Background: Many tRNA synthetases (AARS) have extracellular and nuclear functions. Some of these functions might require the export of their mRNAs.
Results: Packaging in exosomes of mRNAs for AARSs and a splice variant was demonstrated.
Conclusion: Exosomes capture translation-competent AARS-derived mRNAs, which can be regulated by external stimuli.
Significance: Exosomes are a source for extracellular distribution of AARS-derived RNA information.
Keywords: Alternative Splicing, Aminoacyl tRNA Synthetase, Cell Biology, Exosomes, mRNA, Jurkat Cell Exosome mRNAs, tRNA Synthetase Splice Variant
Abstract
Although tRNA synthetases are enzymes that catalyze the first step of translation in the cytoplasm, surprising functions unrelated to translation have been reported. These studies, and the demonstration of novel activities of splice variants, suggest a far broader reach of tRNA synthetases into cell biology than previously recognized. Here we show that mRNAs for most tRNA synthetases can be detected in exosomes. Also detected in exosomes was an mRNA encoding a unique splice variant that others had associated with prostate cancer. The exosomal mRNAs encoding the native synthetase and its cancer-associated splice variant could be translated in vitro and in mammalian cells into stable proteins. Other results showed that selection by exosomes of the splice variant mRNA could be regulated by an external stimulus. Thus, a broad and diverse regulated pool of tRNA synthetase-derived mRNAs is packaged for genetic exchange.
Introduction
The aminoacyl tRNA synthetases (AARSs),2 as a protein family (1), have progressively added novel domains in evolution with a fecundity that is not matched by any other family (2). These domain additions are not needed for catalysis, but rather to endow the synthetases with functions beyond translation. These functions are associated with secreted, extracellular forms and with nuclear forms that are active in many different cell-signaling pathways (3–12). In addition, a recent study demonstrated that a novel domain, appended to SerRS at the time of the invertebrate to vertebrate transition, was essential for development of the closed vascular system (13). Thus, novel domains added to tRNA synthetase are endowed with nontranslation functions that are as essential as the activity of the synthetase in protein synthesis (12).
The discovery and analysis of exosomes revealed yet another mechanism for cells to communicate and to transfer intracellular components. These components include specific mRNAs that can be translated and thus highlight how exosomes implement genetic exchange between cells (14–17). With this perspective, we were especially interested in the idea that, in addition to protein secretion, the exosome-driven intercellular transmission of AARS mRNAs is a genetic communication that brings new functions to the targeted recipient. In addition, we were interested in finding at least one AARS mRNA variant that encoded a novel appended domain, but had ablated at least part of the core enzyme needed for catalysis. We infer that such a variant would have to be redirected to an alternative function. In this connection, a splice variant of human GlnRS was reported to be associated with some cancers (18, 19). This variant (designated here as GlnRSΔiABD) has an internal deletion of 42 amino acids in the anticodon binding domain (ABD). This deletion eliminates anticodon recognition by GlnRSΔiABD and in some ways is reminiscent of the recently reported internally deleted splice variant of human HisRS (HisRSΔCD), which lacks the entire catalytic domain (20). Thus, we were also motivated to search in exosomes for mRNA encoding GlnRSΔiABD and, if successful, to see whether its appearance could be regulated.
EXPERIMENTAL PROCEDURES
Jurkat T Cell Activation and Exosome Purification
Jurkat T cells were grown and maintained in RPMI 1640 medium supplemented with 10% FBS and 0.5% penicillin/streptomycin. Human CD3ϵ monoclonal antibody (clone UCHT1) was obtained from R&D systems (Minneapolis, MN). For activation, culture flasks were coated with 1 μg/ml CD3ϵ antibody in PBS or PBS alone at 37 °C overnight. The flasks were washed once with PBS before use. Jurkat cells suspended in culture medium, which consisted of exosome-free medium cleaned by overnight centrifugation at 100,000 × g (Hitachi CP100, Hitachinaka, Ibaraki, Japan), were added to the CD3ϵ antibody-coated flask for activation or to a PBS-coated flask as a control. After incubation for 6 h, cells were harvested by centrifugation at 300 × g for 10 min. Exosomes were purified from the collected medium by differential centrifugation as described previously (21). Briefly, medium was centrifuged at 2000 × g for 10 min at 4 °C followed by filtration through a 0.22-μm filter. Exosomes were pelleted by ultracentrifugation at 100,000 × g for 90 min at 4 °C. The pellet was washed once with PBS and then resuspended in PBS. The protein content in the purified exosomes was quantified by the BCA protein assay kit (Pierce).
