Background: Melanophilin links myosin Va to Rab27a-melanosomes, a cargo it transports along actin tracks.
Results: Melanophilin recruits more myosin Va motors to actin, slows their speed, and increases their processive run length.
Conclusion: The actin binding domain of melanophilin acts as a tether to enhance processive runs.
Significance: Proteins that link motors to cargo can enhance motor properties in ways that are favorable for their biological role.
Keywords: Gating, Intracellular Trafficking, Myosin, Nanotechnology, Protein Dynamics, Slac-2a, Melanophilin, Processivity, Regulation, Tethering
Abstract
Myosin Va (myoVa) is a molecular motor that processively transports cargo along actin tracks. One well studied cargo in vivo is the melanosome, a pigment organelle that is moved first by kinesin on microtubules and then handed off to myoVa for transport in the actin-rich dendritic periphery of melanocytes. Melanophilin (Mlph) is the adapter protein that links Rab27a-melanosomes to myoVa. Using total internal reflection fluorescence microscopy and quantum dot-labeled full-length myoVa, we show at the single-molecule level that Mlph increases the number of processively moving myoVa motors by 17-fold. Surprisingly, myoVa-Mlph moves ∼4-fold slower than myoVa alone and with twice the run length. These two changes greatly increase the time spent on actin, a property likely to enhance the transfer of melanosomes to the adjacent keratinocyte. In contrast to the variable stepping pattern of full-length myoVa, the myoVa-Mlph complex shows a normal gating pattern between the heads typical of a fully active motor and consistent with a cargo-dependent activation mechanism. The Mlph-dependent changes in myoVa depend on a positively charged cluster of amino acids in the actin binding domain of Mlph, suggesting that Mlph acts as a “tether” that links the motor to the track. Our results provide a molecular explanation for the uncharacteristically slow speed of melanosome movement by myoVa in vivo. More generally, these data show that proteins that link motors to cargo can modify motor properties to enhance their biological role.
Introduction
The correct and timely cellular distribution and positioning of organelles are essential for physiological processes. An intricate transport system has evolved to provide cells with the fine tuning necessary to generate asymmetric distributions of cellular content required for their function and morphology. Whereas long range transport in mammalian cells occurs along microtubule tracks and involves kinesin and dynein motor proteins, the distribution and retention of many vesicles and membranous structures at the cell periphery depend on myosin Va (myoVa)2 driven movement along actin (1–3). One such well studied cellular system is the transport of melanosomes, melanin-containing pigment granules. Melanosomes rely on kinesin-2 for long range transport on microtubules from the perinuclear region outward to the cell periphery. Near the cell periphery, kinesin hands off the melanosome to myoVa, perhaps assisted by the microtubule plus-end tip tracking protein end-binding protein 1 (EB1) (4). MyoVa uses actin tracks to distribute and then capture melanosomes in dendritic extensions near the melanocyte plasma membrane, in preparation for melanosome transfer to the surrounding keratinocytes (2, 3, 5). The melanosome-transport complex on actin consists of myoVa, two adapter proteins that join motor to cargo, and the melanosome. Melanophilin (Mlph, also called Slac2-a) binds to both myoVa and to the small GTPase Rab27a, which is incorporated into the melanosome membrane (Fig. 1A). The absence or dysfunction of any of these three proteins leads to abnormal pigmentation (6, 7).
FIGURE 1.
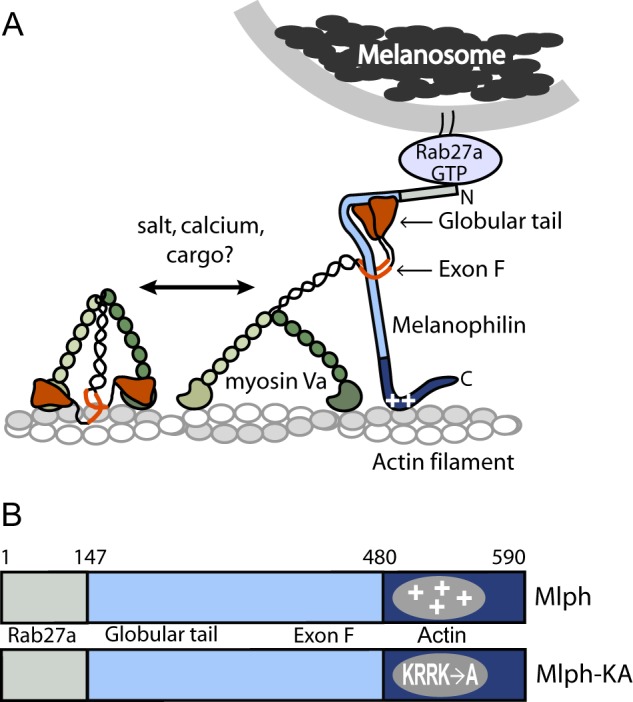
Schematic model of the myoVa-melanosome transport complex and the domain structure of Mlph. A, Mlph linking myoVa to melanosomes via Rab27-GTP. MyoVa exists in an equilibrium between a folded, inhibited state and an extended, active conformation, depending on the ionic strength and calcium concentration in vitro. Binding of cargo has been hypothesized to be the physiologic activator of myoVa. Binding of Mlph to myoVa requires both the globular tail and exon F. Mlph also binds via electrostatic interactions to actin (indicated by plus symbols on Mlph). B, domain structure of Mlph. The N-terminal domain binds to Rab 27a, the intrinsically disordered central portion to myoVa, and the C-terminal region to actin. A mutant Mlph (Mlph-KA) contains four point mutations (K493A/R495A/R496A/K497A) that ablate the positive charge necessary to interact strongly with actin.
