cells among CD8+ or CD4+ T-cells was also determined. (B), Representative FACS profiles of T-cells subsets in the peripheral blood of patients and HDs are shown. *, p<0.05.
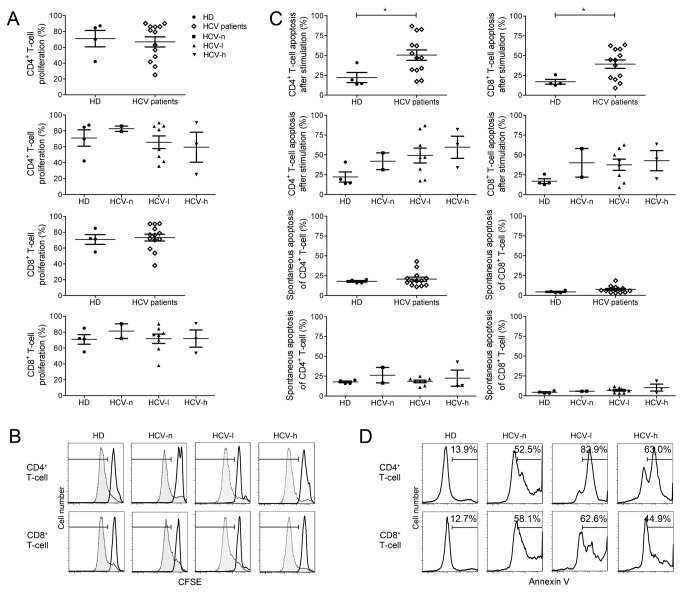
We next examined the proliferation and apoptosis of total CD4+ and CD8+ T-cells from CHC patients. Surprisingly, we did not observe any significant difference in the proliferation of total CD4+ or CD8+ T-cells from CHC patients and HDs (Figure 3A and 3B). However, total T-cells from all CHC groups exhibited significantly enhanced activation-induced apoptosis, with a >2-fold increase in CD8+ T-cells and a ~2-3-fold increase in CD4+ T-cells when compared to T-cells from HDs (Figure 3C and 3D). In contrast, the spontaneous apoptosis rates of total CD4+ and CD8+ T-cells were similar among the CHC patients and HDs (Figure 3C ). These results suggest that changes in the host environment induced by HCV infection may sensitize the T-cells to activation-induced apoptosis.
Figure 3. Enhanced activation-induced apoptosis of global CD4+ and CD8+ T-cells from CHC patients.
Freshly isolated PBMCs were stimulated with plate-bound anti-CD3/CD28 for 3 days. T-cells proliferation was measured by CFSE dilution, and apoptosis was measured by Annexin V staining. (A), Proliferation rates of total CD4+ and CD8+ T-cells from the CHC and HD groups. (B), Representative FACS profiles of T-cells proliferation for each group of CHC patients and HDs. (C), Apoptosis rates of CD4+ and CD8+ T-cells from the CHC patients and HDs with or without anti-CD3/CD28 stimulation. (D), Representative FACS profiles of T-cells apoptosis for each group of CHC patients and HDs. For (A) and (C), each dot represents one individual. *, p<0.05.
The roles of cytokines in activation-induced apoptosis of T-cells from CHC patients
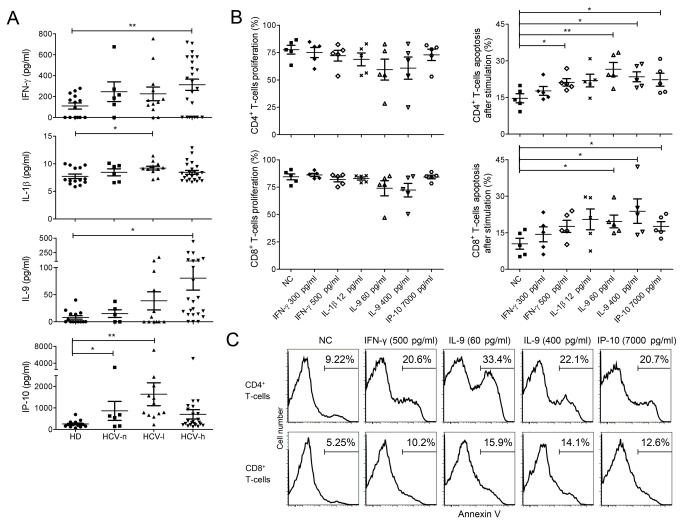
Cytokines play important roles in regulating T-cells survival. We hypothesized that dysregulated cytokine expression in the CHC patients might sensitize CD4+ and CD8+ T-cells to activation-induced apoptosis. Plasma levels of IFN-γ, IL-1β, IL-9, and IP-10 were elevated in the CHC patients (Figure 4A ). Interestingly, the levels of IFN-γ and IL-9 were highest in the HCV-h group, whereas the levels of IL-1β and IP-10 were highest in the HCV-l group (Figure 4A ). The levels of other primary inflammatory cytokines, such as TNF-α, were similar in CHC patients and HDs (data not shown).
Figure 4. Elevated cytokine levels in the plasma of CHC patients and their roles in sensitizing T-cells to activation-induced apoptosis.
