Abstract
A new noninvasive screening tool for colorectal neoplasia detects epigenetic alterations exhibited by gastrointestinal tumor cells shed into stool. There is insufficient existing data to determine temporal associations between colorectal cancer (CRC) progression and aberrant DNA methylation. To evaluate the feasibility of using fecal DNA methylation status to determine CRC progression, we collected stool samples from 14 male SD rats aged six weeks, and administered subcutaneous injections of either 1,2-dimethylhydrazine or saline weekly. p16 DNA methylation statuses in tumorous and normal colon tissue, and from stool samples were determined using methylation-specific PCR. Additionally, p16 methylation was detected in stool DNA from 85.7% of the CRC rats. The earliest change in p16 methylation status in the DMH-treated group stool samples occurred during week nine; repeatabilities were 57.1% in week 19 (p = 0.070) and 85.7% in week 34 (p = 0.005). A temporal correlation was evidenced between progression of CRC and p16 methylation status, as evidenced by DMH-induced rat feces. Using fecal DNA methylation status to determine colorectal tissue methylation status can reveal CRC progression. Our data suggests that p16 promoter methylation is a feasible epigenetic marker for the detection and may be useful for CRC screening.
Keywords: colorectal cancer, DNA methylation, stool test for colorectal cancer
Introduction
Colorectal cancer (CRC) accounts for more than 600,000 deaths each year worldwide (Jemal et al., 2011). In Taiwan in recent years, CRC has become the most common form of cancer occurrence (Promotion, 2010). Effective treatment is possible during the early stages of CRC, but the disease is generally asymptomatic. Unfortunately, the therapy is rarely successful in stage IV after the tumor cells have spread to lymph nodes and other organs (Wong et al., 2004). Thus, an effective screening test and early detection would provide substantial clinical benefits. The present technology CRC screening tools for an average-risk population fall into two categories: (1) physical examination, including colonoscopy, virtual colonoscopy, sigmoidoscopy, double contrast barium enema (DCBE), and digital rectal exam (DRE) (Bretthauer, 2011), and (2) stool tests, including detection of occult blood and exfoliated mutant or epigenetic changes of DNA (Harrison and Benziger, 2011). Colonoscopy and sigmoidoscopy are highly specific and sensitive screening tools; however, these are less acceptable to the public because of invasive medical procedures. Fecal occult blood testing (FOBT), the most efficient noninvasive screening test for colorectal cancer, has a limited impact on survival rate because of its low sensitivity to early stage CRC (Baek et al., 2009).
To provide early detection, it is important to develop a noninvasive and screening tool that is sensitive to CRC characteristics (Krishnan and Wolf, 2011; Labianca and Merelli, 2010). At the molecular level, CRC progression is accompanied by specific genetic and epigenetic changes (Schmid, 2010). Aberrant DNA hypermethylation of CpG islands within promoters is associated with the development and progression of colorectal cancer, which in turn leads to silencing of tumor suppressor genes such as p16INK4a, MGMT and MLH1, SFRP2, and vimentin genes (Chen et al., 2005; Wang and Tang, 2008; Psofaki et al., 2010; Shima et al., 2010).
A novel and non-invasive screening tool for detection of colorectal neoplasia is to assay the methylation alterations present in gastrointestinal tumor cells shed into stool. (Lenhard et al., 2005; Azuara et al., 2010). However, existing data was insufficient to determine temporal correlation between the progression of CRC and aberrant DNA methylation. To address this knowledge gap, we assayed p16 gene methylation changes in the feces of a rat model with chemically induced colorectal cancer. The assay could provide evidence that feces contain a detectable biomarker to diagnose early stage colorectal cancer. The aim of this study was to evaluate the feasibility of using fecal DNA methylation status to predict CRC progression.