MyoVa is a two-headed motor that steps processively along actin filaments, meaning that each head can take many 72-nm hand-over-hand steps along actin without dissociating (for review, see Ref. 8). The long step is made possible by the ∼24-nm elongated α-helical lever arm that is stabilized by binding six calmodulins. Following the neck is a ∼30-nm stretch of coiled-coil that dimerizes the molecule. Truncation of the molecule at this point produces a constitutively active motor called heavy meromyosin (myoVa-HMM). In full-length myoVa, the tail continues as an interrupted span of coiled-coil, which contains six alternatively spliced exons called A–F (9–11). The most C-terminal portion of the molecule is the globular tail domain, which consists solely of α-helices and loops (12). Alternatively spliced tail exons, in combination with the C-terminal globular tail, specify different cargoes by virtue of their ability to bind unique adapter proteins. The melanocyte isoform of myoVa contains exons ACDEF. Both exon F and the globular tail are required for Mlph binding (7, 13).
The globular tail also interacts with the motor domain, stabilizing a folded conformation with inhibited ATPase activity (14–16). In vitro, the folded, inactive conformation can be converted into the active, extended state by increasing the ionic strength or by increasing the calcium concentration at physiologic ionic strength (17) (Fig. 1A). Within the cell, cargo may be the physiological activator by virtue of its ability to displace the head-tail interaction. Although cargo-dependent activation is an attractive mechanism because it would prevent futile processive motion of motors without cargo, it has never been proven.
Recent work has shown that the stepping pattern of full-length myoVa in the absence of cargo is more complex than that of the constitutively active myoVa-HMM (18). Full-length myoVa alternated between two modes of movement. The fast motion was characterized by hand-over-hand stepping similar to that of myoVa-HMM. This was interspersed by periods of very slow movement (<50 nm/s), which showed back steps and short forward steps, suggestive of altered gating between the heads. The slow movement was less frequent at higher salt. This behavior is consistent with the idea that full-length myoVa in the absence of cargo alternates between the active (fast) and inhibited (slow) states during a processive run.
The adapter protein that links myoVa to the Rab27a-melanosome is Mlph, a 65-kDa protein consisting of three domains (Fig. 1B). The N-terminal region of Mlph binds the GTP form of Rab27a (19, 20). This region of Mlph was crystallized in complex with Rab27b, a related member of the small GTPase family (21). The crystal structure showed three subdomains: two helical synaptotagmin-like protein homology domains (SHD1 and SHD2) and a zinc binding motif. In contrast, the central region of Mlph is intrinsically unstructured (22). It has two binding sites for myoVa: residues 147–240 constitute the globular tail binding domain (23), and amino acids 300–400 interact with alternatively spliced exon F (22). The interaction between Mlph and myoVa is strengthened by residues 440–483, which are predicted to have a propensity for coiled-coil formation (24). The C terminus of Mlph (∼amino acids 480–590) contains a positively charged region implicated in actin binding (25, 26), and interaction with EB1, a protein that tracks the microtubule tip (4). Specifically, a cluster of four positively charged residues (Lys-493, Arg-495, Arg-496, and Lys-497) is important for actin binding and essential for melanosome localization in melanocytes (26).
The small GTPase Rab27a is anchored into the melanosome membrane via lipophilic geranylgeranyl groups that are added post-translationally (27). An important feature of Rab proteins is their cycle of association with membranes in the GTP state and their dissociation from membranes in the GDP state (27). A tripartite complex of myoVa, Mlph, and green fluorescent protein (GFP)-tagged Rab27a-GTP has been shown to move processively on actin in vitro, providing proof of principle that a transport complex can be reconstituted from these three proteins (28).
Here we use single-molecule total internal reflection fluorescence (TIRF) microscopy to determine the effect of Mlph on myoVa processivity near physiological ionic strength (150 mm KCl). We show that Mlph binding impacts all aspects of myoVa activity: the stepping pattern, speed, run length, and number of motors recruited to move on actin. The data are consistent with a cargo-dependent activation mechanism. Our results suggest that the efficiency of melanosome transport depends on a Mlph-dependent tethering of myoVa to actin at the expense of speed.
MATERIALS AND METHODS
Expressed Protein
The melanocyte isoform of mouse myoVa with either an N-terminal or C-terminal biotin tag and a C-terminal FLAG-tag (DYKDDDDK) was co-expressed with CaMΔall and purified as described previously (17, 18). CaMΔall is a calcium-insensitive calmodulin due to a point mutation in each of the four calcium binding loops (17). It binds tightly to the heavy chain irrespective of calcium concentration and thus produces a more intact myosin heavy chain following purification of myoVa from Sf9 cells. MyoVa with only a C-terminal FLAG tag was also expressed, as was a myoVa-HMM with a C-terminal biotin tag and C-terminal FLAG tag.