(A), Cytokine levels, including IFN-γ, IL-1β, IL-9, and IP-10, in the plasma of CHC patients and HDs as measured by Luminex assay. (B), Effect of pre-incubation of PBMCs with the indicated cytokines on T-cells proliferation as measured by CFSE dilution and T-cells apoptosis as measured by Annexin-V staining. PBMCs from HDs were pre-incubated with or without (NC) the indicated cytokines for 48 hrs and then stimulated with anti-CD3/CD28 for 3 days before the measurement of proliferation and apoptosis. Data were from 5 independent HDs. (C), Representative FACS profiles of CD4+ and CD8+ T-cells apoptosis after pre-incubation with cytokines followed by stimulation. *, p<0.05; **, p<0.01.
We next examined whether these cytokines sensitized CD4+ and CD8+ T-cells to activation-induced cell death. PBMCs from HDs were pre-cultured with IFN-γ, IL-1β, IL-9, or IP-10 for 48 hours before stimulation with anti-CD3/CD28 antibodies. The concentrations of cytokines used were equivalent to either the median or the maximal value measured in the CHC patients. At these concentrations, these four cytokines did not significantly alter TCR-induced CD4+ and CD8+ T-cell proliferation (Figure 4B ), suggesting that the abnormal levels of these cytokines did not cause gene expression changes related to the cell proliferation machinery. In contrast, pre-incubation of HD T-cells with IFN-γ, IL-9, or IP-10 resulted in significantly enhanced activation-induced CD4+ or CD8+ T-cells apoptosis (Figure 4B and 4C); however, pre-incubation of T-cells with IL-1β did not affect activation-induced cell death (Figure 4B ) (p>0.05). These results demonstrate that IL-9, IFN-γ and IP-10 sensitize T-cells to activation-induced apoptosis and suggest that cytokine levels in CHC patients affect global T-cells function.
Gene expression profiles of CD4+ and CD8+ T-cells in CHC patients
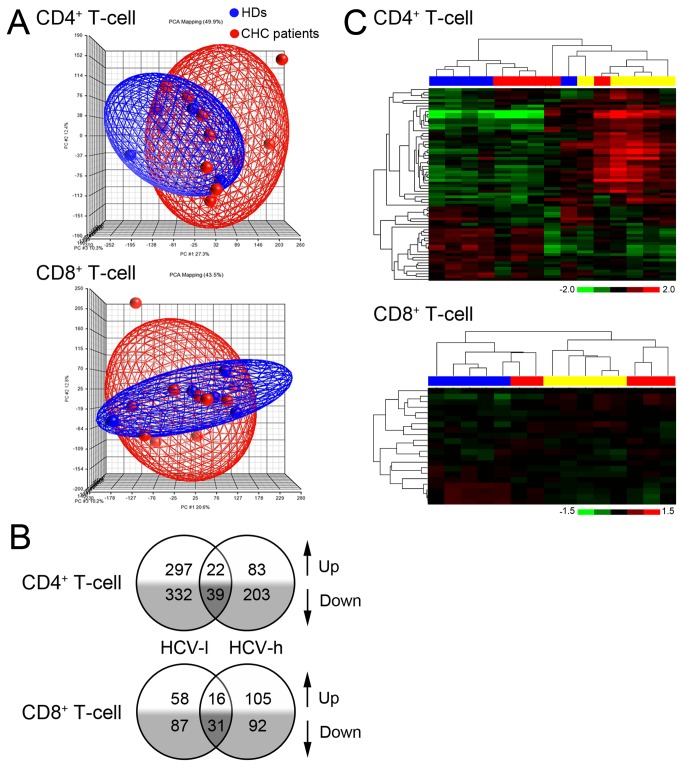
These results suggest that the host environment in CHC patients may reprogram the gene expression profiles of CD4+ and CD8+ T-cells. We thus sought to define the gene expression profiles of total CD4+ and CD8+ T-cells from CHC patients. Surprisingly, the overall gene expression patterns of global CD4+ or CD8+ T-cells from CHC patients were not dramatically different from those from HDs and cannot be used to separate CHC patients from the HDs (Figure 5A and Figure S2 ). Interestingly, T-cells from HCV-h and HCV-l patients shared only a few common changed genes (61 genes for CD4+ T-cells and 47 genes for CD8+ T-cells) (Figure 5B ). Furthermore, no common pathway was altered in T-cells from both HCV-h and HCV-l patients (Figure S3 ). These results suggest that differences in HCV titers influence the gene expression patterns of CD4+ and CD8+ T-cells in CHC patients.
Figure 5. Overall gene expression profiles of CD4+ or CD8+ T-cells from CHC patients.
(A), Scatterplot graph of CD4+ and CD8+ T-cells with PCA (principal component analysis) mapping. CHC patients, n=10; HDs, n=5. (B), The number of genes differentially expressed (fold-change ≥1.5, p<0.05) in CD4+ and CD8+ T-cells of HCV-l and HCV-h group when compared to those in HDs. The microarray data were derived from 5 individuals for each group. The numbers in the overlapping region represent the genes changed in both the HCV-l and HCV-h groups. (C), Clustering analysis of significantly changed apoptosis-related genes in CD4+ and CD8+ T-cells from CHC patients and HDs. Blue blocks represent HDs; yellow blocks represent HCV-l samples; red blocks represent HCV-h samples.
