Abstract
Endive (Cichorium endivia L.) and chicory (C. intybus L.) both have 2n = 18, but until now, there has been no detailed karyomorphological characterization. The present work evaluated five accessions of each species using FISH with rDNA probes and fluorochrome staining with CMA and DAPI. Both species presented distinct banding patterns after fluorochrome staining: while endive had proximal CMA++/DAPI− bands in the short arms of pairs 1, 2 and 3, chicory had proximal CMA-positive bands in chromosomes 1 and 3 and interstitial in the short arm of chromosome 8. Among endive accessions, FISH procedures revealed conserved position and number of 5S and 45S rDNA sites (two and three pairs, respectively), associated with the CMA-positive bands. Notwithstanding, polymorphisms were detected within chicory accessions regarding the number and the distribution of rDNA sites in relation to the most frequent karyotype (two pairs with 45S and one with 5S rDNA). The karyological markers developed allowed karyotypic differentiation between both species, uncovering peculiarities in the number and position of rDNA sites, which suggest chromosome rearrangements, such as translocations in chicory cultivars. The interspecific and intraspecific polymorphisms observed emphasize the potential of karyomorphological evaluations, helping our understanding of the relationships and evolution of the group.
Keywords: compositae, Cichorieae, CMA/DAPI, rDNA sites, chromosome rearrangements
Introduction
The Cichorieae tribe (subfamily Cichorioideae, family Asteraceae) comprises about 70 genera and 1,500 species. The group is morphologically well characterized by the presence of latex and perfect flowers within the capitulum. Among the economically most important members of the tribe, lettuce (Lactuca sativa L.), chicory (Cichorium intybus L.) and endive (C. endivia L.) stand out due to their worldwide use as green salad vegetables (Bremer, 1994; Kilian et al., 2009). While being less used than lettuce, the economic importance of chicory and endive is increasing, with growing incorporation in cooking recipes due to their nutritional value (Lucchin et al., 2008), which impacts the size and distribution of the areas cultivated worldwide.
Controversies regarding the taxonomy of Cichorium have been raised since the 18th century, especially regarding the total number of species and their delimitation (Lucchin et al., 2008). Some authors suggested the existence of three (Tutin et al., 1976), four (Pignatti, 1982) or seven (Wagenitz and Bedarff, 1989) valid species. However, approaches based on AFLP markers (Kiers et al., 2000) and ITS sequences (Kilian and Gemeinholzer, 2007) recognized six species subdivided into three distinct groups: (1) C. bottae A. Deflers., as sister to all the other species, (2) C. intybus and C. spinosum L., and (3) C. endivia, C. pumilum Jacq. and C. calvum Sch. Bip.
Chromosome studies using FISH (fluorescent in situ hybridization) and double fluorochrome staining with CMA (chromomycin A3 - for GC-rich heterochromatic regions) and DAPI (4’,6-diamidino-2-phenylindole - for AT-rich heterochromatic regions) revealed important karyotypic features in Asteraceae, including cultivated (e.g. sunflower, Vanzela et al., 2002; lettuce, Matoba et al., 2007) and wild species (Fregonezi et al., 2004; Garcia et al., 2010). Despite their informativeness, few previous reports are available which have employed these methods, especially considering the size and diversity of the family, as well as their relevance in an evolutionary context within the angiosperms (Watanabe et al., 2007).
Previous cytogenetic evaluations for C. endivia and C. intybus using conventional staining reported the diploid number 2n = 18 for both species and similar chromosome morphologies (e.g.Rick, 1953), thus were unable to effectively differentiate between species as has also been observed for other genera of Cichoriodeae (see Matoba et al., 2007). Therefore, aiming to identify chromosomal differences between these species, 10 Cichorium accessions were analysed - five from C. endivia and five from C. intybus - using FISH with 5S and 45S rDNA probes and CMA/DAPI staining. The results uncovered interesting chromosome polymorphisms, especially in the number of 45S rDNA sites between both species, also revealing chromosome variations within C. intybus accessions, bringing interesting aspects to light for the understanding of their relationships and evolution.
