Abstract
An outbreak of cholera began in Haiti in October of 2010. To understand the progression of epidemic cholera in Haiti, in April of 2012, we initiated laboratory-enhanced surveillance for diarrheal disease in four Haitian hospitals in three departments. At each site, we sampled up to 10 hospitalized patients each week with acute watery diarrhea. We tested 1,616 specimens collected from April 2, 2012 to March 28, 2013; 1,030 (63.7%) specimens yielded Vibrio cholerae, 13 (0.8%) specimens yielded Shigella, 6 (0.4%) specimens yielded Salmonella, and 63 (3.9%) specimens tested positive for rotavirus. Additionally, 13.5% of children < 5 years old tested positive for rotavirus. Of 1,030 V. cholerae isolates, 1,020 (99.0%) isolates were serotype Ogawa, 9 (0.9%) isolates were serotype Inaba, and 1 isolate was non-toxigenic V. cholerae O139. During 1 year of surveillance, toxigenic cholera continued to be the main cause of acute diarrhea in hospitalized patients, and rotavirus was an important cause of diarrhea-related hospitalizations in children.
Introduction
In October of 2010, an outbreak of cholera began in Haiti, a country where cholera cases had not been seen in at least a century. By November of 2010, toxigenic Vibrio cholerae O1 had been confirmed in all 10 of Haiti's administrative departments.1 Cholera treatment facilities (CTFs), semipermanent facilities often associated with a hospital or health center, were quickly constructed and staffed to treat cholera patients, and a National Cholera Surveillance System (NCSS) was established to collect data on the number of cases, hospitalizations, and deaths in CTFs. As of June 30, 2013, 663,134 cholera cases and 8,160 deaths attributed to cholera had been reported to the NCSS.2,3
Since the beginning of the outbreak, the NCSS has used a case definition for cholera based on symptoms rather than laboratory confirmation; the NCSS defines a cholera case as any patient with three or more episodes of acute watery diarrhea within 24 hours. Before March of 2012, the proportion of syndromic cholera cases reported to the NCSS with laboratory-confirmed cholera was not systematically evaluated.
In April of 2012, the Haiti National Public Health Laboratory (LNSP) and the US Centers for Disease Control and Prevention (CDC) Haiti established sentinel laboratory-based surveillance for acute diarrheal illness in Haiti to estimate the proportion of hospitalized patients at CTFs and hospitals with laboratory-confirmed cholera, monitor changes in cholera antimicrobial susceptibility and serotypes, and better understand the burden of other diarrheal diseases in Haiti.
Materials and Methods
We selected four hospitals for surveillance. The first two hospitals, Hôpital Universitaire La Paix (HUP) and Hôpital Foyer Saint Camille (HSC), are located in the capital of Haiti, Port-au-Prince, in West Department. The third site, Hôpital Saint Nicolas (HSN) de Saint Marc, is in Artibonite Department, and the fourth site, Hôpital Saint Michel de Jacmel (SMJ), is in South East Department. HSC is a privately funded Catholic institution; the other three hospitals are public facilities. We chose these four hospitals because they all have associated CTFs, are relatively large facilities, and are located within a 3-hour drive from LNSP in Port-au-Prince, making transport of specimens manageable.
At each site, we used convenience sampling to collect stool specimens from up to 10 hospitalized patients per week with acute watery diarrhea (defined as three or more episodes of acute watery diarrhea within 24 hours, with onset of symptoms within the past 7 days). Nurses were trained to collect one-half of the weekly specimens from patients < 5 years old. Patients who had taken antibiotics either at home or in the health facility were excluded. Hospitalized patients were selected from CTFs, pediatric wards, medicine wards, and emergency rooms. Nurses selected patients at different times of the day and week depending on patient flow and their work load. We also administered a questionnaire to patients to collect demographic and clinical information. In September of 2012, after recognizing that only 18% of patients sampled were children < 5 years old, we increased the maximum number of weekly samples to 15 patients at HUP, the hospital with the highest overall patient volume, in an effort to increase enrollment of younger children.