We further analyzed the gene expression changes using the online DAVID program. Significantly changed genes involved in important biological processes for CD4+ and CD8+ T-cell function were selected from the HCV-h and HCV-l groups (Table S1 and S2 ). Although the inhibitory receptors PD-1, Tim3, 2B4, and CTLA-4 are upregulated in HCV-specific CD8+ T-cells in CHC patients [18–24], the mRNA expression of these receptors was not significantly changed in global CD4+ or CD8+ T-cells of CHC patients, suggesting that the inhibitory receptors do not contribute to global T-cells dysfunction during chronic HCV infection.
Because global CD4+ and CD8+ T-cells from the CHC patients exhibited enhanced activation-induced apoptosis, we analyzed the expression profiles of genes related to apoptosis. The number of significantly changed apoptosis-related genes was greater in CD4+ T-cells from the CHC patients than in CD8+ T-cells (Table S1 and S2 ). Interestingly, the significantly changed apoptosis-related genes in CD4+ T-cells were largely different from those in CD8+ T-cells (Table S1 and S2 ), suggesting that two different sets of genes may be involved in the enhanced activation-induced apoptosis of CD4+ and CD8+ T-cells. Furthermore, genes altered in the HCV-l group were different from those in the HCV-h group for both CD4+ and CD8+ T-cells (Table S1 and S2 ), suggesting that differences in HCV titers have a qualitative effect on T-cells gene expression profiles. Importantly, the expression signatures of the apoptosis-related genes in CD4+ and CD8+ T-cells of CHC patients were distinct from those of HDs and were sufficient to distinguish the CHC patients from HDs (Figure 5C ). These results demonstrate that chronic HCV infection results in altered expression of apoptosis-related genes in global CD4+ and CD8+ T-cells.
Correlation of apoptotic gene expression levels with the pathogenesis of HCV infection
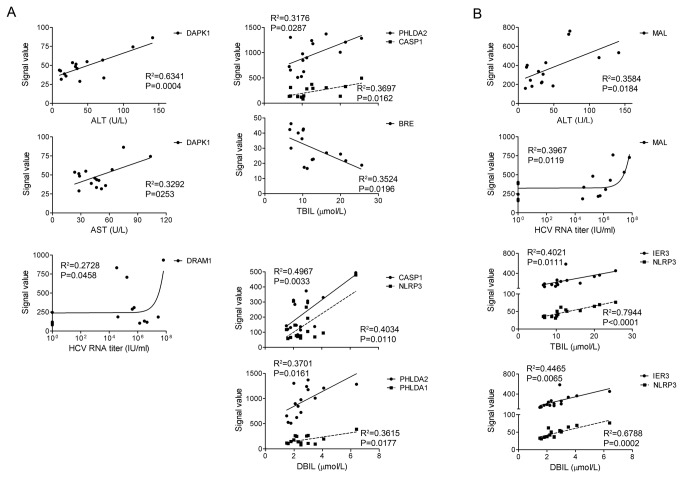
We analyzed the significance of the expression levels of apoptotic genes during HCV infection by correlating them with other clinical parameters. We found that the expression levels of 20 apoptosis-related genes in CD4+ T-cells and 6 apoptosis-related genes in CD8+ T-cells were significantly correlated (p<0.05) with at least one clinical parameter (Figure 6 and data not shown). Interestingly, the expression levels of 5 apoptosis-related genes death-associated protein kinase 1 (DAPK1), pleckstrin homology-like domain family A member 1 (PHLDA1), pleckstrin homology-like domain family A member 2 (PHLDA2), CASP1, and NLR family pyrin domain-containing 3 (NLRP3) in CD4+ T-cells from CHC patients were positively correlated with ALT, AST, TBIL, and DBIL values (Figure 6A ). Expression of the anti-apoptotic gene brain and reproductive organ-expressed (BRE) was negatively correlated with TBIL. DNA damage-regulated autophagy modulator 1 (DRAM1) expression in CD4+ T-cells was correlated with HCV RNA titers in the CHC patients (Figure 6A ). Interestingly, the expression of NLRP3 in CD8+ T-cells from CHC patients was positively correlated with TBIL and DBIL values (Figure 6B ). In addition, the expression levels of mal, T-cell differentiation protein (MAL) in total CD8+ T-cells were positively correlated with the ALT values and HCV RNA titers (Figure 6B ). These significant correlations suggest that dysregulation of T-cells death may be associated with the pathogenesis of chronic HCV infection.
Figure 6. Correlation between the expression levels of apoptosis-related genes and clinical parameters in CHC patients.
(A), Correlation of the expression levels of apoptosis-related genes in CD4+ T-cells from CHC patients with clinical parameters. (B), Correlation of the expression levels of the apoptosis-related genes in CD8+ T-cells from CHC patients with clinical parameters. The original fluorescent signal values for each gene detected in microarray assays were used for the correlation study.