Mouse melanophilin (Mus musculus accession NP_443748) (Mlph) was cloned into baculovirus expression vector pAcSG2 with an N-terminal His6 tag. Another version of Mlph had a C-terminal SNAP tag (New England Biolabs) for labeling with biotin, followed by a FLAG tag for affinity purification. An N-His Mlph-KA was made by introducing four point mutations into the actin binding site: K493A/R495A/R496A/K497A (26).
Melanophilin constructs were expressed in the baculovirus/insect cell system. After 72 h of infection, cells expressing His-tagged Mlph were pelleted and resuspended in buffer A (50 mm NaPi, 5 mm imidazole, pH 8, 0.2 m NaCl, 7% sucrose, 7 mm β-mercaptoethanol, 1 μg/ml leupeptin, 0.2 mm AEBSF, and lysed by sonication. The lysate was clarified in a Beckman Ti70 rotor at 50,000 rpm for 40 min. The supernatant was incubated with a Ni2+-charged resin (His-select; Sigma) for 1 h. The resin was first washed with buffer A and then with buffer A containing 25 mm imidazole to remove nonspecifically bound protein. His-tagged protein was specifically eluted with buffer A containing 200 mm imidazole. Following elution, 1 mm dithiothreitol (DTT) was added. Some preparations were further purified on a Mono Q column equilibrated with 10 mm HEPES, pH 7, 200 mm NaCl, 1 mm EGTA, 1 mm DTT. Protein was eluted with a gradient of 200–600 mm NaCl. Peak fractions were concentrated and dialyzed against 10 mm imidazole, pH 7.8, 0.2 m NaCl, 1 mm EGTA, 1 mm DTT, 50% glycerol, and stored at −20 °C.
Mlph with a C-terminal SNAP tag followed by a FLAG tag was purified on a FLAG affinity column (anti-FLAG M2 affinity gel; Sigma). After 72 h of infection, cells were pelleted and resuspended in 10 mm imidazole, pH 7.4, 0.2 m NaCl, 1 mm EGTA, 7% sucrose, 1 mm DTT, 0.5 mm AEBSF, 0.5 mm TLCK, 5 μg/ml leupeptin (20 ml of buffer/billion cells) and lysed by sonication. The lysate was clarified in a Beckman Ti70 rotor at 50,000 rpm for 40 min. The supernatant was incubated with FLAG affinity resin for 1 h at 4 °C and then washed with 10 mm imidazole, pH 7.4, 0.2 m NaCl, 1 mm EGTA. FLAG-tagged protein was eluted with 0.1 mg/ml FLAG peptide in the wash buffer. One mm DTT was added to the peak fractions, which were then concentrated in a centrifugal filter device (Amicon Ultra 10K; Millipore). The protein was then dialyzed against 10 mm imidazole, pH 7.4, 0.2 m NaCl, 1 mm EGTA, 1 mm DTT, 50% glycerol and stored at −20 °C.
For attachment of SNAP-Biotin (New England Biolabs) SNAP-tagged Mlph was dialyzed against 500 mm Tris-Cl, pH 7.5, 0.2 m NaCl, 1 mm DTT, concentrated in a centrifugal filter device (Amicon Ultra 10K), and clarified (5 min, 10,000 × g). 5 μm SNAP-tagged Mlph and 10 μm SNAP-tagged substrate in 50 mm Tris-Cl, pH 7.5, 0.2 m NaCl, 0.1% Tween 20, 1 mm DTT were incubated for 30 min at 37 °C. Excess reagent was removed by dialysis against 10 mm imidazole, pH 7.4, 0.2 m NaCl, 1 mm DTT. The biotinated protein was further dialyzed against the same buffer containing 50% glycerol and stored at −20 °C.
Analytical Ultracentrifugation
The sedimentation coefficient of myoVa was determined by centrifugation in the Optima XL-I analytical ultracentrifuge (Beckman Coulter). Sedimentation velocity runs were performed in the An60Ti rotor at 35,000 rpm and 20 °C, in 25 mm imidazole, pH 7.4, 4 mm MgCl2, 1 mm EGTA, 1 mm DTT, and KCl concentrations ranging from 75 to 300 mm. Myosin concentration was ∼2 μm. Sedimentation values were determined by curve fitting to one or more species, using the DCDT+ program (29).