Materials and Methods
Seeds of five accessions of C. endivia and five of C. intybus from different provenances (Table 1) were germinated in Petri dishes. Root tips were collected, pre-treated with 8-hydroxyquinoline (2 mM) at 8 °C for 24 h, fixed in 3:1 ethanol:acetic acid (v/v) at room temperature (ca. 25 °C) for 4–24 h and stored at −20 °C as described by Benko-Iseppon and Morawetz (2000). Fixed root tips were digested for 3 h at 37 °C in an enzymatic solution containing 2% (w/v) cellulase (`Onozuka R-10’, Serva) and 20% (v/v) pectinase (Sigma-Aldrich), incubated in 60% acetic acid for 20 min at 37 °C and squashed in a drop of 60% acetic acid. Coverslips were removed after freezing in liquid nitrogen and the slides were air-dried. Slides were aged for three days at room temperature and then stained with CMA (0.5 mg/mL, 1 h) and DAPI (1 μg/mL, 30 min), mounted in McIlvaine’s buffer (pH 7.0):glycerol (1:1, v/v) and stored for three days, according to Schweizer and Ambros (1994). Cell images were acquired using a Leica DMLB epifluorescence microscope and a Leica DFC 340FX camera with the Leica CW4000 software.
Table 1.
Analysed Cichorium endivia and C. intybus accessions with their provenances.
| Accession | ProvenanceA | Additional information |
|---|---|---|
| Cichorium endivia L. | Flora-FreyB | winter endive |
| C. endivia subsp. endivia var. endivia | IPKC (CICH 23 Italy) | - |
| C. endivia subsp. endivia var. crispum Lam. | IPK (CICH 709 China) | Hua Yie Sheng Tsai |
| C. endivia subsp. endivia var. latifolium Lam. | IPK (CICH 388 Netherlands) | Bubikopf |
| C. endivia subsp. divaricatum (Schousb.) P. D. Sell | IPK (CICH 66 Italy) | - |
| C. intybus L. | Flora-Frey | - |
| C. intybus var. intybus Hegi | IPK (CICH 499 Germany) | Wild chicory |
| C. intybus var. foliosum Hegi | IPK (CICH 615 Netherlands) | Liber vo |
| C. intybus var. foliosum cv. Zoom Hegi | KiepenkerlbC | - |
| C. intybus var. sativum Lam. & DC. | IPK (CICH 75 Czechoslovakia) | Slezka |
In parenthesis: registration number and place of origin, if informed.
Commercially available seeds.
Institut für Pflanzengenetik und Kulturpflanzenforschung (Leibniz Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany).
The R2 clone, with an 18S-5.8S-25S rDNA repeat unit (6.5 kb) isolated from Arabidopsis thaliana (L.) Heynh. (Wanzenböck et al., 1997), and the D2 clone, which consists of two 5S rRNA repeats (∼ 400 bp) isolated from Lotus japonicus (Regel) K. Larsen (Pedrosa et al., 2002), were used as probes and were labelled by nick translation (Invitrogen) with digoxigenin-11-dUTP (Roche) and biotin-11-dUTP (Sigma), respectively. The FISH pre-treatment and post-hybridization washes were based on Pedrosa et al. (2002), in which the stringency wash (77%) was performed with 0.1x SSC at 42 °C. Chromosome and probe denaturation and detection were performed according to Heslop-Harrison et al. (1991) and Jiang et al. (1996), respectively, with minor modifications. The slides were denatured in 70% formamide at 90 °C for 10 min. The hybridization mixture, containing 50% formamide (v/v), 2x SSC, 10% dextran sulphate (w/v) and 2.5–5 ng/μL of probe, was denatured at 75 °C for 10 min. Each slide received 10 μL of the hybridization mixture and was hybridized at 37 °C for at least 18 h. Digoxigenin-labelled probes were detected using sheep anti-digoxigenin-FITC (Roche) and amplified with donkey anti-sheep-FITC (Sigma), in 1% (w/v) BSA. Biotin-labelled probes were detected with mouse anti-biotin (Dako) and the signal was visualized with rabbit anti-mouse TRITC conjugate (Dako), in 1% (w/v) BSA. Preparations were counterstained and mounted with 2 μg/mL DAPI in Vectashield (Vector) (1:1; v/v).
Cell images were acquired and optimized for contrast and brightness, while DAPI pictures were pseudo-coloured in grey with the Adobe Photoshop CS4 (Adobe Systems Incorporated) software. Metaphase chromosomes of three cells stained with DAPI from each accession were measured by using the MicroMeasure 3.3 (Reeves, 2001) software and the idiograms were constructed using Adobe Flash CS4 Professional (Adobe Systems Incorporated). Chromosomes were ordered in decreasing order according to their size. The chromosome arm ratio (AR = length of the long arm/length of the short arm) was used to classify chromosome morphologies as metacentric (AR = 1.00–1.49) or submetacentric (AR = 1.50–2.99), according to Guerra (1986).