RNA Isolation
The cellular or exosomal RNA was isolated by the RNeasy mini kit (Qiagen, Hilden, Germany). Cells or exosomes were lysed in the RLT buffer and added with 3.5 volumes of 100% ethanol before passing through the RNeasy mini spin column. The subsequent steps were performed according to the manufacturer's instructions. Residual genomic DNA was removed by TURBO DNase (Invitrogen). To confirm that the extracted RNA was from inside the exosomes, Jurkat exosomes were pretreated with 0.1 μg/μl RNase A (Invitrogen) for 10 min at 37 °C. Then, RNase A was inactivated by 0.4 μg/μl proteinase K (Bioline, London, UK) for 20 min at 37 °C before RNA purification. As a control for RNase activity, 5 μg of cellular RNA were included.
Isolation of Polyribosomal RNA
Polyribosomal mRNA was isolated from Jurkat T cells according to the protocol described by del Prete et al. (22) with slight modifications. Briefly, cells were firstly lysed in RLN lysis buffer (Qiagen, Hilden, Germany) to which was added 0.5% IGEPAL (Sigma-Aldrich), 40 mm dithiothreitol, 500 units/ml RNase inhibitor (ABI Biosystems, Foster City, CA), 150 μg/ml cycloheximide, 650 μg/ml heparin, and 10 mm proteinase inhibitor. After centrifugation at 12,000 × g for 5 min at 4 °C, the supernatant was layered onto a linear sucrose gradient (10–40% sucrose (w/v) supplemented with 10 mm Tris-HCl (pH 7.5, 140 mm NaCl, 1.5 mm MgCl2, 10 mm dithiothreitol, 100 μg/ml cycloheximide, 0.5 mg/ml heparin) and centrifuged at 36,000 rpm (Hitachi CP100) for 6 h at 4 °C. Fourteen fractions (500 μl) were collected. For each fraction, absorbance at 260 nm was determined and plotted. To remove heparin, RNAs in these fractions were then precipitated with 2 m lithium chloride at 4 °C overnight. After centrifugation at 12,000 × g for 15 min at 4 °C, the RNA pellet was washed twice with 70% ice-cold ethanol, air-dried, and resuspended in RNase-free water. Proteins were separated from RNAs using the RNeasy kit.
Quantitative Real-time PCR (qRT-PCR)
All human tissue poly(A)+ RNAs were obtained from Clontech (catalog numbers: 636102, 636106,636170, 636591, 636128, 636105, 636113, 636119, 636121, 636101, 636118, 636146, 636125, 636162, and 636120). Total RNA of human Jurkat T, Raji B, THP-1, IMR-32, MCF-7, or MDA-MB-231 cells was extracted with the PureLinkTM RNA mini kit (Invitrogen). Complementary DNA (cDNA) was synthesized from RNA samples using a SuperScript III first-strand synthesis kit (Invitrogen) and an oligo(dT) primer. The PCR reactions were performed using the SYBR Green master mix (Roche Applied Science, Mannheim, Germany) and with the optimized primer pairs (supplemental Table 1) on a ViiATM 7 real-time PCR system (Life Technologies). The thermal cycling steps were as follows: 2 min at 50 °C, 10 min at 95 °C, and then 45 cycles of 95 °C for 30 s and 60 °C for 30 s. The data were analyzed using the ViiATM 7 RUO software (Life Technologies). For tissue distribution analysis, mRNA expression of QRS1 or GlnRSΔiABD was normalized to two housekeeping genes, RPL9 and RPS11. For analysis of RNA levels in exosomes, U6 snRNA was employed for normalization (17). To determine the copy numbers of QRS1 and GlnRSΔiABD mRNAs in the samples, the standard curve method was employed. Briefly, cDNAs encoding QRS1 or GlnRSΔiABD were cloned into a pET21a vector and confirmed by sequencing. The constructs were quantified with NanoDrop 1000 (Thermo Scientific). The standard curves were constructed using 10, 50, 102, 103, 104, and 105 copies of QRS1 or GlnRSΔiABD as the templates in qPCR reactions, which were run in parallel with the samples. (Copy numbers were calculated using the URI Genomics and Sequencing Center dsDNA copy number calculator.) The standard curves were plotted as cycle threshold (Ct) values versus the logarithm of copy number. The copy number of QRS1 or of GlnRSΔiABD mRNAs in the samples was extrapolated from the standard curve.
Western Blot Analysis
Total proteins were extracted from Jurkat cells with 50 mm Tris buffer (pH 8.0) containing 1% Triton X-100 and 5 mm EDTA. To release exosomal proteins, exosomes were mixed with 1:1 2× SDS loading buffer and incubated at 70 °C for 10 min and at 95 °C for 5 min. Proteins were separated on a NuPAGE 4–12% Bis-Tris gel (Invitrogen) and transferred to a nitrocellulose membrane. The membranes were separately stained with a monoclonal antibody targeted to TSG101 (Abcam, Cambridge, MA) a polyclonal antibody to Bip (Abcam), or a polyclonal antibody to Alix (Santa Cruz Biotechnology, Dallas, TX).
