Abstract
We aim to construct a diagnostic model for bacterial coinfection in dengue patients (Dengue Dual Infection Score [DDIS]); 2,065 adult dengue patients (mean age = 41.9 ± 17.2 years, 58.4% male, 83 patients with bacterial coinfection) seen at a university hospital from January of 2005 to February of 2010 were studied. The DDIS was created by assigning one point to each of five risk factors for bacterial coinfection: pulse rate ≥ 90 beats/minute, total white cell count ≥ 6 × 109/L, hematocrit < 40%, serum sodium < 135 mmol/L, and serum urea ≥ 5 mmol/L. The DDIS identified bacterial coinfection (derivation set area under the curve = 0.793, 95% confidence interval = 0.732–0.854; validation set area under the curve = 0.761, 95% confidence interval = 0.637–0.886). A DDIS of ≥ 4 had a specificity of 94.4%, whereas a DDIS of ≥ 1 had a sensitivity of 94.4% for bacterial coinfection. The DDIS can help to select dengue patients for early bacterial cultures and empirical antibiotics.
Introduction
Dengue is the most common human arboviral disease, with an estimated 390 million infections per year.1 Globally, it is responsible for nearly 500,000 hospitalizations, and 3.6 billion people remain at risk.2–4 The usual form of dengue infection is dengue fever, which is self-limiting and characterized by fever, myalgia, arthralgia, retro-orbital headache, and petechial rash. Dengue hemorrhagic fever and dengue shock syndrome, marked by capillary leakage, are the most serious manifestations of dengue infection, with mortality rates as high as 10–20%.2,5
The clinical course of a viral infection can be adversely affected by bacterial coinfection.6–8 However, bacterial coinfection can be easily overlooked in dengue-endemic or -epidemic settings. A simple clinical rule for the identification of concurrent bacterial infection in dengue patients would be pivotal for triggering timely antibiotic therapy within the usual context of supportive management.
We, therefore, aimed to create a risk score for the identification of bacterial coinfection in adult dengue patients (dengue dual infection). Covariates used for the risk score were deliberately kept simple so that this instrument can be readily and cheaply deployed in the emergency department and lower income countries with constrained diagnostic resources.
Methods
Study population and definitions.
The study took place in a 1,000-bed academic medical center in Singapore, which like neighboring Southeast Asian countries, is endemic for dengue. We analyzed the case records for all patients age > 16 years who had positive laboratory confirmation of dengue infection from January 1, 2005 to February 28, 2010. Patients were seen at either the emergency department or the outpatient clinic, and they were not necessarily admitted. Investigation of possible bacterial coinfection was clinically determined by the attending physicians. We did not exclude patients who had taken antibiotics before hospital attendance, because both clinically and microbiologically diagnosed cases of bacterial coinfection were considered. About 70% of the cohort was randomly chosen for risk score derivation, with the remainder for validation. Our Ethical Review Board waived the need for informed consent (DSRB E/2009/337).
Confirmatory assays for dengue could include antidengue immunoglobulin M (IgM) antibody (enzyme-linked immunosorbent assay [ELISA]), dengue non-structural protein 1 (NS1) antigen, or molecular testing of dengue RNA by polymerase chain reaction (PCR). Like others, we used the IgM assay as one of our confirmatory tests for dengue.9–15 The likelihood of cross-reactivity was low, because other flaviviruses (yellow fever, Japanese encephalitis, tick-borne encephalitis, and West Nile fever) are not endemic in our setting. Also, IgM antibodies are specific, even in Japanese encephalitis and yellow fever vaccinees.16 The dengue NS1 antigen was detected using commercially available assays.17 Molecular testing involved amplification of dengue RNA by real-time reverse transcriptase nested PCR.18
Bacterial coinfection was defined as any clinical diagnosis of bacterial infection (e.g., pneumonia) or any bacteremia or bacteruria from cultures taken within 48 hours of admission. Cases with diagnosis of pneumonia were all verified by a consultant pulmonologist (K.C.S.), who based the assessment on the recorded clinical presentation and radiology. Blood and urine cultures were done conventionally, with the former supplemented by an automated BacT/Alert System (bioMerieux SA, Craponne, France). We chose a 48-hour threshold to increase the likelihood that bacterial infection truly existed at the point of presentation, while allowing a reasonable time window for clinicians to have done microbiological testing. Patients with contaminated blood or urinary cultures (e.g., positive for skin flora, such as coagulase-negative staphylococci) were not considered to have bacterial coinfection. Laboratory test information was also available for patients who were not admitted (for instance, patients who had blood tests and chest radiographs done at the emergency department and were discharged thereafter). Hospital length of stay was counted as 0 days if patients were only managed as outpatients.
Fourteen candidate variables that could be reliably measured and readily available at the point of presentation were selected for the diagnostic model. These covariates included demographics (age and sex), vital signs (temperature, systolic blood pressure, pulse rate, and respiratory rate recorded at presentation), and basic laboratory tests (total leukocyte count, neutrophil percentage, hematocrit, platelet count, sodium, potassium, urea, and creatinine recorded within 24 hours of presentation). Symptoms, including days of fever, were not used for the diagnostic model, because assessment can be subjective; we could not ensure that all the patients were specifically probed for all the symptoms. Comorbidities were not included in the diagnostic model, because their presence or absence may not be firmly established initially. Inflammatory markers (C-reactive protein, erythrocyte sedimentation rate, and procalcitonin) were not routinely checked in our hospital.