Whole stool was collected in a cup, and two swabs were placed into Cary–Blair transport medium (Becton, Dickinson and Company, Sparks, MD). Whole stool and inoculated transport medium were stored at 2–8°C for up to 3 days before transport to LNSP. At LNSP, specimens were inoculated onto Thiosulfate-Citrate-Bile salts-sucrose (TCBS) agar (Thermo Scientific™, Remel™, Lenexa, KS), MacConkey agar (Becton, Dickinson and Company, Sparks, MD), and Hektoen and Xylose Lysine Deoxycholate (XLD) agar (Becton, Dickinson and Company, Sparks, MD). Plates were incubated at 37°C for 24 hours, and the appropriate colonies were selected for additional testing to identify V. cholerae, Shigella, and Salmonella. Whole-stool specimens collected before December of 2012 were also tested for V. cholerae with the Crystal VC Rapid Diagnostic Test (RDT; Span Diagnostics, Surat, India). Pure cultures of V. cholerae were tested by disc diffusion on Mueller–Hinton agar (Becton, Dickinson and Company, Sparks, MD) without blood and with ampicillin, tetracycline (as a marker for doxycycline), and trimethoprim/sulfamethoxazole. Results were interpreted according to Clinical and Laboratory Standards Institute (CLSI) guidelines4; 115 isolates were sent to the Enteric Diseases Laboratory at CDC Atlanta for toxigenicity and additional antimicrobial susceptibility testing.4,5 All stool specimens were also tested for rotavirus using Premier Rotaclone EIA (Meridian Diagnostics, Cincinnati, OH) enzyme immunoassay (EIA).
Data were entered and stored in a Microsoft Access 2010 database (Microsoft Corporation, Redmond, WA), and analyzed using SAS 9.3 (SAS, Cary, NC). Descriptive statistics were generated using frequency procedures, and differences in proportions were tested using χ2 test statistics. Univariate logistic regression was used to assess whether demographic factors (e.g., age and sex) were associated with cholera. Factors that were statistically significant in univariate models were included in a multivariable model, and two-way interactions were assessed. No interactions were significant at P < 0.05, and only the main effects were included in the final model. Sex was not included in the final model, because it was neither significant in a univariate model nor a confounder in the multiple regression model. Although the patient questionnaire collected some clinical variables, in this manuscript, we focus mainly on demographic characteristics of patients; clinical data will be presented in a separate manuscript.
Only samples positive for V. cholerae by culture were considered confirmed. The sensitivity and specificity of the Crystal VC RDT were evaluated using cholera culture as the gold standard. The CDC determined this surveillance activity to be non-research, and therefore, approval was not required from the CDC Institutional Review Board. The Haiti Ethical Review Committee also considered this activity to be non-research. Verbal consent was obtained from adults and the parents or guardians of minors.
Results
From April 2, 2012 to March 28, 2013, we collected stool specimens from 1,616 patients at the four sites. Almost one-half (47.1%) of the specimens were from females. The median age was 25 years (range = < 1 month to 95 years), and 319 (20.0%) patients were under 5 years (Table 1). The highest percentage of patients was from HUP (35.2%), whereas the lowest percentage was from HSC (20.5%). Most patients (84.5%) were treated in CTFs, whereas 12.2% of patients were from pediatric wards; only 3.4% of patients were enrolled from the emergency room or medical wards. Enrolled patients had a mean of 14.4 stools/day, and 56.9% of patients were severely dehydrated.
Table 1.
Demographic characteristics of patients with acute diarrhea in Haiti from April 2, 2012 to March 28, 2013
| Overall (N = 1,616) | Cholera (N = 1,616) | Shigella (N = 1,616) | Salmonella (N = 1,616) | Rotavirus (N = 1,616) | No pathogen detected (N = 1,616) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | n | % | n | % | |
| Total | 1,030 | 63.7 | 13 | 0.8 | 6 | 0.4 | 63 | 3.9 | 517 | 40.0 | ||
| Demographics* | ||||||||||||
| Age (years) | ||||||||||||
| 0–1 | 202 | 12.6 | 27 | 13.4 | 2 | 1.0 | 0 | 0.0 | 31 | 15.4 | 143 | 70.8 |
| 2–4 | 117 | 7.3 | 57 | 48.7 | 0 | 0.0 | 1 | 0.9 | 12 | 10.3 | 50 | 42.7 |
| 5–17 | 231 | 14.5 | 163 | 70.6 | 2 | 0.9 | 1 | 0.4 | 9 | 3.9 | 59 | 25.5 |
| 18–65 | 957 | 59.9 | 707 | 73.9 | 9 | 0.9 | 3 | 0.3 | 8 | 0.8 | 233 | 24.4 |
| 65 and older | 92 | 5.8 | 63 | 68.5 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 29 | 31.5 |
| Sex | ||||||||||||
| Male | 853 | 52.9 | 540 | 63.3 | 7 | 0.8 | 5 | 0.6 | 31 | 3.6 | 276 | 32.4 |
| Female | 759 | 47.1 | 487 | 64.2 | 6 | 0.8 | 1 | 0.1 | 30 | 4.0 | 240 | 31.6 |
| Hospital | ||||||||||||
| HUP | 569 | 35.2 | 402 | 70.7 | 4 | 0.7 | 2 | 0.4 | 16 | 2.8 | 150 | 26.4 |
| HSN | 381 | 23.6 | 219 | 57.5 | 6 | 1.6 | 2 | 0.5 | 19 | 5.0 | 140 | 36.8 |
| SMJ | 334 | 20.7 | 247 | 74.0 | 0 | 0.0 | 1 | 0.3 | 3 | 0.9 | 84 | 25.2 |
| HSC | 332 | 20.5 | 162 | 48.8 | 3 | 0.9 | 1 | 0.3 | 23 | 6.9 | 143 | 43.1 |
| Hospital ward | ||||||||||||
| Pediatrics | 195 | 12.2 | 11 | 5.6 | 2 | 1.0 | 0 | 0.0 | 32 | 16.1 | 151 | 77.4 |
| Emergency/internal medicine | 54 | 3.4 | 14 | 25.9 | 2 | 3.7 | 0 | 0.0 | 2 | 3.7 | 37 | 68.5 |
| Cholera treatment center | 1,356 | 84.5 | 1,000 | 73.8 | 9 | 0.7 | 6 | 0.4 | 28 | 2.1 | 324 | 23.9 |
Missing information: age (17 patients), sex (4 patients), and hospital ward (11 patients).