In Vitro Coupled Transcription/Translation Assay
The cDNA encoding GlnRSΔiABD was cloned into a pcDNA3.1 vector and confirmed by sequencing. The TnT® T7-coupled reticulocyte lysate system was purchased from Promega (Madison, WI). The reaction was performed according to the manufacturer's protocol. The empty vector pcDNA3.1 was included as a negative control. After the reaction, 1% of product was subjected to Western blot analysis with a mouse anti-QRS1 polyclonal antibody (Abcam).
RESULTS
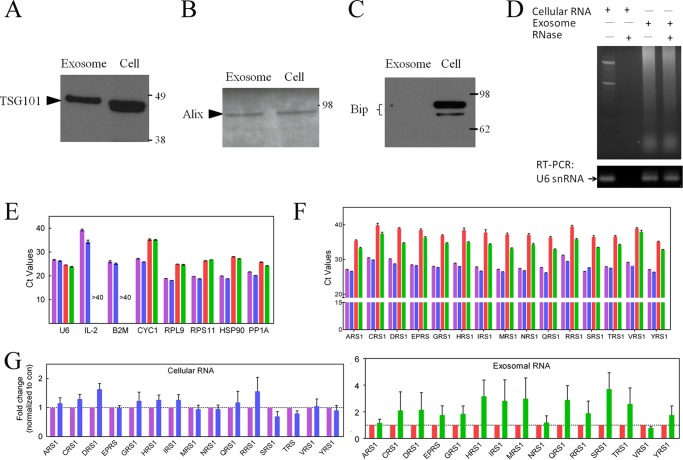
As part of our ongoing investigations of novel functions of tRNA synthetases, we chose Jurkat cells in part because they can be studied in both a quiescent and an activated (by CD3 stimulation) state. Following standard protocols with differential centrifugation methods, about 20 μg of exosomes were purified from 100 ml of culture medium of 108 Jurkat cells. For evaluation of the purity, a sample of the exosomes was subjected to Western blot analysis to detect components known to be markers for exosomes. We also tested for contamination from the endoplasmic reticulum (Fig. 1). This analysis showed clearly the presence of exosomal markers TSG101 (Fig. 1A) and Alix (Fig. 1B) and the absence of the Bip marker of the endoplasmic reticulum (Fig. 1C) (21, 23).
FIGURE 1.
Detection of AARS mRNAs in the exosomal RNA from Jurkat cells. A–C, exosomes were purified from conditioned medium of Jurkat T cells. Four μg of cell lysates and of exosome samples were subjected to Western blot analysis with polyclonal antibodies against TSG101 (A), Alix (B), and Bip (C). D, cellular RNA was degraded after RNase treatment. Jurkat exosomes treated with RNase showed no difference on the agarose gel and RT-PCR (U6 snRNA) as compared with control exosomes. E, representative Ct values of IL-2 mRNAs and seven housekeeping genes (U6 snRNA, B2M, CYC1, RPL9, RPS11, HSP90, and PP1A) were expressed as mean ± S.E. (n = 3). F, the level of mRNAs for 15 genes from the family of cytoplasmic AARSs. The mRNAs for ARS1, CRS1, DRS1, EPRS, GRS1, HRS1, IRS1, MRS1, NRS1, QRS1, RRS1, SRS1, TRS1, VRS1, and YRS1 were analyzed with qRT-PCR. The Ct values for cellular RNA of nonactivated (the control, purple column) or activated (blue) Jurkat cells and for exosomal RNA from control (red) or activated (green) Jurkat cells are presented with a mean ± S.E. (n = 3). G, the expression level in cellular (blue column) or exosomal (green) RNA from activated Jurkat cells was normalized as the -fold change relative to control (con) (nonactivated) cellular (purple) or control exosomal (red) RNA, respectively. The data were expressed as mean ± S.E. of three independent experiments.
