Abstract
The identification of genetically homogeneous groups of individuals is an ancient issue in population genetics and in the case of crops like wheat, it can be valuable information for breeding programs, genetic mapping and germplasm resources. In this work we determined the genetic structure of a set of 102 Argentinean bread wheat (Triticum aestivum L.) elite cultivars using 38 biochemical and molecular markers (functional, closely linked to genes and neutral ones) distributed throughout 18 wheat chromosomes. Genetic relationships among these lines were examined using model-based clustering methods. In the analysis three subpopulations were identified which correspond largely to the origin of the germplasm used by the main breeding programs in Argentina.
Keywords: wheat, genetic structure, molecular markers, genetic diversity
Introduction
Bread wheat (Triticum aestivum L.) is the principal winter crop grown in Argentina for both internal consumption and export. Genetically improved cultivars and agricultural practices have resulted in an increased average wheat yield during the past 41 years, from 1352 kg/ha in 1969 to more than 3500 kg/ha in 2010. To maintain this rate of wheat productivity, exploring the genetic variability at molecular levels in adaptation and yield components and integrating such information with conventional breeding methods will be critical (Chao et al., 2007). The identifications of genomic regions associated with relevant agronomic traits through QTL mapping using bi-parental populations, can now be complemented with alternative genetic mapping strategies like Association Mapping, in which the accurate determination of the population genetic structure is important for the appropriate association between the genotype and the phenotype (Breseghello and Sorrells, 2006; Peng et al., 2009; Le Couviour et al., 2011).
In wheat, the assessment of genetic structure in different populations has been historically based on qualitative and quantitative traits (Spagnoletti-Zeuli and Qualset, 1987; Van Beuningen and Busch, 1997b) and pedigree records (Burkhamer et al., 1998; Van Beuningen and Busch, 1997a; Bered et al., 2002). However, pedigree records are not always available or detailed enough for this type of analysis, especially when large numbers of breeding lines or cultivars are being assessed. More recently, biochemical markers such as variation in storage protein subunits (Metakovsky and Branlard, 1998; Lerner et al., 2009) and/or molecular markers like RAPDs, RFLPs, AFLPs, SSRs, DArTs and SNPs become alternative methods of obtaining a large amount of data for precisely calculating genetic relationship estimates (Mukhtar et al., 2002; Parker et al., 2002; Soleimani et al., 2002; Neumann et al., 2011; Chao et al., 2010). Particularly in Argentina the levels and patterns of genetic diversity among local wheat cultivars have been investigated using SSRs (Manifesto et al., 2001) and storage proteins (Lerner et al., 2009).
Frequently, the biochemical and/or molecular information has been analyzed using tree-based methods that calculate genetic distance between individuals and tree construction algorithms such as UPGMA or neighbor joining to group them in clusters (Sneath and Sokal, 1973; Saitou and Nei, 1987). An alternative model-based method developed recently by Pritchard et al. (2000) and implemented in the software Structure aims at delineating clusters of individuals on the basis of their genotypes at multiple loci using a Bayesian approach (Breseghello and Sorrells, 2006; Chao et al., 2010). However Evanno et al. (2005) demonstrated that in most cases the estimated “log probability of data” used in Structure fails to provide a correct estimation of the number of clusters, (K). Hence, they developed an ad hoc statistic ΔK based on the rate of change in the log probability of data between successive K values, and found that Structure accurately detects the uppermost hierarchical level of structure. Based on these parameters, Earl and von Holdt (2012) developed Structure Harvester, a website and software for visualizing the output based on Evanno et al. (2005).
In this report, we determined the genetic structure of a set of Argentinean wheat cultivars using a model-based approach. 102 cultivars representative of the main breeding companies in Argentina were characterized using a set of 38 biochemical and molecular markers, each mapped to a single chromosome location and distributed over 18 of the 21 wheat chromosomes.
Materials and Methods
Plant material
A set of 102 bread wheat cultivars registered in Argentina was selected to determine its genetic structure based on molecular and biochemical markers. This set included old and recent commercial cultivars selected from the main wheat breeding companies in Argentina. Seed stocks were kindly provided by the Instituto Nacional de Tecnología Agropecuaria (INTA) Marcos Juárez Wheat Germplasm Bank (Marcos Juárez, Argentina).
Genotypic data
For each accession, genomic DNA was extracted from fresh leaves of single plants using a fast, small-scale DNA isolation procedure based on Helguera et al. (2005). Sample genotyping included: (1) functional markers Vrn-A1 (Yan et al., 2004), Vrn-B1, Vrn-D1 (Fu et al., 2005), Ppd-D1 (Beales et al., 2007), Rht-B1, Rht-D1 (Ellis et al., 2002), PinA-D1 (Gautier et al., 1994), Glu-A3 (Zhang et al., 2004), Wx-A1 and Wx-B1 (McLauchlan et al., 2001, Vanzetti et al., 2009), Vp1-B3 (Yang et al., 2007), Ppo-A1 and Ppo-D1 (He et al., 2007), 7oe+8* (Butow et al., 2004); (2) molecular markers closely linked to genes Lr10 (Schachermayr et al., 1997), Lr34 (Lagudah et al., 2006), Lr24 (Schachermayr et al., 1995) and Ppd-B1 (Díaz et al., 2012); and (3) 17 SSR markers selected according to information available in the GrainGenes database (Matthews et al., 2003) and one ISBP marker newly developed for this work. In all cases, polymerase chain reactions (PCR) were performed in 25 μL aliquots in a PTC-100 thermal cycler (MJ Research Inc., Waltham, MA, USA). The reaction contained 100 ng of genomic DNA (template), 1X Taq polymerase buffer (Promega Corp. Madison, WI, USA), 1 U Taq DNA polymerase (Promega), 0.2 mM of each deoxynucleotide, 0.2 μM of each primer, and 1.5 mM of MgCl2. Primers names, sequences and cycling conditions for each molecular marker are detailed in Table S1. SSR markers were run on 6% non-denaturing polyacrylamide gels in 0.5X TBE buffer using a Mega-Gel Dual High-Throughput Vertical Electrophoresis Unit (CBS Scientific Co, Del Mar, CA, USA), stained with ethidium bromide (0.5 g/L) and visualized under UV light. In the case of functional genes and genes closely linked to molecular markers, 10 μL of the PCR products were run on 2% agarose (Promega) gels in SB buffer (Brody and Kern, 2004) at constant power (100 V) for about 30 min. After electrophoresis, the gels were stained with ethidium bromide (0.5 g/L) and visualized under UV light.