Materials and Methods
Animals
All 14 male Sprague-Dawley rats used in the experiment were obtained from BioLASCO Taiwan Co. Ltd. (Taipei, Republic of China), six weeks of age and weighing 250–300 g. They were individually housed in ventilated stainless-steel cages with soft shavings air-conditioned room (temperature 22 ± 1 °C, humidity 50–60% with 12-hour light-dark cycles, lights on at 0700), and were provided free access to food and water throughout the experiment. All procedures were approved by the Institutional Animal Care and Use Committee of National Defense Medical Center (certificate number IACUC-09-047), and were performed in accordance with National Institute of Health guidelines for the treatment of animals.
Generation of DMH induced tumors
Beginning at seven weeks of age, the rats received subcutaneous (s.c.) injections of either 1,2-dimethylhydrazine (DMH) (Sigma Chemical Co) at a dosage of 20 mg/kg body weight (n = 7), or the same volume of saline (n = 7) weekly for 30 weeks. Rats were sacrificed at age 34 weeks through inhaled overdose of CO2.
Collection of samples
Fecal pellets were collected weekly and placed in 2 mL microcentrifuge tubes for DNA extraction. Tissue samples were harvested after the rats were sacrificed. Samples were resected with scissors and inspected for tumors. Tumor, normal tissue close to a tumor, and normal tissue samples were dissected with a razor blade and placed in 2 mL microcentrifuge tubers for DNA extraction. Each tissue sample was sectioned for standard histological preparation and hematoxylin-eosin staining. Tissue and fecal samples were stored at −80 °C.
DNA extraction and sodium bisulfite treatment
Genomic DNA was extracted either from frozen tissue samples using a DNeasy Blood & Tissue kit (QIA-GENE, Germany), or from fecal samples using a QIAamp DNA stool Mini kit (QIAGENE, Germany). The resulting DNA was modified with sodium bisulfite using an EZ DNA Methylation kit (Zymo Research, Orange, County, CA), and a methylated DNA positive control for MSP assays was generated using SssI methylase (Zymo Research, Orange County, CA).
Tissue and fecal DNA methylation studies
The methylation status of tissue samples was determined by methylation-specific PCR (MSP) of bisulfite treated DNA. MSP was carried out in a volume of 15 μL with 7.5 μL of HotStart Taq Premlx (RBC Bioscience), 0.6 μL of each primer, and 0.6 μL of bisulphite treated DNA. PCR conditions included denaturation at 95 °C for 10 min followed by 35 cycles at 95 °C for 30 s, annealing temperature at 55 °C for 35 s, 72 °C for 30 s, and a extension at 72 °C for 4 min.
As only limited quantities of DNA are obtained from fecal samples (Zou et al., 2009) we adopted a nested strategy (Glockner et al., 2009). During the first stage, we carried out amplification by bisulfite-sequencing PCR (BSP) for 35 cycles using primers located in the flanking region of the CDKN2A promoter. In the following stage, MSP amplification was carried out for 30 cycles using 1:100 dilutions of the first-stage products. Table 1 lists the MSP and BSP primer sets, and annealing temperatures.
Table 1.
MSP and BSP primer sets and annealing temperature.
| Forward primer (5′ → 3′) | Annealing temperature (°C) | Product size (bp) | |
|---|---|---|---|
| F* | GGGGAGGGTTTTTTAGATATTTT | 55 | 225 |
| R | TTAAACCACCCATACTCACCTAAAC | ||
| U-F** | GTGAATTTGAGGAGAGTGATTTG | 57 | 129 |
| U-R | CAAAACATTTAATAAAACCCCAA | ||
| M-F | AATTCGAGGAGAGCGATTCG | 62 | 123 |
| M-R | AACGTTTAATAAAACCCCGA | ||
BSP primer: F, forward; R, reverse.
MSP primer: U, unmethylated; M, methylated.