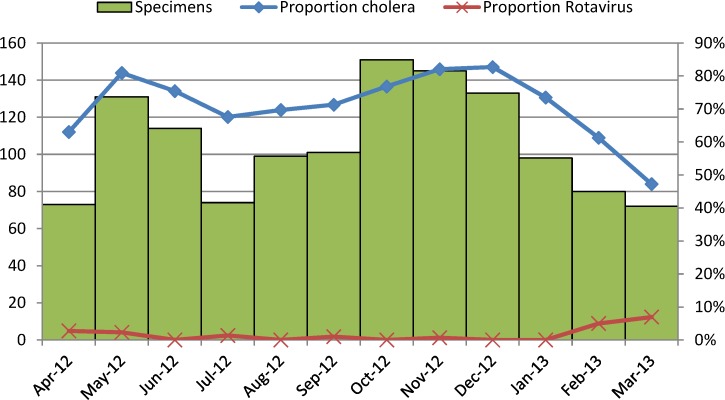
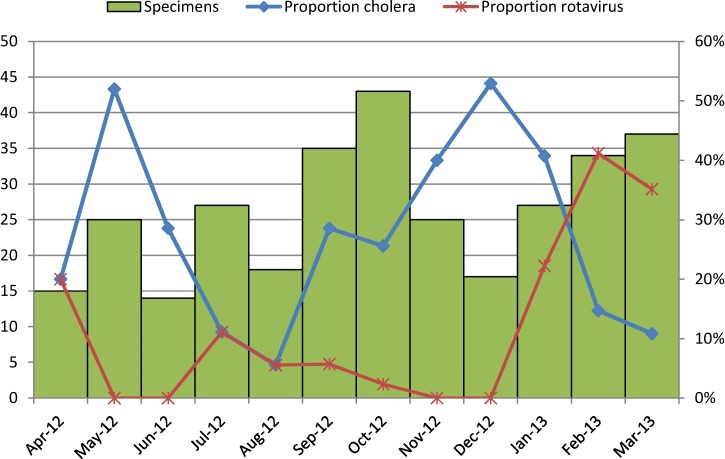
Of 1,616 specimens tested by culture, 1,030 (63.7%) specimens yielded V. cholerae, 6 (0.4%) specimens yielded Salmonella, and 13 (0.8%) specimens yielded Shigella. By EIA, 63 (3.9%) specimens tested positive for rotavirus. Four specimens were positive for both V. cholerae and rotavirus. One specimen yielded V. cholerae, Salmonella, and Shigella. No other coinfections were documented. A greater percentage of patients over 18 years (73.4%) and between 5 and 17 years (70.6%) tested positive for cholera compared with patients 2–4 (48.7%) and under 2 years (13.4%; P < 0.0001) (Table 1). The proportion of children under 5 years who were treated in pediatric wards and tested positive for cholera was lower than the proportion of children under 5 years treated in CTFs who were positive for cholera (6.1% versus 55.6%, P < 0.0001). The monthly proportion of specimens among patients 5 years and older that tested positive for cholera ranged from 47% to 83% (Figure 1). The proportion was highest from May to June and from November to December of 2012. Among patients under 5 years old, the proportion ranged from 6% to 52%, and it was highest in May and December of 2012 (Figure 2).
