Abstract
Cultivars of sugarcane (Saccharum) are hybrids between species S. officinarum (x = 10, 2n = 8x = 80) and S. spontaneum (x = 8, 2n = 5 – 16x = 40 – 128). These accessions have 100 to 130 chromosomes, 80–85% of which are derived from S. officinarum, 10–15% from S. spontaneum, and 5–10% are possible recombinants between the two genomes. The aim of this study was to analyze the repetition of DNA sequences in S. officinarum and S. spontaneum. For this purpose, genomic DNA from S. officinarum was digested with restriction enzymes and the fragments cloned. Sixty-eight fragments, approximately 500 bp, were cloned, sequenced and had their identity analyzed in NCBI, and in the rice, maize, and sorghum genome databases using BLAST. Twelve clones containing partial transposable elements, one single-copy control, one DNA repetitive clone control and two genome controls were analyzed by DNA hybridization on membrane, using genomic probes from S. officinarum and S. spontaneum. The hybridization experiment revealed that six TEs had a similar repetitive DNA pattern in the genomes of S. officinarum and S. spontaneum, while six TEs were more abundant in the genome of S. officinarum. We concluded that the species S. officinarum and S. spontaneum have differential accumulation LTR retrotransposon families, suggesting distinct insertion or modification patterns.
Keywords: sugarcane, dot-blot hybridization, LTR retrotransposons, repetitive DNA
Sugarcane (Saccharum spp.) is a cultivated plant of significant economic importance, as it accounts for 70% of all sugar production in the world. In recent years, due to the global energy crisis, it has also emerged as an excellent source of renewable energy by the production of ethanol. This cultivated plant belongs to the genus Saccharum, family Poaceae, and the main species are S. officinarum, S. spontaneum, S. robustum, S. sinense, S. barberi and S. edule. Modern varieties of sugarcane are a complex of polyploids and aneuploids (Grivet and Arruda, 2001), originating from the recombination of different hybrids derived from two highly polyploid species, S. officinarum (x = 10, 2n = 8x = 80) and S. spontaneum (x = 8, 2n = 5–16x = 40–128). Usually, they have between 100 and 130 chromosomes, 70–80% of which are derived from S. officinarum, 10–20% from S. spontaneum and approximately 10% are recombinants between the two species (D’Hont et al., 1996, Piperidis et al., 2010). Furthermore, it is treated as an extremely large genome, with small chromosomes, which complicates the understanding of their genetic architecture and taxonomy (Pan et al., 2000).
The Saccharum genus presents taxonomic difficulties due to the existence of cross-hybridization producing “synthetic species” and to polymorphisms due to ploidy and aneuploidy, besides high selective pressure caused by genetic improvement. Relationships between the genera Saccharum, Erianthus, Sclerostachya and Narenga have suggested that they constitute an interrelated group involved in the origin of sugarcane, being called the “Saccharum complex” (Mukherjee, 1957). In addition to these genera, Miscanthus Anderss. section Diantra Keng, Erianthus Mickx. section Ripidium Henrard, and Sclerostachya (Hack) were included in the Saccharum complex (Daniels and Roach, 1987). Other phylogenetic analyses performed in Miscanthus, Saccharum and other close genera showed that the many species of Saccharum are closer to the Miscanthus species than to other species of those genera (Hodkinson et al., 2002). However, molecular data have shown only two true species in the genus Saccharum, called S. spontaneum and S. officinarum, which include the wild S. robustum and the races S. barberi, S. sinense and S. edule (Irvine, 1999; Grivet et al., 2004).
