Background: PilA is the major type IVa pilin that forms the conductive nanowires of Geobacter sulfurreducens.
Results: We report the atomic resolution structure of PilA determined with solution state NMR spectroscopy.
Conclusion: The Geobacter sulfurreducens PilA adopts a long, kinked α-helix with a dynamic C-terminal region.
Significance: The structure provides a foundation to build a model of the bacterial nanowire.
Keywords: Bacterial Metabolism, Bioenergetics, Electron Transport, Membrane Proteins, NMR, Structural Biology, Pili
Abstract
Several species of δ proteobacteria are capable of reducing insoluble metal oxides as well as other extracellular electron acceptors. These bacteria play a critical role in the cycling of minerals in subsurface environments, sediments, and groundwater. In some species of bacteria such as Geobacter sulfurreducens, the transport of electrons is proposed to be facilitated by filamentous fibers that are referred to as bacterial nanowires. These nanowires are polymeric assemblies of proteins belonging to the type IVa family of pilin proteins and are mainly comprised of one subunit protein, PilA. Here, we report the high resolution solution NMR structure of the PilA protein from G. sulfurreducens determined in detergent micelles. The protein is >85% α-helical and exhibits similar architecture to the N-terminal regions of other non-conductive type IVa pilins. The detergent micelle interacts with the first 21 amino acids of the protein, indicating that this region likely associates with the bacterial inner membrane prior to fiber formation. A model of the G. sulfurreducens pilus fiber is proposed based on docking of this structure into the fiber model of the type IVa pilin from Neisseria gonorrhoeae. This model provides insight into the organization of aromatic amino acids that are important for electrical conduction.
Introduction
Several species of anaerobic metal-reducing δ proteobacteria utilize various strategies of extracellular electron transport (EET)2 to deliver the electrons produced during respiration to insoluble minerals, containing species such as Fe(III) and Mn(IV), as well as other soluble and insoluble extracellular electron acceptors (1–5). Some of these strategies include direct transfer of electrons via multiheme cytochromes (6), transport via soluble redox mediators (7), and conduction-based transport via filamentous bacterial nanowires or another conductive matrix (8, 9). The ability to carry out EET has enabled these bacteria to play important roles in mineral and nutrient cycling (1), bioremediation of toxic heavy metals (2, 10), and energy production via bacterial fuel cells (6, 11).
Geobacter species represent important and abundant microorganisms that are capable of EET. They putatively employ conductive nanowires to transfer electrons over comparatively long distances to extracellular electron acceptors (8). These nanowires are members of the type IVa family of pili and are comprised of primarily one protein subunit, pilin (12). The fibers can reach lengths up to 20 μm, ∼20 times the length of a typical Geobacter cell (13). In Geobacter sulfurreducens, a model organism capable of EET, the major pilin subunit is encoded by the gene pila, which produces the protein PilA. Similar to other type IVa pilins, PilA is expressed as a prepilin that is cleaved in the inner membrane prior to assembly into the fiber (14). Cleavage is carried out by a prepilin peptidase that cleaves between a conserved glycine and the phenylalanine that becomes the N-terminal residue of the mature protein (15). The cleaved pilin subunits are assembled from the inner membrane into mature fibers (16). The core of these fibers is formed by a highly conserved N-terminal region that G. sulfurreducens PilA shares with other type IVa pilins (Fig. 1).
FIGURE 1.
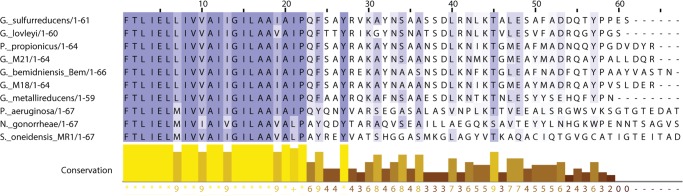
Alignment of various major pilin subunit amino acid sequences. Species whose pilin subunit amino acid sequence length is >66 were truncated to 67 amino acids. Eight species capable of EET and two species that are not capable of EET were included in the alignment.
Atomic resolution structures of several type IVa pilins have been reported (16, 17). These structures share a similar overall architecture, consisting of a conserved N-terminal helical region followed by a divergent globular head domain. However, G. sulfurreducens PilA is considerably shorter (61 amino acids) when compared with the pilins of known structure (∼150 amino acids) (8). This suggests that differences in the sequence or organization of the α-helical region could contribute to electrical conductivity in G. sulfurreducens nanowires.
The mechanism of electrical conduction by G. sulfurreducens nanowires is the focus of intense research. Two models of electron transport have been proposed to explain the conduction. One model suggests that electrical conduction involves electron superexchange between redox active sites such as cytochromes that are bound to the nanowire fibers (18). An alternative hypothesis proposes metallic like conduction along the nanowire (19). This model differs from superexchange in that it envisions the electrons as delocalized across the entire nanowire rather than hopping or tunneling between discrete redox centers (20).