Glutenin analysis
Glutenins were extracted from single seeds and analyzed by SDS-PAGE according to protocols described by Pflüger et al. (2001). Glu-A1, Glu-B1 and Glu-D1 subunits were analyzed by SDS-PAGE in 8% polyacrylamide gels (16×18 cm) in a Hoefer electrophoresis system (Hoefer Inc. Holliston, MA, USA) at 30 mA/gel for approximately 12 h. The gels were stained with 0.2% (w/v) Coomassie Blue R-250 (Promega), in 5% (v/v) ethanol and 12% (w/v) trichloroacetic acid overnight and destained in tap water for 24 h.
Allele diversity
All cultivars were treated as pure lines. A small proportion of heterozygosity was observed, and the following criteria were used to define the working allele. In the case of SSRs, where cultivars displayed two bands with different intensities, only the stronger band was considered. Yet, if the two bands showed similar intensities, then the most frequent allele was considered. If none of these options could be applied, the sample was scored as missing data. In the case of biochemical (glutenins) and functional molecular markers, samples showing heterozygous alleles were scored as missing data. Rare alleles (with frequency lower than 5%) were treated as missing data for population structure. The effective number of alleles per locus was computed on the basis of common alleles as ne = 1/∑pi2 (Hartl and Clark, 1997). The estimate ne represents the number of equally frequent alleles that would result in the same probability observed when randomly drawing two different alleles from the population. It is a measure of variability at the locus that takes into account both allele number and frequency. The polymorphism index content (PIC), a measure of allelic diversity, was calculated according to Nei’s coefficient (Nei, 1973), PIC = 1-∑(pi2), where pi is the frequency of the ith polymorphism detected in the germplasm.
Population structure
Thirty-eight unlinked or distantly linked marker loci, distributed over all the wheat chromosomes, except 6A, 6B and 7B, were used for assessment of population structure. Population structure was investigated using a Bayesian clustering approach to infer the number of clusters (populations) with the softwares Structure v.2.3.3 (Pritchard et al., 2000) and Structure Harvester (Evanno et al., 2005; Earl and von Holdt, 2012). No prior information was used to define the clusters, and the number of subpopulations (K) was set from 1 to 10, without admixture and with correlated allele frequencies, burn-in phase of 105 iterations, and a sampling phase of 2 × 105 replicates, runs with K = 1 to 10 were repeated 10 times (Falush et al., 2003; Breseghello and Sorrells, 2006). This method estimates the proportion of the genomes of each individual derived from the different clusters and assigns individuals to subpopulations based on membership probability. We used the run that assigned all the cultivars to a single cluster at a probability > 0.50. The degree of differentiation of each subpopulation was measured by a modified FST parameter (Falush et al., 2003). The program Genetix (Belkhir et al., 1996–2004) was used to compute an overall FST (Weir and Cockerham, 1984) and to conduct multiple correspondence analysis, with three dimensions.
Results
The genetic diversity of a collection of 102 bread wheat cultivars from Argentina was assessed using 35 molecular markers (13 functional markers, 4 markers closely linked to genes, 17 SSR, and 1 ISBP) and 3 storage proteins (Table 1). A total number of 124 alleles was detected in the panel, including 21 rare alleles (with frequencies lower than 5% in the panel) that were discarded in the population structure studies. The number of alleles per locus varied between two and seven, with an average of 3.26. The average numbers of common (excluding rare alleles in the analysis) and effective alleles (ne) were 2.65 (from 1 to 6) and 2.05 (from 1.01 to 4.76), respectively. Polymorphism information content (PIC) values obtained from the 38 polymorphic markers varied between 0.076 and 0.788, with an average of 0.458. The mean frequency of missing data was 0.43%, or 1.65% when rare alleles were included (Table 1).
Table 1.