Gene expression signatures of total CD4+ and CD8+ T-cells in chronic HCV infection differ from those in chronic HIV or HBV infection
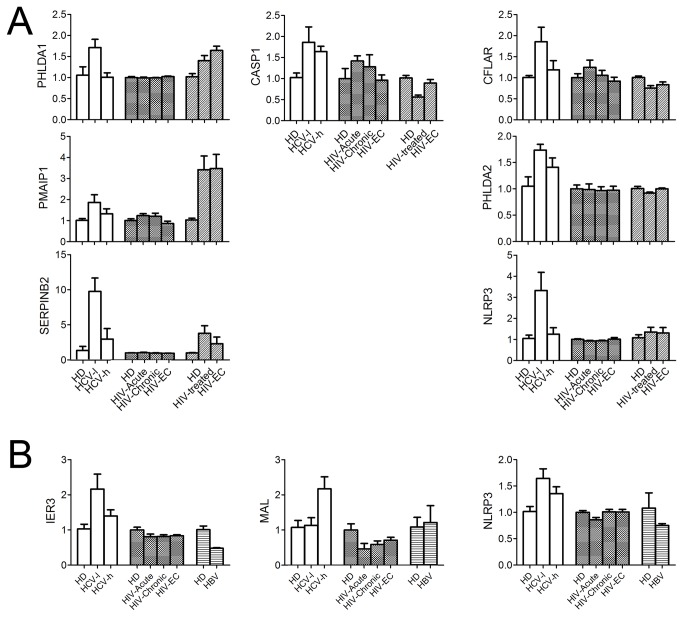
An important feature of untreated HIV infection is the gradual loss of global CD4+ T-cells via apoptosis [25]. We compared the gene expression profiles of CD4+ and CD8+ T-cells from CHC patients to those of HIV or HBV patients using published microarray data [9–11]. Interestingly, the expression of 65 genes was significantly changed in both CHC and HIV patients (Table S3 ). Despite these commonly changed genes, the T-cells gene expression patterns in these two groups of patients differed greatly. First, the expression of interferon-stimulated genes (ISGs) including interferon alpha-inducible protein 6 (IFI6), interferon alpha-inducible protein 27 (IFI27), interferon-inducible protein 44 (IFI44), 2'-5'-oligoadenylate synthetase 1 (OAS1), and myxovirus resistance protein 1 (MX1), was upregulated early during the acute phase of HIV-1 infection and maintained through the chronic phase but was not upregulated in CD4+ T-cells from CHC patients. Second, many apoptosis-related gene expression changes in CD4+ T-cells from CHC patients were not observed in CD4+ T-cells from HIV-1 infected patients (Figure 7A and Table S3 ). These results suggest that HCV and HIV infection have different impacts on the expression profiles of apoptosis-related genes in total CD4+ T-cells.
Figure 7. Expression patterns of apoptosis-related genes in T-cells from patients infected with HCV, HIV-1, and HBV.
(A), Relative expression levels of apoptosis-related genes in CD4+ T-cells during HCV or HIV-1 infection. (B), Relative expression levels of apoptosis-related genes in CD8+ T-cells during HCV, HIV-1, or HBV infection. The gene expression levels in T-cells from HDs were defined as 1, and fold changes are shown.
Like chronic HCV infection, chronic HBV (CHB) infection also causes functional exhaustion of both antigen-specific and global CD8+ T-cells [13,15,26–28]. We previously identified gene changes in total CD8+ T-cells from CHB patients [9]. Only 15 genes were altered during both CHC and CHB infection, and 6 of these genes displayed opposite changes in CHC and CHB patients (Table S4 ). The ISG signature associated with CD8+ T-cells during acute and chronic HIV-1 infection [10,11] was not shared with CD8+ T-cells from CHC patients. Importantly, the expression levels of the majority of the apoptosis-related genes that were altered in CD8+ T-cells from CHC patients either were not changed or displayed opposite changes in CD8+ T-cells from patients chronically infected with HBV or HIV-1 (Figure 7B and Table S4 , Table S5 ). Taken together, these results demonstrate that global CD8+ T-cells express a unique apoptosis-related gene signature during chronic HCV infection that differs from that observed during chronic HBV or HIV-1 infection.
Discussion
Although the functional exhaustion of HCV-specific T-cells during CHC is well established, the functional status of total CD4+ and CD8+ T-cells in CHC patients remains poorly understood. In this study, we examined the functional status and gene expression profiles of total CD4+ and CD8+ T-cells in a group of long-term treatment-naïve CHC patients. Total CD4+ and CD8+ T-cells from CHC patients exhibited enhanced activation-induced apoptosis but proliferated normally. The plasma levels of IFN-γ, IL-9, and IP-10 in these patients were significantly elevated, and pre-incubation of primary T-cells from healthy donors with these cytokines sensitized T-cells to activation-induced apoptosis. Furthermore, although the overall gene expression pattern in CD4+ and CD8+ T-cells from CHC patients was indistinguishable from that from HDs, CHC patients’ T-cells expressed unique apoptosis-related gene signatures that differed from those in T-cells from patients chronically infected with HIV-1 or HBV. Our data provide important insight into the functional status of global T-cells during chronic HCV infection and suggest that the host environment during chronic HCV infection alters T-cells function.
The enhanced activation-induced apoptosis of total CD4+ and CD8+ T-cells from CHC patients is likely due to the combined effects of several cytokines, including IFN-γ, IP-10, and IL-9. The role of IFN-γ in priming T-cells for activation-induced death has been firmly established [29,30]. Moreover, elevated IFN-γ in CHC patients modulates macrophage activation, and IFN-γ can directly act on T-cells or indirectly induce T-cells death by promoting the production of galectin-9 by monocytes and macrophages during chronic HCV infection [31]. Galectin-9 induces the apoptosis of HCV-specific CTLs by interacting with its cellular receptor, T cell immunoglobulin mucin-3 (Tim-3) [31]. Elevated IP-10 in CHC patients is a predictor of poor therapeutic outcome [32,33]. A previous report also showed that in combination with IL-2 and IFN-α, IP-10 induces the activation-independent apoptosis of primary human T-cells [34]. Our data clearly demonstrate that IP-10 also sensitizes primary human T-cells to activation-induced apoptosis.
