Abstract
We have modified an existing semi-nested multiplex polymerase chain reaction (PCR) by adding one Plasmodium knowlesi-specific nested PCR, and validated the latter against laboratory and clinical samples. This new method has the advantage of being relatively affordable in low resource settings while identifying the five human Plasmodium species with a three-step PCR.
Since the first Malaysian reports and the recognition of Plasmodium knowlesi as the fifth human Plasmodium species,1–5 there have been continuous attempts to improve the molecular detection of this Plasmodium species of simian origin.6–11 Initially, a nested 18S small subunit (ssu) ribosomal DNA (rDNA)-based polymerase chain reaction (PCR) identified the first large cohort of naturally acquired P. knowlesi infections in humans in Borneo.1 However, the primers used to detect the P. knowlesi DNA (Pmk8-Pmkr9) can cross-react with the Plasmodium vivax ribosomal RNA (rRNA) gene, requiring systematic confirmation by either a secondary PCR (1) or sequencing.6,12 Since then, more specific methods using nested6 or non-nested PCR,10,11 loop-mediated isothermal amplification,8 or real time PCR7,9 have been developed.
Plasmodium knowlesi infections in humans were reported in Vietnam12 and this species needs to be accounted for in the newly defined national malaria elimination strategies.13 Applying the multiplex semi-nested (SnM) PCR published by Rubio and others14 for the detection of the four previously described human Plasmodium species, we recently showed that the malaria parasite reservoir in Central Vietnam was more complex than previously reported by standard microscopy.15 We therefore added another secondary PCR, specific for P. knowlesi, into the existing SnM-PCR to enable the detection of the five Plasmodium species within a three-step PCR.
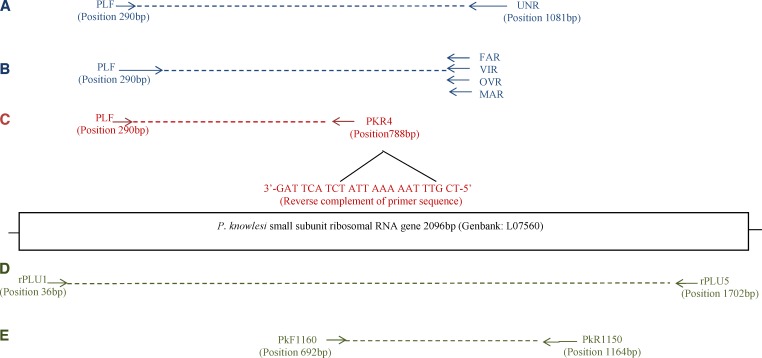
The existing SnM-PCR protocol was adapted from Rubio and others,14 and consists of a first reaction Plasmodium genus specific (Figure 1A) followed by a semi-nested PCR including the Plasmodium-specific forward (PLF) primer used in the primary PCR, plus species-specific reverse primers for Plasmodium falciparum, P. vivax, Plasmodium malaria, and Plasmodium ovale (Figure 1B). We hereby describe the newly added semi-nested (Sn-) PCR for the identification of P. knowlesi (Figure 1C). The analysis of existing oligonucleotide primers for P. knowlesi showed that the reverse complement of the PkF1140 primer designed and validated by Imwong and others6 was compatible with the PLF primer described by Rubio and others.14 Therefore, the Sn-PCR of P. knowlesi includes the PLF forward primer, and a species-specific reverse primer (PKR4: 3′-GAT TCA TCT ATT AAA AAT TTG CT-5′), the latter being the reverse complement of the PkF1140 primer shortened by two base pairs (bp). The specificity of the PKR4 was subsequently assessed in silico by aligning sequences for the 18S ssu rRNA genes of different human and non-human Plasmodium species (P. knowlesi, P. coatney, P. cynomolgi, P. inui, P. fragile, P. reichenowi, P. lophurae, P. gallinacae) as well as 29 P. knowlesi sequences available on GenBank (http://www.ncbi.nlm.nih.gov/genbank/index.html).
Figure 1.
Primers position for the modified SnM-PCR amplification are shown for the single primary reaction (A), followed by two secondary PCRs: one for the four usual human Plasmodium species (B), following protocol by Rubio and others,15 and the new secondary PCR with the PkR4 primer (C). The relative positions of the different primers used for the nested PCR amplification to detect P. knowlesi by Imwong and others6 are indicated below the gene: primary PCR (D) and nested PCR (E).