Biochemical and molecular markers used in the study, chromosome location, number of alleles, number of common alleles, effective allele number and polymorphic information content (for details see Supplementary Tables S2 and S3).
| N | Locus | Marker1 | Chr. | AN | NCA | ne | PIC |
|---|---|---|---|---|---|---|---|
| 1 | Glu-A1 | B | 1A | 4 | 3 | 2.19 | 0.554 |
| 2 | Glu-B1 | B | 1B | 7 | 6 | 3.30 | 0.696 |
| 3 | Glu-D1 | B | 1D | 3 | 2 | 1.01 | 0.162 |
| 4 | Vrn-A1 | F | 5A | 3 | 3 | 2.18 | 0.541 |
| 5 | Vrn-B1 | F | 5B | 2 | 2 | 1.98 | 0.495 |
| 6 | Vrn-D1 | F | 5D | 2 | 2 | 1.95 | 0.487 |
| 7 | Ppd-D1 | F | 2D | 2 | 2 | 2.01 | 0.512 |
| 8 | Rht-B1 | F | 4B | 2 | 2 | 1.88 | 0.467 |
| 9 | Rht-D1 | F | 4D | 2 | 2 | 1.71 | 0.415 |
| 10 | PinA-D1 | F | 5D | 2 | 2 | 1.99 | 0.496 |
| 11 | Glu-A3 | F | 1A | 7 | 6 | 4.76 | 0.788 |
| 12 | Wx-A1 | F | 7A | 2 | 2 | 1.10 | 0.093 |
| 13 | Wx-B1 | F | 4A | 3 | 2 | 1.34 | 0.304 |
| 14 | Vp1-B3 | F | 3B | 4 | 4 | 2.94 | 0.555 |
| 15 | Ppo-D1 | F | 2A | 2 | 2 | 1.77 | 0.488 |
| 16 | Ppo-A1 | F | 2D | 2 | 2 | 1.54 | 0.364 |
| 17 | Lr10 | CL | 1A | 3 | 3 | 2.36 | 0.584 |
| 18 | Lr24 | CL | 3D | 2 | 2 | 1.17 | 0.144 |
| 19 | Lr34 | CL | 7D | 2 | 2 | 1.46 | 0.315 |
| 20 | Ppd-B1 | CL | 2B | 2 | 2 | 1.54 | 0.349 |
| 21 | Xgwm124 | N | 1B | 4 | 1 | 1.08 | 0.076 |
| 22 | Xgwm493 | N | 3B | 4 | 3 | 2.68 | 0.641 |
| 23 | Xgwm295 | N | 7D | 4 | 2 | 1.27 | 0.274 |
| 24 | Xgwm265 | N | 2A | 3 | 2 | 1.74 | 0.436 |
| 25 | Barc174 | N | 7A | 5 | 5 | 2.78 | 0.646 |
| 26 | Xgwm261 | N | 2D | 4 | 4 | 1.88 | 0.468 |
| 27 | Xgwm219 | N | 6B | 3 | 3 | 2.51 | 0.602 |
| 28 | Xgwm374 | N | 2B | 2 | 2 | 1.56 | 0.359 |
| 29 | Xgwm156 | N | 5A | 4 | 4 | 2.39 | 0.581 |
| 30 | Barc70 | N | 7A | 6 | 4 | 3.86 | 0.747 |
| 31 | Xwmc147 | N | 1D | 2 | 2 | 2.00 | 0.508 |
| 32 | Xgwm111 | N | 7D | 5 | 2 | 1.39 | 0.386 |
| 33 | Xgwm113 | N | 4B | 2 | 2 | 1.61 | 0.379 |
| 34 | Xgwm264 | N | 3B | 3 | 3 | 2.58 | 0.612 |
| 35 | Xgwm533 | N | 3D | 4 | 2 | 1.35 | 0.315 |
| 36 | Wms5 | N | 3A | 4 | 4 | 3.39 | 0.704 |
| 37 | Xwmc44 | N | 1B | 5 | 3 | 2.07 | 0.561 |
| 38 | Cs37 | N | 4D | 2 | 2 | 1.44 | 0.303 |
B: Biochemical, F: Functional, CL: Closely linked to genes, N: Neutral.
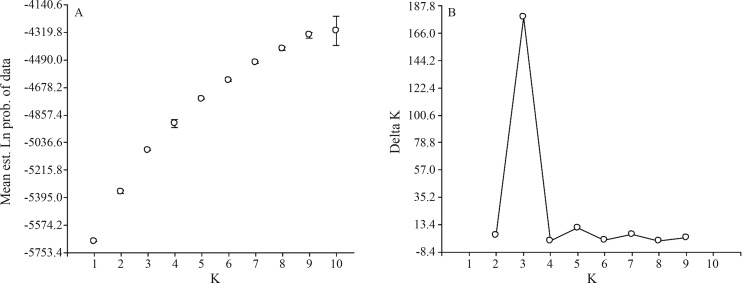
We explored the population genetic structure among the accessions using a model-based method. A model-based method is a cluster analysis that evaluates genetic similarity among genotypes without using prior information. After a first analysis with the Structure program we could not determine precisely the K number (number of subgroups or subpopulations) in the population, as the curve of the Ln probability of data [L(K)], did not generate a plateau after K = 10 (Figure 1A). Therefore, we used the output of Structure as input data for Structure Harvester, now obtaining a clear peak with the highest ΔK value at K = 3 (Figure 1B). The analysis showed that three subpopulations were optimal for assigning all except 13 cultivars into one among the three clusters, with an a posteriori probability > 0.80. The 13 genotypes assigned to individual clusters with an a posteriori probability > 0.50 (but < 0.80) are underlined in Table 2. The three subpopulations K1, K2 and K3 included 17, 48 and 37 cultivars, respectively, with FST averages slightly higher in K1 (0.1939) than in K2 and K3 (0.1279 and 0.1218, respectively), this evidencing a moderated differentiation within subgroups. Furthermore, the FST value across subpopulations was 0.1485, indicating a moderate differentiation also between subgroups.
Figure 1.
Population genetic structure analysis of Argentinian wheat germplasm. (A) Mean L(K) (±SD) over 10 runs for each K value. The model considered here is a hierarchical island model using all 102 individuals per population and 38 loci. (B) ΔK calculated as ΔK = m|L″(K)| /s[L(K)]. The modal value of this distribution is the true K or the uppermost level of structure, here 3 clusters.