Surprisingly, we found that a high concentration of IL-9 promotes activation-induced apoptosis in T-cells. IL-9, a common γc-sharing cytokine, is a T-cells growth factor [35,36]. Neutralization or genetic deletion of IL-9 in mouse models dramatically reduced the effector T-cells population, suggesting a role for IL-9 in promoting T-cells survival and proliferation [37,38]. However, in our study, T-cells cultured in high concentrations of IL-9 were more sensitive to activation-induced apoptosis. Two possible mechanisms might account for this observation. First, high concentrations of IL-9 might induce an altered expression pattern of pro- versus anti-apoptotic genes in human T-cells. IL-4, another γc-sharing cytokine, was recently shown to induce the upregulation of the pro-apoptotic genes Bcl-2-like protein 11 (Bim) and Bcl-2-associated X (Bax) in T-cells [39]. Second, IL-9 may act indirectly by inducing pro-apoptotic cytokines in other PBMC populations. The precise mechanisms by which IL-9 primes human T-cells for activation-induced apoptosis require further investigation. Nevertheless, high plasma IL-9 levels are associated with poor treatment outcomes in HIV/HCV-coinfected patients [40] and in patients with chronic heart failure [41]. Together, these data suggest that high plasma IL-9 levels are pathogenic during chronic HCV infection. Importantly, our results strongly suggest that these cytokines act together on both global T-cells and HCV-specific T-cells. Thus, to restore the impaired survival of antigen-specific T-cells in CHC patients, it will be essential to neutralize the pro-apoptotic activities of these cytokines.
Although the overall gene expression patterns of CD4+ and CD8+ T-cells in CHC patients were largely indistinguishable from those of HD T-cells, the expression pattern of apoptosis-related genes in T-cells from CHC patients was dramatically altered and was sufficient to distinguish CHC patients from HDs. Consistent with the normal lymphocyte compartment but enhanced activation-induced apoptosis observed in the CHC patients, these results suggest that CHC-induced host inflammation primarily affects apoptosis. Furthermore, the expression of multiple apoptosis-related genes was dysregulated in CD4+ T-cells from CHC patients. The expression of some of these genes, such as BCL2A1, was also perturbed in global CD8+ T-cells from CHC patients. Interestingly, one-third of these genes were significantly correlated with clinical parameters in CHC patients. For example, the expression of DAPK1 was significantly positively correlated with ALT and AST values. DAPK1 is induced after T-cell stimulation and inhibits T-cells activation [42]. The expression of superoxide dismutase 2 (SOD2), a protein that inhibits radiation-induced apoptosis [43], was strongly correlated with ALT, TBIL and DBIL values, suggesting that apoptosis-related genes may contribute to pathology during HCV infection.
Importantly, the gene signature in T-cells from CHC patients differed from those in T-cells from HIV-infected and HBV-infected patients. The gene signature of CD4+ T-cells from HIV-infected patients is characterized by the induction of ISGs [10,11]. We recently demonstrated that three apoptosis-related genes –Apoptotic protease-activating factor 1 (APAF1), mitogen-activated protein kinase 4 (MAP2K4), and tumor protein p53 (TP53) – were upregulated in PBMCs from HIV rapid progressors [44]. Similarly, CD8+ T-cells from HIV-1 infected patients also exhibit upregulation of ISGs. Although apoptosis is enhanced in T-cells from HIV-1 patients, the changes in the expression of apoptosis-related genes are different from those observed in CHC patients, suggesting that these changes are virus-specific. Thus, specific strategies are needed to prevent virus-induced T-cells loss in chronic HCV infection.
In conclusion, our data suggest that activation-induced apoptosis is a primary functional failure for global CD4+ and CD8+ T-cells in long-term chronic HCV infection, accompanying with unique apoptosis-related gene expression signature. Several cytokines elevated in the HCV patients’ plasma, including IFN-γ, IP-10 and IL-9, are responsible for sensitization of global T-cells to activation-induced cell death. However, it remains to be investigated whether these cytokines directly lead antigen-specific T-cells to activation-induced apoptosis.
Supporting Information
Correlations between the HCV RNA titer and other clinical parameters among the CHC patients. Shown are the correlations between HCV RNA titers and ALT, AST, DBIL, TBIL, age and gender. None has achieved statistic significance.
(TIF)
Clustering analysis of genes with CV>0.5. (A), Clustering result for CD4+ T-cells is presented. (B), Clustering result for CD8+ T-cells is presented. The blocks in blue represent HD samples, the blocks in yellow represent HCV-l samples, and the block in red represents HCV-h samples.
(TIF)
List of altered gene pathways in CD4+ and CD8+ T lymphocytes of CHC patients. Shown are the results of pathway analysis by GeneGo for significantly changed expression of genes (fold change ≥1.5, p<0.05) of CD4+ (A) and CD8+ (B) T-cells. Note that the pathways in HCV-l and HCV-h patients differed from each other.
(TIF)
Genes involved in critical biological processes with up- or down-regulated expression from CD4+ T-cells in HCV-h or HCV-l groups compared to healthy donors.
(DOCX)
Genes involved in critical biological processes with up- or down-regulated expression from CD8+ T-cells in HCV-h or HCV-l groups compared to healthy donors.
(DOCX)
Common genes of CD4+ T-cells shared by HCV and HIV-1infection.