There are several strategies for analyzing these relationships, one of which is the differential amplification of repetitive DNA sequences, widely studied in several organisms (Ugarkovic and Plohl, 2002). Thus, determining the distribution of repetitive DNA sequences in species of genus Saccharum and close genera will permit a better understanding of the relationships between the genomes of these species and of the taxonomy of the group, besides the sequencing of the genome. The transposable elements (TEs) have been reported to be responsible for improving the genome. They show great diversity, with different families in plants, and with differences among individuals of the same species (reviewed by Morgante et al., 2007). In sugarcane, there are reports on TEs such as the one of Domingues et al. (2012) who described 35 families within four Copia and Gypsy lineages, and the one of Kajihara et al. (2012), showing that TEs in sugarcane are transcriptionally active, however, to our best knowledge, no analysis based on DNA hybridization showing differences between species of genus Saccharum has been published so far. In this work, we aimed to evaluate the abundance of transposable elements in S. officinarum and S. spontaneum.
Buds of S. officinarum and S. spontaneum accessions were obtained from the Serra do Ouro germplasm bank and germinated in pots. Young leaves were collected from each accession, genomic DNA was extracted using the CTAB method, as described by Saghai-Maroof et al. (1984), and quantified in 1% agarose gel. 10 μg of DNA from S. officinarum (cultivar Lousier) was digested using the restriction enzyme MboI (Fermentas), following its protocol. The digestion products were separated in a 2% agarose gel, and a region of 200 to 500 bp of the gel was excised and DNA purified. The fragments were then cloned into the vector CloneJET (Fermentas) and transformed in E. coli. The clones were confirmed by PCR, using 50 μL of reaction solution containing 50 ng of DNA, 1x enzyme buffer, 1.5 mM of MgCl2, 0.2 mM of dNTPs, 0.5 U of Taq-polymerase (Fermentas) and 0.2 μM of each primer. The DNA was then submitted to 30 amplification cycles at 94 °C for 1 min, 55 °C for 1 min, and 72 °C for 1 min. A DNA standard of 1 kb molecular weight was used for the determination of the molecular weight of the respective fragments of amplified DNA. The amplification products were separated in a 1% agarose gel and visualized using ethidium bromide.
The clones were amplified by PCR, the products were visualized on 1% agarose gels and sent to Macrogen (South Korea) for purification and DNA sequencing using primers from vector. The sequences were analyzed by BLAST against GenBank sequences and the genomes of rice, maize and sorghum. The max target sequences (100), which automatically adjust parameters for short input sequences, and the expected number of chance matches in a random model (expected threshold = 10) were used as BLAST general parameter.
DNA from clones containing partial TEs was placed (dots) onto a nylon membrane (Hybond N+, Amersham Biosciences) and fixed at 120 °C for 30 min. The membrane was pre-hybridized in a solution of DIG Easy Hyb (DIG High Prime DNA Labeling and Detection Starter Kit II (Roche Applied Science) for 30 min. Probes were produced from genomic DNA of S. officinarum and S. spontaneum digested with MobI and labeled with digoxigenin-AP by “Random Primer” following the manufacturer’s instructions (DIG High Prime DNA Labeling and Detection Starter Kit II). The probes were denatured at 100 °C for 5 min, added to the hybridization solution at 37 °C, and the mix was then placed onto the membrane and hybridized for 12 hours at 37 °C. After hybridization, the membrane was washed for 2x 5 min in 2x SSC and 0.1% SDS, followed by two washes of 15 min in 0.1x SSC and 0.1% SDS. Hybridization was visualized in a reaction with CSPD (Disodium 3-(4-methoxyspiro {1,2-dioxetane-3,2’-(5’-chloro)tricyclo [3.3.1.13,7]decan}-4-yl)phenylphosphate) (Roche Applied Science) and the signals evidenced by exposure to X-ray film. DNA hybridization was performed with three replicates, and the experimental controls were: single copy clone (JX101456 - partial tubulin-specific chaperone E), repetitive clone (Scent7 - described as repetitive by Nagaki et al., 1998), and the genomic DNA controls.