Results
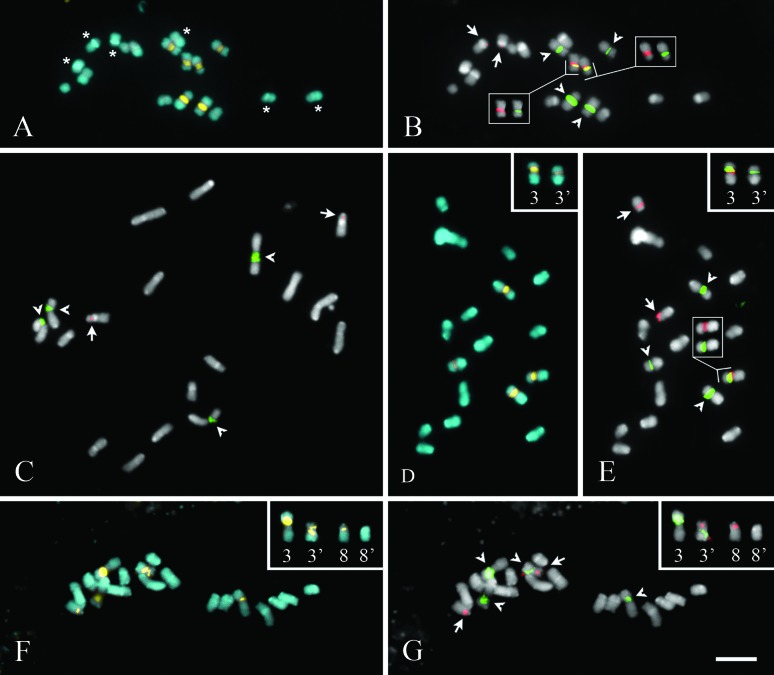
The chromosome numbers of all C. endivia (Figure 1A and 1B) and C. intybus (Figure 1C–G) accessions were stable (2n = 18), with gradually decreasing chromosome sizes. Size and morphology of the chromosomes, in combination with the cytogenetic markers allowed for the easily identification of all chromosome pairs in C. endivia and most of them in C. intybus (except for the pairs 5, 6 and 7). Chromosome complements showed averages of 2.84 μm per chromosome for C. endivia (ranging from 1.67 μm to 4.01 μm) and 3.20 μm for C. intybus (ranging 2.30 μm from to 4.29 μm). The average size of the whole diploid complement for C. endivia was 51.13 μm, and 57.58 μm for C. intybus.
Figure 1.
Metaphase chromosomes of Cichorium endivia subsp. endivia (A and B), C. intybus var. foliosum (C), C. intybus var. intybus (D and E), and C. intybus var. foliosum cv. Zoom. (F and G). Superposition of chromosomes stained with DAPI (blue) and CMA (yellow) (A, D and F). Chromosomes hybridized with 45S (green) and 5S (red) rDNA probes, counterstained with DAPI (grey) (B, C, E and G). Asterisks indicate DAPI+ bands; arrows and arrowheads indicate 5S and 45S rDNA sites, respectively; inside inserts in B and E indicate chromosomes with both 5S and 45S rDNA sites; and right corner inserts in D, E, F and G indicate heteromorphic chromosome pairs. Scale bar in G corresponds to 5 μm.