(DOCX)
Common genes of CD8+ T-cells shared by HCV and HBV infection.
(DOCX)
Common genes of CD8+ T-cells shared by HCV and HIV-1 infection.
(DOCX)
Acknowledgments
We thank Dr. Claire Gordy for critically reading this manuscript. We thank the patients and healthy donors for their support of this study.
Funding Statement
This study is supported by the National Grand Program on Key Infectious Diseases (No. 2012ZX10002-006, No.2013ZX10004-601, No.2013ZX10002002-006; No.2012ZX10002004-006) from the Ministry of Health of China, the Capital Health Development Research Project (No.2011-2018-04) from Beijing Municipal Bureau of Health and the Institute of Pathogen Biology (No.2009IPB108), Chinese Academy of Medical Science & Peking Union Medical College. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pawlotsky JM (2004) Pathophysiology of hepatitis C virus infection and related liver disease. Trends Microbiol 12: 96-102. doi: 10.1016/j.tim.2003.12.005. PubMed: 15036326. [DOI] [PubMed] [Google Scholar]
- 2. Shepard CW, Finelli L, Alter MJ (2005) Global epidemiology of hepatitis C virus infection. Lancet Infect Dis 5: 558-567. doi: 10.1016/S1473-3099(05)70216-4. PubMed: 16122679. [DOI] [PubMed] [Google Scholar]
- 3. Rehermann B (2009) Hepatitis C virus versus innate and adaptive immune responses: a tale of coevolution and coexistence. J Clin Invest 119: 1745-1754. doi: 10.1172/JCI39133. PubMed: 19587449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guidotti LG, Chisari FV (2006) Immunobiology and pathogenesis of viral hepatitis. Annu Rev Pathol 1: 23-61. doi: 10.1146/annurev.pathol.1.110304.100230. PubMed: 18039107. [DOI] [PubMed] [Google Scholar]
- 5. Dustin LB, Rice CM (2007) Flying under the radar: the immunobiology of hepatitis C. Annu Rev Immunol 25: 71-99. doi: 10.1146/annurev.immunol.25.022106.141602. PubMed: 17067278. [DOI] [PubMed] [Google Scholar]
- 6. Wedemeyer H, He XS, Nascimbeni M, Davis AR, Greenberg HB et al. (2002) Impaired effector function of hepatitis C virus-specific CD8+ T cells in chronic hepatitis C virus infection. J Immunol 169: 3447-3458. PubMed: 12218168. [DOI] [PubMed] [Google Scholar]
- 7. Gruener NH, Lechner F, Jung MC, Diepolder H, Gerlach T et al. (2001) Sustained dysfunction of antiviral CD8+ T lymphocytes after infection with hepatitis C virus. J Virol 75: 5550-5558. doi: 10.1128/JVI.75.12.5550-5558.2001. PubMed: 11356962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Radziewicz H, Ibegbu CC, Hon H, Osborn MK, Obideen K et al. (2008) Impaired hepatitis C virus (HCV)-specific effector CD8+ T cells undergo massive apoptosis in the peripheral blood during acute HCV infection and in the liver during the chronic phase of infection. J Virol 82: 9808-9822. doi: 10.1128/JVI.01075-08. PubMed: 18667503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jin WJ, Chen CF, Liao HY, Gong LL, Yuan XH et al. (2012) Downregulation of the AU-rich RNA-binding protein ZFP36 in chronic HBV patients: implications for anti-inflammatory therapy. PLOS ONE 7: e33356. doi: 10.1371/journal.pone.0033356. PubMed: 22428029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hyrcza MD, Kovacs C, Loutfy M, Halpenny R, Heisler L et al. (2007) Distinct transcriptional profiles in ex vivo CD4+ and CD8+ T cells are established early in human immunodeficiency virus type 1 infection and are characterized by a chronic interferon response as well as extensive transcriptional changes in CD8+ T cells. J Virol 81: 3477-3486. doi: 10.1128/JVI.01552-06. PubMed: 17251300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vigneault F, Woods M, Buzon MJ, Li C, Pereyra F et al. (2011) Transcriptional profiling of CD4 T cells identifies distinct subgroups of HIV-1 elite controllers. J Virol 85: 3015-3019. doi: 10.1128/JVI.01846-10. PubMed: 21177805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ogg GS, Jin X, Bonhoeffer S, Dunbar PR, Nowak MA et al. (1998) Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science 279: 2103-2106. doi: 10.1126/science.279.5359.2103. PubMed: 9516110. [DOI] [PubMed] [Google Scholar]
- 13. Maini MK, Boni C, Lee CK, Larrubia JR, Reignat S et al. (2000) The role of virus-specific CD8(+) cells in liver damage and viral control during persistent hepatitis B virus infection. J Exp Med 191: 1269-1280. doi: 10.1084/jem.191.8.1269. PubMed: 10770795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Edwards BH, Bansal A, Sabbaj S, Bakari J, Mulligan MJ et al. (2002) Magnitude of functional CD8+ T-cell responses to the gag protein of human immunodeficiency virus type 1 correlates inversely with viral load in plasma. J Virol 76: 2298-2305. doi: 10.1128/jvi.76.5.2298-2305.2002. PubMed: 11836408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Webster GJ, Reignat S, Brown D, Ogg GS, Jones L et al. (2004) Longitudinal analysis of CD8+ T cells specific for structural and nonstructural hepatitis B virus proteins in patients with chronic hepatitis B: implications for immunotherapy. J Virol 78: 5707-5719. doi: 10.1128/JVI.78.11.5707-5719.2004. PubMed: 15140968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Seeff LB (2002) Natural history of chronic hepatitis C. Hepatology 36: S35-S46. doi: 10.1016/S0168-8278(02)80104-0. PubMed: 12407575. [DOI] [PubMed] [Google Scholar]