To start our search for mRNAs encoding for AARSs in exosomes from Jurkat cells, we first investigated exosomal RNA from Jurkat cells. In contrast to added cellular RNA, which was degraded by RNase, there was no difference between control and RNase-treated exosomes (Fig. 1D), suggesting that exosomal RNA was protected within the exosomes. Then, we used qRT-PCR to annotate the number of cycles above the background threshold (Ct) that is needed to detect a specific mRNA. U6 snRNA, an RNA commonly found in exosomes, was used as a positive marker and was easily detected. As expected, expression of U6 snRNA in the cellular RNA was relatively stable upon activation (Ct for quiescent = 26.7 ± 0.1 versus 26.2 ± 0.2 for activated) and in exosomal RNA (Ct for quiescent 24.6 ± 0.1 versus 23.8 ± 0.2 in activated) (Fig. 1E). To search for a negative control, we also checked the expression levels of six other housekeeping genes (Fig. 1E). Importantly, of these six genes, β-2 microglobulin (B2M) had a similar level of expression as U6 snRNA inside the cell. In contrast, B2M mRNA was not detected in the exosomes. Thus, exosomal RNAs appear not to be contaminated by cellular RNAs. We also investigated the expression level of IL-2 mRNA in quiescent and activated Jurkat cells and in exosomes. As expected (24), IL-2 mRNA was up-regulated after activation (Ct value changed from 39.3 ± 0.4 to 34.2 ± 0.8). In contrast, after 45 cycles of qPCR, IL-2 mRNA was not detected in exosomal RNA samples from either quiescent or activated Jurkat cells (Fig. 1E).
Next, we investigated the presence of mRNAs encoding 15 different cytoplasmic tRNA synthetases. These included mRNAs for alanyl-tRNA synthetase (AlaRS, ARS1), ArgRS (RRS1), AsnRS (NRS1), AspRS (DRS1), CysRS (CRS1), GlnRS (QRS1), Glu-ProRS (EPRS), GlyRS (GRS1), HisRS (HRS1), IleRS (IRS1), MetRS (MRS1), SerRS (SRS1), ThrRS (TRS1), ValRS (VRS1), and TyrRS (YRS1). (The mRNAs encoding cytoplasmic histidyl-tRNA synthetase (HRS1), cytoplasmic tryptophanyl-tRNA synthetase (WRS1), and mitochondrial asparaginyl-tRNA synthetase (NRS2) were annotated in exosomes extruded from two mast cell lines (MC/9 and HMC-1) (14, 25). Our work also showed the presence of HRS1 in exosomes from Jurkat cells.) The primers used for this analysis are listed in supplemental Table 1. (The average length of the coding sequences for 15 AARS genes was around 2400 bp. The average distance between two pairs of primers for each AARS mRNA was 1050 bp.) To assure and validate that we covered a large portion of the mRNA, these primers were designed to detect exon junctions at two well separated regions of the mRNAs.
In most cases, two pairs of primers were used for confirmation. Because in this and other work (20) we found that the abundance of splice variants (such as those that might contain an internal deletion) was less than 1% of that of the full-length mRNAs, the primers essentially yield Ct values that monitor the amount of the full mRNA.
The mRNAs for all 15 cytoplasmic tRNA synthetases were found in both whole cell lysates and exosomal fractions. The Ct values for the whole cells lysates were in the range of Ct = 23.1–29.4. For the exosomes, the Ct values ranged from 29.9 to 40.9. Thus, Ct values for the exosomal mRNAs for the cytoplasmic AARSs suggest a level that is about 1% or less of that found in whole cells (23.1 versus 29.9, for example, is a difference of 26.8 = 111.4 (Fig. 1F)).
The data were further analyzed, with normalization to control cellular or exosomal RNA, to evaluate regulation upon Jurkat cell activation. For either cellular or exosomal RNAs, expression levels of 15 cytoplasmic AARS genes were not altered (after activation) in a statistically significant way as compared with the controls (Fig. 1G).