Genomic DNA of the S. officinarum accessions (Lousier and IJ76-530) was digested for 20 hours with the enzyme EcoRI (Fermentas), after which the fragments were separated in a 2% agarose gel at 30 V for 6 h. Then the gel was washed in depurination, denaturation and neutralization solutions. After 24 h, the DNA was fixed on a nylon membrane (Hybond N+, Amersham Biosciences) at 80 °C for 2 h. The membrane was pre-hybridized in a solution of DIG Easy Hyb (DIG High Prime DNA Labeling and Detection Starter Kit II) for 30 min. The probes were labeled with a random primer using PCR clones Soffa.4, Soff.e4 and Soff.f2 DNA and then denatured at 100 °C for 5 min and added to the hybridization solution at 37 °C for 12 hours. After hybridization, the membrane was washed for 2x 5 min in 2x SSC and 0.1% SDS, followed by two 15 min washes in 0.1x SSC and 0.1% SDS. The hybridization was visualized in a reaction with CSPD and the signals evidenced by exposure to X-ray film.
The sequences were analyzed using the NCBI sequence database for maize mobile elements, in the genomes of rice, maize and sorghum. Twelve clones showed analogies with TEs deposited in the databases, suggesting that they are abundant in the genome of S. officinarum (Table 1). Clone Soff.a4, which is highly repeated in the genome of S. officinarum and S. spontaneum, has a similarity with a centromeric sequence belonging to the SCEN family (Table 1), described by Nagaki et al. (1998). The same clone had also high similarity to the LTR retrotransposon Maximus family, reported in sugarcane by Domingues et al. (2012). For the grass species, BLAST showed 11 repetitive clones in sorghum, seven in maize and four in rice (Table 1).
Table 1.
BLAST results for repetitive DNA sequences of Saccharum officinarum
| Gene bank N. | Clone name | Repetitive in: sorghum/maize/rice | Order | Superfamily | Family | e-value |
|---|---|---|---|---|---|---|
| JX101444 | Soff.a2 | y/n/n | LTR | Copia | Maximus/Sire | 3e-125 |
| JX101454 | Soff.a4 | *y/n/n | LTR | Copia | Maximus/Sire | 1e-08 |
| JX101446 | Soff.a9 | y/n/n | LTR | Copia | Maximus/Sire | 2e-37 |
| JX101450 | Soff.b7 | y/y/y | LTR | CopiaGypsy | Maximus/SireDEL/TEKAY | 5e-48 |
| JX101445 | Soff.b11 | n/n/y | LTR | Copia | Maximus/Sire | 8e-66 |
| JX101451 | Soff.b12 | **y/y/n | LTR | Copia | Maximus/Sire | 9e-125 |
| JX101443 | Soff.c3 | n/y/n | LTR | Gypsy | DEL/TEKAY | 4e-59 |
| JX101452 | Soff.d8 | y/y/n | LTR | Copia | Maximus/Sire | 5e-28 |
| JX101453 | Soff.d11 | y/y/n | LTR | Copia | Maximus/Sire | 1e-63 |
| JX101448 | Soff.e4 | y/y/y | LTR | Copia | Maximus/Sire | 8e-180 |
| JX101455 | Soff.e6 | y/n/n | LTR | Copia | Maximus/Sire | 1e-48 |
| JX101442 | Soff.f2 | y/y/y | LTR | Gypsy | DEL/TEKAY | 1e-77 |
| JX101456 | Soff.a10 | n/n/n | - | - | - |
Only chromosome #3;
only chromosomes #3, 5, 6 and 7.