Figure 1.
Proportion of samples from patients 5 years and older positive for V. cholerae by stool culture and rotavirus by month from April 2, 2012 to March 28, 2013.
Figure 2.
Proportion of samples from patients under 5 years positive for V. cholerae by stool culture and rotavirus by month from April 2, 2012 to March 28, 2013.
Results from unadjusted logistic regression models showed that age, hospital site, and hospital ward were all associated with culture-confirmed cholera (Table 2). In an adjusted model including age, hospital site, and hospital ward, patients 0–2 years were less likely to have cholera (odds ratio [OR] = 0.27, 95% confidence interval [95% CI] = 0.16–0.46) compared with patients 5 years and older (Table 2). In a model that adjusted for both age and hospital ward, compared with patients from HUP, patients from HSN had lower odds of testing positive for cholera (OR = 0.54, 95% CI = 0.40–0.73). Diarrheal patients treated in pediatric wards (OR = 0.05, 95% CI = 0.02–0.10) and emergency and internal medicine wards (OR = 0.14, 95% CI = 0.07–0.26) were much less likely to have cholera compared with patients treated in CTFs, even after adjusting for patient age and hospital.
Table 2.
Unadjusted and adjusted results of logistic regression models of culture-confirmed cholera in Haiti from 2012 to 2013
| Unadjusted ORs (95% CI) cholera | Adjusted ORs (95% CI) cholera | |
|---|---|---|
| Age (years) | ||
| 0–2 | 0.06 (0.04–0.09) | 0.27 (0.16–0.46) |
| 2–4 | 0.36 (0.24–0.52) | 0.64 (0.41–1.02) |
| ≥ 5 | Reference | Reference |
| Hospital | ||
| HUP | Reference | Reference |
| HSN | 0.56 (0.43–0.74) | 0.54 (0.40–0.73) |
| SMJ | 1.18 (0.87–1.60) | 1.00 (0.71–1.38) |
| HSC | 0.40 (0.30–0.52) | 0.93 (0.65–1.34) |
| Hospital ward | ||
| Pediatrics | 0.02 (0.01–0.04) | 0.05 (0.02–0.10) |
| Emergency/internal medicine | 0.13 (0.07–0.23) | 0.14 (0.07–0.26) |
| Cholera treatment center | Reference | Reference |
Of 1,029 V. cholerae serogroup O1 toxigenic isolates, 1,020 (99.0%) isolates were identified as serotype Ogawa, and 9 (0.9%) isolates were identified as serotype Inaba. One non-toxigenic isolate was identified as V. cholerae serogroup O139 by RDT and standard serogroup O139 antisera testing at LNSP. It was later confirmed to be a non-toxigenic isolate of V. cholerae O139. One-hundred fifteen isolates were tested for antimicrobial susceptibility at CDC Atlanta. All isolates were susceptible to ampicillin, and all but six isolates were resistant to trimethoprim-sulfamethoxazole. One isolate from a patient at HSN showed intermediate resistance to tetracycline; all others were fully susceptible to tetracycline.
In total, 741 specimens were tested for cholera by the Crystal VC RDT. The sensitivity and specificity of the RDT were 95.6% and 79.5%, respectively, and the positive and negative predictive values were 89.4% and 91.0%, respectively (Table 3).
Table 3.
Sensitivity, specificity, negative predictive value, and positive predictive value of Crystal VC RDT compared with stool culture for V. cholerae
| RDT result | Culture result | ||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 457 | 54 | 511 |
| Negative | 21 | 209 | 230 |
| Total | 478 | 263 | 741 |
Sensitivity: 457/478 = 95.6%. Specificity: 209/263 = 79.5%. Positive predictive value: 457/511 = 89.4%. Negative predictive value: 209/230 = 91.0%.
Rotavirus was identified more frequently in children 0–1 (15.4%) and 2–4 years (10.3%) compared with patients 5–18 (3.9%) and > 18 years (0.8%; P < 0.0001). The proportion of children under 5 years who were treated in pediatric wards and tested positive for rotavirus was similar to the proportion of children under 5 years treated in CTFs who were positive for rotavirus (15.6% versus 10.2%, P = 0.38). The monthly proportion of rotavirus among children under 5 years of age ranged from 0% to 41% (Figure 2). The proportion was highest in February and March of 2013 and lowest from May to June of 2012 and from November to December of 2012, when the percentage of children with V. cholerae was highest.