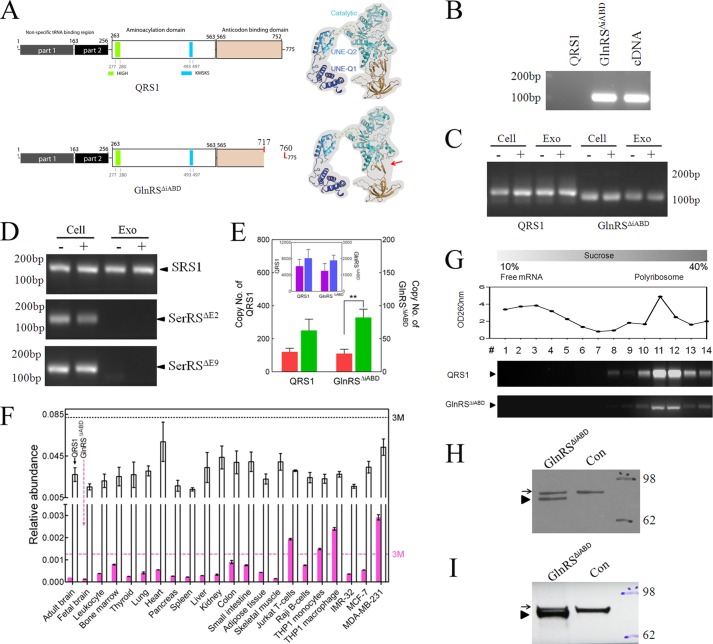
As mentioned in a recent publication on HisRSΔCD (20), an amplification-based transcriptome sequencing method was developed for the discovery of splicing variants of mRNAs for AARSs. Because of a prior study indicating an association of a specific splice variant (designated as GlnRSΔiABD) of GlnRS mRNA with cancer (19), first reported in a prostate cancer cell line, we were particularly interested in screening for the presence of this variant (Fig. 2). The splice variant mRNA skips exon 23 and results in deletion of a region encoding a 42-amino acid segment in the ABD that is needed for recognition of the anticodon of tRNAGln (26) (Fig. 2A). Based on the location of the exon-skipping event, we designed a pair of GlnRSΔiABD mRNA-specific primers that do not amplify mRNA for QRS1 (Fig. 2B). Interestingly, GlnRSΔiABD was found in the exosomal RNA of both quiescent and activated Jurkat cells (Fig. 2C). In contrast, two splicing variants of SerRS expressed in the cell were not detected in the exosomal RNA (Fig. 2D). To support the conclusion that the exosomal RNA preparation was not contaminated by cellular RNA, we reconfirmed the presence of U6 snRNA and absence of mRNA encoding B2M and IL-2.
FIGURE 2.
GlnRSΔiABD was present in exosomes extruded from Jurkat cells. A, the consequence of the internal deletion in the ABD of GlnRS (to give GlnRSΔiABD) is modeled based on the structure of several lower homologs (Protein Data Bank (PDB) numbers 3tl4, 2hz7, and 1exd). The location of the splice junction in the structure is marked with a red arrow. B, The specific primers for GlnRSΔiABD were tested in a qPCR reaction with the templates of 106 copies of QRS1, 106 copies of GlnRSΔiABD, and cDNA generated from RNA of Jurkat cells. C, using RT-PCR, expression of GlnRSΔiABD was tested using cellular (cell) and exosomal (exo) RNA samples. (The notation − is for nonactivated Jurkat cells, and + is for activated Jurkat cells.) D, two SerRS splicing variants were present in the cells but not in the exosome. Using RT-PCR, expression of SRS1, SerRSΔE2, and SerRSΔE9 was tested using cellular (cell) and exosomal (exo) RNA samples. SerRSΔE2 and SerRSΔE9 are SerRS splicing variants with deletion of the second and ninth exons, respectively. E, the copy numbers of QRS1 and GlnRSΔiABD were determined by qRT-PCR and using a standard curve. The data represent mean ± S.E. of three independent experiments. ** designates p < 0.01 in a Student's t test. F, the tissue distribution across 22 samples of QRS1 (empty column) and GlnRSΔiABD (pink column) was determined by qRT-PCR, using primers that specifically target QRS1 or GlnRSΔiABD. The data are expressed as mean ± S.E. (n = 3). A light magenta dashed line marks 3 times the median (3M) from the expression levels in 22 samples. G, endogenous GlnRSΔiABD mRNA was detected in the polyribosomal fractions but not free mRNA fractions isolated from Jurkat cells. Fourteen fractions were obtained from across a 10–40% sucrose gradient. The mRNA for QRS1 and GlnRSΔiABD was detected with specific PCR primers. H, a recombinant gene encoding GlnRSΔiABD in pcDNA3.1 and an empty vector control (con) were subjected to a rabbit T7 RNA polymerase-coupled translation-transcription reticulocyte lysate system, and the products were analyzed by Western blotting. The black arrow points to rabbit QRS1, whereas the arrowhead marks human GlnRSΔiABD. I, The HEK 293T cells were transfected with a recombinant gene encoding GlnRSΔiABD in a pcDNA3.1 vector or with an empty vector (Con). Cell lysates were analyzed by Western blot analysis. The position of QRS1 is indicated by a black arrow, and an arrowhead marks the position of GlnRSΔiABD.
A qRT-PCR analysis was used to determine the levels of the transcripts for QRS1 and GlnRSΔiABD. The levels of mRNAs for QRS1 and GlnRSΔiABD were not significantly changed after activation of Jurkat cells (copy number = 6170.8 ± 1672.9 and 8124.6 ± 2167.9 and copy number = 1256.1 ± 462.0 and 1898.9 ± 314.2, respectively, for mRNAs of QRS1 and GlnRSΔiABD). In contrast, transcripts for GlnRSΔiABD were most clearly enhanced in the exosomes from activated Jurkat cells (copy number = 120.2 ± 21.7 versus 249.7 ± 69.1 (p = 0.1, quiescent versus activated) for QRS and copy number = 27.3 ± 6.6 versus 82.0 ± 12.6 for GlnRSΔiABD (p = 0.009) (Fig. 2E)).