Here we report the heterologous expression and structure determination of G. sulfurreducens PilA (GSu PilA). To our knowledge, this solution NMR structure is the first atomic resolution structure of a type IVa pilin that is involved in the formation of conductive bacterial nanowires. The structure was determined in the presence of 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC) detergent micelles, revealing the interactions of a full-length type IVa pilin with a lipid environment.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The PilA protein was expressed as a fusion with TrpLE in C41(DE3) Escherichia coli using the expression vector pTCLE (21, 22). Fusion to TrpLE results in the formation of insoluble inclusion bodies, facilitating the expression and purification of the protein. The plasmid also encodes a His6 tag between the TrpLE and the cleavage site. The pila gene from G. sulfurreducens KN400 was optimized for expression in E. coli, purchased from IDT, and inserted into the pTCLE plasmid using the existing Nde1 and Xho1 restriction sites. Cells transformed with the plasmid were grown to an A600 of ∼0.6 and then induced for 4 to 6 h with the addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside. The cells were harvested by centrifugation at 9000 × g for 30 min and stored at −20 °C.
Thawed cells were lysed by incubating in Bugbuster reagent (Pierce Scientific) with 100 μg/ml lysozyme and 200 μg/ml DNase for up to 1 h. The insoluble inclusion bodies were separated from the soluble cellular components by centrifugation at 15,000 × g for 30 min. Then the inclusion bodies were solubilized in loading buffer containing 6 m guanidine-HCl, 10 mm imidazole, and 50 mm sodium phosphate at pH 8.0.
The fusion protein was separated using a nickel-Sepharose column (GE Healthcare) and eluted from the column using a step gradient from 10 mm imidazole to 250 mm imidazole. β-Mercaptoethanol was added to a final concentration of 1% (v/v), and the protein was stored at 4 °C overnight. The fusion protein was precipitated by dialysis against MilliQ-purified water and pelleted by centrifugation at 9000 × g for 30 min. The pelleted protein was further washed with MilliQ-purified water and dried under vacuum.
The fusion protein was dissolved in 70% trifluoroacetic acid and cleaved for 2 h with 1 m cyanogen bromide. The cleavage reaction was stopped by lyophylization. Cleaved PilA was dissolved in 6 m guanidine-HCl and separated from uncleaved fusion protein using a 2.6 cm by 60 cm S-100 gel filtration column. Cleaved PilA was concentrated and diluted into 1% β-octylglucopyranoside, 50 mm sodium phosphate pH 6.0. Insoluble protein was removed by centrifugation for 15 min at 9000 × g. The purified PilA was extensively dialyzed against MilliQ-purified water to remove β-octylglucopyranoside, lyophilized, and dissolved in 200 mm DHPC.
NMR Data Collection
NMR experiments were conducted on 750 and 800 MHz (1H frequencies) Agilent (Varian) VNMRS NMR spectrometers. All NMR spectra were acquired at 35 °C. The experiments used for backbone assignments were HNCA, HNCO, HNCACB, and CBCA(CO)NH. Side chain assignments were made using an HCCH-TOCSY and CCH-TOCSY. Some 3J-coupling constants were acquired from a three-dimensional HNHA experiment. 1H 1H NOE-based distance restraints were derived from three-dimensional 15N-edited NOESY-HSQC and three-dimensional 13C-edited NOESY-HSQC experiments, both with 200-ms mixing times. Micelle localization was assessed using a titration with Gd(III)-diethylenetriaminepentaacetic acid (Gd-DTPA). The Gd-DTPA was prepared as a 250 mm stock solution in MilliQ-purified water, and the pH was adjusted to 5.0 using 6 m NaOH. Gd-DTPA was added to a 1 mm solution of PilA in 200 mm DHPC over a concentration range of 0 to 8 mm. All spectra were processed using nmrPipe and visualized using CARA (Rochus Keller, ETH Zurich), nmrviewJ (23), or Sparky (24).
Structure Calculation
Backbone dihedral angles were obtained using the program TALOS (25). H-H NOE peak volumes were calculated using Gaussian peak fitting in the program Sparky. These volumes were converted into distance constraints using CYANA 2.1 (26). NOESY peak lists were partially assigned manually. The TALOS dihedral angles, 3J coupling constants, and NOESY peak lists were used to perform automated assignment and structure calculation in CYANA (version 2.1). Following initial structure calculation, hydrogen bond restraints were added based on chemical shifts, temperature coefficients (27), and initial structures. Temperature coefficients were determined by acquiring 15N HSQC spectra at 25° to 45 °C. Hydrogen bonds were assigned where the change in proton chemical shift was more positive than −4.6 ppb/K (27). The automated assignment and structure calculations were repeated with these hydrogen bond restraints. The 20 lowest energy structures were subjected to further refinement in explicit water using CNS-Solve (version 1.1) (28, 29). We found that refinement in explicit water resulted in overall improvements in geometry and structural statistics, such as clashscore. Of these refined structures, 18 were selected that did not contain any distance restraint violations >0.5 Å or dihedral angle violations >5°. For figures where a single model from the ensemble is shown, the conformer with the best clashscore and MolProbity score was used (30). A model of the G. sulfurreducens nanowire was produced by superimposing our structure (residues 2–50) onto the homologous region of Neisseria gonorrhoeae fimbrial protein (Protein Data Bank code 2HIL) (12) using the software Chimera (31). The atomic coordinates for the structural ensemble have been deposited in the Protein Data Bank under code 2M7G.