Discussion
We found that, from 18 to 30 months after the beginning of the cholera epidemic in Haiti, nearly two-thirds of hospitalized diarrheal patients who were sampled at four large health facilities in Haiti had laboratory-confirmed V. cholerae. These results improve our understanding of the cholera epidemic in Haiti; although 104,295 syndromically identified cholera cases were reported to the NCSS during the 1-year period covered by this analysis, including 17,677 children under 5 years,2 if national patterns mirror our findings, 27% of the reported cases in adults and 74% of the reported cases in children under 5 years may have been caused by other diarrheal diseases. Nevertheless, the fact that the percentage of monthly adult cholera cases among all sampled diarrheal cases was almost always over 50% during our 12-month surveillance period illustrates that sustained cholera transmission continues to occur in Haiti. The consistently high proportion of culture-positive cholera among diarrheal patients in this 12-month period in Haiti differs from findings from countries in which cholera has been endemic for a much longer time, where lower proportions of diarrheal disease are attributable to cholera during long periods of the year.6,7 Some of this difference, especially in adults, may be the result of repeated exposures over a longer period of endemicity.
In multivariable analysis, we found that patients at HSN had lower odds of being positive for cholera. Because the cholera outbreak began in the Artibonite, where HSN is located, it is possible that higher population immunity in this area resulted in the lower proportion of cholera cases.
In Haiti, healthcare providers have been instructed to refer suspected cholera patients to external CTFs, which are constructed and maintained to reduce the risk of environmental contamination and nosocomial cholera transmission and ensure safe waste removal. However, we found that approximately one-tenth of diarrheal patients treated in the pediatric wards, medical wards, and emergency rooms of the four hospitals had cholera. This finding illustrates that efforts to triage cholera patients to external CTFs based on symptoms alone are not perfect; previous studies have also shown that cholera cases are difficult to diagnose on clinical grounds alone.8,9 Because some cholera patients will likely continue to be admitted to regular hospital wards in Haiti, ensuring appropriate infection control practices in all hospital wards is necessary to prevent cholera transmission within facilities.
In March of 2012, the Haitian National Laboratory characterized two V. cholerae isolates as non-Ogawa serotypes; these isolates were subsequently confirmed to be serotype Inaba by CDC Atlanta.10 Previous infection with V. cholerae serotype Ogawa, the prevailing serotype before March of 2012, confers relatively less immunity to reinfection with the V. cholerae Inaba serotype. This less-effective cross-protective immunity raised concerns for an increase in incidence of cholera among previously infected individuals. However, we did not find evidence of broad serotype conversion in our sample; only 9 of 1,030 isolates were serotype Inaba. Additionally, of the isolates tested for antimicrobial susceptibility, only one isolate was intermediately resistant to tetracycline, a marker for susceptibility to doxycycline, the most commonly used antibiotic for treatment of cholera in Haiti. The close monitoring of antimicrobial susceptibility can continue to inform clinical cholera treatment guidelines in Haiti.
In our study, nearly all isolates were serogroup O1, and we did not identify any non-O1/non-O139 isolates. Our findings differ from a previous study of cholera patients from November of 2010, a month after the beginning of the cholera outbreak in Haiti, in which non-Ol/non-O139 V. cholerae was isolated from 21% of patients.11 However, our findings are consistent with other studies of samples collected from patients identified later in the Haiti outbreak, which found only serogroup O1 isolates.12,13 These results are not unexpected; non-O1/O139 serogroups generally cause a diarrheal disease that is less severe than cholera and does not have epidemic potential, whereas toxigenic O1 and O139 serogroups have epidemic potential.14
The V. cholerae O139 isolate was initially identified by culture and agglutination in O139 antiserum. Then, a suspension of an isolated colony was tested using the Crystal VC RDT and found to be positive for O139. The isolate was later confirmed as a non-toxigenic V. cholerae serogroup O139 by CDC Atlanta. Non-toxigenic O139 is uncommon in the United States but has been documented in CDC's Cholera and Other Vibrio Illness Surveillance System.15 Toxigenic O139 cholera has been imported to the United States by tourists and seafood, but transmission in the Western Hemisphere has never been documented.16
In our study, like a previous study in Haiti,17 the Crystal VC RDT for cholera performed well compared with culture, the gold standard for cholera diagnosis. In addition to high sensitivity and relatively high specificity, the test had high positive and negative predictive values. Others studies that have evaluated the Crystal VC have found similarly high sensitivity but lower specificity estimates (49–73%).18–20 In our study, the rapid test for cholera was performed in a laboratory by trained technicians. The location and training of the staff performing the RDT may partly explain the improved test performance relative to other studies. In settings where the diagnostic gold standard of culture is not readily available, a rapid test for cholera can be a reliable alternative for early confirmation of cholera outbreaks and a trigger for the rapid resource mobilization necessary for a successful public health response. Although the Crystal VC RDT can identify both serogroup O1 and O139 isolates, final confirmation of these new strains should be done by culture and cholera toxin testing; although the RDT is highly sensitive and very specific, culture remains the gold standard.