- 17. Simmonds P, Bukh J, Combet C, Deléage G, Enomoto N et al. (2005) Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42: 962-973. doi: 10.1002/hep.20819. PubMed: 16149085. [DOI] [PubMed] [Google Scholar]
- 18. Urbani S, Amadei B, Tola D, Massari M, Schivazappa S et al. (2006) PD-1 expression in acute hepatitis C virus (HCV) infection is associated with HCV-specific CD8 exhaustion. J Virol 80: 11398-11403. doi: 10.1128/JVI.01177-06. PubMed: 16956940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Penna A, Pilli M, Zerbini A, Orlandini A, Mezzadri S et al. (2007) Dysfunction and functional restoration of HCV-specific CD8 responses in chronic hepatitis C virus infection. Hepatology 45: 588-601. doi: 10.1002/hep.21541. PubMed: 17326153. [DOI] [PubMed] [Google Scholar]
- 20. Radziewicz H, Ibegbu CC, Fernandez ML, Workowski KA, Obideen K et al. (2007) Liver-infiltrating lymphocytes in chronic human hepatitis C virus infection display an exhausted phenotype with high levels of PD-1 and low levels of CD127 expression. J Virol 81: 2545-2553. doi: 10.1128/JVI.02021-06. PubMed: 17182670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nakamoto N, Cho H, Shaked A, Olthoff K, Valiga ME et al. (2009) Synergistic reversal of intrahepatic HCV-specific CD8 T cell exhaustion by combined PD-1/CTLA-4 blockade. PLOS Pathog 5: e1000313 PubMed: 19247441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vali B, Jones RB, Sakhdari A, Sheth PM, Clayton K et al. (2010) HCV-specific T cells in HCV/HIV co-infection show elevated frequencies of dual Tim-3/PD-1 expression that correlate with liver disease progression. Eur J Immunol 40: 2493-2505. doi: 10.1002/eji.201040340. PubMed: 20623550. [DOI] [PubMed] [Google Scholar]
- 23. McMahan RH, Golden-Mason L, Nishimura MI, McMahon BJ, Kemper M et al. (2010) Tim-3 expression on PD-1+ HCV-specific human CTLs is associated with viral persistence, and its blockade restores hepatocyte-directed in vitro cytotoxicity. J Clin Invest 120: 4546-4557. doi: 10.1172/JCI43127. PubMed: 21084749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bengsch B, Seigel B, Ruhl M, Timm J, Kuntz M et al. (2010) Coexpression of PD-1, 2B4, CD160 and KLRG1 on exhausted HCV-specific CD8+ T cells is linked to antigen recognition and T cell differentiation. PLOS Pathog 6: e1000947 PubMed: 20548953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cummins NW, Badley AD (2010) Mechanisms of HIV-associated lymphocyte apoptosis: 2010. Cell Death Dis 1: e99. doi: 10.1038/cddis.2010.77. PubMed: 21368875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boni C, Fisicaro P, Valdatta C, Amadei B, Di Vincenzo P et al. (2007) Characterization of hepatitis B virus (HBV)-specific T-cell dysfunction in chronic HBV infection. J Virol 81: 4215-4225. doi: 10.1128/JVI.02844-06. PubMed: 17287266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Das A, Hoare M, Davies N, Lopes AR, Dunn C et al. (2008) Functional skewing of the global CD8 T cell population in chronic hepatitis B virus infection. J Exp Med 205: 2111-2124. doi: 10.1084/jem.20072076. PubMed: 18695005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lopes AR, Kellam P, Das A, Dunn C, Kwan A et al. (2008) Bim-mediated deletion of antigen-specific CD8 T cells in patients unable to control HBV infection. J Clin Invest 118: 1835-1845. doi: 10.1172/JCI33402. PubMed: 18398508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu Y, Janeway CA Jr. (1990) Interferon gamma plays a critical role in induced cell death of effector T cell: a possible third mechanism of self-tolerance. J Exp Med 172: 1735-1739. doi: 10.1084/jem.172.6.1735. PubMed: 2147950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Refaeli Y, Van Parijs L, Alexander SI, Abbas AK (2002) Interferon gamma is required for activation-induced death of T lymphocytes. J Exp Med 196: 999-1005. doi: 10.1084/jem.20020666. PubMed: 12370261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mengshol JA, Golden-Mason L, Arikawa T, Smith M, Niki T et al. (2010) A crucial role for Kupffer cell-derived galectin-9 in regulation of T cell immunity in hepatitis C infection. PLOS ONE 5: e9504. doi: 10.1371/journal.pone.0009504. PubMed: 20209097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dolganiuc A, Norkina O, Kodys K, Catalano D, Bakis G et al. (2007) Viral and host factors induce macrophage activation and loss of toll-like receptor tolerance in chronic HCV infection. Gastroenterology 133: 1627-1636. doi: 10.1053/j.gastro.2007.08.003. PubMed: 17916356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lagging M, Romero AI, Westin J, Norkrans G, Dhillon AP et al. (2006) IP-10 predicts viral response and therapeutic outcome in difficult-to-treat patients with HCV genotype 1 infection. Hepatology 44: 1617-1625. doi: 10.1002/hep.21407. PubMed: 17133471. [DOI] [PubMed] [Google Scholar]