Statistical analysis.
Univariate comparisons of proportions, means, and medians were done using Fisher exact, Student t, and Wilcoxon rank sum tests, respectively. Using the derivation cohort, a univariate screen of 14 candidate variables was first performed with bacterial coinfection as the dependent variable. To maintain a familywise α=0.05 after multiple comparisons, a threshold P value of 0.003 was computed using Bonferroni adjustment. Age, pulse rate, respiratory rate, total leukocyte count, neutrophil percentage, hematocrit, platelet count, sodium, urea, and creatinine met this threshold. Creatinine was excluded because of collinearity with urea (r = 0.72, P < 0.001).
Using logistic regression, the remaining nine variables were analyzed. Five variables (pulse rate, total leukocyte count, hematocrit, sodium, and urea) with multivariate P values < 0.10 were retained in the final diagnostic model. These five variables were each dichotomized by maximizing the area under individual receiver operating characteristic (ROC) curves. Using logistic regression, standardized coefficients for the dichotomized variables were computed, and each variable was then weighted equally for the final risk score (Dengue Dual Infection Score [DDIS]). The DDIS was a summation of the five variables (each scored as 0 or 1) and could range from 0 to 5. Logistic regression models were calibrated using the Hosmer–Lemeshow χ2 test. Model discrimination was tested in both the derivation and validation sets using ROC curves. We also used a separate method of internal validation, where our derived model was repeatedly fit in 1,000 bootstrap samples (bootstrap resampling) using all patients with complete data. The mean area under the curve (AUC) for 1,000 bootstrap models then represented the estimated model performance for future samples.
Results
Patient characteristics.
In total, 2,065 patients age > 16 years were studied (Table 1). Patients presented after a median of 4 days of self-reported fever (interquartile range = 2–5 days); 65 (3.15%) of 2,065 patients had taken antibiotics before hospital attendance, and 110 (5.67%) patients were on β-blockers (mean heart rate did not differ by β-blocker use; P = 0.86). The patient numbers and percentages analyzed by year were 796 (38.5% in 2005), 227 (11.0% in 2006), 515 (24.9% in 2007), 203 (9.8% in 2008), 287 (13.9% in 2009), 37 (1.8% in January and February of 2010), reflecting outbreaks in 2005 and 200719–21; 1,775 patients were tested for dengue IgM antibodies, and 1,589 (89.5%) patients were positive. Eight hundred patients were tested for dengue NS1 antigen, and one hundred forty-three (17.9%) patients were positive; 1,236 patients were tested for dengue RNA, and 662 (53.6%) patients were positive. Of 662 positive PCR results, the following serotypes were recorded: type 1 = 217 (32.8%), type 2 = 320 (48.3%), type 3 = 109 (16.5%), type 4 = 13 (1.9%), types 1 and 2 simultaneously present = 1 (0.2%), and types 1 and 3 simultaneously present = 2 (0.3%).
Table 1.
Patient characteristics
| Characteristics | Full cohort (N = 2,065) | Cohort A (N = 1,835)* | Cohort B (N = 1,157)† |
|---|---|---|---|
| Age (years) | 41.9 ± 17.2 | 41.8 ± 17.2 | 41.4 ± 17.0 |
| Male (%) | 1,205 (58.4) | 1,064 (58.0) | 696 (60.2) |
| Ethnicity (%) | |||
| Chinese | 1,471 (71.2) | 1,311 (71.4) | 843 (72.9) |
| Malay | 255 (12.3) | 229 (12.5) | 135 (11.7) |
| Indian | 165 (8.0) | 144 (7.9) | 84 (7.3) |
| Other | 174 (8.4) | 151 (8.2) | 95 (8.2) |
| Comorbid conditions (%) | |||
| Stroke | 7 (0.3) | 6 (0.3) | 5 (0.4) |
| Dementia | 8 (0.4) | 6 (0.3) | 4 (0.4) |
| Diabetes mellitus | 209 (10.1) | 191 (10.4) | 112 (9.7) |
| Hypertension | 327 (15.8) | 296 (16.1) | 178 (15.4) |
| Hyperlipidemia | 114 (5.5) | 97 (5.3) | 66 (5.7) |
| Coronary artery disease | 101 (4.9) | 94 (5.1) | 53 (4.6) |
| Chronic heart failure | 19 (0.9) | 18 (1.0) | 14 (1.2) |
| Asthma | 84 (4.1) | 74 (4.0) | 42 (3.6) |
| Chronic obstructive lung disease | 12 (0.6) | 9 (0.5) | 8 (0.7) |
| Bronchiectasis | 1 (0.1) | 1 (0.1) | 1 (0.1) |
| Chronic renal failure | 34 (1.7) | 34 (1.7) | 15 (1.3) |
| Cancer | 21 (1.0) | 18 (1.0) | 11 (1.0) |
| Arthritis | 11 (0.5) | 11 (0.6) | 11 (0.5) |
| Presence of bacterial coinfection (%) | 83 (4.0) | 78 (4.3) | 52 (4.5) |
Cohort A: Cohort of patients with complete data for the DDIS, including both the derivation and validation sets.