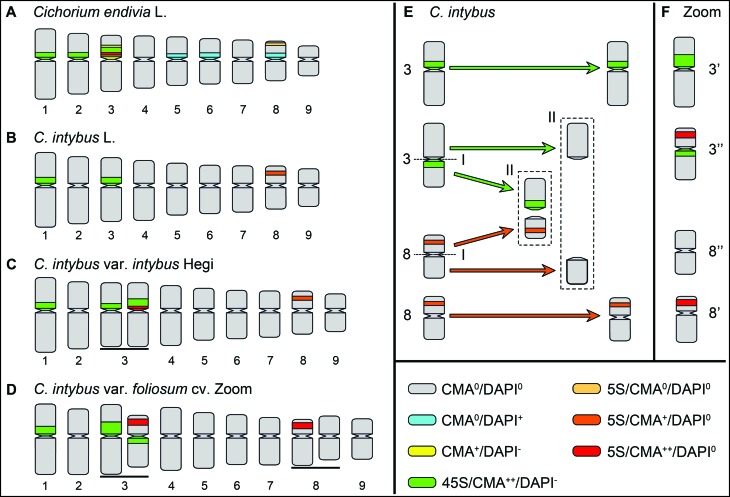
Nucleolus organizing regions (NORs) were observed (usually not distended) in the three larger chromosome pairs (1, 2 and 3) of C. endivia (Figure 2A), while only pairs 1 and 3 of C. intybus presented satellites (Figures 2B–D). Chromosome morphology was metacentric, except for C. intybus var. foliosum cv. Zoom that displayed a submetacentric pair (chromosome 7) and two heteromorphic pairs (3 and 8), each with a submetacentric and a metacentric chromosome (Figure 2D). Considering the similarity regarding the chromosome measurements for C. endivia (Figure 2A) and C. intybus (Figure 2B) accessions, the sampled mean values for each species were grouped into single idiograms, except for C. intybus var. intybus Hegi (Figure 2C) and C. intybus var. foliosum cv. Zoom (Figure 2D) that were processed in exclusive idiograms.
Figure 2.
Schematic representation of Cichorium endivia and C. intybus chromosomes: idiograms of C. endivia (A), C. intybus (B), C. intybus var. intybus (C) and C. intybus var. foliosum cv. Zoom (D). Heteromorphic pairs in C. intybus var. intybus and C. intybus var. foliosum cv. Zoom accessions are underlined (C and D). Representation of possible reciprocal translocation with breakages in the centromeres (centric fission - I, and fusion - II), involving a homologue of pairs 3 and 8 of C. intybus (E), in the genesis of the heteromorphic chromosomes pairs of C. intybus var. foliosum cv. Zoom cytotype (F).
The CMA/DAPI staining revealed the prevalence of CMA++/DAPI− heterochromatin for both species. In C. endivia, bands were visible in the short arm of pairs 1, 2 and 3 (Figures 1A and 2A), while in C. intybus the same distribution was observed in pairs 1 and 3 (Figures 1D and 2B), with an additional faint CMA+/DAPI− band in the intercalary region of the short arm of pair 8 (data not shown; Figure 2B). Additionally, two C. intybus accessions showed heteromorphic chromosomes, regarding both size and distribution of CMA bands. In C. intybus var. intybus, a larger CMA++/DAPI− band was observed in one chromosome of pair 3 (Figures 1D and 2C). On the other hand, C. intybus var. foliosum cv. Zoom displayed two heteromorphic chromosome pairs. In pair 3, one of the homologues exhibited a proximal band in the short arm, while the other chromosome had two CMA++/DAPI− bands (one intercalary in the short arm and the other in the proximal region of the long arm) (Figures 1F and 2D). Additionally, in pair 8, besides a small difference between chromosome sizes, one homologue displayed an intercalary CMA++/DAPI− band in the short arm, while the other presented no bands (Figures 1F and 2D).
Additional proximal CMA0/DAPI+ bands were visible in the short arms of chromosome pairs 5, 6 and 8 in C. endivia (Figures 1A and 2A). Nevertheless, small terminal CMA0/DAPI+ bands were observed in C. intybus, which were not always visible, especially when the chromosomes were fully condensed (Figure 1D, F). Therefore, these small marks were not represented in the idiogram. In both cases, these additional bands were enhanced after FISH procedures, as well as other marks that became visible in terminal and proximal positions (noticeable when comparing the DAPI staining in Figures 1A and D - before FISH, with C, E, F - after FISH).
The FISH assay using the 45S rDNA probe uncovered signals in the proximal region of the short arms of the three larger chromosome pairs of C. endivia, which were co-localized with the CMA++/DAPI− bands (Figures 1B and 2A). Only in pair 3, did the 45S rDNA site occupied a part of the CMA++/DAPI− band (Figure 2A). Regarding the distribution of the 5S rDNA, two chromosome pairs had hybridization signals: one site was observed in a proximal position of the short arm in pair 3 (co-localized with a CMA++/DAPI− band), adjacent to the distal 45S rDNA site, while the other was visible in the subterminal region of the short arm in pair 8, with no associated fluorochrome band (Figures 1B and 2A).