In our sample, 13.5% of diarrheal patients < 5 years old tested positive for rotavirus. This percentage is lower than the percentages described in previous studies from other countries in the region; a surveillance network from Latin America and the Caribbean region found that, among children < 5 years hospitalized for acute diarrhea, the median percentage positive for rotavirus was 31.5% (range = 24–47%).21 However, in Haiti, a country in the midst of a cholera epidemic, the proportion of diarrheal specimens positive for rotavirus might be expected to be lower, because cholera adds considerably to the denominator of hospitalized children with diarrhea. Indeed, in our surveillance system, almost one-half of specimens from 2- to 4-year-old children were positive for cholera.
Our findings have several limitations. First, we only included four hospital sites in three departments of Haiti, which limits our ability to generalize results to the rest of the country. Additionally, although we included 10–15 patients weekly from each site, patients were selected by convenience sampling and came predominately from CTFs. Therefore, our results may have been subject to selection bias and may not be representative of all diarrheal patients at each site or patients treated at health facilities without CTFs. Furthermore, we included hospitalized patients only; the NCSS includes hospitalized and non-hospitalized patients. Therefore, our patient population is not representative of all patients reported to the NCSS. We were not able to estimate the overall burden of cholera, because we were unable to obtain denominator data for the catchment area of the hospitals and overall hospital admissions. Also, although we excluded patients who had received antibiotics, we failed to identify the etiology of disease for 35% of patients sampled. To strengthen surveillance in the future, we plan to add identification of other enteric pathogens, such as enterotoxigenic Escherichia coli. Although all four hospitals have backup generators and samples were transported in cold boxes with freezer packs, it is possible that, in some instances, some specimens were not consistently maintained at the correct temperature, which may have influenced pathogen recovery. Additionally, we did not collect data on patient outcomes in our study, and we are, therefore, not able to analyze how outcomes differed by pathogen or other factors. Finally, although cholera was more commonly identified during Haiti's two rainy seasons and although rotavirus was more common during the cooler months of the year, data from several years of surveillance are needed to better understand the seasonality of rotavirus and cholera, both of which have been shown to vary by season in previous studies in other countries.22,23
More than 2 years after the start of the cholera epidemic in Haiti, in four hospitals in different parts of the country, cholera remained the predominant cause of diarrheal-related hospitalizations among adults and an important cause of hospitalizations for diarrhea among children. Despite lower case-fatality rates and decreased international attention since the beginning of the epidemic, cholera continues to impose significant demands on human and financial resources from Haiti's health system. Rotavirus seems to be a smaller but significant cause of diarrhea in children < 5 years old. The introduction of rotavirus vaccine planned for later this year could help reduce diarrheal disease among children in Haiti. In areas at risk for cholera outbreaks, as the World Health Organization has recommended, oral cholera vaccine could be used.24 Ultimately, broad access to safe drinking water and sanitation in Haiti will be necessary to reduce the large burden of cholera and other waterborne diseases on the Haitian population.
ACKNOWLEDGMENTS
We thank Roc Magloire, Josiane Buteau, Rossignol Emmanuel, Marc-Covens Junior Jean-Baptiste, Sherly Morisseau, Renette Anseime, Rossignol Emmanuel, Chedelène Riviere, Lourdy Narcisse, Marie Géanne Ulysse, Fabiola Charles, Finelia Saint Louis, Dieunane Nikechta Cherisier, Mirlène Alix, Chrismène Cyprien, the administrators and staff of Hôpital Universitaire la Paix, Hôpital Foyer Saint Camille, Hôpital Saint Nicolas, and Hôpital Saint Michel de Jacmel, Cheryl Bopp, Laura Dickmeyer, Nancy Garrett, Peter Gerner-Smidt, Jean Whichard, Joan Brunkard, Janell Routh, Nick Schaad, Barbara J. Marston, Manish Patel, Umesh Parashar. and Jon Gentsch.