Statistical analyses
All statistical analyses were performed using the SPSS 21.0 software package for Windows (SPSS Taiwan Corp.). We computed the sensitivity and specificity of the fecal DNA methylation assay by using the McNemar test to compare methylation statuses between fecal and tissue samples for rats treated with DMH (n = 7) and those injected with saline (n = 7); the Fisher’s exact test was used to evaluate the differences of p16 hypermethylation between the two groups in stool samples. A p value ≤ 0.05 was considered statistically significant.
Results
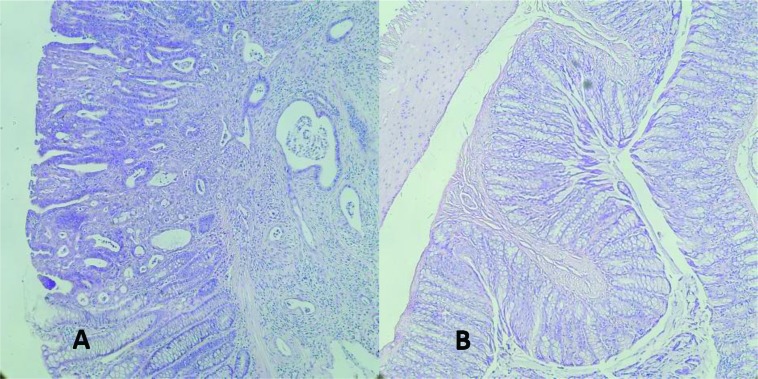
There were no significant differences in body weight between experimental and control rats at euthanasia, and both groups exhibited a healthy appearance. All of the DMH-treated, but none of the control group rates developed colorectal adenomatous cells (Figure 1).
Figure 1.
Histological analysis of (A) DMH-induced rat colonic adenoma and (B) normal tissue. Magnification: x 20; Hematoxylin-eosin staining.
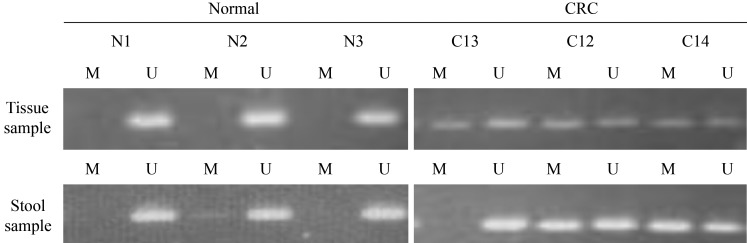
We analyzed the methylation status of the p16 gene in DNA from tissue samples. A methylated p16 gene was detected in all intestinal adenoma tissue samples from the DMH-treated group. By contrast, there was no hypermethylation of the p16 gene in the control group tissue samples (Figure 2). We then assessed the p16 gene methylation status of tissue and stool samples from corresponding donor rats to ascertain the consistency of p16 hypermethylation between the two sample types. After euthanasia in week 34, p16 hypermethylation was found in the stool of 85.7% (6/7) of the DMH-treated rats but in none (0/7) of the control group. The McNemar test p-value was 1.000 (Table 2).
Figure 2.
Methylation status of the p16 gene in fecal specimens and in CRC tissue taken from the same donor rat. N: normal tissue; C: CRC tissue; M: Methylated; U: Unmethylated.
Table 2.
Consistency of p16 hypermethylation between tissue and stool samples.
| Tissue samples
|
p-value* | ||||
|---|---|---|---|---|---|
| Methylated | Unmethylated | ||||
| Stool samples | Week 9 | Methylated | 5 | 0 | 0.500 |
| Unmethylated | 2 | 7 | |||
|
| |||||
| Week 19 | Methylated | 4 | 0 | 0.250 | |
| Unmethylated | 3 | 7 | |||
|
| |||||
| Week 34 | Methylated | 6 | 0 | 1.000 | |
| Unmethylated | 1 | 7 | |||
McNemar test.