Cohort B: Cohort of patients with complete data for the DDIS used for the derivation set.
Of 2,065 patients, 83 (4.0%) patients had 110 instances of bacterial coinfection marked by positive blood culture (25/110; 22.7%), positive urine culture (43/110; 39.1%), or physician-diagnosed bacterial pneumonia (42/110; 38.2%). Among 68 positive blood and urine cultures, 47 (69.1%) cultures grew gram-negative bacteria, and 21 cultures grew gram-positive bacteria (Table 2). Cases with pneumonia were diagnosed when radiological features were present in an appropriate clinical context (fever, cough, phlegm, and dyspnea) and treated with antibiotics. No other infections apart from pneumonia were diagnosed on clinical/radiological grounds alone. Patients with bacterial coinfections had worse outcomes compared with patients without bacterial coinfections, and they were more likely to have diabetes mellitus, hypertension, hyperlipidemia, chronic renal failure, and cancer (Table 3).
Table 2.
Microbiological isolates
| Sources of infection | N | |
|---|---|---|
| Positive blood cultures | ||
| Methicillin-sensitive Staphylococcus aureus | Infective endocarditis | 2 |
| Infected arteriovenous fistula | 1 | |
| Infected vascular catheter | 1 | |
| Primary bacteremia | 1 | |
| Methicillin-resistant Staphylococcus aureus | Infective endocarditis | 1 |
| Primary bacteremia | 2 | |
| Streptococcus agalactiae | Primary bacteremia | 1 |
| Group A Streptococcus | Lower limb cellulitis | 1 |
| Aeromonas maltophilia | Cholangitis | 1 |
| Escherichia coli | Urinary tract | 3 |
| Cholecystitis | 3 | |
| Klebsiella pneumoniae | Cholangitis | 2 |
| Urinary tract | 1 | |
| Primary bacteremia | 1 | |
| Kluyvera cryocrescens | Primary bacteremia | 1 |
| Salmonella enteritidis | Primary bacteremia | 1 |
| Salmonella typhi | Primary bacteremia | 2 |
| Total | 25 | |
| Positive urinary cultures | ||
| Enterococcus faecalis | Urinary tract | 11 |
| Escherichia coli | Urinary tract | 20 |
| Klebsiella pneumoniae | Urinary tract | 7 |
| Pseudomonas aeruginosa | Urinary tract | 3 |
| Pantoea species | Urinary tract | 1 |
| Citrobacter koseri | Urinary tract | 1 |
| Total | 43 | |
Table 3.
Comorbid conditions and clinical outcomes for dengue patients
| Comorbid conditions and clinical outcomes | Patients with bacterial infections (N = 83) | Patients without bacterial infections (N = 1,982) | P value |
|---|---|---|---|
| Comorbid conditions (%) | |||
| Stroke | 0/83 (0.0) | 7/1,982 (0.4) | 1.000 |
| Dementia | 1/83 (1.2) | 7/1,982 (0.4) | 0.280 |
| Diabetes mellitus | 21/83 (25.3) | 188/1,982 (9.5) | < 0.001* |
| Hypertension | 30/83 (36.1) | 297/1,982 (15.0) | < 0.001* |
| Hyperlipidemia | 9/83 (10.8) | 105/1,982 (5.3) | 0.044* |
| Coronary artery disease | 6/83 (7.2) | 95/1,982 (4.8) | 0.295 |
| Chronic heart failure | 0/83 (0.0) | 19/1,982 (1.0) | 1.000 |
| Asthma | 1/83 (1.2) | 83/1,982 (4.2) | 0.265 |
| Chronic obstructive lung disease | 2/83 (2.4) | 10/1,982 (0.5) | 0.081 |
| Bronchiectasis | 0/83 (0.0) | 1/1,982 (0.1) | 1.000 |
| Chronic renal failure | 4/83 (4.8) | 30/1,982 (1.5) | 0.045* |
| Cancer | 3/83 (3.6) | 18/1,982 (0.9) | 0.049* |
| Arthritis | 1/83 (1.2) | 10/1,982 (0.5) | 0.364 |
| Admitted to ICU (%) | 6/83 (7.2) | 8/1,982 (0.4) | < 0.001* |
| Hospital mortality (%) | 16/83 (19.3) | 32/1,982 (1.6) | < 0.001* |
| Hospital LOS (days) | 8.7 ± 7.5 | 3.9 ± 3.8 | < 0.001* |
| Required platelet transfusion (%) | 3/83 (3.6) | 55/1,982 (2.8) | 0.650 |
| Required blood transfusion (%) | 3/83 (3.6) | 29/1,982 (1.4) | 0.120 |
ICU = intensive care unit; LOS = length of stay.
P < 0.05.
Derivation and validation of diagnostic model.