The Sn-PCR was performed in 25 μL reaction mixture with final concentration for a 1× reaction of 1× Qiagen loading buffer (10× Qiagen buffer, Hilden, Germany), 100 μM of each dNTP (Eurogentec, Seraing, Belgium), 0.5 μM PLF, 0.2 μM PKR4, 1 Unit of Qiagen HotstarTaq plus polymerase, and 2 μL DNA (a 1/500 dilution of the primary PCR product was used as the template). Cycling conditions were as follows: a 5 min denaturation and activation of the Qiagen HotstarTaq plus polymerase step at 94°C, followed by 30 cycles of 20 sec at 51°C for annealing, followed by elongation at 72°C for 1 min, and a 30 sec denaturation at 94°C. The final cycle was followed by an extension time of 10 min at 72°C. The PCR products were detected by 2% agarose gel stained with ethidium bromide, and visualized under UV light.
Reference blood samples from P. knowlesi-infected patients (N = 13) were kindly provided by the IMR, Kuala Lumpur. Furthermore, 80 blood samples (filter paper) from malaria-infected patients collected during a previous survey carried out in Vietnam15 were used to further validate the specificity of the PLF-PKR4 primers. The DNA extraction was done using the QIAamp DNA Micro Kit (Qiagen, Hilden, Germany).
Primary and nested PCR products were cloned into plasmid vectors (pCR4-TOPO vector) and transformed into Mach1 T1B Escherichia coli cells using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA). Thirty bacterial colonies, 10 from two distinct P. knowlesi samples harboring the amplified primary PCR fragment, and 10 harboring the secondary PCR fragment (five colonies for each of the two P. knowlesi samples) were purified and sequenced. In addition, DNA extracts from the 13 P. knowlesi reference samples were subjected to the new Sn-PCR and the amplified PCR fragments of expected size (actual size amplicon = 498 bp), were cut out of the agarose gel, purified, cloned, and sequenced. The sequences obtained were analyzed with the BioEdit program (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and compared with available sequences in GenBank. All sequences obtained from plasmids with the primary and nested PCR inserts were confirmed as P. knowlesi.
The sensitivity and specificity of our Sn-PCR protocol was assessed using two previously published nested PCR protocols for the detection of P. knowlesi: one using Pmk8-Pmkr9 primers1 (hereafter called PCR1) and the one using the PkF1150-PkR15560 primers6 (hereafter called PCR2). The relative sensitivity was determined by a 10-fold serial dilution of the P. knowlesi H strain (Plasmodium knowlesi Genomic DNA-P. knowlesi H strain, MRA-456G obtained at the MR4 [http://www.mr4.org/]) using DNA extract from human blood collected from healthy donors. The analytic sensitivity was determined using a 10-fold serial dilution of the Primary PCR product of PCR 1 and 2 (using primers rPLU1 and 5). This PCR product was first cloned, extracted, and sequenced. The extracted plasmid DNA concentration was determined using three measurements of the Nanodrop (Thermo Scientific, Landsmeer, The Netherlands). A 10-fold serial dilution of the plasmid DNA containing the P. knowlesi fragment was made to determine the sensitivity of the different PCRs. The PCR1 showed the highest in both experiments with an analytical sensitivity at dilutions as low as 1 fg/μL compared with the 100 fg/μL for PCR2 and our Sn-PCR. In both panels the P. knowlesi MR4 H strain as well as the plasmid dilution series, our Sn-PCR and the PCR2 showed the same sensitivity.
The specificity of the three protocols was tested using different genomic DNA controls from Plasmodium species originating from monkeys (1 P. cynomolgy, 1 P. simium, 1 P. fragile; reference strains from NIMPE, Hanoi), and humans (4 P. vivax, 2 P. falciparum, 1 P. ovale, and 1 P. malariae; reference strains from ITM, Antwerp), 1 P. knowlesi (IMR, Kuala Lumpur) as well as genomic DNA (gDNA) of non-Plasmodium origin (1 Leishmania donovani, 1 Schistosoma mansoni, 1 Trypanosoma cruzi, 1 HIV provirus, 1 Mycobacterium tuberculosis, and 1 Mycobacterium ulcerans). Our Sn-PCR and PCR2 showed no cross-reaction, whereas PCR1 showed a false positive P. knowlesi with a 153 bp band for one of the four P. vivax controls.