Sequence Alignment
The amino acid sequences of major pilin subunits from 10 species of bacteria were aligned using Clustal Omega on the EMBL-EBI website (32, 33). Eight species of bacteria capable of EET were selected, Geobacter sulfurreducens KN400, Geobacter lovleyi, Pelobacter propionicus, Geobacter M21, Geobacter bemidjiensis bem, Geobacter M18, Geobacter metallireducens, and Shewanella oneidensis. Two control bacterial species that do not carry out EET were also included, Pseudomonas aeruginosa and N. gonorrhoeae. These strains were chosen based on the availability of full-length high resolution structures of the pilin subunits. The alignment was visualized using Jalview (version 2.8) (34).
RESULTS
Type IVa pilins are expressed in vivo as pre-pilins that are cleaved in the inner membrane prior to assembly into filaments. To study the cleaved pilin, we expressed only the coding sequence for mature GSu PilA as a fusion protein with TrpLE (21, 22). Following purification, cleavage of the TrpLE-PilA fusion with cyanogen bromide yielded sufficient PilA protein from 1 liter of cells to produce a 1 mm NMR sample in 350 μl of 200 mm DHPC.
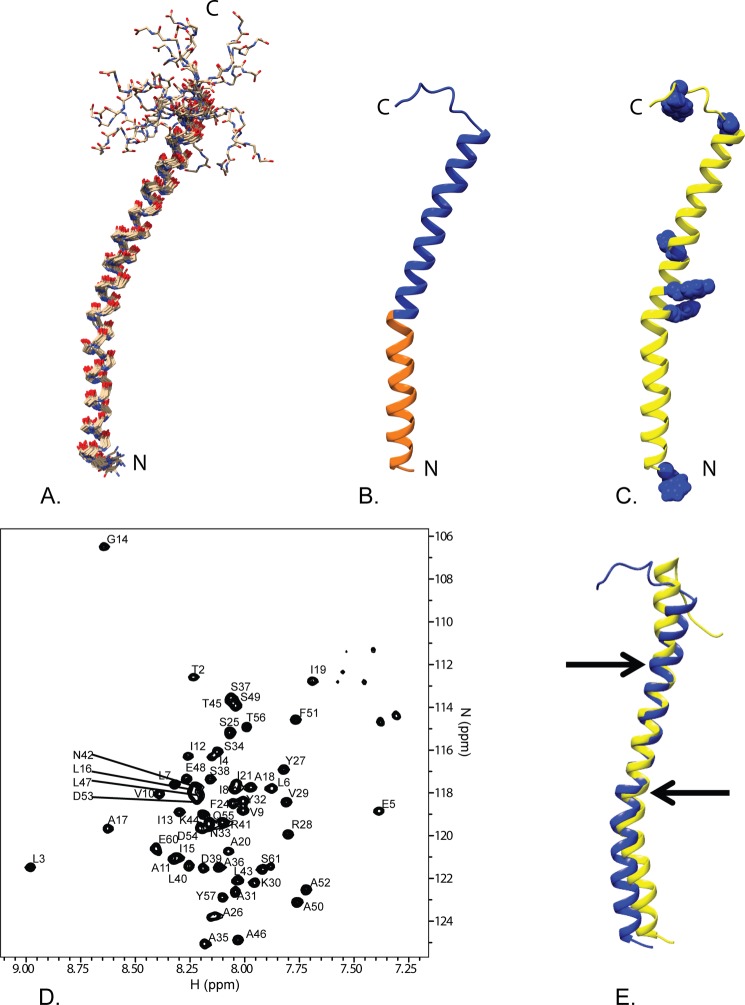
Fig. 2 shows an overview of the GSu PilA structure determined using solution state NMR spectroscopy. The backbone r.m.s.d. to the mean of the structural ensemble is 0.6 Å for the ordered residues. The 15N TROSY spectrum of the protein in DHPC micelles is shown in Fig. 2D. A summary of the structural statistics and restraints is provided in Table 1. Overall, the structure adopts a bent α-helix from residue 1 to residue 52 that is ∼75 Å long. The bend is located at proline 22, which is highly conserved throughout type IVa pilin proteins, as shown in Fig. 1. The structure is poorly restrained from residue 53 to the C terminus, resulting in a number of divergent and extended structures for this region.
FIGURE 2.
