Abstract
Floodwater samples (N = 110) collected during the 2011 Bangkok floods were tested for Leptospira using culture and polymerase chain reaction (PCR); 65 samples were PCR-positive for putatively non-pathogenic Leptospira species, 1 sample contained a putatively pathogenic Leptospira, and 6 samples contained Leptospira clustering phylogenetically with the intermediate group. The low prevalence of pathogenic and intermediate Leptospira in floodwater was consistent with the low number of human leptospirosis cases reported to the Bureau of Epidemiology in Thailand. This study provides baseline information on environmental Leptospira in Bangkok together with a set of laboratory tests that could be readily deployed in the event of future flooding.
Leptospirosis is an endemic infection throughout the tropics, where outbreaks are well-described, often in the context of heavy freshwater flooding.1,2 Flooding of the Bangkok Metropolitan Region between late October of 2011 and January of 2012 posed a potential risk for a leptospirosis outbreak, and case reports were monitored by the Bureau of Epidemiology of Thailand. The purpose of this article is to describe two adjunctive methods (direct polymerase chain reaction [PCR] and culture) to determine whether pathogenic Leptospira spp. was present in the floodwater.
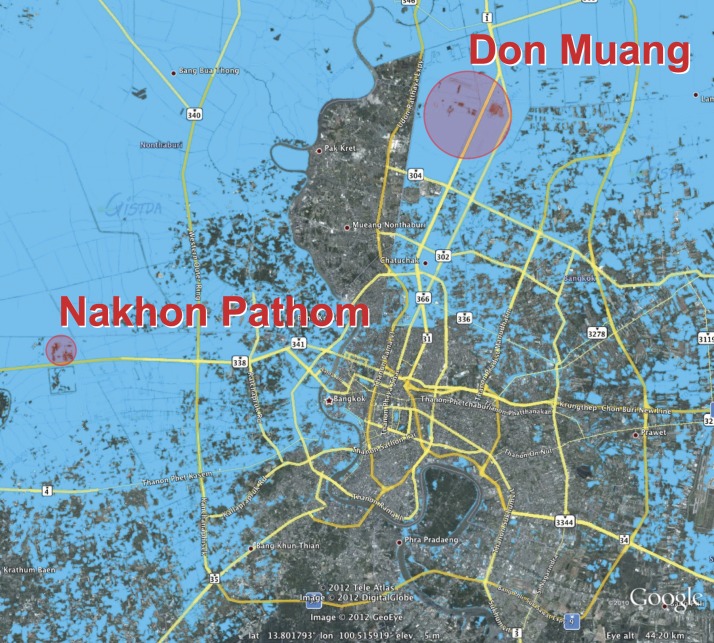
A total of 110 floodwater samples was collected between November 14 and December 6 of 2011; 70 samples were taken within a radius of approximately 2 km around the Salaya Campus of Mahidol University in Nakhon Pathom province (sample code NP), and 40 samples were taken within a radius of approximately 8 km in Don Muang district, Bangkok province (sample code DM) (Figure 1). At each sampling point, 100 mL floodwater were collected at a depth of 20 cm into a sterile glass bottle. Samples were maintained at ambient temperature (25–32°C) during transportation to the laboratory; 50 mL sample were used for direct PCR assay, and the remaining 50 mL sample were used for culture.
Figure 1.
Map of the Bangkok Metropolitan Region showing the extent of flooding in 2011 and the two sampling zones. Flood data were obtained from the Geo-Informatics and Space Technology Development Agency in Thailand, and the map was generated using Google Earth (Google Inc.). The area flooded in 2011 is shown in blue, and the sampling zones are shown as red circles.
A published PCR assay based on amplification and sequencing of a region of the 16S rRNA gene (rrs)3 was used to test all water samples. This testing was performed on a 50-mL sample that was first centrifuged at 3,000 × g for 30 minutes. The deposit was resuspended in 140 μL sterile water, and DNA was extracted from the suspension using the QIAamp Viral RNA Mini Kit (QIAGEN, Santa Clarita, CA) Five microliters DNA extract were used in the reaction, which was performed as described previously.3 The 433-bp amplicons were sequenced, and the species of Leptospira was inferred based on position in a maximum likelihood (ML) phylogenetic tree, which included 36 rrs sequences from GenBank for Leptospira species belonging to the pathogenic, intermediate (of intermediate pathogenicity), non-pathogenic, or unculturable groups Supplemental Table 1. The tree was constructed using the algorithm implemented in PhyML version 3.0.1,4 and the model of sequence evolution used was the generalized time-reversible (GTR) model with γ-distributed rate variation. An ML tree was constructed using the nearest neighbor interchange (NNI) method.4 The MEGA program5 was used to display and edit the tree. The rrs sequences generated during this study were submitted to GenBank, and they are provided in Supplemental Table 2.