The FISH assay using the 45S rDNA probe in C. intybus revealed two pairs of hybridization sites in the proximal region of the short arms in pairs 1 and 3, and, as observed for C. endivia, those sites were co-localized with CMA++/DAPI− bands (Figures 1C and 2B). The FISH procedures with the 5S rDNA probe showed evidence for a single site in the intercalary position of the short arm in pair 8, co-localized with a CMA+/DAPI− band (Figures 1C and 2B). In C. intybus var. intybus, an extra 5S rDNA site was observed in one chromosome of pair 3 (Figures 1D and 2C).
In C. intybus var. foliosum cv. Zoom, a heteromorphism related to the distribution of the rDNA sites could be detected in pairs 3 and 8 (Figures 1G and 2D). Regarding the 45S rDNA site of pair 3, one of the homologues displayed a proximal site in the short arm, similar to the other accessions, but slightly larger. However, the other site was in a proximal region of the long arm at the homologue of this chromosome pair. For 5S rDNA, an intercalary site was observed in the short arm of a homologue of pair 3 (carrier of 45S rDNA) while the other was intercalary in the short arm of a homologue of pair 8. The second homologue of pair 8 exhibited no marks. In this accession, all rDNA sites co-localized with the CMA++/DAPI− bands (Figures 1F–G and 2D).
Discussion
Previous karyotypic analyses of Cichorium species were restricted to chromosome counts regarding the species C. endivia (Rick, 1953), C. intybus (Dobes et al., 1997), C. intybus var. intybus (Lövkvist and Hultgård, 1999) and C. spinosum (Montmollin, 1986), all of them with 2n = 2x = 18. This chromosome number is very frequent within the Cichorioidae, as observed in Lactuca, Sonchus and Tolpis, being considered conserved throughout the subfamily (Bremer, 1994).
Contrary to the classic cytogenetic methods, which could not elucidate the chromosome evolution within other Cichorioideae genera, the distribution of heterochromatic blocks has been shown to be a very informative approach (e.g.Matoba et al., 2007). Thus, the presence of repetitive DNA clusters (rDNA sites and fluorochrome bands) observed in both C. endivia and C. intybus, as well as differences in their distribution patterns, suggest the great importance of such DNA elements during the karyotype evolution within the genus.
In general, 45S rDNA sites occur either in terminal or sub-terminal position of chromosomes within subfamilies of Asteraceae, as observed in Chaptalianutans (subfamily Mutisioideae) (Fregonezi et al., 2004) and in the genera Lactuca (Matoba et al., 2007), Tragopogon (Garcia et al., 2010) and Vernonia (Salles-de-Melo et al., 2010) from the subfamily Cichorioideae. However, despite the prevalence of terminal NORs in karyotypes of angiosperms as a whole (Roa and Guerra, 2012), both C. endivia and C. intybus had only proximal NORs. Similarly, proximal NORs have been reported in karyotypes of Achyrocline spp. (subfamily Asteroideae) Hypochaeris spp. (subfamily Cichorioideae), which may also be evidences of structural changes (such as paracentric inversions) throughout chromosome evolution within these plant groups (Cerbah et al., 1998; Ruas et al., 2005; Mazzella et al., 2010).
The co-localization of CMA/DAPI bands and 45S rDNA sites in the analysed species revealed the presence of CMA++ heterochromatin associated with NORs. This association has also been previously described for Asteraceae (Fregonezi et al., 2004; Mazzella et al., 2010; Salles-de-Melo et al., 2010) and has been shown to be quite frequent within angiosperms (Guerra, 2000). On the other hand, the occurrence of CMA+ bands in association with 5S rDNA sites in the family has been mainly associated with the co-localization of 45S and 5S rDNA sites, as observed for species of Achyrocline (Mazzella et al., 2010) and Artemisia (Pellicer et al., 2008; Konowalik et al., 2010). Moreover, as occurred for both species herein analysed, Hypochaeris catharinensis Cabrera also showed co-localization of CMA+ bands and 5S rDNA (Reck et al., 2011), a feature described for other higher plants (e.g.Cabral et al., 2006; Vasconcelos et al., 2010).