Overview of the solution NMR structure of G. sulfurreducens PilA. A, overlay of the ensemble of 18 structures that did not contain NOE violations >0.5 Å or dihedral angle violations >5°. B, ribbon diagram of the selected conformer (see “Materials and Methods”) with the highly conserved core domain corresponding to amino acids 1–22 colored in orange and the rest of the protein colored blue. C, ribbon diagram of the selected conformer with the aromatic residues shown in blue space filling. The amino terminus and carboxyl terminus are indicated by N and C, respectively. D, 15N TROSY spectrum of GSu PilA with backbone amide assignments. Data were collected at 750 MHz on an Agilent VNMR spectrometer. E, overlay of the homology model of GSu PilA (38) and the experimentally determined GSu PilA structure. The arrows indicate where the degree of bend differs between the structures near residues 22 and 42. To emphasize the differences, the structures were aligned using residues 23–41.
TABLE 1.
Summary of structural statistics and restraints
Structural statistics were calculated using the Protein Structure Validation Suite (43).
| Distance restraints | |
| Total NOE | 755 |
| Intra-residue | 172 |
| Inter-residue | 583 |
| Sequential (|i − j| = 1) | 253 |
| Medium-range (|i − j|<5) | 329 |
| Long-range (|i − j|>5) | 1 |
| H-Bond Restraints (|i − j|<5) | 27 |
| Total dihedral angle restraints | 104 |
| φ | 51 |
| ψ | 53 |
| 3J HNHA coupling constant restraints | 36 |
| Total restraints | 922 |
| Total no. of restricting constraints per restrained residuea | 15.4 |
| Structure statisticsb | |
| Violations | |
| r.m.s.d. of distance violation/constraint (Å) | 0.06 |
| r.m.s.d. of dihedral angle violation/constraint | 0.22° |
| Max dihedral angle violation | 3.3° |
| Max distance constraint violation (Å) | 0.47 |
| r.m.s.d. from ideal geometry | |
| Bond length (Å) | 0.021 |
| Bond angles | 1.1° |
| Average pairwise r.m.s.d (Å) | |
| Ordered backbonec | 0.6 |
| All Backbone | 3.6 |
| Ordered heavy atomsc | 1.1 |
| All heavy atoms | 4.0 |
| Ramachandran statisticsd | |
| Most favored regions | 99.8% |
| Allowed regions | 0.2% |
| Disallowed regions | 0% |
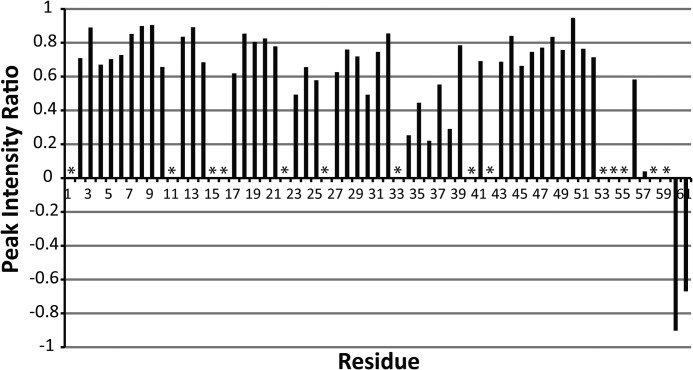
To determine whether the poorly restrained C terminus of the protein is a result of increased dynamics, we performed an H-N heteronuclear NOE experiment. The results of this experiment are shown in Fig. 3. The H-N heteronuclear NOE ratios near and below zero at the C-terminal residues (56–61) indicate increased dynamics compared with the rest of the PilA protein. We also observed multiple peaks in the 15N HSQC spectrum that correspond to single residues in the C-terminal region, suggesting that the C terminus may be adopting multiple conformations. In addition to the clearly dynamic C-terminal region, there is a second region of increased dynamics located at residues 34–38. Flexibility in this helix has been suggested to contribute to proper packing of other type IVa pilins into fibers (35).
FIGURE 3.
Summary of H-N heteronuclear NOE data. Spectra with and without proton saturation (3s) were acquired. The plot shows the ratio of cross-peak intensities with and without proton saturation. The data were collected at 800 MHz. Asterisks indicate resonances that could not be clearly identified and quantitatively characterized.