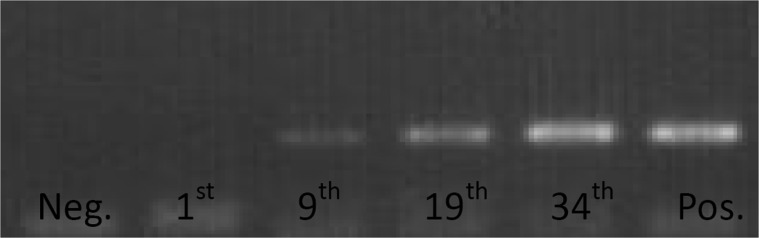
When retrospectively assaying stool samples to assess the temporal association between CRC progression and aberrant DNA methylation we detected the earliest instance of p16 hypermethylation in week nine. Samples from the DMH-treated group showed a p16 hypermethylation ratio of 71.4% (5/7) against 0% (0/7) from the control group (McNemar test p = 0.500, Table 2); the repeatability of using fecal DNA methylation status for identifying CRC in our rat samples was 57.1% (4/7) during week 19 and 85.7% (6/7) in week 34 (McNemar test p = 0.250 and 1.000, respectively) (Table 2).
From the two groups of this study (DMH-treat and saline control), the methylation status in stool samples of periods of time was significantly different in week nine and 34 (Fisher’s exact test p-value were 0.021 and 0.005, respectively) and it was borderline significant in week 19 (p = 0.070) (Table 3). The repeatability of using fecal DNA methylation status was used as an indicator for CRC progression (Figure 3). Samples were taken at the indicated rat age.
Table 3.
Differences in p16 hypermethylation between DMH-treated and saline-control groups in stool samples.
| Week | DMH-treated | Saline-control | p-value* | |
|---|---|---|---|---|
| 9 | Methylated | 5(71.4) | 0(0) | 0.021 |
| Unmethylated | 2(28.6) | 7(100) | ||
|
| ||||
| 19 | Methylated | 4(57.1) | 0(0) | 0.070 |
| Unmethylated | 3(42.9) | 7(100) | ||
|
| ||||
| 34 | Methylated | 6(85.7) | 0(0) | 0.005 |
| Unmethylated | 1(14.3) | 7(100) | ||
Fisher’s exact test.
The sensitivities in weeks 9, 19 and 34 were 71.4% (5/7), 57.1% (4/7) and 85.7% (6/7), respectively, in the DMH-treated group stool samples; the specificity of each week was 100% (7/7) in the normal saline group.
Figure 3.
The repeatability of using fecal DNA methylation status to identify CRC progression. Samples were taken at the indicated rat age.
Discussion
The development and progression of colorectal cancer follows the “adenoma-carcinoma” sequence, in which colorectal tumor cells develop via a worsening dysplasia of normal colonic mucosa (Fearon and Vogelstein, 1990). An uncomplicated surgical procedure can provide an effective treatment if diagnosis is made during the early stages before metastasis occurs (Kim et al., 2010). Thus, it is desirable to find biomarkers that have high sensitivity and specificity towards CRC.
There is abundant evidence supporting the efficacy of FOBT in decreasing colorectal cancer mortality. However, the impact of FOBT on CRC incidence is lessened by the method’s low sensitivity (Heitman et al., 2010; Levi et al., 2007). Mandel et al. (2000) reported that FOBT decreases CRC incidence by 17–20%. Conversely, Heresbach et al. (2006) provided evidence that FOBT has an insignificant effect on CRC incidence. The incidence of CRC and its mortality rates are expected to increase due to our aging population, particularly if there are no new screening tools developed to replace FOBT.(Mariotto et al., 2006)
In a normal intestinal tract, the epithelium is constantly and rapidly renewed by the turnover of 5 × 1010 epithelial cells per day (Mehl, 1991). The shedding rate of carcinoma colonocytes is faster than that of normal cells, and we can use this characteristic to identify tumor cells and assay the status of genetic or epigenetic abnormalities. Promoter hypermethylation analysis of stool DNA is a promising noninvasive test for early diagnosis of CRC, and this area has received much research interest. An increasing number of genes is found to undergo promoter region hypermethylation in the tissue and stool of CRC patients, and Glockner et al. (2009) found a higher sensitivity to TFPI2 methylation in patients with CRC (73–89%), in patients with adenomas (21–43%), and a high specificity toward TFPI2 methylation by patients with either CRC or adenomas (93–100%). Other reports observed high sensitivity (> 68%) and specificity (> 84%) towards SFRP2 and GATA4 methylation (Oberwalder et al., 2008; Wang and Tang, 2008; Hellebrekers et al., 2009)
There is substantial evidence supporting the value of using methylation analysis of stool DNA as a screening tool, although we cannot exclude the possibility of bias due to the nature of these cross-sectional studies (Thomas et al., 2005); the literature contains no reports of a sequential association between CRC and changes in gene methylation status in stool. Our findings answer questions about the consistency and temporal association between the progression of CRC and aberrant DNA methylation.