- 34. Sidahmed AM, León AJ, Bosinger SE, Banner D, Danesh A et al. (2012) CXCL10 contributes to p38-mediated apoptosis in primary T lymphocytes in vitro. Cytokine 59: 433-441. doi: 10.1016/j.cyto.2012.05.002. PubMed: 22652417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goswami R, Kaplan MH (2011) A brief history of IL-9. J Immunol 186: 3283-3288. doi: 10.4049/jimmunol.1003049. PubMed: 21368237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. He YW, Malek TR (1998) The structure and function of gamma c-dependent cytokines and receptors: regulation of T lymphocyte development and homeostasis. Crit Rev Immunol 18: 503-524. doi: 10.1615/CritRevImmunol.v18.i6.20. PubMed: 9862091. [DOI] [PubMed] [Google Scholar]
- 37. Li H, Nourbakhsh B, Cullimore M, Zhang GX, Rostami A (2011) IL-9 is important for T-cell activation and differentiation in autoimmune inflammation of the central nervous system. Eur J Immunol 41: 2197-2206. doi: 10.1002/eji.201041125. PubMed: 21674475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li H, Nourbakhsh B, Ciric B, Zhang GX, Rostami A (2010) Neutralization of IL-9 ameliorates experimental autoimmune encephalomyelitis by decreasing the effector T cell population. J Immunol 185: 4095-4100. doi: 10.4049/jimmunol.1000986. PubMed: 20805418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dunkle A, Dzhagalov I, He YW (2011) Cytokine-dependent and cytokine-independent roles for Mcl-1: genetic evidence for multiple mechanisms by which Mcl-1 promotes survival in primary T lymphocytes. Cell Death Dis 2: e214. doi: 10.1038/cddis.2011.95. PubMed: 21975296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Guzmán-Fulgencio M, Jiménez JL, Berenguer J, Fernández-Rodríguez A, López JC et al. (2012) Plasma IL-6 and IL-9 predict the failure of interferon-alpha plus ribavirin therapy in HIV/HCV-coinfected patients. J Antimicrob Chemother 67: 1238-1245. doi: 10.1093/jac/dkr595. PubMed: 22294644. [DOI] [PubMed] [Google Scholar]
- 41. Cappuzzello C, Di Vito L, Melchionna R, Melillo G, Silvestri L et al. (2011) Increase of plasma IL-9 and decrease of plasma IL-5, IL-7, and IFN-gamma in patients with chronic heart failure. J Transl Med 9: 28. doi: 10.1186/1479-5876-9-28. PubMed: 21418620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chuang YT, Fang LW, Lin-Feng MH, Chen RH, Lai MZ (2008) The tumor suppressor death-associated protein kinase targets to TCR-stimulated NF-kappa B activation. J Immunol 180: 3238-3249. PubMed: 18292548. [DOI] [PubMed] [Google Scholar]
- 43. Epperly MW, Sikora CA, DeFilippi SJ, Gretton JA, Zhan Q et al. (2002) Manganese superoxide dismutase (SOD2) inhibits radiation-induced apoptosis by stabilization of the mitochondrial membrane. Radiat Res 157: 568-577. doi:10.1667/0033-7587(2002)157[0568:MSDSIR]2.0.CO;2. PubMed: 11966323. [DOI] [PubMed] [Google Scholar]
- 44. Zhang ZN, Xu JJ, Fu YJ, Liu J, Jiang YJ et al. (2013) Transcriptomic Analysis of Peripheral Blood Mononuclear Cells in Rapid Progressors in Early HIV Infection Identifies a Signature Closely Correlated with Disease Progression. Clin Chem (. (2013)) PubMed: 23592504. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlations between the HCV RNA titer and other clinical parameters among the CHC patients. Shown are the correlations between HCV RNA titers and ALT, AST, DBIL, TBIL, age and gender. None has achieved statistic significance.
(TIF)
Clustering analysis of genes with CV>0.5. (A), Clustering result for CD4+ T-cells is presented. (B), Clustering result for CD8+ T-cells is presented. The blocks in blue represent HD samples, the blocks in yellow represent HCV-l samples, and the block in red represents HCV-h samples.
(TIF)
List of altered gene pathways in CD4+ and CD8+ T lymphocytes of CHC patients. Shown are the results of pathway analysis by GeneGo for significantly changed expression of genes (fold change ≥1.5, p<0.05) of CD4+ (A) and CD8+ (B) T-cells. Note that the pathways in HCV-l and HCV-h patients differed from each other.
(TIF)
Genes involved in critical biological processes with up- or down-regulated expression from CD4+ T-cells in HCV-h or HCV-l groups compared to healthy donors.
(DOCX)
Genes involved in critical biological processes with up- or down-regulated expression from CD8+ T-cells in HCV-h or HCV-l groups compared to healthy donors.
(DOCX)
Common genes of CD4+ T-cells shared by HCV and HIV-1infection.
(DOCX)
Common genes of CD8+ T-cells shared by HCV and HBV infection.
(DOCX)
Common genes of CD8+ T-cells shared by HCV and HIV-1 infection.
(DOCX)