Table 2.
Distribution of 102 Argentinean wheat cultivars in the three subpopulations detected using a model-based approach.
| K | Cultivar name* |
|---|---|
| 1 | Baguette 10, Baguette 17, Baguette 18, Baguette 19, Baguette 21, Baguette 30, Baguette 31, Baguette 9, Baguette P. 11, BSYN 100, BSYN 200, BSYN 300, Klein 32, Klein Atlas, Klein Centauro, Sinvalocho, SRN Nogal |
| 2 | ACA 223, ACA 801, ACA 901, ACA 906, ACA 907, BIOINTA 1001, BIOINTA 1003, BIOINTA 1004, BIOINTA 1005, BIOINTA 1006, BIOINTA 3004, Buck AGP-FAST, Buck Huanchen, Buck Puelche, Don Mario Arex, Don Mario Atlax, Don Mario Cronox, Don Mario Onix, INIA Centinela, INIA Churrinche, INIA Torcaza, Klein Brujo, Klein Cacique, Klein Capricornio, Klein Carpincho, Klein Castor, Klein Don Enrique, Klein Escorpión, Klein Gavilán, Klein Gladiador, Klein Guerrero, Klein León, Klein Pantera, Klein Proteo, Klein Rayo, Klein Tauro, Klein Tigre, Klein Yarará, Klein Zorro, LE 2330, LE 2331, LE 2333, LE 2341, ProINTA Elite, ProINTA Guazú, ProINTA Isla Verde, ProINTA Oasis, Relmo Sirirí |
| 3 | ACA 201, ACA 202, ACA 320, ACA 903B, Barletta 77, BIOINTA 1000, BIOINTA 1002, BIOINTA 2001, BIOINTA 2004, BIOIN-TA 3003, BIOINTA 3005, Buck 55CL, Buck Aniversario, Buck Baqueano, Buck Bigua, Buck Brasil, Buck Chacarero, Buck Guapo, Buck Malevo, Buck Mangrullo, Buck Meteoro, Buck Napostá, Buck Norteño, Buck Pingo, Buck Pronto, Buck Ranquel, Buck Taita, Don Mario Themix, INIA Cóndor, Klein Chajá, Klein Impacto, Klein Nutria, Klein Rendidor, Marcos Juárez INTA, Olaeta Artillero, ProINTA Gaucho, ProINTA Granar |
Underlined cultivars were assigned to individual clusters with a p > 0.50, cultivars not underlined were assigned with a p > 0.80.
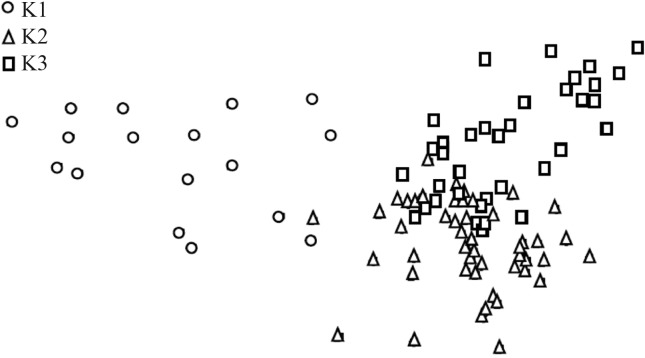
Figure 2 shows the projection of the multiple correspondence analysis (MCA) cloud on one orthogonal plane, with different symbols identifying each subpopulation according to the classification from Structure. The cloud was continuous, with three protrusions corresponding to the three subpopulations. In agreement with FST estimates, sub-populations 2 and 3 were less dispersed than 1.
Figure 2.
Orthogonal projection of the cloud of points representing the genetic distance among cultivars, based on 38 unlinked molecular markers analyzed by multiple correspondence analysis. Subpopulations K1-K3 were inferred using Structure harvester.
Discussion
Genetic variability of the panel
The means for allele numbers and PIC values observed across all markers used in this work were 3.26 and 0.458 respectively (Table 1). These values were considerably lower than those observed for SSR loci (9.4 and 0.720; Manifesto et al., 2001) and for storage proteins (5.00 and 0.544; Lerner et al., 2009) for different but overlapping collections of wheat cultivars from Argentina. The mean allele number detected herein was also significantly lower than those revealed by SSR analysis in US wheat germplasm (4.8 and 7.2 alleles), reported by Breseghello and Sorrells (2006) and Chao et al. (2007) respectively. It was also lower than the 6.20 and 7.49 alleles observed by Plaschke et al. (1995) and Le Couviour et al. (2011) in European wheat germplasm, and the 5.4 alleles detected by Dreisigacker et al. (2004) in CIMMYT germplasm. Finally, Balfourier et al. (2007) working with a worldwide wheat collection of 3,942 entries from 73 countries detected the very high average value of 23.9 alleles per locus. These large differences in the number of alleles detected may be due to differences in the technologies used to detect polymorphism, as well as the type of molecular markers selected for the characterization (and/or the rate and amount of the germplasm evaluated).