Fifty milliliters each water sample were passed through a sterile 0.2-μm filter (Sartorius AG, Gottingen, Lower Saxony, Germany); 0.5 mL filtrate were inoculated into a tube containing 3 mL Leptospira Vanaporn Wuthikanun (LVW) solid agar slant containing 1% Noble agar base and 10% rabbit serum.6 Slants were incubated at 30°C in 5% CO2 for 2 days followed by 30°C in air for a total of 28 days.6 The surface fluid on each agar slant was examined two times weekly by dark-field microscopy. If spirochetes were observed, 100 μL surface fluid were spread-plated onto LVW agar6 supplemented with 2,6-dichlorophenolindophenol (10 mg/mL; Sigma-Aldrich) to enhance visibility of Leptospira colonies and incubated at 30°C in 5% CO2 for 2 days followed by 30°C in air for up to 28 days. Plates were examined two times weekly for visible colonies. We hypothesized that water samples might contain more than one strain of Leptospira species, which may manifest as different colony morphologies on solid agar. In view of this possibility, a single colony of each morphology type on a given plate was picked for additional analysis. These colonies were inoculated into 3 mL Ellinghausen-McCullough-Johnson-Harris (EMJH) broth and incubated at 30°C in air for 5–7 days to achieve a Leptospira concentration of 108 cfu/mL. DNA was then extracted using a boiling method and screened using a multiplex PCR assay developed during this study (using both rrs and lipL32 genes as targets) (Supplemental Text 1). Colonies were assigned to pathogenic, intermediate, or non-pathogenic groups (Supplemental Text 1). Samples that were positive for pathogenic and intermediate species were further evaluated using the rrs assay.3
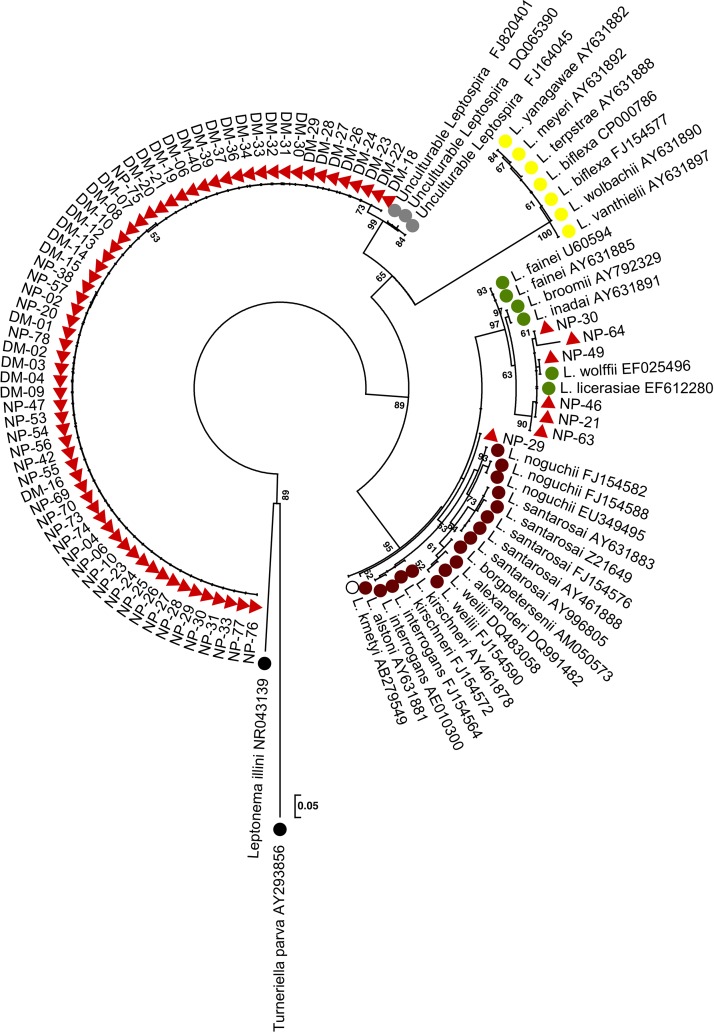
Direct rrs PCR of 110 floodwater samples yielded an amplicon that could be sequenced in 65 cases (59%), of which 45 cases were from Nakhon Pathom and 20 cases were from Bangkok. These 65 sequences were resolved into eight rrs alleles with two polymorphic sites. Analysis of eight alleles using the blastn algorithm implemented in BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) showed that the highest matches (nucleotide identities ranging from 98% to 99%) were rrs sequences cloned from unculturable bacteria in freshwater samples from several countries, including the United States, Korea, and China in 2003, 2008, and 2010, respectively (GenBank ID codes DQ065390, FJ164045, and FJ820401, respectively). On phylogenetic analysis, all eight alleles resided in the non-pathogenic Leptospira spp. cluster (Figure 2).
Figure 2.
Phylogenetic analysis of partial rrs sequences. An ML tree was based on a 443-nt region of rrs. Thirty-six reference sequences from GenBank are shown as color-coded circles that denote their species group: pathogenic (brown), intermediate pathogenicity (green), non-pathogenic (yellow), uncertain pathogenicity (white), or unculturable (grey). Black circles denote two members of the family Leptospiracae. Leptospira species sequences obtained from floodwater samples are shown as red triangles. Two samples are shown two times, because they each contained two different species.