Processivity Assay
MyoVa or myoVa-HMM was diluted to 0.35 μm in buffer B-300 (10 mm imidazole, pH 7.4, 300 mm KCl, 4 mm MgCl2, 1 mm EGTA, 10 mm DTT), mixed with a 2-fold molar excess of actin and 1 mm MgATP, and centrifuged for 15 min at 400,000 × g to remove any myosin unable to dissociate from actin in the presence of ATP. The protein in the supernatant was diluted to 0.1–0.2 μm. Assays were performed at room temperature (22 °C) in 25-μl flow chambers. Flow cells were incubated with 0.1 mg/ml N-ethylmaleimide-modified myosin in buffer B-300 for 4 min, blocked with buffer B-25 (10 mm imidazole, pH 7.4, 25 mm KCl, 4 mm MgCl2, 1 mm EGTA, 10 mm DTT) plus 1 mg/ml BSA, rinsed with 3 volumes of buffer B-25, incubated with 0.05 μm rhodamine-phalloidin-labeled actin filaments in buffer B-25 for 3 min, and blocked with buffer B-25 plus 1 mg/ml BSA. The flow chamber was then rinsed with buffer B-150 (10 mm imidazole, pH 7.4, 150 mm KCl, 4 mm MgCl2, 1 mm EGTA, 50 mm DTT, 0.1 mg/ml CaMΔall, 2 mm MgATP, an oxygen scavenger system (3 mg/ml glucose, 0.1 mg/ml glucose oxidase, 0.18 mg/ml catalase), and an ATP-regenerating system (0.5 mm phosphoenolpyruvate and 100 units/ml pyruvate kinase)). To determine step sizes, experiments were performed at 2–4 μm ATP.
For labeling of myoVa only, motors were mixed with a 4-fold excess of 655-nm streptavidin-coated quantum dots (Qdots; Invitrogen) to ensure that most moving Qdots had single motors bound. Motors were mixed with a 10-fold molar excess of Mlph and incubated for 2 min.
To attach Qdots to both the His tag of Mlph and biotinated myoVa, the protein mixture contained equal concentrations of myoVa, Mlph, and 655-nm anti-His Qdots (gift from Paul Selvin), and a 4-fold excess of streptavidin 565-nm Qdots (Invitrogen). Because the addition of Mlph led to a very high density of motors on actin, a 3-fold molar excess of myoVa which did not contain a biotin tag was added to the flow cell, to prevent cross-linking of adjacent motors on actin via Qdots with multiple streptavidin moieties. This further guaranteed that data were obtained from single motors.
To label only the C terminus of Mlph with a Qdot, a 2-fold molar excess of SNAP-biotin-Mlph was mixed with myoVa lacking a biotin tag and incubated for 5 min. A 2-fold molar excess of streptavidin 655-nm Qdots was added and incubated for 5 min. The protein complex was diluted to a final motor concentration of 0.3–5.0 nm in buffer B-150 and added to the flow cell.
Data Acquisition and Analysis
The microscope setup has been described previously (18). Images were taken at 10-Hz frame rate using 2 × 2 pixel binning. Total acquisition time was 100 s. All image stacks were analyzed using ImageJ v1.44k (National Institutes of Health). We required motors to move continuously for five consecutive frames to be analyzed. Qdot position versus time was tracked with subpixel resolution using the SpotTracker plugin for ImageJ (30) for the duration of the movement. Speeds were calculated as the displacement from origin/time. Run length was calculated as total displacement from origin between first and last tracked frame. Kaplan-Meier analysis (31) of run lengths included all tracks. For characteristic run lengths analysis, traces were not analyzed if they started outside the time and space coordinates of the movie or if they terminated at the end of a filament or at the edge of the field of view. Statistical analysis was performed in GraphPad Prism (GraphPad Software, Inc.) All errors are given as S.D. unless otherwise noted. The characteristic run lengths and speeds were estimated by least squares fitting to cumulative distribution functions. Step sizes were calculated in MATLAB using the Kerssemakers step-finding algorithm (32). Average step sizes were determined by least squares fitting to histograms.
Binding of Mlph to Actin
Chicken skeletal actin was prepared from acetone powder (33). Actin (4 μm) was incubated with either WT-Mlph or Mlph-KA (2 μm) in 10 mm imidazole, pH 7.4, 150 mm NaCl, 4 mm MgCl2, 1 mm EGTA, and 1 mm DTT. The complex was spun at 4 °C for 20 min at 393,000 × g. Supernatant and pellet fractions were run on 12% SDS-gels.
RESULTS
MyoVa Is Poised to Be Activated at Physiological Salt
We expressed and purified the melanocyte isoform of myoVa, with alternatively spliced exons ACDEF in the tail for these studies (Fig. 2A). The transition from the folded, inhibited to extended, active conformation of full-length myoVa was followed by analytical ultracentrifugation over a range of ionic strengths (Fig. 3). Consistent with earlier findings, myoVa adopted a ∼14 S folded conformation at low salt (<100 mm KCl), and a ∼10 S extended conformation at salt concentrations >300 mm KCl (15, 17, 34). The transition between the two conformations is fairly steep and occurs near physiological ionic strength (∼150–200 mm KCl). Under cellular conditions the motor is therefore poised to switch between the active and inhibited states. All experiments described here used 150 mm KCl where myoVa is predominantly in the inhibited conformation.
FIGURE 2.