In C. intybus var. foliosum cv. Zoom, an additional, more intense band (CMA++/DAPI−) associated with the 5S rDNA site was observed, suggesting an amplification of this site, a fact confirmed by a higher intensity of both CMA staining and FISH signals. On the other hand, the lack of CMA-positive bands in the 5S rDNA site of the chromosome 8 of C. endivia may probably be related to either a considerably lower number of rDNA repetitions or a distinct GC content in the intergenic sequences Additionally, epigenetic changes in chromatin conformation due to cytosine methylation or post-translational histone changes could affect the CMA association (see Cabral et al., 2006, and references therein).
In contrast to the published results for Lactuca species in which the number of rDNA sites were very stable (two pairs with 45S and one with 5S rDNA; Matoba et al., 2007), we observed a slight difference regarding the number of chromosome pairs bearing 45S and 5S rDNA sites between the analysed species (three pairs with 45S and two with 5S rDNA for C. endivia, and two pairs with 45S and one with 5S rDNA for C. intybus). Similarly to the Cichorium species, Tragopogon species (Cichorieae; with 2n = 2x = 12) also exhibited variation regarding the number of 45S rDNA sites among T. dubius Scop. and T. pratensis L. (both with one pair of sites) and T. porrifolius L. (two pairs) (Pires et al., 2004).
Contrary to the similarity among different C. endivia accessions, the two distinct karyotypes noticed among C. intybus accessions indicate the occurrence of recent chromosome changes involving rDNA sites in cultivated varieties. The occurrence of an extra 5S rDNA site in one chromosome of pair 3 of C. intybus var. intybus may be an indication of non-homologous recombination between chromosome pairs 3 and 8, followed by amplification of 5S rDNA repetitive motifs at the new location. On the other hand, the presence of two heteromorphic pairs (3 and 8) in the karyotype of C. intybus var. foliosum cv. Zoom seem to be the result of a reciprocal translocation with breakages in the centromeres (centric fission/fusion) involving a homologue of each pair (Figure 2E,F). Besides the detected changes, other rearrangements may have occurred, including the amplification of the heterochromatin that was co-localized with the 45S rDNA in the first homologue of the pair 3 and the 5S rDNA sites, and the reduction of part of the second homologue chromosome of pair 8, possibly by means of deletion related or not to the translocation process. Additionally, a different morphology was observed in the submetacentric pair 7 and the first homologue of pair 8 (both with a larger long arm).
The present results uncovered interspecific differences between C. endivia and C. intybus, besides the occurrence of some unexpected intraspecific karyotypic variations among C. intybus accessions. The identified features represent the first pieces of evidence regarding the distribution of the main chromosome markers within this genus, revealing high diversity in the apparently homogeneous karyotypes previously described after standard staining.
Acknowledgments
The authors thank to the researchers Brunno Leite, Ana Rafaela Oliveira, Geyner Alves and Kyria Bortoleti for relevant suggestions during the execution of this work. For the concession of valuable germplasm accessions we thank the Institut für Pflanzengenetik und Kulturpflanzen-forschung Gatersleben (IPK), Germany. The present study was financially supported by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil) and FACEPE (Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco, Brazil).
Footnotes
Associate Editor: Marcelo Guerra
References
- Benko-Iseppon AM, Morawetz W. Cytological comparison of Calyceraceae and Dipsacaceae with special reference to their taxonomic relationships. Cytologia. 2000;65:123–128. [Google Scholar]