Markers associated with traits of agronomic interest
In our study we selected a set of markers (biochemical, functional markers and closely linked to genes markers) related with relevant traits for breeding, like growth habit and/or vernalization response (Vrn-A1, Vrn-B1 and Vrn-D1), photoperiod sensitivity (Ppd-D1), plant height (Rht-B1, Rht-D1), grain texture (PinA-D1), starch waxy proteins variants (Wx-A1 and Wx-B1), PPO activity (Ppo-A1, Ppo-D1), variants of the Viviparous-1B gene (Vp1-B3) associated with pre-harvest tolerance (Yang et al., 2007), low molecular weight glutenins (Glu-A3) and high molecular weight glutenins (Glu-A1, Glu-B1, Glu-D1)
The natural variation scanned with markers based on winter/spring allelic variants from Vrn-A1, Vrn-B1 and Vrn-D1 loci confirmed the spring growth habit as the best adapted for the wheat production area of Argentina (91 of the tested cultivars carried at least one spring allele, considering Vrn-A1, Vrn-B1 and Vrn-D1 loci, vs. 11 cultivars with the triple winter alleles combination). These data agree with previous phenotypic (Appendino et al., 2003) and molecular data (Fu et al., 2005). We also noticed a higher frequency of the photoperiod insensitive (PI) alleles Ppd-D1a and/or Ppd-B1a alleles (74 cultivars) than the combination of Ppd-D1b and Ppd-B1b alleles associated with photoperiod sensitivity (PS) (28 cultivars), this suggesting a better adaptation of photoperiod insensitivity to the environmental conditions in Argentina (between 27° and 38° S). A high frequency of PI alleles was also observed in low latitude regions of Japan (36° N), associated also in this case with early flowering to avoid rains at harvest and preharvest sprouting (Seki et al., 2011). Unlike this situation, Lanning et al. (2012) evaluated PS and PI spring near-isogenic lines (NILs) and observed better agronomic perfomance in PS NILs planted at higher latitudes (between 45° and 54° N) and considering early planting dates, as no difference between PS and PI lines occurred for the latest planting date. In the case of the dwarfing genes Rht-B1 and Rht-D1, the most frequent allelic combinations were semi-dwarf phenotypes (37 Rht-B1b/Rht-D1a and 29 Rht-B1a/Rht-D1b) followed by tall phenotypes (35 Rht-B1a/Rht-D1a) and finally, only one dwarf phenotype (Rht-B1b/Rht-D1b). The reduced height alleles Rht-B1b and Rht-D1b are incorporated in breeding programs to reduce lodging (Flintham et al., 1997) and increase the harvest index (Gooding et al., 2012) under favorable environments, including problably, most fertile environments in Argentina.
These data would support a better adaptation of spring, photoperiod insensitive and semidwarf wheats to dominant environments in Argentina, however, a fine tuning evaluation of spring NILs carrying different combinations of vernalization, photoperiod insensitivity, as well as plant height alleles is still a pending issue.
In the case of markers closely linked to Lr genes, the most valuable information is perhaps, the relatively high number of cultivars that probably possess the adult plant leaf rust resistance gene Lr34 (20 cultivars), a finding which agrees with Vanzetti et al. (2011). This gene has supported resistance to leaf rust in wheat for more than fifty years and is extensively used in breeding programs worldwide (Krattinger et al., 2009).
Relevant information for bread-making quality can be the presence of the Glu-B1 7oe subunit associated with improved dough strength of wheat (Butow et al., 2004) in ten cultivars. Valuable alleles for the development of cultivars with superior bread quality, partial waxy wheats, low PPO activity, and pre-harvest sprouting tolerance were also detected in the panel.
K = 3 is associated with the main breading programs in Argentina
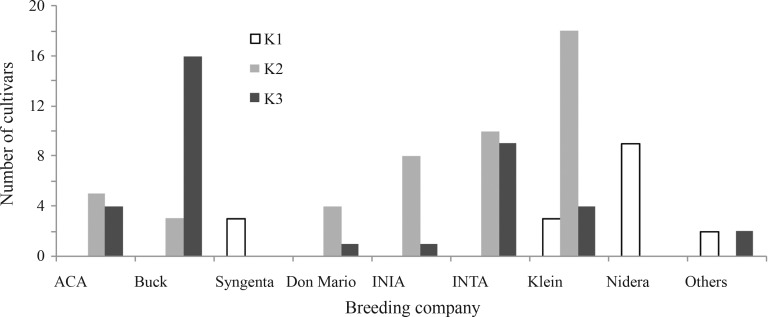
In this work, using a model-based approach we detected three subpopulations in the collection of 102 Argentinean wheat cultivars. Our hypothesis is that this subpopulation division actually reflects the origin of the germplasm used by the main breeding programs in Argentina. For example, K1 is composed mainly (70.58%) by cultivars from Nidera and Syngenta breeding programs, as 100% of the cultivars tested and released by these companies were grouped only in K1. It is worthy of note that this germplasm (at least early materials released by Nidera (Bulos et al., 2006), has a European origin, mainly from France, and was introduced gradually to Argentina since 1999. The K1 subpopulation also includes old cultivars from the Klein breeding program, like Klein 32 (released in 1932), Klein Atlas (1963) and Klein Centauro (1989) (Figure 3). K2 is mainly composed (60.41%) by cultivars from Klein, INIA and Don Mario breeding programs, the 72% of Klein, 87.5% of INIA and 80% of Don Mario tested cultivars were included in K2 (Figure 3). The cultivars grouped in K2 are basically (1) introductions and selections made in CIMMYT, as well as crosses made in Argentina, including CIMMYT material like Bobwhite, Kavkaz, Pastor, Seri, Veery and Weebill, (2) introductions from Brazil (some materials from Don Mario Breeding Company), and (3), to a lesser degree, materials selected from crosses including traditional germplasm from Argentina. Finally, K3 is composed mostly by cultivars belonging to the Buck Breeding Program (43.24%). INTA and ACA Breeding Programs have an even distribution of their tested cultivars between the K1 and K2 subpopulations (Figure 3). The cultivars grouped in K3 are mostly derived from traditional germplasm from Argentina and, to a lesser degree, from CIMMYT.