There are currently two animal models commonly employed to investigate the rules of DNA methylation in progression of CRC: (1) using genetic manipulation of animals to overexpress or down-regulate specific genes that are direct regulators of DNA methylation and methylation-related gene expression; and (2) carcinogen-treated animal models with assessment of somatic epigenetic alterations that arise in the resulting tumors (Conerly and Grady, 2010). Our study design adopted the latter category; we used DMH to change the genome methylation status and induce CRC. DMH-induced colon apoptosis in the rat model exhibits a characteristic aberrant crypt foci-adenoma-carcinoma sequence and associated increase in cellular proliferation of colonic epithelial cells similar to that observed in humans (Robertis et al., 2011).
Borinstein et al. (2010) used an AOM-induced mice model to investigate the relationship between methylation of candidate genes, CRC, and normal tissues, and these authors reported 25% p16 methylation in CRC tissue, whereas we found 100% methylation to occur. We attribute the difference between Borinstein’s and our own findings to the use of different carcinogens and rat species. In human studies, the extent of p16 hypermethylation could be as high as 70% (Psofaki et al., 2010), which is in accordance with our results, indicating that p16 methylation is a high frequency CRC marker.
Although this study reveals a temporal correlation between progression of colorectal cancer and p16 methylation status in rat feces, the observed high frequency of p16 methylation in tumor samples may be due to the DMH carcinogenic mechanism (Robertis et al., 2011). Thus, our findings may not represent the true temporal causal relation between the natural history of colorectal cancer and p16 methylation status in human feces.
Our study demonstrates that p16 hypermethylation in stool occurs before CRC develops and that changes in the methylation status of the p16 gene in stool are statistically significant or borderline significant. The consistency of hypermethylated p16 between stool sample and tumor tissue demonstrates that detecting the hypermethylated p16 gene in stool to identify colorectal neoplasia provides a sensitive early detection, consistent and noninvasive screening tool.
Acknowledgments
This study was funded by a grant from Zuoying Branch of Kaohsiung Armed Forces General Hospital, Taiwan, Republic of China. Grant number ZAFGH9901.