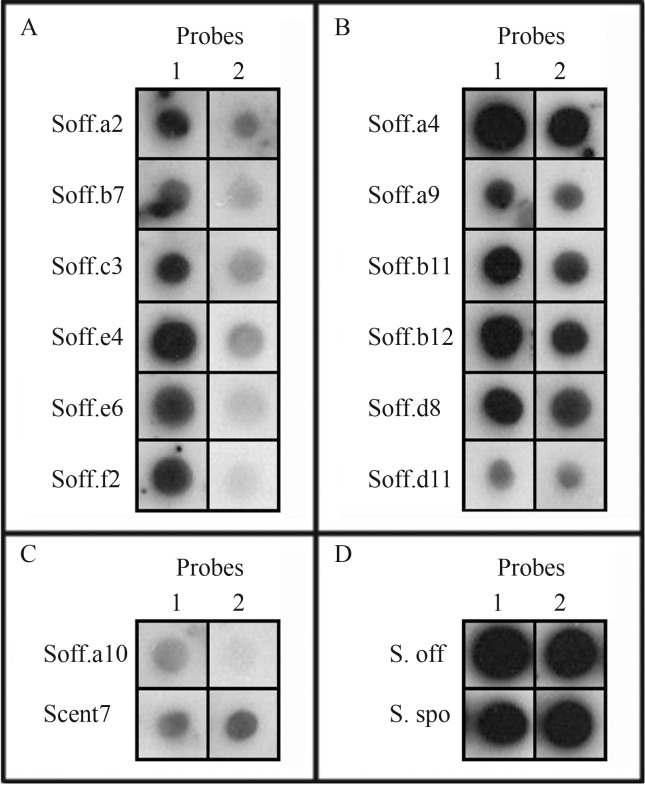
Two membranes composed of 14 clones (12 clones containing partial TEs and two controls) and two genomes were hybridized with genomic probes from S. officinarum and S. spontaneum, respectively. A stringency of 80% was used, to allow high specificity of the sequences with the genomes analyzed. The results showed clear dots when compared with the genomic controls (genomic DNA from S. officinarum and S. spontaneum), the DNA repetitive control and the single copy control (Figure 1).
Figure 1.
Hybridization profile for Saccharum officinarum (1) and S. spontaneum (2) genomic probes. (A) clones with greater intensity in the genome of S. officinarum; (B) highly repetitive clones in the two genomes; (C) repetition control (Scent7) and clone as a single copy (Soff.a10); and (D) positive control with probe from genomic DNA of S. officinarum (S.off) and S. spontaneum (S. spo).
Some clones showed greater signal intensity in the genome of S. officinarum (Soff.a2, Soff.b7, Soff.c3, Soff.e4, Soff.e6 and Soff.f2) (Figure 1A), whereas other clones had similar signal intensity in the two genomes (Soff.a4, Soff.a9, Soff.b11, Soff.b12, Soff.d8 and Soff.d11) (Figure 1B). A clone of repetitive DNA (Scent7) for the genus Saccharum, described by Nagaki et al. (1998), was added as a repetition control, and the signal showed similarity to Soff.a9 and Soff.d11, indicating consistency in the hybridizations and confirming that the sequences are repetitive (Figure 1C).
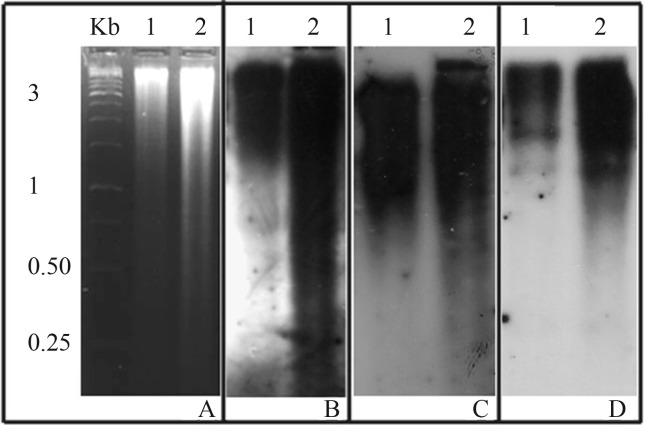
The Soff.a4, Soff.e4 and Soff.f2 (JX101454, JX101448 and JX101442 sequences, respectively) were analyzed by Southern blot in two S. officinarum accessions, in order to obtain their repetition patterns in the genome, using the same conditions as for dot blot hybridization. The results showed that these sequences were dispersed in the genome of S. officinarum, appearing as a smear in the two accessions analyzed (Figure 2).
Figure 2.
Southern blot of Saccharum officinarum, accession Lousier (1) and IJ76-530 (2). Genomic DNA was digested with EcoRI (A) and hybridized with the probes Soff.e4 (JX101448) (B), Soff.a4 (JX101454) (C), and Soff.f2 (JX101442) (D).