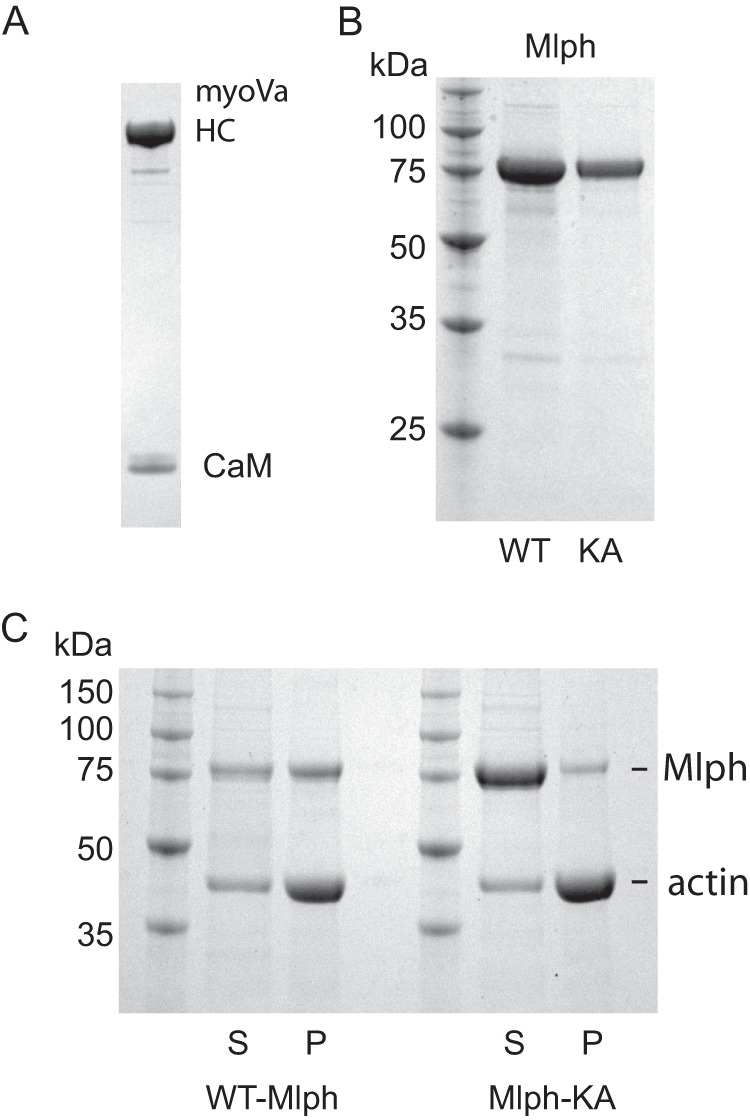
SDS-gels of purified proteins and ability of Mlph to bind actin. A, purified full-length myoVa with an N-terminal biotin tag showing the myoVa heavy chain (HC) and calmodulin (CaM); 4–12% gradient SDS-gel. B, left lane, marker proteins with indicated molecular masses. Middle lane, WT-Mlph. Right lane, mutant Mlph-KA, 12% SDS-gel. C, co-sedimentation of actin with WT-Mlph or Mlph-KA showing that Mlph-KA has a decreased ability to interact with actin. S, supernatant; P, pellet; 12% SDS-gel.
FIGURE 3.
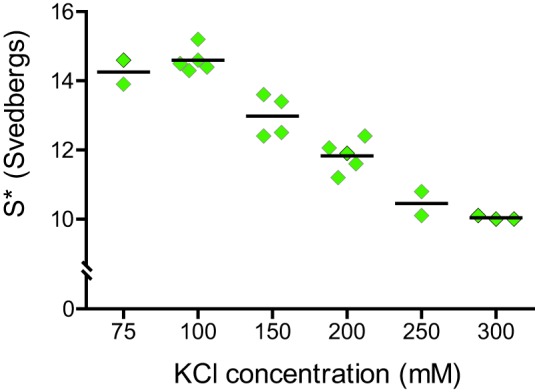
Salt-dependent conformational transition of myoVa. Analytical ultracentrifugation was used to follow the ionic strength dependence of the folded, inhibited conformation (∼14 S) to the active, extended (∼10 S) form. Intermediate S values represent an equilibrium between bent and extended molecules. Individual data points represent separate experiments. Horizontal bar indicates the average value.
Mlph Recruits More Motors to the Actin Track
Mlph was expressed using the baculovirus/insect cell expression system and purified by affinity chromatography (Fig. 2B). To show that Mlph binds to myoVa under the ionic conditions and protein concentrations used for the single-molecule experiments, a biotin tag on the motor domain of myoVa was labeled with a 565-nm streptavidin-Qdot, and N-His-Mlph was labeled with a 655-nm anti-His-Qdot (Fig. 4). Simultaneous movement of both red and green Qdots along rhodamine-phalloidin-labeled actin filaments was observed by TIRF microscopy (Fig. 4 and supplemental Movie S1). Unexpectedly, the speed at which the myoVa-Mlph complex moved (∼30 nm/s for the complex shown in Fig. 4) was much slower than in the absence of Mlph, which averaged ∼500 nm/s.
FIGURE 4.