The specificity was further tested with the 80 clinical samples, previously analyzed by SnM-PCR for the identification of the four human Plasmodium species.15 Filter paper dried blood spots were then tested for the presence of P. knowlesi using the three nested PCR protocols (Table 1). Results showed that PCR1 identified six P. knowlesi infections, whereas our Sn-PCR and PCR 2 did not detect any. Sequencing confirmed the absence of P. knowlesi in all six samples, and the cross-reactivity of PCR1 primers with human DNA (97% homology to region 6853624 bp to 6853772 bp on the Human chromosome 16 genomic contig GenBank accession number: NW 001838290.1). No evidence of contamination of the PCR products was found.
Table 1.
Specificity of the three nested protocols using 80 clinical samples from Vietnam
| Mono- and mixed malaria infections* | Total tested | Positive result for P. knowlesi by protocol: | ||
|---|---|---|---|---|
| n | Nested PCR1† (Pmk8-Pmkr9) | Nested PCR2‡ (PkF1160-PkR1150) | Sn-PCR (PLF-PKR4) | |
| P. falciparum (P.f.) | 38 | – | – | – |
| P. vivax (P.v.) | 14 | – | – | – |
| P. malariae (P.m.) | 5 | – | – | – |
| P. ovale (P.o.) | 1 | – | – | – |
| P.f. + P.v. | 7 | 2 | – | – |
| P.f. + P.m. | 3 | – | – | – |
| P.m. + P.o. | 1 | – | – | – |
| P.f. + P.v. + P.m. | 5 | 2 | – | – |
| P.f. + P.m. + P.o. | 2 | 1 | – | – |
| P.v. + P.m. + P.o. | 3 | 1 | – | – |
| P.f. + P.v. + P.m. + P.o. | 1 | – | – | – |
| Total (N) | 80 | 6 | 0 | 0 |
Further simplification of the protocol into a five-species SnM-PCR should be feasible as preliminary data have shown that the P. knowlesi-specific primer was compatible in silico with the other four human Plasmodium-specific primers. This requires additional optimization efforts, however if successful, will further enhance the value of the current protocol.
Our modified SnM-PCR, offers the advantage of being feasible and affordable for poor income settings where real time PCR is not available yet, while reliably detecting the five human Plasmodium species within a three-step assay.
Footnotes
Financial support: This study was financed by the UBS Optimus Foundation and the Belgian Cooperation within the Framework Agreement III.
Disclosure: The P. knowlesi H strain was obtained through the MR4 as part of the BEI Resources Repository, NIAID, NIH: Plasmodium knowlesi Genomic DNA - P. knowlesi H strain, MRA-456G, deposited by A. W. Thomas.
Authors' addresses: Nguyen Van Hong, National Institute of Malariology, Parasitology and Entomology - Clinical Research Department, Hanoi, Viet Nam, E-mail: nvhong1982@yahoo.com. Peter van den Eede, Chantal Van Overmeir, Anna Rosanas-Urgell, Umberto D'Alessandro, and Annette Erhart, Institute of Tropical Medicine, Antwerp, Belgium, E-mails: petervandeneede@gmail.com, cvoverm@itg.be, arosanas@itg.be, udalessandro@mrc.gm, and aerhart@itg.be. Indra Vythilingham, Faculty of Medicine, University of Malaya - Parasitology Department, Kuala Lumpur, Malaysia, E-mail: Indra.vythilingham@gmail.com. Pham Vinh Thanh, Ngo Duc Thang, Nguyen Manh Hung, and Le Xuan Hung, National Institute of Malariology, Parasitology and Entomology - Epidemiology Department, Hanoi, Viet Nam, E-mails: phamvinht@gmail.com, thangnimpevn@yahoo.com, drmanhhung@gmail.com, and xuanhungvsr@ yahoo.com.