These results prompted us to investigate the expression of mRNA for GlnRSΔiABD in additional cell types. Across 15 tissues and seven cell lines, expression levels of mRNAs for QRS1 were not significantly different. In contrast, for the mRNA for GlnRSΔiABD, the expression level in Jurkat T, THP-1 monocyte, macrophage, and MDA-MB-231 cells stood out from the panel of the others (>3 median) (Fig. 2F). The expression level of GlnRSΔiABD mRNA was significantly up-regulated in the more advanced and invasive MDA-MB-231 cells as compared with MCF-7 cells.
We next investigated the potential for translation of the GlnRSΔiABD mRNA. First, we used specific PCR primers to identify the mRNA for endogenous GlnRSΔiABD in polyribosomal RNA. Using sucrose gradient centrifugation to separate polyribosomes, we were able by RT-PCR to identify the GlnRSΔiABD mRNA in the polyribosome but not free RNA fractions, which was similar to the result with QRS1 mRNA (Fig. 2G). We then constructed a recombinant cDNA gene for the GlnRSΔiABD mRNA detected in exosomes. The idea was to test whether the mRNA could be translated into a stable protein. First, we used the rabbit reticulocyte-coupled transcription-translation system (27, 28). As shown in Fig. 2H, a protein product of the expected size for GlnRSΔiABD was easily detected. This result showed that exosomal mRNA for GlnRSΔiABD is active for translation. It also demonstrated that the GlnRSΔiABD-encoded protein is internally deleted in a way that allows it to fold into a stable structure.
Next, we transfected HEK 293T cells with the vector encoding the GlnRSΔiABD gene. Cell lysates were then analyzed by Western blot methods. As shown in Fig. 2I, proteins corresponding in size to endogenous QRS1 and GlnRSΔiABD were easily detected. These results further support the conclusion that the exosomal mRNA sequences for tRNA synthetases and their splice variants are functional in protein synthesis.
DISCUSSION
Given the expansion of tRNA synthetase functions into areas of biology beyond translation, the question of whether exosome-mediated export of their mRNAs could play a role in the distribution of synthetase-derived information was of some interest. Genetic material consisting of mRNA was first identified in exosomes extruded from two human mast cell lines (MC/9 and HMC-1) (14). Significantly, the exosome mRNA from these mast cell lines was functional in both in vitro and in vivo translation assays (14). Although several components of the translational apparatus, such as ribosomal proteins and elongation factors, were also found in the exosomes (29), no evidence supported the presence of rRNAs (14). The absence of rRNAs suggests that translation is not actively taking place in exosomes. Thus, although tRNA synthetases are essential components of the translation apparatus, the capture of their mRNAs is not for translation nor to implement translation in exosomes. In that connection, several recent studies document a regulatory role for specific mRNAs, which is distinct from a role in translation (30–32).
The presence of mRNAs for specific proteins in exosomes and the discovery of microRNAs in exosomes have enlarged the perspective for how RNAs can play a role in genetic exchange between cells. In that connection, recent work by Montecalvo et al. (33) has reinforced the concept of the functional significance of exosome-based export of RNAs. Our work shows that most, if not all, mRNAs encoding human cytoplasmic tRNA synthetases, and the mRNA for a specific splice variant, are captured by exosomes. Interestingly, the absence of detectable mRNA in exosomes for B2M and IL-2 (even when expression of IL-2 was up-regulated (Fig. 1E)) highlights a mechanism for mRNA selection that is yet to be understood. However, the observation that the relative abundance of the exosomal GlnRSΔiABD mRNA splice variant was regulated by an external stimulus (for activating Jurkat cells (Fig. 2E)) shows that the selection mechanism is sensitive to specific exogenous signals. In addition, the abundance of the GlnRSΔiABD mRNA variant was tissue-specific (Fig. 2F), suggesting that parameters determining cell type also play a role in mRNA selection. Thus, exosomes provide a versatile format for selection and distribution of tRNA synthetase-derived genetic information. Also, the observation that an mRNA encoding a catalytically impaired splice variant is packaged by exosomes further highlights the potential for the broad reach of alternative noncatalytic forms of tRNA synthetases into areas of biology that are outside of translation.
Acknowledgments
We thank Drs. Min Guo, Kyle P. Chiang, and Melissa Ashlock for helpful comments and advice.