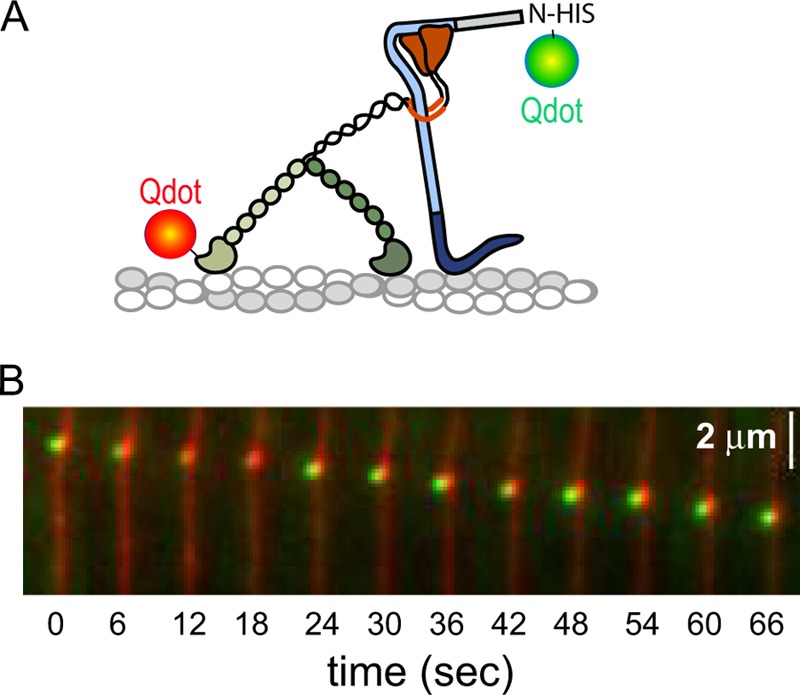
Mlph binds to myoVa in single-molecule assays. A, myoVa with a biotin tag on the motor domain was labeled with a 565-nm streptavidin-Qdot, and Mlph with an N-terminal His tag with a 655-nm anti-HIS Qdot. B, simultaneous movement of both red and green Qdots along rhodamine-phalloidin-labeled actin filaments was observed by TIRF microscopy. Speed, ∼30 nm/s. Conditions: 150 mm KCl, pH 7.4, 2 mm MgATP.
The speed, run length, and frequency of motile events in the presence or absence of Mlph were investigated in more detail using a Qdot on the myoVa motor domain and a 10-fold molar excess of unlabeled Mlph (Fig. 5A, upper). A visually striking change caused by addition of Mlph was a 17 ± 2-fold increase in the number of myoVa motors moving per time and length of actin filament, compared with myoVa alone (t test, p < 0.001) (Fig. 5B). In the absence of Mlph, the run frequency of the constitutively active, truncated myoVa-HMM, was only 3.3 ± 0.9 times higher than that observed with full-length myoVa (t test, p < 0.001) (Fig. 6A). The increase in run frequency due to motor activation alone is thus not sufficient to explain the 17-fold increase seen when Mlph is added to full-length myoVa.
FIGURE 5.
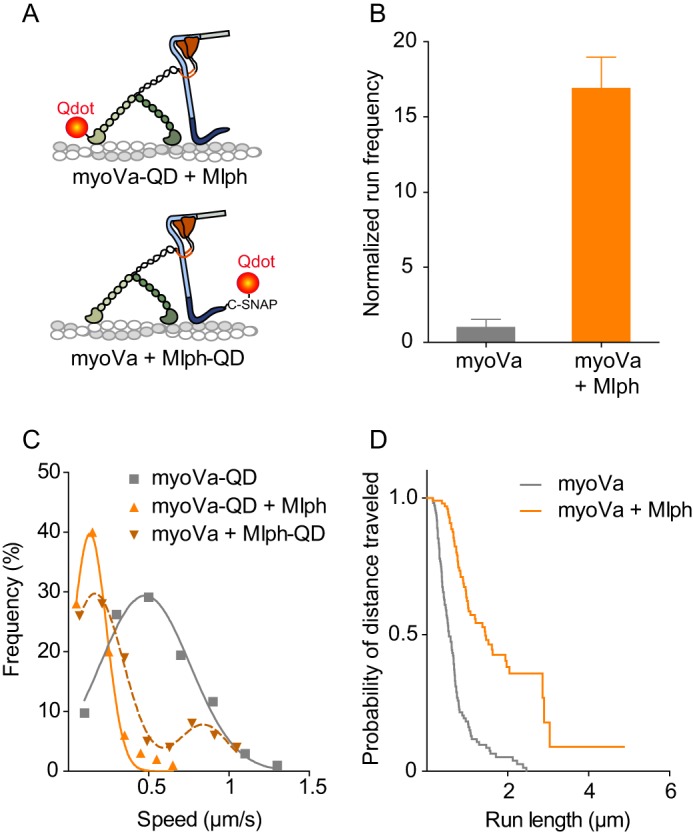
Comparison of myoVa single-molecule kinetics in the presence or absence of Mlph. A, illustration of labeling approaches for the myoVa-Mlph complex. Upper panel, myoVa with an N-terminal biotin tag was labeled with a 655-nm streptavidin-Qdot. Mlph was unlabeled. Lower panel, Mlph with a C-terminal SNAP-biotin tag was labeled with a 655-nm streptavidin-Qdot. MyoVa was unlabeled. B, normalized frequency of motors starting to move within a fixed time frame and area. Values are normalized to actin length and the number of runs with myoVa under the same conditions. Error bars are S.D. (n = 11 for myoVa, and n = 5 for myoVa-Mlph) (t test, p < 0.001). C, histogram and Gaussian fit of speeds for