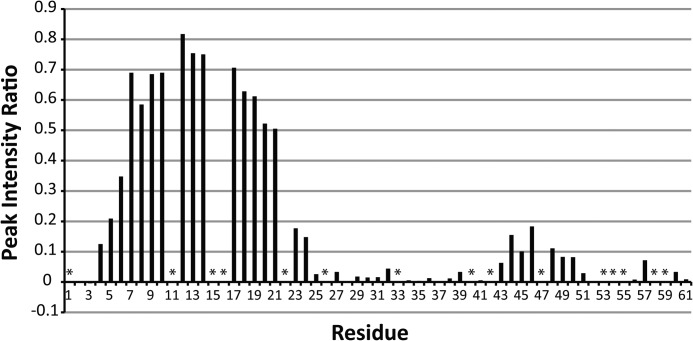
Our structure was determined in the presence of DHPC detergent micelles. The interaction between the micelle and protein was analyzed using the polar probe Gd-DTPA. Residues that are located inside of the detergent micelle will be protected from relaxation by Gd-DTPA, whereas residues located outside of the micelle will experience significant paramagnetic relaxation. The results of this experiment are shown in Fig. 4. The data clearly show substantial protection from paramagnetic relaxation in the N-terminal region of GSu PilA from residue 5 to residue 21. These data indicate that the N terminus of PilA is associated with the detergent micelle.
FIGURE 4.
Summary of paramagnetic relaxation by Gd-DTPA. The site-specific reduction in peak intensity upon addition of 4 mm Gd-DTPA to an 15N-labeled sample of GSu PilA is shown. Values near 1 indicate no loss of intensity and protection from Gd-DTPA. No correction was made for the small sample dilution (∼3% maximum) that occurred during the titration. The data were collected at 750 MHz. Asterisks indicate resonances that could not be clearly identified and quantitatively characterized.
DISCUSSION
GSu PilA and the pilins of many related species are atypical members of the type IVa family of pilins. The previously reported structures of type IVa pilins have shown that they generally consist of a long α-helical domain followed by a globular domain (16). The globular domains of these pilins make extensive contact along the N-terminal helix, leading to the distinction of two subdomains within the N-terminal helix, α1-N (amino acids 1–28) and α1-C (amino acids 29–52). However, GSu PilA is 61 amino acids long, 80 or more amino acids shorter than other type IVa pilins for which structures are available. Our structure shows that GSu PilA consists of only the N-terminal α-helix combined with a short and flexible C-terminal region. Thus, the α1-C subdomain of GSu PilA lacks stabilization from a C-terminal globular domain, probably contributing to the increased dynamics observed in this region. In other type IVa pilins, a proline or glycine at residue 42 gives rise to a second bend in the N-helix (12, 36). G. sulfurreducens and related species (see Fig. 1) lack this Pro/Gly and instead have an asparagine at this position. In our structure, the bend at residue 42 is not evident, likely due to the asparagine substitution at this location.
Our NMR data show that the C-terminal region of GSu PilA is highly dynamic in solution. Interestingly, the amino acid sequence in this region is poorly conserved, except for residues Tyr-57 and Pro-58. These residues are highly conserved among EET capable bacteria with short pilins, but are poorly conserved in other species (see Fig. 1). The conservation of these residues in an otherwise divergent region suggests that they play an important role in the function of this type of pilin. Thus, it is possible that this region gains structure upon fiber formation.
In addition to the flexibility in the C-terminal region of GSu PilA, we also observed significant dynamics in the middle of the PilA helix (see Fig. 3, residues 34–38). A principal feature of type IVa pili is flexibility that allows these fibers to bend and stretch without breaking (37). It has been suggested that the inter-subunit interactions in a bent fiber may be different from those in a straight fiber (16). The increased dynamics that we observe in the central region of the helix may also contribute to fiber flexibility by allowing the subunits to accommodate distortions that might occur as the fiber bends in response to its local environment or as it is assembled.
A homology model of GSu PilA was recently published based upon the x-ray crystal structure of P. aeruginosa full-length pilin (38). This homology model is also α-helical; however, the homology model differs from our structure with an average pairwise r.m.s.d. of 2.20 Å for residues 3–50. The structures differed in the degree of bend around proline 22 and the homology model exhibited the bend at residue 42 that is not present in our structure. This is not surprising because these are the primary sites where the structure of the P. aeruginosa pilin diverges from our experimental structure of Gsu PilA. In the case of the bend at residue 42, this is likely due to the presence of an asparagine at this position instead of the proline or glycine normally found in other pilins, including the template structure from P. aeruginosa. The difference in the bend at proline 22 could be due to differences in amino acid sequence 2 residues C-terminal of proline 22. In addition, the template structure from P. aeruginosa was determined using x-ray crystallographic methods and these differences may be caused by the protein adopting slightly different conformations in solution versus the crystal. The differences between the homology model and our experimental structure are illustrated in Fig. 2E.
Assembly of pilin fibers is thought to occur from a reservoir of pilin subunits in the bacterial inner membrane (16). Our structural data are consistent with this model. The N-terminal region of GSu PilA is associated with the membrane-mimicking detergent micelle, suggesting that this region is likely inserted into the bacterial inner membrane prior to polymerization. Its length of 22 amino acids (including proline 22) is consistent with the average length of other membrane spanning α-helices (39). Importantly, this length would position the N terminus of GSu PilA, and consequently the prepilin cleavage site, at the edge of the hydrophobic membrane where it could be easily accessed by the prepilin peptidase. Thus, the membrane may play an additional role in positioning the prepilin cleavage site for efficient cleavage by the prepilin peptidase.