Cultivated sugarcane accessions are formed by two main genomes which correspond to the pure species S. officinarum and S. spontaneum. A process of backcrossing associated with “nobilization” allowed to combine a fraction of 80–85% from S. officinarum with 10–15% from S. spontaneum. Both species are polyploid, S. officinarum with 2n = 80 and S. spontaneum with 2n = 40–128 (DHont et al., 1998), and recent phylogenetic studies have shown that the genus Saccharum is monophyletic, comprising only two true species (S. officinarum and S. spontaneum). Their speciation is relatively recent, as it dates back to approximately 1.5–2 million years (Jannoo et al., 2007), suggesting similar DNA sequences. In the present study, we used repetitive DNA sequences to identify differential abundance of the TEs in the genomes of S. officinarum and S. spontaneum, using DNA hybridization. As both species are polyploid, differential accumulation of repetitive DNA sequences may occur since speciation. Indeed, differentially amplified sequences were detected indirectly by D’Hont et al. (1996) and Piperidis et al. (2010), using GISH to identify the genomes.
The fraction of dispersed repetitive DNA is the major component in many eukaryotic genomes, being the largest contributor to variation in DNA content between similar organisms (Zhao et al., 1998). Mobile DNA elements have contributed significantly to this increase, by being selfish DNA and by multiplying in a disorderly way in the genomes (Bowen and Jordan, 2002). TEs described by Domingues et al. (2012) in the sugarcane genome from the R570 (BAC hybrid) are transcriptionally active (Araujo et al., 2005), and a search of repetitive DNA has identified species-specific repeated DNA in Saccharum (Kim et al., 2011). However, there is no information about differential accumulation of TEs between the Saccharum species. In the present study, the repetitive DNA sequences were similar to TEs, suggesting that these elements are highly abundant in the genomes of the Saccharum species and must have contributed to the expansion of the genomes. Additionally, six of the DNA sequences analyzed were more abundant in the genome of S. officinarum, suggesting differential accumulation between genomes.
Studies of repetitive DNA in Saccharum have shown a large number of repetitive sequences in this group (Alix et al., 1998, 1999; Nagaki and Murata, 2005) in which some sequences are common to several species, and differential amplification of repetitive DNA was observed by Nakayama (2004), showing accumulation of a specific sequence in the genome of S. officinarum and suggesting that repetitive DNA amplification is a common evolutionary mechanism in the genomes. Indeed, collinearity between sugarcane and sorghum was described by Wang et al. (2010) for 20 genomic regions, and unaligned regions between sugarcane and sorghum sequences are occupied mostly by repeats. Garsmeur et al. (2011) showed that sugarcane and sorghum genomes are collinear in the genic regions and present high structure and sequence conservation. These results suggest that the main mechanism of divergence among species is the repetitive component of their genomes. In other species, as well as in the genus Phaseolus, the differences between species are probably due to differential amplification of the repetitive DNA (Pedrosa-Harand et al., 2006). Furthermore, Zhang et al. (2006) showed species-specific accumulation repetitive DNA in Triticum.
Repetitive sequences of the SCEN family were found to occupy 0.6% of the sugarcane genome, or 2.6 × 105 copies per haploid genome (Nagaki et al., 1998). Our results showed that SCEN sequences are distributed within the genome of S. officinarum and S. spontaneum with a high number of repetitions, confirming the findings of Nagaki et al. (1998). The SCENT7 sequence (SCEN family) was used as repetitive control DNA. Its signal was less than that of SCENM9 (similar to Soff.a4), however the SCEN family showed a large divergence in the sequence, suggesting that members of this family were differentially amplified in the genomes of S. officinarum and S. spontaneum. We conclude that the species S. officinarum and S. spontaneum have differential repetitive DNA, belonging to LTR retro-transposons families, suggesting a distinct pattern of insertion or modification, which is the important factor for increasing the differences between genomes in the genus Saccharum.