References
- 1.Singh B, Kim Sung L, Matusop A, Radhakrishnan A, Shamsul SS, Cox-Singh J, Thomas A, Conway DJ. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–1024. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- 2.Cox-Singh J, Singh B. Knowlesi malaria: newly emergent and of public health importance? Trends Parasitol. 2008;24:406–410. doi: 10.1016/j.pt.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox-Singh J, Davis TM, Lee KS, Shamsul SS, Matusop A, Ratnam S, Rahman HA, Conway DJ, Singh B. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin Infect Dis. 2008;46:165–171. doi: 10.1086/524888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vythilingam I, Noorazian YM, Huat TC, Jiram AI, Yusri YM, Azahari AH, Norparina I, Noorrain A, Lokmanhakim S. Plasmodium knowlesi in humans, macaques and mosquitoes in peninsular Malaysia. Parasites & Vectors. 2008;1:26. doi: 10.1186/1756-3305-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White NJ. Plasmodium knowlesi: the fifth human malaria parasite. Clin Infect Dis. 2008;46:172–173. doi: 10.1086/524889. [DOI] [PubMed] [Google Scholar]
- 6.Imwong M, Tanomsing N, Pukarittayakemee S, Day NP, White NJ, Snounou G. Spurious amplification of P. vivax small subunit RNA gene using the primers currently used to detect P. knowlesi. J Clin Microbiol. 2009;47:4173–4175. doi: 10.1128/JCM.00811-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babady NE, Sloan LM, Rosenblatt JE, Pritt BS. Detection of Plasmodium knowlesi by real-time polymerase chain reaction. Am J Trop Med Hyg. 2009;81:516–518. [PubMed] [Google Scholar]
- 8.Iseki H, Kawai S, Takahashi N, Hirai M, Tanabe K, Yokoyama N, Igarashi I. Evaluation of a loop-mediated isothermal amplification (LAMP) method as a diagnostic tool of zoonotic simian malaria parasite Plasmodium knowlesi infection. J Clin Microbiol. 2010;48:2509–2514. doi: 10.1128/JCM.00331-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Divis PCS, Shokoples SE, Singh B, Yanow SK. A TaqMan real-time PCR assay for the detection and quantitation of Plasmodium knowlesi. Malar J. 2010;9:344. doi: 10.1186/1475-2875-9-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucchi NW, Poorak M, Oberstaller J, DeBarry J, Srinivasamoorthy G, Goldman I, Xayavong M, da Silva AJ, Peterson DS, Barnwell JW, Kissinger J, Udhayakumar V. A new single-step PCR assay for the detection of the zoonotic malaria parasite Plasmodium knowlesi. PLoS ONE. 2012;7:e31848. doi: 10.1371/journal.pone.0031848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chew CH, Lim YAL, Lee PC, Mahmud R, Chua KH. A hexaplex PCR detection system for the identification of five human Plasmodium species with internal control. J Clin Microbiol. 2012;50:4012–4019. doi: 10.1128/JCM.06454-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van den Eede P, Nguyen Van H, Van Overmeir C, Vythilingam I, Ngo Duc T, Xuan Hung L, Nguyen Manh H, Anne J, D'Alessandro U, Erhart A. Human Plasmodium knowlesi infections in southern Vietnam. Malar J. 2009;8:249. doi: 10.1186/1475-2875-8-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ministry of Health . National Institute of Malariology, Parasitology, and Entomology; Hanoi: 2012. National malaria control and elimination programme. National strategy for malaria control and elimination for the period of 2012–2015. December 2011. Internal Report. [Google Scholar]
- 14.Rubio JM, Post RJ, van Leeuwen WM, Henry MC, Lindergard G, Hommel M. Alternative polymerase chain reaction method to identify Plasmodium species in human blood samples: the semi-nested multiplex malaria PCR (SnM-PCR) Trans R Soc Trop Med Hyg. 2002;96:S199–S204. doi: 10.1016/s0035-9203(02)90077-5. [DOI] [PubMed] [Google Scholar]
- 15.Hong Nguyen V, Van den Eede P, Van Overmeir C, Thang Ngo D, Hung Le X, D'Alessandro U, Erhart A. The distribution of human Plasmodium species in central Vietnam is complex with marked age-dependent prevalence of symptomatic and patent infections. Am J Trop Med Hyg. 2012;87:989–995. doi: 10.4269/ajtmh.2012.12-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]