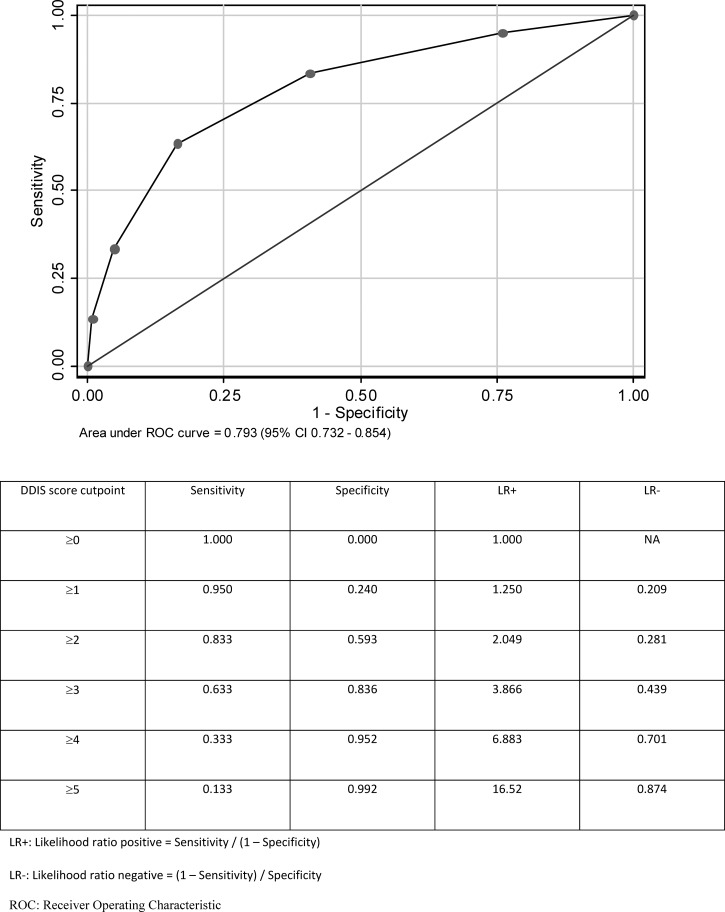
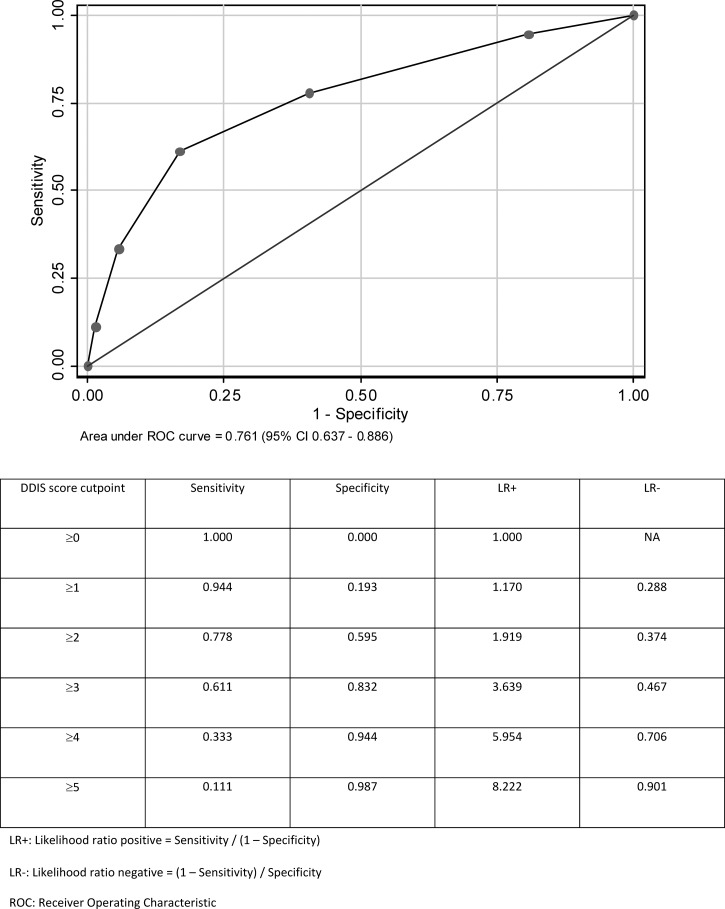
Using the derivation cohort and a univariate screen, logistic regression analysis was performed on complete data for 1,157 patients (52 patients with concurrent bacterial infection) (Table 4). Five variables (pulse rate, total leukocyte count, hematocrit, sodium, and urea) were dichotomized and used to construct the DDIS (Table 5). Logistic regression model calibration was satisfactory (Hosmer–Lemeshow P > 0.05). Using the derivation set, the AUC was 0.793 (95% confidence interval [95% CI] = 0.732–0.854) (Figure 1). Using the validation set, the AUC was slightly reduced to 0.761 (95% CI = 0.637–0.886) (Figure 2). Using bootstrap resampling on all patients with complete data (N = 1,835), the resulting mean AUC of 0.779 (95% CI = 0.696–0.860) was only marginally lower than the AUC for model derivation. Overall, among 1,835 patients in the entire cohort with complete data for the DDIS, 57.60% scored 0 or 1, 36.13% scored 2 or 3, and 6.27% scored 4 or 5.
Table 4.