Acknowledgments
This work received support from the Breeding Program for Sugarcane at the Federal University of Alagoas and from the National Council for Scientific and Technological Development (Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq, Brazil).
Footnotes
Associate Editor: Marcia Pinheiro Margis
References
- Alix K, Baurens FC, Paulet F, Glaszmann JC, D’Hont A. Isolation and characterization of a satellite DNA family in the Saccharum complex. Genome. 1998;41:854–864. [PubMed] [Google Scholar]
- Alix K, Pualet F, Glaszmann JC, D’Hont A. Inter-Alu-like species-specific sequences in the Saccharum complex. Theor Appl Genet. 1999;99:962–968. [Google Scholar]
- Araujo PG, Rossi M, de Jesus EM, Saccaro NL, Kajihara D, Massa R, de Felix JM, Drummond RD, Falco MC, Chabregas SM, et al. Transcriptionally active transposable elements in recent hybrid sugarcane. Plant J. 2005;44:707–717. doi: 10.1111/j.1365-313X.2005.02579.x. [DOI] [PubMed] [Google Scholar]
- Bowen NJ, Jordan K. Transposable elements and the evolution of eukaryotic complexity. Curr Issues Mol Biol. 2002;4:65–76. [PubMed] [Google Scholar]
- D’Hont A, Grivet L, Feldmann P, Rao S, Berding N, Glaszmann JC. Characterisation of the double genome structure of modern cultivars (Saccharum spp.) by molecular cytogenetics. Mol Gene Genet. 1996;250:405–413. doi: 10.1007/BF02174028. [DOI] [PubMed] [Google Scholar]
- Daniels J, Roach BT. Taxonomy and evolution. In: Heinz DJ, editor. Sugarcane Improvement Though Breeding. Elsevier; Amsterdam: 1987. pp. 7–84. [Google Scholar]
- Domingues D, Cruz GMQ, Metcalfe CJ, Nogueira FTS, Vicentini R, Alves C de S, Van Sluys MA. Analysis of plant LTR-retrotransposons at the finescale family level reveals individual molecular patterns. BMC Genomics. 2012;13:e137. doi: 10.1186/1471-2164-13-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsmeur O, Charron C, Bocs S, Jouffe V, Samain S, Couloux A, Droc G, Zini C, Glaszmann JC, Van Sluys MA, et al. High homologous gene conservation despite extreme auto-polyploid redundancy in sugarcane. New Phytol. 2011;189:629–642. doi: 10.1111/j.1469-8137.2010.03497.x. [DOI] [PubMed] [Google Scholar]
- Grivet L, Arruda P. Sugarcane genomics: Depicting the complex genome of an important tropical crop. Curr Opin Plant Biol. 2001;5:122–127. doi: 10.1016/s1369-5266(02)00234-0. [DOI] [PubMed] [Google Scholar]
- Grivet L, Daniels C, Glaszmann JC, D’Hont A. A review of recent molecular genetics evidence for sugarcane evolution and domestication. Ethnobot Res Appl. 2004;2:9–17. [Google Scholar]
- Hodkinson TR, Chase MW, Lledó MD, Salamin N, Renvoiza SA. Phylogenetics of Miscanthus, Saccharum and related genra (Saccharinae, Andropogoneae, Poacea) based on DNA sequences from ITS nuclear ribosomal DNA and plastid trnL intro and trnL-F intergenic spacers. J Plant Res. 2002;115:381–392. doi: 10.1007/s10265-002-0049-3. [DOI] [PubMed] [Google Scholar]
- Irvine JE. Saccharum species as horticultural classes. Theor Appl Genet. 1999;98:186–194. [Google Scholar]
- Jannoo N, Grivet L, Chantret N, Garsmeur O, Glaszmann JC, Arruda P, D’Hont A. Orthologous comparison in a gene-rich region among grasses reveals stability in the sugarcane polyploidy genome. Plant J. 2007;50:574–585. doi: 10.1111/j.1365-313X.2007.03082.x. [DOI] [PubMed] [Google Scholar]