This work was supported in part by the Innovation and Technology Fund of Hong Kong (UIM181, UIM192, and UIM199), Grant CA092577 from the NCI (to P. S.) and Grant GM 088278 from the National Institutes of Health (to Y.-X. L.), aTyr Pharma, and by a fellowship from the National Foundation for Cancer Research. Professors Mingjie Zhang, Xiang-Lei Yang, and Paul Schimmel have a financial benefit from aTyr Pharma in the form of compensation or stock ownership or both. The partial support of this work by aTyr Pharma is stated in this footnote. The rest of the authors have affiliations that are annotated, and these affiliations include aTyr Pharma and Pangu Biopharma.

This article contains supplemental Table 1.
- AARS
- aminoacyl tRNA synthetase
- QRS
- GlnRS, glutaminyl-tRNA synthetase
- ABD
- anticodon binding domain
- qRT-PCR
- quantitative real-time PCR
- qPCR
- quantitative PCR
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol
- B2M
- β-2 microglobulin
- CD
- catalytic domain.
REFERENCES
- 1. Ibba M., Soll D. (2000) Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69, 617–650 [DOI] [PubMed] [Google Scholar]
- 2. Guo M., Yang X. L., Schimmel P. (2010) New functions of aminoacyl-tRNA synthetases beyond translation. Nat. Rev. Mol. Cell Biol. 11, 668–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wakasugi K., Schimmel P. (1999) Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science 284, 147–151 [DOI] [PubMed] [Google Scholar]
- 4. Martinis S. A., Plateau P., Cavarelli J., Florentz C. (1999) Aminoacyl tRNA synthetases: a family of expanding functions. EMBO J. 18, 4591–4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yannay-Cohen N., Carmi-Levy I., Kay G., Yang C. M., Han J. M., Kemeny D. M., Kim S., Nechushtan H., Razin E. (2009) LysRS serves as a key signaling molecule in the immune response by regulating gene expression. Mol. Cell 34, 603–611 [DOI] [PubMed] [Google Scholar]
- 6. Kim S., You S., Hwang D. (2011) Aminoacyl-tRNA synthetases and tumorigenesis: more than housekeeping. Nat. Rev. Cancer 11, 708–718 [DOI] [PubMed] [Google Scholar]
- 7. Bonfils G., Jaquenoud M., Bontron S., Ostrowicz C., Ungermann C., De Virgilio C. (2012) Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol. Cell 46, 105–110 [DOI] [PubMed] [Google Scholar]
- 8. Han J. M., Jeong S. J., Park M. C., Kim G., Kwon N. H., Kim H. K., Ha S. H., Ryu S. H., Kim S. (2012) Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 149, 410–424 [DOI] [PubMed] [Google Scholar]
- 9. Sajish M., Zhou Q., Kishi S., Valdez D. M., Jr., Kapoor M., Guo M., Lee S., Kim S., Yang X. L., Schimmel P. (2012) Trp-tRNA synthetase bridges DNA-PKcs to PARP-1 to link IFN-γ and p53 signaling. Nat. Chem. Biol. 8, 547–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yao P., Fox P. L. (2013) Aminoacyl-tRNA synthetases in medicine and disease. EMBO. Mol. Med. 5, 332–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ofir-Birin Y., Fang P., Bennett S. P., Zhang H. M., Wang J., Rachmin I., Shapiro R., Song J., Dagan A., Pozo J., Kim S., Marshall A. G., Schimmel P., Yang X. L., Nechushtan H., Razin E., Guo M. (2013) Structural switch of lysyl-tRNA synthetase between translation and transcription. Mol. Cell 49, 30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo M., Schimmel P. (2013) Essential nontranslational functions of tRNA synthetases. Nat. Chem. Biol. 9, 145–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu X., Shi Y., Zhang H. M., Swindell E. C., Marshall A. G., Guo M., Kishi S., Yang X. L. (2012) Unique domain appended to vertebrate tRNA synthetase is essential for vascular development. Nat. Commun. 3, 681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., Lötvall J. O. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 [DOI] [PubMed] [Google Scholar]
- 15. Zhang Y., Liu D., Chen X., Li J., Li L., Bian Z., Sun F., Lu J., Yin Y., Cai X., Sun Q., Wang K., Ba Y., Wang Q., Wang D., Yang J., Liu P., Xu T., Yan Q., Zhang J., Zen K., Zhang C. Y. (2010) Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol. Cell 39, 133–144 [DOI] [PubMed] [Google Scholar]