Disclaimer: The findings and conclusions in this article are the findings and conclusions of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Authors' addresses: Maria W. Steenland, Benjamin L. Nygren, Michele B. Parsons, Deborah F. Talkington, and Eric D. Mintz, Division of Foodborne, Waterborne, and Environmental Diseases, Centers for Disease Control and Prevention, Atlanta, GA, E-mails: mws475@mail.harvard.edu, ghz8@cdc.gov, zcp9@cdc.gov, dft1@cdc.gov, and edm1@cdc.gov. Gerard A. Joseph, Mentor Ali Ber Lucien, Nicole Freeman, and Jacques Boncy, Laboratoire National de Santé Publique (National Public Health Laboratory), Port-au-Prince, Haiti, E-mails: gerardajo944@gmail.com, drmabl@yahoo.com, nicolemfreeman@gmail.com, and jacques_boncy@hotmail.com. Marisa Hast and S. Arunmozhi Balajee, Health Systems Reconstruction Office, Centers for Disease Control and Prevention, Atlanta, GA, E-mails: ium1@cdc.gov and fir3@cdc.gov. Eyal Leshem, Division of Viral Diseases, Centers for Disease Control and Prevention, Atlanta, GA, E-mail: wgp9@cdc.gov. Stanley Juin, John Vertefeuille, and Mark A. Katz, Centers for Disease Control and Prevention, Haiti, E-mails: Juins@ht.cdc.gov, dki4@cdc.gov, and cfq6@cdc.gov.
References
- 1.Tappero JW, Tauxe RV. Lessons learned during public health response to cholera epidemic in Haiti and the Dominican Republic. Emerg Infect Dis. 2011;17:2087–2093. doi: 10.3201/eid1711.110827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ministere de la Sante Publique et de la Population . Rapports journaliers du MSPP sur l'évolution du choléra en Haiti. 2012. http://www.mspp.gouv.ht/site/index.php?option=com_content&view=article&id=120&Itemid=1 Available at. Accessed July 29, 2013. [Google Scholar]
- 3.Barzilay EJ, Schaad N, Magloire R, Mung KS, Boncy J, Dahourou GA, Mintz ED, Steenland MW, Vertefeuille JF, Tappero JW. Cholera surveillance during the Haiti epidemic—the first 2 years. N Engl J Med. 2013;368:599–609. doi: 10.1056/NEJMoa1204927. [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute . Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria; Proposed Guideline. Wayne, PA: Clinical and Laboratory Standards Institute; 2005. [Google Scholar]
- 5.Fields PI, Popovic T, Wachsmuth K, Olsvik O. Use of polymerase chain reaction for detection of toxigenic Vibrio cholerae O1 strains from the Latin American cholera epidemic. J Clin Microbiol. 1992;30:2118–2121. doi: 10.1128/jcm.30.8.2118-2121.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manna B, Niyogi SK, Bhattacharya MK, Sur D, Bhattacharya SK. Observations from diarrhoea surveillance support the use of cholera vaccination in endemic areas. Int J Infect Dis. 2005;9:117–119. doi: 10.1016/j.ijid.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Chowdhury F, Rahman MA, Begum YA, Khan AI, Faruque AS, Saha NC, Baby NI, Malek MA, Kumar AR, Svennerholm AM, Pietroni M, Cravioto A, Qadri F. Impact of rapid urbanization on the rates of infection by Vibrio cholerae O1 and enterotoxigenic Escherichia coli in Dhaka, Bangladesh. PLoS Negl Trop Dis. 2011;5:e999. doi: 10.1371/journal.pntd.0000999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siddique AK, Ahmed S, Iqbal A, Sobhan A, Poddar G, Azim T, Sack DA, Rahman M, Sack RB. Epidemiology of rotavirus and cholera in children aged less than five years in rural Bangladesh. J Health Popul Nutr. 2011;29:1–8. doi: 10.3329/jhpn.v29i1.7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sur D, Deen JL, Manna B, Niyogi SK, Deb AK, Kanungo S, Sarkar BL, Kim DR, Danovaro-Holliday MC, Holliday K, Gupta VK, Ali M, von Seidlein L, Clemens JD, Bhattacharya SK. The burden of cholera in the slums of Kolkata, India: data from a prospective, community based study. Arch Dis Child. 2005;90:1175–1181. doi: 10.1136/adc.2004.071316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention Notes from the field: Identification of Vibrio cholerae serogroup O1, serotype Inaba, biotype El Tor strain—Haiti, March 2012. MMWR Morb Mortal Wkly Rep. 2012;61:309. [PubMed] [Google Scholar]