Footnotes
Editor: Carlos F.M. Menck
References
- Azuara D, Rodriguez-Moranta F, de Oca J, Soriano-Izquierdo A, Mora J, Guardiola J, Biondo S, Blanco I, Peinado MA, Moreno V, et al. Novel methylation panel for the early detection of colorectal tumors in stool DNA. Clin Colorectal Cancer. 2010;9:168–176. doi: 10.3816/CCC.2010.n.023. [DOI] [PubMed] [Google Scholar]
- Baek YH, Chang E, Kim YJ, Kim BK, Sohn JH, Park DI. Stool methylation-specific polymerase chain reaction assay for the detection of colorectal neoplasia in Korean patients. Dis Colon Rectum. 2009;52:1452–1459. doi: 10.1007/DCR.0b013e3181a79533. discussion 1459–1463. [DOI] [PubMed] [Google Scholar]
- Borinstein SC, Conerly M, Dzieciatkowski S, Biswas S, Washington MK, Trobridge P, Henikoff S, Grady WM. Aberrant DNA methylation occurs in colon neoplasms arising in the azoxymethane colon cancer model. Mol Carcinog. 2010;49:94–103. doi: 10.1002/mc.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretthauer M. Colorectal cancer screening. J Intern Med. 2011;270:87–98. doi: 10.1111/j.1365-2796.2011.02399.x. [DOI] [PubMed] [Google Scholar]
- Chen WD, Han ZJ, Skoletsky J, Olson J, Sah J, Myeroff L, Platzer P, Lu S, Dawson D, Willis J, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97:1124–1132. doi: 10.1093/jnci/dji204. [DOI] [PubMed] [Google Scholar]
- Conerly M, Grady WM. Insights into the role of DNA methylation in disease through the use of mouse models. Dis Model Mech. 2010;3:290–297. doi: 10.1242/dmm.004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- Glockner SC, Dhir M, Yi JM, McGarvey KE, Van Neste L, Louwagie J, Chan TA, Kleeberger W, de Bruine AP, Smits KM, et al. Methylation of TFPI2 in stool DNA: A potential novel biomarker for the detection of colorectal cancer. Cancer Res. 2009;69:4691–4699. doi: 10.1158/0008-5472.CAN-08-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S, Benziger H. The molecular biology of colorectal carcinoma and its implications: A review. Surgeon. 2011;9:200–210. doi: 10.1016/j.surge.2011.01.011. [DOI] [PubMed] [Google Scholar]
- Heitman SJ, Hilsden RJ, Au F, Dowden S, Manns BJ. Colorectal cancer screening for average-risk North Americans: An economic evaluation. PLoS Medicine. 2010;7:e1000370. doi: 10.1371/journal.pmed.1000370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellebrekers DM, Lentjes MH, van den Bosch SM, Melotte V, Wouters KA, Daenen KL, Smits KM, Akiyama Y, Yuasa Y, Sanduleanu S, et al. GATA4 and GATA5 are potential tumor suppressors and biomarkers in colorectal cancer. Clin Cancer Res. 2009;15:3990–3997. doi: 10.1158/1078-0432.CCR-09-0055. [DOI] [PubMed] [Google Scholar]
- Heresbach D, Manfredi S, D’Halluin PN, Bretagne JF, Branger B. Review in depth and meta-analysis of controlled trials on colorectal cancer screening by faecal occult blood test. Eur J Gastroenterol Hepatol. 2006;18:427–433. doi: 10.1097/00042737-200604000-00018. [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Kim MS, Lee J, Sidransky D. DNA methylation markers in colorectal cancer. Cancer Metastasis Rev. 2010;29:181–206. doi: 10.1007/s10555-010-9207-6. [DOI] [PubMed] [Google Scholar]
- Krishnan S, Wolf JL. Colorectal cancer screening and prevention in women. Womens Health (Lond Engl) 2011;7:213–226. doi: 10.2217/whe.11.7. [DOI] [PubMed] [Google Scholar]
- Labianca R, Merelli B. Screening and diagnosis for colorectal cancer: Present and future. Tumori. 2010;96:889–901. [PubMed] [Google Scholar]
- Lenhard K, Bommer GT, Asutay S, Schauer R, Brabletz T, Goke B, Lamerz R, Kolligs FT. Analysis of promoter methylation in stool: A novel method for the detection of colorectal cancer. Clin Gastroenterol Hepatol. 2005;3:142–149. doi: 10.1016/s1542-3565(04)00624-x. [DOI] [PubMed] [Google Scholar]