The structured domain of GSu PilA is structurally similar to the homologous region in other type IVa pilin structures. Fig. 5A illustrates this structural similarity. Only the domain that is thought to interact with the membrane in the native protein showed significant interaction with the DHPC detergent micelle, consistent with a properly folded membrane associated protein. Taken together, these observations suggest that the protein is adopting a biologically relevant conformation.
FIGURE 5.
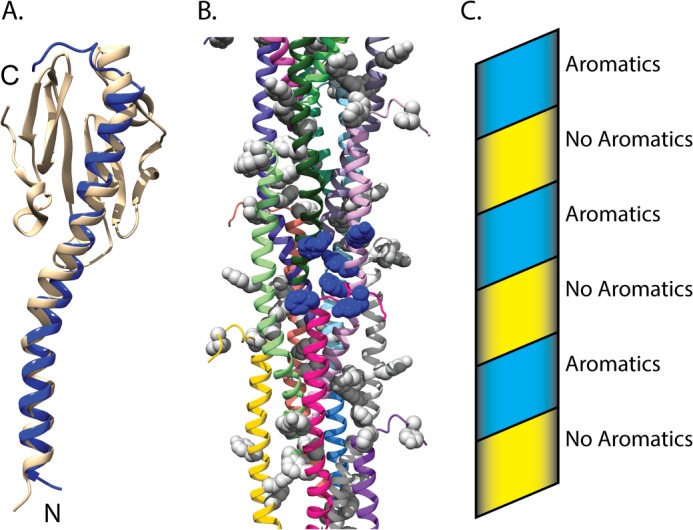
Model of a nanowire fiber based on the structure of G. sulfurreducens PilA. A, superimposition of PilA from G. sulfurreducens on to the homologous type IV pilin from N. gonorrhoeae (Protein Data Bank code 2HIL) (12). The amino terminus and carboxyl terminus are indicated by N and C, respectively. B, model of the bacterial nanowire, based on the pilus assembly of N. gonorrhoeae (Protein Data Bank code 2HIL) (12). Aromatic side chains are shown in space filling. A single cluster of aromatic side chains is shown in blue space filling, whereas all others are shown in gray. Ribbons of each subunit were colored individually. C, schematic diagram showing the progression of the aromatic clusters up the pilus structure. The aromatic band is colored blue, and the aromatic devoid band is colored yellow.
To better understand the possible interchain interactions between GSu PilA subunits, we superimposed the GSu PilA structure onto the pilin in the model of the N. gonorrhoeae pilus (Protein Data Bank code 2HIL) (12, 36). The N. gonorrhoeae pilus core is formed by the helical packing of the α-1 helices into staggered three-helix bundles with a rise of 10.5 Å per pilin subunit (12). The model generated by this superimposition is shown in Fig. 5. The backbone r.m.s.d. for the residues used for the docking (3–50) is 2.6 Å. Fig. 5A shows an individual subunit from the superimposition. The 3.5 to 4.0 nm overall width of the model fiber is consistent with previous electron microscopy studies of G. sulfurreducens conductive nanowires (8, 19). Interestingly, the aromatic residues of neighboring GSu PilA subunits are clustered within a sphere of radius 15 Å, shown in blue in Fig. 5B. These clusters arise from the helical packing of the individual subunits, which results in close contact between the N terminus (Phe-2), the center of the subunit (Phe-24, Tyr-27, and Tyr-32), and the C-terminal region (Phe-51 and Tyr-57) of neighboring subunits. The clustering results in an aromatic rich band and an aromatic devoid band that coil along the pilus structure, as shown in the schematic in Fig. 5C.
A recent study reported that mutation of the aromatic residues C-terminal of proline 22 resulted in substantial loss of electrical conduction by G. sulfurreducens nanowires. These aromatic residues are highly conserved (shown in Fig. 1) in species thought to utilize a similar EET mechanism to that of G. sulfurreducens (40). Taken together with our model, these data suggest that clustering of aromatic amino acids likely plays an important role in conduction, probably by bringing aromatic side chains close enough to contribute to electron transfer through delocalized orbitals (41) and/or promoting electron hopping (18) through tyrosines within or between clusters. The aromatic clusters could also facilitate electron transfer between c-type cytochromes or other redox-active proteins bound to the nanowire (42).
This model provides valuable information about the structure of GSu PilA and G. sulfurreducens nanowires. However, it does not address the possible conformational changes that could occur upon subunit polymerization. Dynamics observed in the middle of the helical region could indicate a possible location of conformational changes upon polymerization. The C-terminal region may be stabilized upon polymerization, altering the interaction between Tyr-57 and other aromatic side chains. Refinement of the model as new experimental data are acquired and further structural characterization of fully assembled G. sulfurreducens nanowires will be critical to our further understanding of bacterial nanowires and the changes that take place upon polymerization. The model also provides a framework for docking of other proteins, such as c-type cytochromes, that may be involved in EET by these nanowires.