Figure 3.
Bar chart representing the frequency of cultivars belonging to each breeding company distributed in the subpopulations K1 to K3.
A similar type of grouping of cultivars by geographic origin and breeding history using a model-based approach was observed by Le Couviour et al. (2011) working with an elite wheat panel from Europe. They identified four sub-populations, including cultivars from UK, Germany and France divided into two subgroups and proposed that the separation between French, German and UK cultivars can be explained by the geographic origin and, in the case of France, the conformation of two subgroups as being due to the breeding history. Furthermore, Chao et al. (2007), when using a similar approach to analyze the genetic structure of U.S. wheat cultivars and breeding lines, found four sub-populations and suggested that the genetic diversity existing among these U.S. wheat germplasm was influenced by regional adaptation. Our data would suggest that in Argentina the most important factor explaining the genetic variability of adapted commercial bread cultivars would be the different core collections of germplasm used by the main breeding programs instead of geographic adaptation, as observed in Europe and US.
The results obtained in this paper are a very valuable source of information for breeding programs for the creation of novel combinations of alleles from genes involved in adaptation, disease resistance and bread-making quality between other traits. Additionally, the genetic structure of the panel of cultivars analyzed in this study is being used as the starting point of association studies considering additional phenotypic traits of interest for the breeding like drought tolerance and yield components like kernel weight between others.
Acknowledgments
This work is part of the project AEBIO241351 “Association mapping for agronomic interest traits” supported by the Instituto Nacional de Tecnología Agropecuaria, Argentina.
Footnotes
Associate Editor: Everaldo Gonçalves de Barros
Supplementary Material
The following online material is available for this article:
Table S1 - Molecular markers used in the study.
Table S2 - Biochemical and functional molecular markers in Argentinean wheat cultivars.
Table S3 - Neutral molecular markers in Argentinean wheat cultivars.
This material is available as part of the online article from http://www.scielo.br/gmb.
References
- Appendino M, Bartoloni N, Slafer G. Vernalization response and earliness per se in cultivars representing different eras of wheat breeding in Argentina. Euphytica. 2003;130:61–69. [Google Scholar]
- Balfourier F, Roussel V, Strelchenko P, Exbrayat-Vinson F, Sourdille P, Boutet G, Koenig J, Ravel C, Mitrofanova O, Beckert M, et al. A worldwide bread wheat core collection arrayed in a 384-well plate. Theor Appl Genet. 2007;114:1265–1275. doi: 10.1007/s00122-007-0517-1. [DOI] [PubMed] [Google Scholar]
- Beales J, Turner A, Griffiths S, Snape JW, Laurie DA. A Pseudo-Response Regulator is misexpressed in the photo-period insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.) Theor Appl Genet. 2007;115:721–733. doi: 10.1007/s00122-007-0603-4. [DOI] [PubMed] [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4.05, logiciel sous Windows pour la génétique des populations. Université de Montpellier II; Montpellier, France: 1996. [Google Scholar]
- Bered F, Barbosa-Neto JF, de Carvalho FIF. Genetic variability in common wheat germplasm based on coefficients of parentage. Genet Mol Biol. 2002;25:211–215. [Google Scholar]
- Breseghello F, Sorrells MS. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics. 2006;172:1165–1177. doi: 10.1534/genetics.105.044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody JR, Kern SE. Sodium boric acid: A Trisfree, cooler conductive medium for DNA electrophoresis. Bio-Techniques. 2004;36:214–216. doi: 10.2144/04362BM02. [DOI] [PubMed] [Google Scholar]
- Bulos M, Echarte M, Sala C. Occurrence of the rust resistance gene Lr37 from Aegilops ventricosa in Argentine cultivars of wheat. Electr J Biotechnol. 2006;9:1–17. [Google Scholar]
- Burkhamer RL, Lanning SP, Martens RJ, Martin JM, Talbert LE. Predicting progeny variance from parental divergence in Hard Red Spring wheat. Crop Sci. 1998;38:243–248. [Google Scholar]
- Butow BJ, Gale KR, Ikea J, Juhász A, Bedö Z, Tamás L, Gianibelli MC. Dissemination of the highly expressed Bx7 glutenin subunit (Glu-B1al allele) in wheat as revealed by novel PCR markers and RP-HPLC. Theor Appl Genet. 2004;109:1525–1535. doi: 10.1007/s00122-004-1776-8. [DOI] [PubMed] [Google Scholar]
- Chao S, Zhang W, Dubcovsky J, Sorrells M. Evaluation of genetic diversity and genome-wide linkage disequilibrium among U.S. wheat (Triticum aestivum L.) germplasm representing different market classes. Crop Sci. 2007;47:1018–1030. [Google Scholar]