Culture yielded colonies from 87 of 110 samples (79.1%), which on dark-field microscopy, consisted of spirochetes (Supplemental Table 4). Picking one representative colony for each morphology type on a given plate yielded 140 single colonies for evaluation by the multiplex PCR assay. Results are presented using sample as the denominator, with additional information provided for samples containing more than one species. A single sample (sample code NP-29) was positive for pathogenic Leptospira species, and phylogenetic analysis of rrs placed this sample in the pathogenic clade on a discrete branch that did not contain any other isolates (Figure 2). Six samples were positive for intermediate Leptospira species. Using BLAST analysis, two samples were designated as L. licerasiae (NP-21 and NP-46), and two samples were designated as L. wolffii (NP-49 and NP-63). The remaining two samples (NP-30 and NP-64) were most closely related to L. licerasiae, with 99% and 97% nucleotide identity, respectively Supplemental Table 3, but they resided on their own branch in the intermediate Leptospira cluster (Figure 2). Samples NP-30 and NP-49 also contained non-pathogenic Leptospira along with an additional 74 samples. Six samples contained non-Leptospiraceae spirochetes. There was overlap between samples that were both culture-positive and positive on direct PCR for unculturable non-pathogenic Leptospira (N = 47).
In summary, we found a low prevalence of pathogenic and intermediate Leptospira species. The culture and PCR assays used here require additional validation to determine their accuracy for environmental use, but our findings are consistent with the low number of human leptospirosis cases reported to the Bureau of Epidemiology of Thailand, with only 11 notified cases from the Bangkok Metropolitan Region between November of 2011 and January of 2012 (Annual Epidemiological Surveillance Report 2011; http://www.boe.moph.go.th/Annual/AESR2011/index.html). Although outbreaks of leptospirosis during periods of flooding are well-documented in Thailand and elsewhere in the world,1,2 our evidence indicates that an outbreak was not the case during the 2011 Bangkok flood.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the staff at the Mahidol-Oxford Tropical Medicine Research Unit and the Faculty of Tropical Medicine, Mahidol University for support.
Footnotes
Financial support: This work was supported by Wellcome Trust Grant 089275/Z/09/Z and the Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand. S.J.P. receives support from the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre.
Authors' addresses: Janjira Thaipadungpanit, Vanaporn Wuthiekanun, Premjit Amornchai, Siriphan Boonsilp, and Nicholas P. J. Day, Mahidol-Oxford Tropical Medicine Research Unit, Mahidol University, Bangkok, Thailand, E-mails: janjira@tropmedres.ac, lek@tropmedres.ac, premjit@tropmedres.ac, boonsilp.s@gmail.com, and nickd@tropmedres.ac. Narisara Chantratita, Surapon Yimsamran, Wanchai Maneeboonyang, Prapin Tharnpoophasiam, Natnaree Saiprom, Yuvadee Mahakunkijcharoen, Pratap Singhasivanon, and Direk Limmathurotsakul, Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand, E-mails: narisara@tropmedres.ac, tmpbw@mahidol.ac.th, surapon.yim@mahidol.ac.th, tmwmn@mahidol.ac.th, ningkhub@hotmail.com, yuvadee.mah@mahidol.ac.th, pratap.sin@mahidol.ac.th, and direk@tropmedres.ac. Sharon J. Peacock, Department of Medicine, University of Cambridge, Addenbrooke's Hospital, Cambridge, UK, E-mail: sjp97@medschl.cam.ac.uk.
References
- 1.Trevejo RT, Rigau-Perez JG, Ashford DA, McClure EM, Jarquín-González C, Amador JJ, de los Reyes JO, Gonzalez A, Zaki SR, Shieh WJ, McLean RG, Nasci RS, Weyant RS, Bolin CA, Bragg SL, Perkins BA, Spiegel RA. Epidemic leptospirosis associated with pulmonary hemorrhage-Nicaragua, 1995. J Infect Dis. 1998;178:1457–1463. doi: 10.1086/314424. [DOI] [PubMed] [Google Scholar]
- 2.Amilasan AS, Ujiie M, Suzuki M, Salva E, Belo MC, Koizumi N, Yoshimatsu K, Schmidt WP, Marte S, Dimaano EM, Villarama JB, Ariyoshi K. Outbreak of leptospirosis after flood, the Philippines, 2009. Emerg Infect Dis. 2012;18:91–94. doi: 10.3201/eid1801.101892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boonsilp S, Thaipadungpanit J, Amornchai P, Wuthiekanun V, Chierakul W, Limmathurotsakul D, Day NP, Peacock SJ. Molecular detection and speciation of pathogenic Leptospira spp. in blood from patients with culture-negative leptospirosis. BMC Infect Dis. 2011;11:338. doi: 10.1186/1471-2334-11-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 5.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wuthiekanun V, Amornchai P, Paris DH, Langla S, Thaipadunpanit J, Chierakul W, Smythe LD, White NJ, Day NP, Limmathurotsakul D, Peacock SJ. Rapid isolation and susceptibility testing of Leptospira spp. using a new solid medium (LVW agar) Antimicrob Agents Chemother. 2012;57:297–302. doi: 10.1128/AAC.01812-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.