Univariate screen and multivariate analysis of risk factors for bacterial infection in dengue patients within the derivation cohort
| Risk factor and group | N | Univariate result | Univariate P value | Multivariate OR* | Multivariate P value |
|---|---|---|---|---|---|
| Age (years) | |||||
| 1 | 63 | 53.3 ± 21.3 | < 0.001 | 1.010 (0.991–1.030) | 0.292 |
| 2 | 1,415 | 41.3 ± 16.8 | |||
| Male (%; reference level: male) | |||||
| 1 | 63 | 30 (47.6) | 0.054 | – | – |
| 2 | 1,415 | 846 (59.9) | |||
| Temperature at ED (°C) | |||||
| 1 | 59 | 37.4 ± 1.2 | 0.810 | – | – |
| 2 | 1,264 | 37.4 ± 1.0 | |||
| Systolic blood pressure at ED (mmHg) | |||||
| 1 | 60 | 125 ± 26 | 0.048 | – | – |
| 2 | 1,330 | 120 ± 19 | |||
| Pulse rate at ED (beats/minute) | |||||
| 1 | 60 | 98 ± 21 | < 0.001 | 1.026 (1.008–1.044) | 0.004† |
| 2 | 1,315 | 86 ± 16 | |||
| Respiratory rate at ED (breaths/minute) | |||||
| 1 | 52 | 20 ± 4 | < 0.001 | 1.069 (0.974–1.173) | 0.162 |
| 2 | 1,178 | 18 ± 2 | |||
| Total white cell count within 24 hours of admission (109/L) | |||||
| 1 | 63 | 8.0 ± 6.2 | < 0.001 | 1.062 (0.995–1.134) | 0.070† |
| 2 | 1,409 | 4.1 ± 3.2 | |||
| Neutrophil percentage within 24 hours of admission (%) | |||||
| 1 | 63 | 69 ± 21 | <0.001 | 1.014 (0.995–1.033) | 0.150 |
| 2 | 1,410 | 55 ± 19 | |||
| Hematocrit within 24 hours of admission (%) | |||||
| 1 | 63 | 37.1 ± 7.0 | <0.001 | 0.923 (0.880–0.970) | 0.001† |
| 2 | 1,408 | 41.8 ± 5.6 | |||
| Platelets within 24 hours of admission (109/L) | |||||
| 1 | 63 | 112 ± 69 | <0.001 | 0.998 (0.993–1.003) | 0.410 |
| 2 | 1,408 | 82 ± 60 | |||
| Sodium within 24 hours of admission (mmol/L) | |||||
| 1 | 63 | 134 ± 7 | < 0.001 | 0.946 (0.894–1.001) | 0.056† |
| 2 | 1,330 | 136 ± 4 | |||
| Potassium within 24 hours of admission (mmol/L) | |||||
| 1 | 62 | 3.8 ± 0.8 | 0.608 | – | – |
| 2 | 1,323 | 3.8 ± 0.6 | |||
| Urea within 24 hours of admission (mmol/L) | |||||
| 1 | 63 | 9.7 ± 13.4 | < 0.001 | 1.038 (0.998–1.080) | 0.063† |
| 2 | 1,329 | 4.7 ± 4.3 | |||
| Creatinine within 24 hours of admission (μmol/L) | |||||
| 1 | 62 | 156 ± 245 | < 0.001 | – | – |
| 2 | 1,322 | 85 ± 72 | |||
Hosmer–Lemeshow χ2 = 6.92, P = 0.5455. ED = emergency department; group 1 = patients with bacterial infections; group 2 = patients without bacterial infections; OR = odds ratio (95% CI).
Logistic regression analysis based on complete data for N = 1,157 patients (N = 52 patients with concurrent bacterial infection).
Risk factors chosen for DDIS.
Table 5.
Logistic regression of dichotomized risk factors for bacterial infection in dengue patients within the derivation cohort
| Risk factor | OR* | Standardized coefficients | P value |
|---|---|---|---|
| Pulse rate at ED ≥ 90 (beats/minute) | 2.19 (1.24–3.85) | 0.78 (0.22–1.35) | 0.007 |
| Total white cell count within 24 hours of admission ≥ 6 (109/L) | 2.13 (1.15–3.94) | 0.76 (0.14–1.37) | 0.016 |
| Hematocrit within 24 hours of admission < 40 (%) | 3.52 (2.00–6.18) | 1.26 (0.69–1.82) | < 0.001 |
| Sodium within 24 hours of admission < 135 (mmol/L) | 2.44 (1.39–4.28) | 0.89 (0.33–1.45) | 0.002 |
| Urea within 24 hours of admission ≥ 5 (mmol/L) | 2.65 (1.44–4.87) | 0.98 (0.37–1.58) | 0.002 |
Hosmer–Lemeshow χ2 = 5.40, P = 0.6113. ED = emergency department; OR = odds ratio (95% CI).
Logistic regression analysis based on complete data for N = 1,299 patients (N = 60 patients with concurrent bacterial infection).
Figure 1.
Using DDIS for the identification of bacterial infection (N = 1,299 patients in the derivation set with complete data and 60 patients with bacterial infection).
Figure 2.
Using DDIS for the identification of bacterial infection (N = 536 patients in the validation set with complete data and 18 patients with bacterial infection).
Discussion
Our results showed that a small but clinically significant proportion of dengue-infected patients who presented to the hospital also had bacterial coinfection. We developed and validated a DDIS, which had an AUC of 0.793 in the derivation cohort and 0.761 in the validation cohort. Only about 5% of patients with DDIS < 1 had bacterial coinfection, whereas nearly 95% of patients with DDIS ≥ 4 had bacterial coinfections. It is, thus, possible to identify patients who are unlikely to need empirical antibiotics and select patients who will benefit most from early therapy.