- 11.Hasan NA, Choi SY, Eppinger M, Clark PW, Chen A, Alam M, Haley BJ, Taviani E, Hine E, Su Q, Tallon LJ, Prosper JB, Furth K, Hoq MM, Li H, Fraser-Liggett CM, Cravioto A, Huq A, Ravel J, Cebula TA, Colwell RR. Genomic diversity of 2010 Haitian cholera outbreak strains. Proc Natl Acad Sci USA. 2012;109:E2010–E2017. doi: 10.1073/pnas.1207359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katz LS, Petkau A, Beaulaurier J, Tyler S, Antonova ES, Turnsek MA, Guo Y, Wang S, Paxinos EE, Orata F, Gladney LM, Stroika S, Folster JP, Rowe L, Freeman MM, Knox N, Frace M, Boncy J, Graham M, Hammer BK, Boucher Y, Bashir A, Hanage WP, Van Domselaar G, Tarr CL. Evolutionary dynamics of Vibrio cholerae O1 following a single-source introduction to Haiti. MBio. 2013;4:e00398-13. doi: 10.1128/mBio.00398-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talkington D, Bopp C, Tarr C, Parsons MB, Dahourou G, Freeman M, Joyce K, Turnsek M, Garrett N, Humphrys M, Gomez G, Stroika S, Boncy J, Ochieng B, Oundo J, Klena J, Smith A, Keddy K, Gerner-Smidt P. Characterization of toxigenic Vibrio cholerae from Haiti, 2010–2011. Emerg Infect Dis. 2011;17:2122–2129. doi: 10.3201/eid1711.110805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menon M, Mintz ED, Tauxe R. Bacterial Infections of Humans. 4th Ed. Berlin: Springer; 2009. p. 255. [Google Scholar]
- 15.Centers for Disease Control and Prevention Cholera and Other Vibrio Illness Surveillance System. 2013. http://www.cdc.gov/nationalsurveillance/cholera_vibrio_surveillance.html Available at. Accessed July 29, 2013.
- 16.Boyce TG, Mintz ED, Greene KD, Wells JG, Hockin JC, Morgan D, Tauxe RV. Vibrio cholerae O139 Bengal infections among tourists to Southeast Asia: an intercontinental foodborne outbreak. J Infect Dis. 1995;172:1401–1404. doi: 10.1093/infdis/172.5.1401. [DOI] [PubMed] [Google Scholar]
- 17.Boncy J, Rossignol E, Dahourou G, Hast M, Buteau J, Stanislas M, Moffett D, Bopp C, Balajee SA. Performance and utility of a rapid diagnostic test for cholera: notes from Haiti. Diagn Microbiol Infect Dis. 2013;76:521–523. doi: 10.1016/j.diagmicrobio.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 18.Page AL, Alberti KP, Mondonge V, Rauzier J, Quilici ML, Guerin PJ. Evaluation of a rapid test for the diagnosis of cholera in the absence of a gold standard. PLoS One. 2012;7:e37360. doi: 10.1371/journal.pone.0037360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ley B, Khatib AM, Thriemer K, von Seidlein L, Deen J, Mukhopadyay A, Chang NY, Hashim R, Schmied W, Busch CJ, Reyburn R, Wierzba T, Clemens JD, Wilfing H, Enwere G, Aguado T, Jiddawi MS, Sack D, Ali SM. Evaluation of a rapid dipstick (Crystal VC) for the diagnosis of cholera in Zanzibar and a comparison with previous studies. PLoS One. 2012;7:e36930. doi: 10.1371/journal.pone.0036930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukherjee P, Ghosh S, Ramamurthy T, Bhattacharya MK, Nandy RK, Takeda Y, Nair GB, Mukhopadhyay AK. Evaluation of a rapid immunochromatographic dipstick kit for diagnosis of cholera emphasizes its outbreak utility. Jpn J Infect Dis. 2010;63:234–238. [PubMed] [Google Scholar]
- 21.de Oliveira LH, Danovaro-Holliday MC, Andrus JK, de Fillipis AM, Gentsch J, Matus CR, Widdowson MA, Rotavirus Surveillance N. Sentinel hospital surveillance for rotavirus in Latin American and Caribbean countries. J Infect Dis. 2009;200((Suppl 1)):S131–S139. doi: 10.1086/605060. [DOI] [PubMed] [Google Scholar]
- 22.Patel MM, Pitzer VE, Alonso WJ, Vera D, Lopman B, Tate J, Viboud C, Parashar UD. Global seasonality of rotavirus disease. Pediatr Infect Dis J. 2013;32:e134–e147. doi: 10.1097/INF.0b013e31827d3b68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pascual M, Bouma MJ, Dobson AP. Cholera and climate: revisiting the quantitative evidence. Microbes Infect. 2002;4:237–245. doi: 10.1016/s1286-4579(01)01533-7. [DOI] [PubMed] [Google Scholar]
- 24.World Health Organization . Cholera Vaccines: WHO Position Paper. Geneva: World Health Organization; 2010. pp. 117–128. [Google Scholar]