- Levi Z, Rozen P, Hazazi R, Vilkin A, Waked A, Maoz E, Birkenfeld S, Leshno M, Niv Y. A quantitative immunochemical fecal occult blood test for colorectal neoplasia. Ann Intern Med. 2007;146:244–255. doi: 10.7326/0003-4819-146-4-200702200-00003. [DOI] [PubMed] [Google Scholar]
- Mandel JS, Church TR, Bond JH, Ederer F, Geisser MS, Mongin SJ, Snover DC, Schuman LM. The effect of fecal occult-blood screening on the incidence of colorectal cancer. N Engl J Med. 2000;343:1603–1607. doi: 10.1056/NEJM200011303432203. [DOI] [PubMed] [Google Scholar]
- Mariotto AB, Yabroff KR, Feuer EJ, De Angelis R, Brown M. Projecting the number of patients with colorectal carcinoma by phases of care in the US: 2000–2020. Cancer Causes Control. 2006;17:1215–1226. doi: 10.1007/s10552-006-0072-0. [DOI] [PubMed] [Google Scholar]
- Mehl LE. A mathematical computer simulation model for the development of colonic polyps and colon cancer. J Surg Oncol. 1991;47:243–252. doi: 10.1002/jso.2930470409. [DOI] [PubMed] [Google Scholar]
- Oberwalder M, Zitt M, Wontner C, Fiegl H, Goebel G, Kohle O, Muhlmann G, Ofner D, Margreiter R, Muller HM. SFRP2 methylation in fecal DNA - A marker for colorectal polyps. Int J Colorectal Dis. 2008;23:15–19. doi: 10.1007/s00384-007-0355-2. [DOI] [PubMed] [Google Scholar]
- Promotion BOH. Cancer Registry Annual Report, 2007. Department of Health; Taiwan: 2010. [Google Scholar]
- Psofaki V, Kalogera C, Tzambouras N, Stephanou D, Tsianos E, Seferiadis K, Kolios G. Promoter methylation status of hMLH1, MGMT, and CDKN2A/p16 in colorectal adenomas. World J Gastroenterol. 2010;16:3553–3560. doi: 10.3748/wjg.v16.i28.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertis M, Massi E, Poeta M, Carotti S, Morini S, Cecchetelli L, Signori E, Fazio V. The AOM/DSS murine model for the study of colon carcinogenesis: From pathways to diagnosis and therapy studies. J Carcinog. 2011;10:e9. doi: 10.4103/1477-3163.78279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid G. The use of molecular markers in the diagnosis of colorectal cancer screening. Dig Dis. 2010;28:625–628. doi: 10.1159/000320064. [DOI] [PubMed] [Google Scholar]
- Shima K, Nosho K, Baba Y, Cantor M, Meyerhardt JA, Giovannucci EL, Fuchs CS, Ogino S. Prognostic significance of CDKN2A (p16) promoter methylation and loss of expression in 902 colorectal cancers: Cohort study and literature review. Int J Cancer. 2010;128:1080–1094. doi: 10.1002/ijc.25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JR, Nelson JK, Silverman SJ. Research methods in physical activity. Human Kinetics; Champaign: 2005. p. 454. [Google Scholar]
- Wang DR, Tang D. Hypermethylated SFRP2 gene in fecal DNA is a high potential biomarker for colorectal cancer noninvasive screening. World J Gastroenterol. 2008;14:524–531. doi: 10.3748/wjg.14.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JM, Yen MF, Lai MS, Duffy SW, Smith RA, Chen TH. Progression rates of colorectal cancer by Dukes’ stage in a high-risk group: Analysis of selective colorectal cancer screening. Cancer J. 2004;10:160–169. doi: 10.1097/00130404-200405000-00005. [DOI] [PubMed] [Google Scholar]
- Zou H, Taylor WR, Harrington JJ, Hussain FT, Cao X, Loprinzi CL, Levine TR, Rex DK, Ahnen D, Knigge KL, et al. High detection rates of colorectal neoplasia by stool DNA testing with a novel digital melt curve assay. Gastroenterology. 2009;136:459–470. doi: 10.1053/j.gastro.2008.10.023. [DOI] [PubMed] [Google Scholar]