Dengue dual infections, which are shown by our patient cohort and others, are associated with high mortality and morbidity compared with dengue without concurrent bacterial infections.22 In a prior series of 100 patients with dengue hemorrhagic fever or dengue shock syndrome, 7 (7%) patients had concurrent bacteremia.22 In contrast, among all our patients with laboratory-confirmed dengue, 25 (1.2%) patients had concurrent bacteremia, and 83 (4.0%) patients had bacterial coinfection, which agrees more closely with the 3.6% bacterial infection rate among 606 adult dengue patients in another study.14 Like others have shown, the majority of bacteremia isolates were gram-negative.22 Primary gram-negative bacteremia was postulated to be caused by the breakdown of the intestinal mucosal barrier in severe dengue infections.22 We additionally observed a broad range of other infections, such as cellulitis and cholecystitis (with potential secondary infection), that have been highlighted in case reports.23,24 However, we cannot exclude the possibility that bacterial infection may also increase susceptibility to dengue infection.
We identified several clinical risk factors for dengue dual infection. These variables were clinically relevant and easily and reliably measured at the point of presentation (or shortly thereafter). We showed that the pulse rate was significantly higher in dengue with bacterial coinfections, congruent with the finding of relative bradycardia in isolated dengue fever.25,26 We also showed that total leukocyte count was significantly higher in dengue with bacterial coinfections. Similarly, other studies have shown higher leukocyte levels in febrile illnesses that were not caused by dengue infection, although levels were similar between dengue fever and dengue hemorrhagic fever.10,27 Other risk factors, like hematocrit, sodium, and urea, have not been well-studied.
Although several comorbidities proved to be significantly more prevalent in patients with bacterial coinfection, we did not include these comorbidities in our diagnostic model for two reasons. First, we had decided a priori to only use objective and reliable risk factors in the diagnostic model. The presence of comorbidities may not always be accurate or available from the clinical history, and hence, they were omitted. Second, age was correlated with both increased comorbidity and bacterial coinfection. The point biserial correlation coefficients were 0.28, 0.41, 0.29, 0.07, and 0.13 (P values ≤ 0.002) when analyses of age by diabetes mellitus, hypertension, hyperlipidemia, chronic renal failure, and cancer were done. Therefore, each of the comorbidities (when added to the logistic derivation model) became non-significant, and they did not affect the significance levels or coefficient sizes of the other risk factors.
Interestingly, on the univariate screen, we found no difference in initial temperature for patients with and without bacterial coinfection, showing that fever was not a reliable sign for bacterial infection.28 Thrombocytopenia at presentation was also less pronounced in patients with bacterial coinfection. Because platelet counts in dengue fall progressively until a nadir at days 5 and 6 of illness,29,30 this finding can be explained by patients with more severe illness presenting earlier. In our study, patients with bacterial coinfection presented after a median of 2 days of self-reported fever, whereas patients without bacterial coinfection presented after a median of 4 days of self-reported fever (P value < 0.001).
Strengths of the study include large sample size and use of multivariable analysis for relatively uncommon events, such as dengue dual infection. Prior studies were relatively smaller,22,24 which would preclude such an investigation. All diagnoses of dengue infection were laboratory-confirmed. The small reduction in the AUC between the derivation and validation cohorts suggests that the DDIS score is generalizable. Furthermore, risk-scoring using routinely available clinical and laboratory parameters allows for broad application in most healthcare settings.
There are several limitations. Our study was done in a single center, but we had a varied multiracial patient population that would improve the generalizability of our results. Our study was retrospective, but such data have been used to develop other important risk scores, like the CURB-65 index for pneumonia.31 We studied adult patients, and the results may not be applicable to children. Not all patients had bacterial cultures done, but we used a combination of clinical and microbiological criteria to detect bacterial infection, which can exist in the absence of positive cultures.32 It is unlikely that many bacterial infections were missed, because patients not diagnosed with bacterial coinfections had good clinical outcomes. For this reason, it is also unlikely that many delayed bacterial infections developed in the hospital from dengue-induced neutropenia or nosocomial infections. The ideal study would be to prospectively perform serial multiple site cultures for all suspected dengue patients, but such research would be exceedingly difficult. We did not use convalescent sera to detect additional dengue cases, but such testing is rarely done. We also did not restrict our analyses to only NS1 antigen- and PCR-confirmed cases, because our target patient population included all dengue patients who presented to hospital, regardless of day of dengue illness. Finally, our study design did not allow us to determine if fatalities in coinfected patients could have been avoided through earlier recognition, because we did not have information on other aspects of patient care, such as compliance to sepsis bundles. However, this limitation did not affect the primary aim of the study, which was to determine early indicators of bacterial coinfection.
Although the overall proportion of patients with coinfection may be low, the absolute numbers can be high during periods of epidemics or in areas of high endemicity. The DDIS can be readily built into a protocol to alert the attending physician if a patient suspected of dengue might have a high risk of bacterial coinfection, which increases the chances of early antibiotic therapy. Nonetheless, prospective validation in other cohorts will be needed before we can be more confident of its wider application. With the advent of newer inflammatory markers, their addition to the risk score can be done to see if a beneficial net reclassification of cases exists.