- Chao S, Dubcovsky J, Dvorak J, Luo MC, Baenziger S, Matnyazov R, Clark D, Talbert L, Anderson J, Dreisigacker S, et al. Population- and genome-specific patterns of linkage disequilibrium and SNP variation in spring and winter wheat (Triticum aestivum L.) BMC Genomics. 2010;11:e727. doi: 10.1186/1471-2164-11-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz A, Zikhali M, Turner AS, Isaac P, Laurie DA. Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum) PLoS One. 2012;7:e33234. doi: 10.1371/journal.pone.0033234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreisigacker S, Zhang P, Warburton ML, Van Ginkel M, Hoisington D, Melchinger AE. SSR and pedigree analyses of genetic diversity among CIMMYT wheat lines targeted to different megaenvironments. Crop Sci. 2004;44:381–388. [Google Scholar]
- Earl DA, von Holdt BM. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012;4:359–361. [Google Scholar]
- Ellis MH, Spielmeyer W, Gale KR, Rebetzke GJ, Richards RA. “Perfect” markers for the Rht-B1b and Rht-D1b dwarfing genes in wheat. Theor Appl Genet. 2002;105:1038–1042. doi: 10.1007/s00122-002-1048-4. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flintham JE, Börner A, Worland AJ, Gale MD. Optimizing wheat grain yield: Effects of Rht (gibberellin-insensitive) dwarfing genes. J Agric Sci. 1997;128:11–25. [Google Scholar]
- Fu D, Szücs P, Yan L, Helguera M, Skinner JS, von Zitzewitz J, Hayes PM, Dubcovsky J. Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Genet Genom. 2005;273:54–65. doi: 10.1007/s00438-004-1095-4. [DOI] [PubMed] [Google Scholar]
- Gautier MF, Aleman ME, Guirao A, Marion D, Joudrier P. Triticum aestivum puroindolines, two cystine-rich seed proteins: cDNA sequence analysis and developmental gene expression. Plant Mol Biol. 1994;25:43–57. doi: 10.1007/BF00024197. [DOI] [PubMed] [Google Scholar]
- Gooding MJ, Addisu M, Uppal RK, Snape JW, Jones HE. Effect of wheat dwarfing genes on nitrogen-use efficiency. J Agric Sci. 2012;150:3–22. [Google Scholar]
- Hartl D, Clark A. Principles of Population Genetics. Sinauer Associates; Sunderland: 1997. p. 635. [Google Scholar]
- He XY, He ZH, Zhang LP, Sun DJ, Morris CF, Fuerst EP, Xia XC. Allelic variation of polyphenol oxidase (PPO) genes located on chromosomes 2A and 2D and development of functional markers for the PPO genes in common wheat. Theor Appl Genet. 2007;115:47–58. doi: 10.1007/s00122-007-0539-8. [DOI] [PubMed] [Google Scholar]
- Helguera M, Vanzetti L, Soria M, Khan IA, Kolmer J, Dubcovsky J. PCR markers for Triticum speltoides leaf rust resistance gene Lr51 and their use to develop isogenic hard red spring wheat lines. Crop Sci. 2005;45:728–734. [Google Scholar]
- Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFadden H, Bossolini E, Selter LL, Keller B. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science. 2009;323:1360–1363. doi: 10.1126/science.1166453. [DOI] [PubMed] [Google Scholar]
- Lagudah ES, McFadden H, Singh RP, Huerta-Espino J, Bariana HS, Spielmeyer W. Molecular genetic characterization of the Lr34/Yr18 slow rusting resistance gene region in wheat. Theor Appl Genet. 2006;114:21–30. doi: 10.1007/s00122-006-0406-z. [DOI] [PubMed] [Google Scholar]
- Lanning SP, Hucl P, Pumphrey M, Carter AH, Lamb PF, Carlson GR, Wichman DM, Kephart KD, Spaner D, Martin JM, et al. Agronomic performance of spring wheat as related to planting date and photoperiod response. Crop Sci. 2012;52:1633–1639. [Google Scholar]
- Le Couviour F, Faure S, Poupard B, Flodrops Y, Dubreuil P, Praud S. Analysis of genetic structure in a panel of elite wheat varieties and relevance for association mapping. Theor Appl Genet. 2011;123:715–727. doi: 10.1007/s00122-011-1621-9. [DOI] [PubMed] [Google Scholar]
- Lerner SE, Kolman MA, Rogers WJ. Quality and endosperm storage protein variation in Argentinean grown bread wheat. I. Allelic diversity and discrimination between cultivars. J Cereal Sci. 2009;49:337–345. [Google Scholar]
- Manifesto MM, Schlatter AR, Hopp HE, Suarez EY, Dubcovsky J. Quantitative evaluation of genetic diversity in wheat germplasm using molecular markers. Crop Sci. 2001;41:682–690. [Google Scholar]
- Matthews DE, Carollo VL, Lazo GR, Anderson OD. GrainGenes, the genome database for small-grain crops. Nucleic Acids Res. 2003;31:183–186. doi: 10.1093/nar/gkg058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan A, Ogbonnaya FC, Hollingsworth B, Carter M, Gale KR, Henry RJ, Holton TA, Morell MK, Rampling LR, Sharp PJ, et al. Development of robust PCR-based DNA markers for each homoeo-allele of granule-bound starch synthase and their application in wheat breeding programs. Austr J Agric Res. 2001;52:1409–1416. [Google Scholar]
- Metakovsky EV, Branlard G. Genetic diversity of French common wheat germplasm based on gliadin alleles. Theor Appl Genet. 1998;96:209–218. [Google Scholar]
- Mukhtar MS, Rahman M, Zafar Y. Assessment of genetic diversity among wheat (Triticum aestivum L.) cultivars from a range of localities across Pakistan using random amplified polymorphic DNA (RAPD) analysis. Euphytica. 2002;128:417–425. [Google Scholar]