myoVa alone (v = 502 ± 269 nm/s, n = 103; gray squares and line) or in the presence of a 10-fold molar excess of unlabeled Mlph (v = 135 ± 103 nm/s, n = 100; orange triangles and solid line) (t test, p < 0.001). Labeling strategy is depicted in A, upper. Alternatively, an unlabeled motor was used, and the Qdot was attached to the C terminus of Mlph and myoVa was unlabeled (brown dashed line and inverted triangles). This labeling strategy is depicted in A, lower. These data best fit two Gaussians (v1 = 155 ± 188 nm/s; v2 = 860 ± 149 nm/s, n = 100). The small fraction of runs with a higher velocity implied that the tethering capacity of some labeled Mlph was compromised by the labeling. D, Kaplan-Meier plot of run lengths showing the percent of motors that have traveled at least a certain distance. MyoVa alone (median run length 550 nm, n = 116; gray line) or in the presence of a 10-fold molar excess of Mlph (median run length 1440 nm, n = 100; orange) (log-rank test, p < 0.001). Conditions: 150 mm KCl, pH 7.4, 2 mm MgATP.
FIGURE 6.
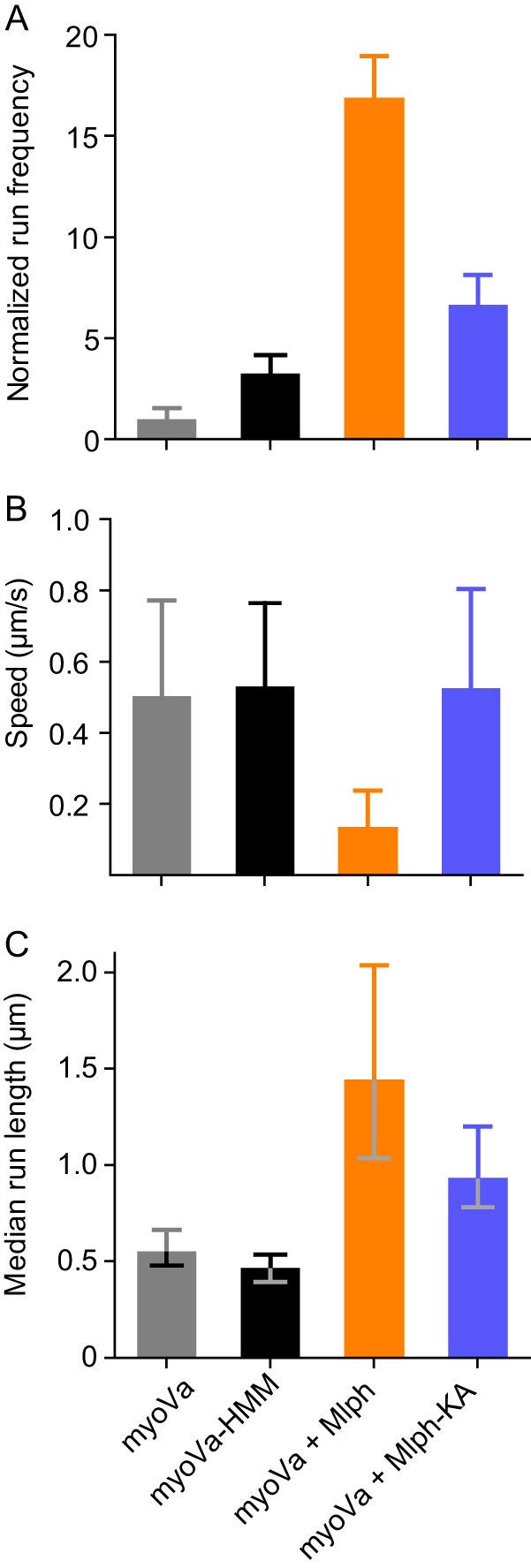
Comparison of single-molecule properties with different motors and different Mlph constructs. A, normalized frequency of motors starting to move within a fixed time frame and area. Values are normalized to actin length and number of runs with myoVa under the same conditions. Error bars are S.D. For myoVa-HMM versus myoVa-Mlph-KA, p < 0.01; all other pairs, p < 0.001. B, mean speed as determined by the fit of a Gaussian distribution to the histogram. Error bars are S.D. Differences between myoVa-Mlph and all other constructs are statistically significant (t test, p < 0.001). C, median run length as determined by Kaplan-Meyer survival statistics. Error bars are 95% confidence intervals. Difference between myoVa-HMM and myoVa is not significant. All other differences are statistically significant (log-rank test). For myoVa-Mlph versus myoVa-Mlph-KA, p < 0.01; all other pairs are p < 0.001.