In conclusion, we developed and validated a risk score for the identification of bacterial coinfection in dengue patients. The DDIS is simple and easy to use in most clinical settings, including resource-limited settings.
ACKNOWLEDGMENTS
The authors are grateful to the nursing students and lecturers from Ngee Ann Polytechnic, Singapore, for helping with the data collection.
Footnotes
Authors' addresses: Kay C. See, Jason Phua, Hwee S. Yip, Leong L. Yeo, and Tow K. Lim, University Medicine Cluster, National University Hospital, Singapore and Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, E-mails: Kay_Choong_SEE@nuhs.edu.sg, Jason_PHUA@nuhs.edu.sg, Hwee_Seng_YIP@nuhs.edu.sg, Leonard_LL_YEO@nuhs.edu.sg, and Tow_Keang_LIM@nuhs.edu.sg.
References
- 1.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilder-Smith A, Schwartz E. Dengue in travelers. N Engl J Med. 2005;353:924–932. doi: 10.1056/NEJMra041927. [DOI] [PubMed] [Google Scholar]
- 3.Halstead SB. Dengue. Lancet. 2007;370:1644–1652. doi: 10.1016/S0140-6736(07)61687-0. [DOI] [PubMed] [Google Scholar]
- 4.Wilder-Smith A, Renhorn KE, Tissera H, Abu Bakar S, Alphey L, Kittayapong P, Lindsay S, Logan J, Hatz C, Reiter P, Rocklov J, Byass P, Louis VR, Tozan Y, Massad E, Tenorio A, Lagneau C, L'Ambert G, Brooks D, Wegerdt J, Gubler D. DengueTools: innovative tools and strategies for the surveillance and control of dengue. Glob Health Action. 2012;2012:5. doi: 10.3402/gha.v5i0.17273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rigau-Perez JG, Clark GG, Gubler DJ, Reiter P, Sanders EJ, Vorndam AV. Dengue and dengue haemorrhagic fever. Lancet. 1998;352:971–977. doi: 10.1016/s0140-6736(97)12483-7. [DOI] [PubMed] [Google Scholar]
- 6.Rice TW, Rubinson L, Uyeki TM, Vaughn FL, John BB, Miller RR, 3rd, Higgs E, Randolph AG, Smoot BE, Thompson BT. Critical illness from 2009 pandemic influenza A virus and bacterial coinfection in the United States. Crit Care Med. 2012;40:1487–1498. doi: 10.1097/CCM.0b013e3182416f23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palacios G, Hornig M, Cisterna D, Savji N, Bussetti AV, Kapoor V, Hui J, Tokarz R, Briese T, Baumeister E, Lipkin WI. Streptococcus pneumoniae coinfection is correlated with the severity of H1N1 pandemic influenza. PLoS One. 2009;4:e8540. doi: 10.1371/journal.pone.0008540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purcell K, Fergie J. Concurrent serious bacterial infections in 2396 infants and children hospitalized with respiratory syncytial virus lower respiratory tract infections. Arch Pediatr Adolesc Med. 2002;156:322–324. doi: 10.1001/archpedi.156.4.322. [DOI] [PubMed] [Google Scholar]
- 9.Ratnam I, Black J, Leder K, Biggs BA, Matchett E, Padiglione A, Woolley I, Panagiotidis T, Gherardin T, Pollissard L, Demont C, Luxemburger C, Torresi J. Incidence and seroprevalence of dengue virus infections in Australian travelers to Asia. Eur J Clin Microbiol Infect Dis. 2012;31:1203–1210. doi: 10.1007/s10096-011-1429-1. [DOI] [PubMed] [Google Scholar]
- 10.Gregory CJ, Santiago LM, Arguello DF, Hunsperger E, Tomashek KM. Clinical and laboratory features that differentiate dengue from other febrile illnesses in an endemic area–Puerto Rico, 2007–2008. Am J Trop Med Hyg. 2010;82:922–929. doi: 10.4269/ajtmh.2010.09-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang CC, Wu CC, Liu JW, Lin AS, Liu SF, Chung YH, Su MC, Lee IK, Lin MC. Chest radiographic presentation in patients with dengue hemorrhagic Fever. Am J Trop Med Hyg. 2007;77:291–296. [PubMed] [Google Scholar]
- 12.Lee IK, Liu JW, Yang KD. Clinical and laboratory characteristics and risk factors for fatality in elderly patients with dengue hemorrhagic fever. Am J Trop Med Hyg. 2008;79:149–153. [PubMed] [Google Scholar]