The solution structure of full-length GSu PilA is representative of a unique class of truncated type IVa pilins that is common among EET-capable bacteria. Our structure of GSu PilA enables construction of a model of a G. sulfurreducens pilin fiber. This model provides initial insights into the overall architecture of these structures, supporting a key role and likely arrangement of mechanistically important aromatic amino acid side chains. These structural insights provide an important foundation, which will support further efforts to improve our understanding of extracellular electron transport by G. sulfurreducens and related δ proteobacteria, as well as develop new biological nanomaterials.
Acknowledgments
We thank Dr. Ryan Renslow and Dr. John Cort for helpful conversations and advice.
This work was supported by the William Wiley Post-Doctoral Fellowship from the Environmental Molecular Sciences Laboratory. All data were collected at the Environmental Molecular Sciences Laboratory, a national scientific user facility supported by the Department of Energy's Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory.
The atomic coordinates and structure factors (code 2M7G) have been deposited in the Protein Data Bank (http://wwpdb.org/).
The NMR chemical shifts and restraints have been deposited in the Biological Magnetic Resonance Data Bank under accession no. 19185.
- EET
- extracellular electron transport
- GSu PilA
- G. sulfurreducens PilA
- DHPC
- 1,2-dihexanoyl-sn-glycero-3-phosphocholine
- Gd-DTPA
- Gd(III)-diethylenetriaminepentaacetic acid
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Lovley D. R., Holmes D. E., Nevin K. P. (2004) Dissimilatory Fe(III) and Mn(IV) reduction. Adv. Microb. Physiol. 49, 219–286 [DOI] [PubMed] [Google Scholar]
- 2. Lovley D. R., Phillips E. J. P., Gorby Y. A., Landa E. R. (1991) Microbial reduction of uranium. Nature 350, 413–416 [Google Scholar]
- 3. Lovley D. R., Phillips E. J. (1988) Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 54, 1472–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Myers C. R., Nealson K. H. (1988) Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240, 1319–1321 [DOI] [PubMed] [Google Scholar]
- 5. Lovley D. R., Coates J. D., Blunt-Harris E. L., Phillips E. J. P., Woodward J. C. (1996) Humic substances as electron acceptors for microbial respiration. Nature 382, 445–448 [Google Scholar]
- 6. Shi L., Squier T. C., Zachara J. M., Fredrickson J. K. (2007) Respiration of metal (hydr)oxides by Shewanella and Geobacter: a key role for multihaem c-type cytochromes. Mol. Microbiol. 65, 12–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brutinel E. D., Gralnick J. A. (2012) Shuttling happens: soluble flavin mediators of extracellular electron transfer in Shewanella. Appl. Microbiol. Biotechnol. 93, 41–48 [DOI] [PubMed] [Google Scholar]
- 8. Reguera G., McCarthy K. D., Mehta T., Nicoll J. S., Tuominen M. T., Lovley D. R. (2005) Extracellular electron transfer via microbial nanowires. Nature 435, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 9. Bond D. R., Strycharz-Glaven S. M., Tender L. M., Torres C. I. (2012) On electron transport through Geobacter biofilms. ChemSusChem 5, 1099–1105 [DOI] [PubMed] [Google Scholar]
- 10. Lovley D. R. (2001) Bioremediation. Anaerobes to the rescue. Science 293, 1444–1446 [DOI] [PubMed] [Google Scholar]
- 11. Logan B. E., Hamelers B., Rozendal R., Schröder U., Keller J., Freguia S., Aelterman P., Verstraete W., Rabaey K. (2006) Microbial fuel cells: methodology and technology. Environ. Sci. Technol. 40, 5181–5192 [DOI] [PubMed] [Google Scholar]
- 12. Craig L., Volkmann N., Arvai A. S., Pique M. E., Yeager M., Egelman E. H., Tainer J. A. (2006) Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol. Cell 23, 651–662 [DOI] [PubMed] [Google Scholar]
- 13. Reguera G., Nevin K. P., Nicoll J. S., Covalla S. F., Woodard T. L., Lovley D. R. (2006) Biofilm and nanowire production leads to increased current in Geobacter sulfurreducens fuel cells. Appl. Environ. Microbiol. 72, 7345–7348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arts J., van Boxtel R., Filloux A., Tommassen J., Koster M. (2007) Export of the pseudopilin XcpT of the Pseudomonas aeruginosa type II secretion system via the signal recognition particle-Sec pathway. J. Bacteriol. 189, 2069–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nunn D. N., Lory S. (1991) Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc. Natl. Acad. Sci. U.S.A. 88, 3281–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Giltner C. L., Nguyen Y., Burrows L. L. (2012) Type IV pilin proteins: versatile molecular modules. Microbiol. Mol. Biol. Rev. 76, 740–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Craig L., Pique M. E., Tainer J. A. (2004) Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2, 363–378 [DOI] [PubMed] [Google Scholar]