- Kajihara D, de Gogoy F, Hamaji TA, Blanco SR, Van Sluys MA, Rossi M. Functional characterization of sugarcane mustang domesticated transposases and comparative diversity in sugarcane, rice, maize and sorghum. Genet Mol Biol. 2012;35:632–639. doi: 10.1590/S1415-47572012005000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Robertson JS, Paterson A. Inference of sub-genomic origin of BACs in an interspecific hybrid sugarcane cultivar by overlapping oligonucleotide hybridizations. Genome. 2011;54:727–737. doi: 10.1139/g11-038. [DOI] [PubMed] [Google Scholar]
- Morgante M, Paoli E, Radovic S. Transposable elements and the plant pan-genomes. Curr Opin Plant Biol. 2007;10:149–155. doi: 10.1016/j.pbi.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Murkherjee SK. Origin and distribution of Saccharum. Bot Gas. 1957;119:55–61. [Google Scholar]
- Nagaki K, Murata M. Characterization of CENH3 and centromere-associated DNA sequences in sugarcane. Chromosome Res. 2005;13:195–203. doi: 10.1007/s10577-005-0847-2. [DOI] [PubMed] [Google Scholar]
- Nagaki K, Tsujimoto H, Sasakuma T. A novel repetitive sequence of sugar cane, SCEN family, locating on centromeric regions. Chromosome Res. 1998;6:295–302. doi: 10.1023/a:1009270824142. [DOI] [PubMed] [Google Scholar]
- Nakayama S. Species-specific accumulation of interspersed sequences in genus Saccharum. Genes Genet Syst. 2004;79:361–365. doi: 10.1266/ggs.79.361. [DOI] [PubMed] [Google Scholar]
- Pan YB, Burner DM, Lendendrei BL. An assessment of the phylogenetic relationship among sugarcane and related taxa based on the nucleotide sequence of 5S rRNA intergenic spacers. Genetica. 2000;108:285–295. doi: 10.1023/a:1004191625603. [DOI] [PubMed] [Google Scholar]
- Pedrosa-Harand A, Almeida CCS, Mosiolek M, Blair MW, Schweizer D, Guerra M. Extensive ribosomal DNA amplification during Andean common bean (Phaseolus vulgaris L.) evolution. Theor Appl Genet. 2006;112:924–933. doi: 10.1007/s00122-005-0196-8. [DOI] [PubMed] [Google Scholar]
- Piperidis G, Piperidis N, D’Hont A. Molecular cytogenetic investigation of chromosome composition and transmission in sugarcane. Mol Genet Genomics. 2010;284:65–73. doi: 10.1007/s00438-010-0546-3. [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population-dynamics. Proc Natl Acad Sci USA. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarkovic D, Plohl M. Variation in satellite DNA profiles - Causes and effects. EMBO J. 2002;21:5955–5959. doi: 10.1093/emboj/cdf612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Roe B, Macmil S, Yu Q, Murray JE, Tang H, Chen C, Najar F, Wiley G, Bower J, et al. Microcollinearity between autopolyploid sugarcane and diploid sorghum genomes. BMC Genomics. 2010;11:e261. doi: 10.1186/1471-2164-11-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Li W, Friebe B, Gill BS. Simultaneous painting of three genomes in hexaploid wheat by BAC-FISH. Genome. 2006;47:979–987. doi: 10.1139/g04-042. [DOI] [PubMed] [Google Scholar]
- Zhao X, Si Y, Hanson RE, Crane CF, Prince HJ, Stelly DM, Wendel JF, Paterson AH. Dispersed repetitive DNA has spread to new genomes since polyploid formation in cotton. Genome Res. 1998;8:479–492. doi: 10.1101/gr.8.5.479. [DOI] [PubMed] [Google Scholar]
Internet Resources
- Rice genome http://rice.genomics.org.cn/rice (accessed September 22, 2012).
- Maize genome http://www.maizegdb.org (accessed September 22, 2012).
- Sorghum genome http://www.phytozome.net (accessed September 22, 2012).