- 16. Eldh M., Ekström K., Valadi H., Sjöstrand M., Olsson B., Jernås M., Lötvall J. (2010) Exosomes communicate protective messages during oxidative stress; possible role of exosomal shuttle RNA. PLoS One 5, e15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhu H., Fan G. C. (2011) Extracellular/circulating microRNAs and their potential role in cardiovascular disease. Am. J. Cardiovasc. Dis. 1, 138–149 [PMC free article] [PubMed] [Google Scholar]
- 18. Lukk M., Kapushesky M., Nikkilä J., Parkinson H., Goncalves A., Huber W., Ukkonen E., Brazma A. (2010) A global map of human gene expression. Nat. Biotechnol. 28, 322–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Griffith M., Griffith O. L., Mwenifumbo J., Goya R., Morrissy A. S., Morin R. D., Corbett R., Tang M. J., Hou Y. C., Pugh T. J., Robertson G., Chittaranjan S., Ally A., Asano J. K., Chan S. Y., Li H. I., McDonald H., Teague K., Zhao Y., Zeng T., Delaney A., Hirst M., Morin G. B., Jones S. J., Tai I. T., Marra M. A. (2010) Alternative expression analysis by RNA sequencing. Nat. Methods 7, 843–847 [DOI] [PubMed] [Google Scholar]
- 20. Xu Z., Wei Z., Zhou J. J., Ye F., Lo W. S., Wang F., Lau C. F., Wu J., Nangle L. A., Chiang K. P., Yang X. L., Zhang M., Schimmel P. (2012) Internally deleted human tRNA synthetase suggests evolutionary pressure for repurposing. Structure 20, 1470–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Théry C., Amigorena S., Raposo G., Clayton A. (2006) Isolation and characterization of exosomes from cell culture supernatants and biological fluids. in Current Protocols in Cell Biology (Bonifacino J. S., Dasso M., Harford J. B., Lippincott-Schwartz J., Yamada K. M., eds) Chapter 3, Unit 3.22, John Wiley & Sons, Inc., New York: [DOI] [PubMed] [Google Scholar]
- 22. del Prete M. J., Vernal R., Dolznig H., Müllner E. W., Garcia-Sanz J. A. (2007) Isolation of polysome-bound mRNA from solid tissues amenable for RT-PCR and profiling experiments. RNA 13, 414–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ohshima K., Inoue K., Fujiwara A., Hatakeyama K., Kanto K., Watanabe Y., Muramatsu K., Fukuda Y., Ogura S., Yamaguchi K., Mochizuki T. (2010) Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS One 5, e13247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ledbetter J. A., Gentry L. E., June C. H., Rabinovitch P. S., Purchio A. F. (1987) Stimulation of T cells through the CD3/T-cell receptor complex: role of cytoplasmic calcium, protein kinase C translocation, and phosphorylation of pp60c-src in the activation pathway. Mol. Cell Biol. 7, 650–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mathivanan S., Simpson R. J. (2009) ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 9, 4997–5000 [DOI] [PubMed] [Google Scholar]
- 26. Perona J. J., Rould M. A., Steitz T. A. (1993) Structural basis for transfer RNA aminoacylation by Escherichia coli glutaminyl-tRNA synthetase. Biochemistry 32, 8758–8771 [DOI] [PubMed] [Google Scholar]
- 27. Pelham H. R. B., Jackson R. J. (1976) An efficient mRNA-dependent translation system from reticulocyte lysates. Eur. J. Biochem. 67, 247–256 [DOI] [PubMed] [Google Scholar]
- 28. King R. W., Lustig K. D., Stukenberg P. T., McGarry T. J., Kirschner M. W. (1997) Expression cloning in the test tube. Science 277, 973–974 [DOI] [PubMed] [Google Scholar]
- 29. Mathivanan S., Ji H., Simpson R. J. (2010) Exosomes: extracellular organelles important in intercellular communication. J. Proteomics 73, 1907–1920 [DOI] [PubMed] [Google Scholar]
- 30. Leygue E. (2007) Steroid receptor RNA activator (SRA1): unusual bifaceted gene products with suspected relevance to breast cancer. Nucl. Recept. Signal. 5, e006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wadler C. S., Vanderpool C. K. (2007) A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc. Natl. Acad. Sci. U.S.A. 104, 20454–20459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Candeias M. M., Malbert-Colas L., Powell D. J., Daskalogianni C., Maslon M. M., Naski N., Bourougaa K., Calvo F., Fåhraeus R. (2008) P53 mRNA controls P53 activity by managing Mdm2 functions. Nat. Cell Biol. 10, 1098–1105 [DOI] [PubMed] [Google Scholar]
- 33. Montecalvo A., Larregina A. T., Shufesky W. J., Stolz D. B., Sullivan M. L., Karlsson J. M., Baty C. J., Gibson G. A., Erdos G., Wang Z., Milosevic J., Tkacheva O. A., Divito S. J., Jordan R., Lyons-Weiler J., Watkins S. C., Morelli A. E. (2012) Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 119, 756–766 [DOI] [PMC free article] [PubMed] [Google Scholar]