- 18. Strycharz-Glaven S. M., Snider R. M., Guiseppi-Elie A., Tender L. M. (2011) On the electrical conductivity of microbial nanowires and biofilms. Energy Environ. Sci. 4, 4366–4379 [Google Scholar]
- 19. Lovley D. R. (2012) Long-range electron transport to Fe(III) oxide via pili with metallic-like conductivity. Biochem. Soc. Trans. 40, 1186–1190 [DOI] [PubMed] [Google Scholar]
- 20. Malvankar N. S., Lovley D. R. (2012) Microbial nanowires: a new paradigm for biological electron transfer and bioelectronics. ChemSusChem 5, 1039–1046 [DOI] [PubMed] [Google Scholar]
- 21. Calderone T. L., Stevens R. D., Oas T. G. (1996) High-level misincorporation of lysine for arginine at AGA codons in a fusion protein expressed in Escherichia coli. J. Mol. Biol. 262, 407–412 [DOI] [PubMed] [Google Scholar]
- 22. Yansura D. G. (1990) Expression as trpE fusion. Methods Enzymol. 185, 161–166 [DOI] [PubMed] [Google Scholar]
- 23. Johnson B. A. (2004) Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278, 313–352 [DOI] [PubMed] [Google Scholar]
- 24. Goddard T. D., Kneller D. G. (2009) SPARKY 3, University of California, San Francisco, CA [Google Scholar]
- 25. Cornilescu G., Delaglio F., Bax A. (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 26. Güntert P., Mumenthaler C., Wüthrich K. (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 27. Cierpicki T., Otlewski J. (2001) Amide proton temperature coefficients as hydrogen bond indicators in proteins. J. Biomol. NMR 21, 249–261 [DOI] [PubMed] [Google Scholar]
- 28. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 29. Nederveen A. J., Doreleijers J. F., Vranken W., Miller Z., Spronk C. A., Nabuurs S. B., Güntert P., Livny M., Markley J. L., Nilges M., Ulrich E. L., Kaptein R., Bonvin A. M. (2005) RECOORD: a recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins 59, 662–672 [DOI] [PubMed] [Google Scholar]
- 30. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 32. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., Lopez R. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. (2009) Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Craig L., Taylor R. K., Pique M. E., Adair B. D., Arvai A. S., Singh M., Lloyd S. J., Shin D. S., Getzoff E. D., Yeager M., Forest K. T., Tainer J. A. (2003) Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol. Cell 11, 1139–1150 [DOI] [PubMed] [Google Scholar]
- 36. Hartung S., Arvai A. S., Wood T., Kolappan S., Shin D. S., Craig L., Tainer J. A. (2011) Ultrahigh resolution and full-length pilin structures with insights for filament assembly, pathogenic functions, and vaccine potential. J. Biol. Chem. 286, 44254–44265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Biais N., Higashi D. L., Brujic J., So M., Sheetz M. P. (2010) Force-dependent polymorphism in type IV pili reveals hidden epitopes. Proc. Natl. Acad. Sci. U.S.A. 107, 11358–11363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Feliciano G. T., da Silva A. J., Reguera G., Artacho E. (2012) Molecular and electronic structure of the peptide subunit of Geobacter sulfurreducens conductive pili from first principles. J. Phys. Chem. A 116, 8023–8030 [DOI] [PubMed] [Google Scholar]
- 39. Eyre T. A., Partridge L., Thornton J. M. (2004) Computational analysis of alpha-helical membrane protein structure: implications for the prediction of 3D structural models. Protein Eng. Des. Sel. 17, 613–624 [DOI] [PubMed] [Google Scholar]
- 40. Vargas M., Malvankar N. S., Tremblay P. L., Leang C., Smith J. A., Patel P., Synoeyenbos-West O., Nevin K. P., Lovley D. R. (2013) Aromatic Amino Acids Required for Pili Conductivity and Long-Range Extracellular Electron Transport in Geobacter sulfurreducens. MBio 4, e00105–e00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Malvankar N. S., Vargas M., Nevin K. P., Franks A. E., Leang C., Kim B. C., Inoue K., Mester T., Covalla S. F., Johnson J. P., Rotello V. M., Tuominen M. T., Lovley D. R. (2011) Tunable metallic-like conductivity in microbial nanowire networks. Nat. Nanotechnol. 6, 573–579 [DOI] [PubMed] [Google Scholar]
- 42. Gray H. B., Winkler J. R. (2010) Electron flow through metalloproteins. Biochim Biophys. Acta 1797, 1563–1572 [DOI] [PubMed] [Google Scholar]
- 43. Bhattacharya A., Tejero R., Montelione G. T. (2007) Evaluating protein structures determined by structural genomics consortia. Proteins 66, 778–795 [DOI] [PubMed] [Google Scholar]