- Bremer K. Asteraceae: Cladistics and Classification. Timber Press; Portland: 1994. p. 752. [Google Scholar]
- Cabral JS, Felix LP, Guerra M. Heterochromatin diversity and its co-localization with 5S and 45S rDNA sites in chromosomes of four Maxillaria species (Orchidaceae) Genet Mol Biol. 2006;29:659–664. [Google Scholar]
- Cerbah M, Souza-Chies T, Jubier MF, Lejeune B, Siljak-Yakovlev S. Molecular phylogeny of the genus Hypochaeris using internal transcribed spacers of nuclear rDNA: Inference for chromosomal evolution. Mol Biol Evol. 1998;15:345–354. doi: 10.1093/oxfordjournals.molbev.a025931. [DOI] [PubMed] [Google Scholar]
- Dobes C, Hahn B, Morawetz W. Chromosomenzahlen zur Gefäβpflanzen-Flora Österreichs. Linzer Biol Beitr. 1997;29:5–43. [Google Scholar]
- Fregonezi JN, Torezan JMD, André LL, Vanzela ALL. A karyotypic study of three southern Brazilian Asteraceae species using fluorescence in situ hybridization with a 45S rDNA probe and C-CMA3 banding. Genet Mol Biol. 2004;27:223–227. [Google Scholar]
- Garcia S, Panero JL, Siroky J, Kovarik A. Repeated reunions and splits feature the highly dynamic evolution of 5S and 35S ribosomal RNA genes (rDNA) in the Asteraceae family. BMC Plant Biol. 2010;10:e176. doi: 10.1186/1471-2229-10-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra MS. Reviewing the chromosome nomenclature of Levan et al. Braz J Genet. 1986;9:741–743. [Google Scholar]
- Guerra M. Patterns of heterochromatin distribution in plant chromosomes. Genet Mol Biol. 2000;23:1029–1041. [Google Scholar]
- Heslop-Harrison JS, Schwarzacher T, Anamthawat-Jonsson K, Leitch AR, Shi M, Leitch IJ. In situ hybridization with automated chromosome denaturation. Technique. 1991;3:109–116. [Google Scholar]
- Jiang J, Hulbert SH, Gill BS, Ward DC. Interphase fluorescence in situ hybridization mapping: A physical mapping strategy for plant species with large complex genomes. Mol Gen Genet. 1996;252:497–502. doi: 10.1007/BF02172395. [DOI] [PubMed] [Google Scholar]
- Kiers AM, Mes THM, Meijden RVD, Bachmann K. A search for diagnostic AFLP markers in Cichorium species with emphasis on endive and chicory cultivar groups. Genome. 2000;43:470–476. doi: 10.1139/g00-024. [DOI] [PubMed] [Google Scholar]
- Kilian N, Gemeinholzer B. Studies in the Compositae of the Arabian Peninsula and Socotra - 7. Erythroseris, a new genus and the previously unknown sister group of Cichorium (Cichorieae subtribe Cichoriinae) Willdenowia. 2007;37:283–296. [Google Scholar]
- Kilian N, Gemeinholzer B, Lack HW. Cichorieae. In: Funk VA, Susanna A, Stuessy TE, Bayer RJ, editors. Systematics, Evolution and Biogeography of Compositae. IAPT; Vienna: 2009. pp. 343–383. [Google Scholar]
- Konowalik K, Garcia S, Pellicer J, Kreitschitz A, Vallès J. Cytogenetic characterisation of Artemisia absinthium (Asteraceae, Anthemidae) and its Polish endemic var. calcigena. Ann Bot Fenn. 2010;47:477–488. [Google Scholar]
- Lövkvist B, Hultgård UM. Chromosome numbers in south Swedish vascular plants. Opera Bot. 1999;137:1–42. [Google Scholar]
- Lucchin M, Varotto S, Barcaccia G, Parrini P. Chicory and endive. In: Prohen J, Nuez F, editors. Vegetables I. Springer; New York: 2008. pp. 3–48. [Google Scholar]
- Matoba H, Mizutani T, Nagano K, Hoshi Y, Uchiyama H. Chromosomal study of lettuce and its allied species (Lactuca spp., Asteraceae) by means of karyotype analysis and fluorescence in situ hybridization. Hereditas. 2007;144:235–243. doi: 10.1111/j.2007.0018-0661.02012x. [DOI] [PubMed] [Google Scholar]
- Mazzella C, Rodríguez M, Vaio M, Gaiero P, López-Carro B, Santiñaque FF, Folle GA, Guerra M. Karyological features of Achyrocline (Asteraceae, Gnaphalieae): Stable karyotypes, low DNA content variation and linkage of rRNA genes. Cytogenet Genome Res. 2010;128:169–176. doi: 10.1159/000290689. [DOI] [PubMed] [Google Scholar]
- Montmollin BD. Etude cytotaxonomique de la flore de la Crète. III. Nombres chromosomiques. Candolle. 1986;41:431–439. [Google Scholar]