- Nei M. Analyses of gene diversity in subdivided populations. Proc Natl Acad Sci USA. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann K, Kobiljski B, Dencic S, Varshney RK, Börner A. Genome-wide association mapping: A case study in bread wheat (Triticum aestivum L.) Mol Breed. 2011;27:37–58. [Google Scholar]
- Parker GD, Fox PN, Langridge P, Chalmers K, Whan B, Ganter PF. Genetic diversity within Australian wheat breeding programs based on molecular and pedigree data. Euphytica. 2002;124:293–306. [Google Scholar]
- Peng JH, Bai Y, Haley SD, Lapitan NLV. Microsatellite-based molecular diversity of bread wheat germplasm and association mapping of wheat resistance to the Russian wheat aphid. Genetica. 2009;135:95–122. doi: 10.1007/s10709-008-9262-x. [DOI] [PubMed] [Google Scholar]
- Pflüger LA, D’Ovidio R, Margiotta B, Peña R, Mujeeb-Kazi A, Lafiandra D. Characterisation of high- and low-molecular weight glutenin subunits associated to the D genome of Aegilops tauschii in a collection of synthetic hexaploid wheats. Theor Appl Genet. 2001;103:1293–1301. [Google Scholar]
- Plaschke J, Ganal M, Röder M. Detection of genetic diversity in closely related bread wheat using microsatellite markers. Theor Appl Genet. 1995;91:1001–1007. doi: 10.1007/BF00223912. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens P, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schachermayr G, Messmer MM, Feuillet C, Winzeler H, Winzeler M, Keller B. Identification of molecular markers linked to the Agropyron elongatum-derived leaf rust resistance gene Lr24 in wheat. Theor Appl Genet. 1995;90:982–990. doi: 10.1007/BF00222911. [DOI] [PubMed] [Google Scholar]
- Schachermayr G, Feuillet C, Keller B. Molecular markers for the detection of the wheat leaf rust resistance gene Lr10 in diverse genetic backgrounds. Mol Breed. 1997;3:65–74. [Google Scholar]
- Seki M, Chono M, Matsunaka H, Fujita M, Oda S, Kubo K, Kiribuchi-Otobe C, Kojima H, Nishida H, Kato K. Distribution of photoperiod-insensitive alleles Ppd-B1a and Ppd-D1a and their effect on heading time in Japanese wheat cultivars. Breed Sci. 2011;61:405–412. doi: 10.1270/jsbbs.61.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneath pH, Sokal RR. Numerical Taxonomy. W.H. Freeman; San Francisco: 1973. p. 573. [Google Scholar]
- Soleimani VD, Baum BR, Johnson DA. AFLP and pedigree-based genetic diversity estimates in modern cultivars of durum wheat [Triticum turgidum L. subsp. durum (Desf.) Husn.] Theor Appl Genet. 2002;104:350–357. doi: 10.1007/s001220100714. [DOI] [PubMed] [Google Scholar]
- Spagnoletti-Zeuli PLS, Qualset CO. Geographical diversity for quantitative spike characters in a world collection of durum wheat. Crop Sci. 1987;27:235–241. [Google Scholar]
- Van Beuningen LT, Busch RH. Genetic diversity among North American spring wheat cultivars: I. Analysis of the coefficient of parentage matrix. Crop Sci. 1997a;37:570–579. [Google Scholar]
- Van Beuningen LT, Busch RH. Genetic diversity among North American spring wheat cultivars: III. Cluster analysis based on quantitative morphological traits. Crop Sci. 1997b;37:981–988. [Google Scholar]
- Vanzetti L, Pflüger L, Rodríguez-Quijano M, Carrillo JM, Helguera M. Genetic variability of Waxy genes in hexaploid wheats. Electron J Biotechnol. 2009;12:e4–5. [Google Scholar]
- Vanzetti L, Campos P, Demichelis M, Lombardo L, Vaschetto LM, Aurelia PR, Bainotti C, Helguera M. Identification of leaf rust resistance genes in Argentinean hexaploid wheat (Triticum aestivum L.) using molecular markers and traditional gene postulation. Electron J Biotechnol. 2011;14:e9–9. [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Yan L, Helguera M, Kato K, Fukuyama S, Sherman J, Dubcovsky J. Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor Appl Gen. 2004;109:1677–1686. doi: 10.1007/s00122-004-1796-4. [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhao XL, Xia LQ, Chen XM, Xia XC, Yu Z, He Z, Röder MS. Development and validation of Viviparous-1 STS marker for pre-harvest sprouting tolerante in Chinese wheats. Theor Appl Genet. 2007;115:971–980. doi: 10.1007/s00122-007-0624-z. [DOI] [PubMed] [Google Scholar]
- Zhang W, Gianibelli MC, Rampling LR, Gale KR. Characterisation and marker development for low molecular weight glutenin genes from Glu-A3 alleles of bread wheat (Triticum aestivum. L) Theor Appl Genet. 2004;108:1409–1419. doi: 10.1007/s00122-003-1558-8. [DOI] [PubMed] [Google Scholar]
Internet Resources
- GrainGenes database. http://wheat.pw.usda.gov/GG2/index.shtml (September 15, 2012).
- Structure software. http://pritch.bsd.uchicago.edu/structure.html (September 15, 2012).
- Structure harvester. http://taylor0.biology.ucla.edu/structureHarvester/ (September 15, 2012).
- Genetix software. http://kimura.univ-montp2.fr/genetix/ (September 15, 2012).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following online material is available for this article:
Table S1 - Molecular markers used in the study.
Table S2 - Biochemical and functional molecular markers in Argentinean wheat cultivars.
Table S3 - Neutral molecular markers in Argentinean wheat cultivars.