- 13.Ong A, Sandar M, Chen MI, Sin LY. Fatal dengue hemorrhagic fever in adults during a dengue epidemic in Singapore. Int J Infect Dis. 2007;11:263–267. doi: 10.1016/j.ijid.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 14.Wang CC, Lee IK, Su MC, Lin HI, Huang YC, Liu SF, Wu CC, Lin MC. Differences in clinical and laboratory characteristics and disease severity between children and adults with dengue virus infection in Taiwan, 2002. Trans R Soc Trop Med Hyg. 2009;103:871–877. doi: 10.1016/j.trstmh.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 15.Anders KL, Nguyet NM, Chau NV, Hung NT, Thuy TT, Lien le B, Farrar J, Wills B, Hien TT, Simmons CP. Epidemiological factors associated with dengue shock syndrome and mortality in hospitalized dengue patients in Ho Chi Minh City, Vietnam. Am J Trop Med Hyg. 2011;84:127–134. doi: 10.4269/ajtmh.2011.10-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz E, Mileguir F, Grossman Z, Mendelson E. Evaluation of ELISA-based sero-diagnosis of dengue fever in travelers. J Clin Virol. 2000;19:169–173. doi: 10.1016/s1386-6532(00)00114-1. [DOI] [PubMed] [Google Scholar]
- 17.Pok KY, Lai YL, Sng J, Ng LC. Evaluation of nonstructural 1 antigen assays for the diagnosis and surveillance of dengue in Singapore. Vector Borne Zoonotic Dis. 2010;10:1009–1016. doi: 10.1089/vbz.2008.0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30:545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher D. To the vector borne……mosquito-transmitted diseases in Singapore. Singapore Med J. 2005;46:596–597. [PubMed] [Google Scholar]
- 20.Ooi EE, Wilder-Smith A, Ng LC, Gubler DJ. The 2007 dengue outbreak in Singapore. Epidemiol Infect. 2010;138:958–959. doi: 10.1017/S0950268810000026. [DOI] [PubMed] [Google Scholar]
- 21.Hii YL, Rocklov J, Ng N, Tang CS, Pang FY, Sauerborn R. Climate variability and increase in intensity and magnitude of dengue incidence in Singapore. Glob Health Action. 2009;2009:2. doi: 10.3402/gha.v2i0.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee IK, Liu JW, Yang KD. Clinical characteristics and risk factors for concurrent bacteremia in adults with dengue hemorrhagic fever. Am J Trop Med Hyg. 2005;72:221–226. [PubMed] [Google Scholar]
- 23.Wu KL, Changchien CS, Kuo CM, Chuah SK, Lu SN, Eng HL, Kuo CH. Dengue fever with acute acalculous cholecystitis. Am J Trop Med Hyg. 2003;68:657–660. [PubMed] [Google Scholar]
- 24.Chai LY, Lim PL, Lee CC, Hsu LY, Teoh YL, Lye DC, Krishnan P, Leo YS. Cluster of Staphylococcus aureus and dengue co-infection in Singapore. Ann Acad Med Singapore. 2007;36:847–850. [PubMed] [Google Scholar]
- 25.Lateef A, Fisher DA, Tambyah PA. Dengue and relative bradycardia. Emerg Infect Dis. 2007;13:650–651. doi: 10.3201/eid1304.061212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wittesjo B, Bjornham A, Eitrem R. Relative bradycardia in infectious diseases. J Infect. 1999;39:246–247. doi: 10.1016/s0163-4453(99)90063-4. [DOI] [PubMed] [Google Scholar]
- 27.Kalayanarooj S, Vaughn DW, Nimmannitya S, Green S, Suntayakorn S, Kunentrasai N, Viramitrachai W, Ratanachu-eke S, Kiatpolpoj S, Innis BL, Rothman AL, Nisalak A, Ennis FA. Early clinical and laboratory indicators of acute dengue illness. J Infect Dis. 1997;176:313–321. doi: 10.1086/514047. [DOI] [PubMed] [Google Scholar]
- 28.Seigel TA, Cocchi MN, Salciccioli J, Shapiro NI, Howell M, Tang A, Donnino MW. Inadequacy of temperature and white blood cell count in predicting bacteremia in patients with suspected infection. J Emerg Med. 2012;42:254–259. doi: 10.1016/j.jemermed.2010.05.038. [DOI] [PubMed] [Google Scholar]
- 29.Tai DY, Chee YC, Chan KW. The natural history of dengue illness based on a study of hospitalised patients in Singapore. Singapore Med J. 1999;40:238–242. [PubMed] [Google Scholar]
- 30.Wichmann O, Hongsiriwon S, Bowonwatanuwong C, Chotivanich K, Sukthana Y, Pukrittayakamee S. Risk factors and clinical features associated with severe dengue infection in adults and children during the 2001 epidemic in Chonburi, Thailand. Trop Med Int Health. 2004;9:1022–1029. doi: 10.1111/j.1365-3156.2004.01295.x. [DOI] [PubMed] [Google Scholar]
- 31.Lim WS, van der Eerden MM, Laing R, Boersma WG, Karalus N, Town GI, Lewis SA, Macfarlane JT. Defining community acquired pneumonia severity on presentation to hospital: an international derivation and validation study. Thorax. 2003;58:377–382. doi: 10.1136/thorax.58.5.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Limmathurotsakul D, Jamsen K, Arayawichanont A, Simpson JA, White LJ, Lee SJ, Wuthiekanun V, Chantratita N, Cheng A, Day NP, Verzilli C, Peacock SJ. Defining the true sensitivity of culture for the diagnosis of melioidosis using Bayesian latent class models. PLoS One. 2010;5:e12485. doi: 10.1371/journal.pone.0012485. [DOI] [PMC free article] [PubMed] [Google Scholar]