- Pedrosa A, Sandal N, Stougaard J, Schweizer D, Bachmair A. Chromosomal map of the model legume Lotus japonicus. Genetics. 2002;161:1661–1672. doi: 10.1093/genetics/161.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicer J, Garcia S, Garnatje T, Hidalgo O, Siljak-Yakovlev S, Vallès J. Molecular cytogenetic characterization of some representatives of the subgenera Artemisia and Absinthium (genus Artemisia, Asteraceae) Collect Bot. 2008;27:19–27. [Google Scholar]
- Pignatti S. Flora d’Italia. Vol. 3. Edagricole; Bologna: 1982. p. 780. [Google Scholar]
- Pires JC, Lim KY, Kovarik A, Matyasek R, Boyd A, Leitch AR, Leitch IJ, Bennett MD, Soltis PS, Soltis DE. Molecular cytogenetic analysis of recently evolved Tragopogon (Asteraceae) allopolyploids reveal a karyotype that is additive of the diploid progenitors. Am J Bot. 2004;91:1022–1035. doi: 10.3732/ajb.91.7.1022. [DOI] [PubMed] [Google Scholar]
- Reck M, Benício L, Ruas EA, Rodrigues LA, Ruas PM, Ortiz MA, Talavera S, Urtubey E, Stuessy T, Weiss-Schneeweiss H, et al. Karyotype and AFLP data reveal the phylogenetic position of the Brazilian endemic Hypochaeris catharinensis (Asteraceae) Plant Syst Evol. 2011;296:231–243. [Google Scholar]
- Reeves A. MicroMeasure: A new computer program for the collection and analysis of cytogenetic data. Genome. 2001;44:439–443. [PubMed] [Google Scholar]
- Rick CM. Hybridization between chicory and endive. P Am Soc Hortic Sci. 1953;61:459–466. [Google Scholar]
- Roa F, Guerra M. Distribution of 45S rDNA sites in chromosomes of plants: Structural and evolutionary implications. BMC Evol Biol. 2012;12:e225. doi: 10.1186/1471-2148-12-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruas CF, Vanzela ALL, Santos MO, Fregonezi JN, Ruas MP, Matzenbacher NI, Aguiar-Perecin MLR. Chromosomal organization and phylogenetic relationships in Hypochaeris species (Asteraceae) from Brazil. Genet Mol Biol. 2005;28:129–139. [Google Scholar]
- Salles-de-Melo MRC, Lucena RM, Semir J, Carvalho R, Pereira RCA, Benko-Iseppon AM. Karyological features and cytotaxonomy of the tribe Vernonieae (Asteraceae) Plant Syst Evol. 2010;285:189–199. [Google Scholar]
- Schweizer D, Ambros PF. Chromosome banding. In: Godsen JR, editor. Methods in Molecular Biology. Vol. 29. Humana Press; Totowa: 1994. pp. 97–112. [DOI] [PubMed] [Google Scholar]
- Tutin TG, Heywood VH, Burges NA, Moore DM, Valentine DH, Walters SM, Webb DA. Flora Europaea. Vol. 4. Cambridge University Press; Cambridge: 1976. p. 534. [Google Scholar]
- Vanzela ALL, Ruas CF, Oliveira MF, Ruas PM. Characterization of diploid, tetraploid and hexaploid Helianthus species by chromosome banding and FISH with 45S rDNA probe. Genetica. 2002;114:105–111. doi: 10.1023/a:1015171625890. [DOI] [PubMed] [Google Scholar]
- Vasconcelos S, Souza AA, Gusmão CLS, Milani M, Benko-Iseppon AM, Brasileiro-Vidal AC. Heterochromatin and rDNA 5S and 45S sites as reliable cytogenetic markers for castor bean (Ricinus communis, Euphorbiaceae) Micron. 2010;41:746–753. doi: 10.1016/j.micron.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Wagenitz G, Bedarff U. Taxonomic notes on some species of the genus Cichorium (Compositae-Lactuceae) In: Tan K, editor. Plant Ttaxonomy, Phytogeography and Related Subjects. The Davis & Hedge Festschrift; Edinburgh: 1989. pp. 11–21. [Google Scholar]
- Wanzenböck EM, Schöfer C, Schweizer D, Bachmair A. Ribosomal transcription units integrated via T-DNA transformation associate with the nucleolus and do not require upstream repeat sequences for activity in Arabidopsis thaliana. Plant J. 1997;11:1007–1016. doi: 10.1046/j.1365-313x.1997.11051007.x. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Yahara T, Hashimoto G, Nagatani Y, Soejima A, Kawahara T, Nakazawa M. Chromosome numbers and karyotypes in Asteraceae. Ann Mo Bot Gard. 2007;94:643–654. [Google Scholar]