Mlph Slows Motor Movement
In the absence of Mlph, the speed distribution of myoVa was symmetrical and fitted a Gaussian distribution with an average speed of 502 ± 269 nm/s (Fig. 5C). The constitutively active, truncated myoVa-HMM moved at similar speeds (530 ± 234 nm/s, t test, p > 0.05) (Fig. 6B). Addition of a 10-fold molar excess of unlabeled Mlph decreased the speed of myoVa to 135 ± 103 nm/s, which was slower than either myoVa alone or myoVa-HMM (t test, p < 0.001) (Figs. 5C and 6B).
The experiment was repeated with Mlph that was labeled at its C terminus with a Qdot and unlabeled motor (Fig. 5A, lower). Most runs again had a slow velocity, centered at 167 ± 179 nm/s (Fig. 5C, dashed line), consistent with data obtained when myoVa was labeled with a Qdot. The minor population of fast motors (840 ± 156 nm/s) in this experiment is likely due to impaired binding of some Mlph molecules to actin when the Qdot is attached at the C terminus near the actin binding region. As a control to ensure that the slowing of speed was not due to Mlph simply acting as a roadblock, we compared the speed of myoVa-HMM in the presence of Mlph (468 ± 178 nm/s) with that of myoVa-HMM alone (530 ± 234 nm/s) and found no significant difference (t test, p > 0.05).
Mlph Enhances Run Length
In the absence of Mlph, myoVa traveled over relatively short distances with a median run length of 550 nm (95% CI, 479–664 nm) (Fig. 5D), similar to the median run length of myoVa-HMM (471 nm, 95% CI, 393–536 nm, log-rank test, p > 0.05) (Fig. 6C). Addition of a 10-fold molar excess of Mlph increased the median run length >2.5-fold to 1440 nm (95% CI, 1034–2037 nm), which is significantly longer than runs of either myoVa alone or myoVa-HMM (log-rank test, p < 0.001) (Fig. 5D and supplemental Movies S2 and S3). Run lengths were analyzed using Kaplan-Meier survival statistics (31) because most motors that bound Mlph ran to the end of the actin filament. The survival curve analysis allowed us to include all traces and calculate a median run length that was reached by half of the motors tracked.
For traditional characteristic run length analysis, long runs that terminate at the end of an actin filament are excluded because only traces whose start and stop coordinates are known can be analyzed. This biases the results toward shorter run lengths, Nonetheless, traditional run length analysis showed the same trend but with the expected lower values (myoVa, 472 nm, 95% CI, 448–497 nm; myoVa-HMM, 440 nm, 95% CI, 411–474 nm; myoVa plus a 10-fold excess of Mlph, 862 nm, 95% CI, 788–952 nm).
The Effects of Mlph Are Minimized with Less Positive Charge in the Actin Binding Region
The increased frequency of motile runs, increased run length, and slower speeds of the myoVa-Mlph complex could be the result of Mlph tethering the motor to actin. To test this idea, we used a previously characterized Mlph mutant (Mlph-KA) which binds actin weakly. In this mutant, four positively charged residues in the actin binding region were mutated to Ala (K493A/R495A/R496A/K497A) (26) (Figs. 1B and 2B). These four basic residues have the potential to interact with the negatively charged N-terminal region of actin. To show that Mlph-KA has reduced affinity for actin, actin was co-sedimented with either wild-type Mlph or Mlph-KA. More than half of WT-Mlph pelleted with actin, but only a small fraction of Mlph-KA bound to actin under the same conditions (Fig. 2C). Mlph-KA did, however, retain its ability to bind myoVa because it co-localized and moved with myoVa in the processivity assay (data not shown).
At the single-molecule level, myoVa-Mlph-KA had a significantly lower run frequency (t test, p < 0.001), faster speed (t test, p < 0.001), and shorter median run length (log-rank test, p < 0.01) than seen with WT-Mlph (Fig. 6). Mlph-KA thus muted all the large effects on motor properties caused by WT-Mlph, consistent with a decreased tethering ability. Speeds reverted to that seen with myoVa-HMM. Run frequency and run length were intermediate between values obtained with myoVa-Mlph and myoVa-HMM, perhaps due to some residual actin binding ability that was not abolished by the four point mutations.
Motion of Mlph during Processive Walking
To determine whether Mlph occupies discrete positions on actin or if it is continually dragged along the actin filament when bound to myoVa, a Qdot was attached to the C terminus of Mlph, which is near the actin binding region. MyoVa was unlabeled and did not contain a biotin tag. Interestingly, displacement versus time traces showed step-like behavior (Fig. 7A). The distance between Mlph-Qdot attachment points fit a single Gaussian with a mean of 106 ± 31 nm (Fig. 7B), indicating that Mlph bound to myoVa does not dissociate from actin with every step of the head. This distance is statistically different from all myosin step sizes (t test, p < 0.001).
FIGURE 7.
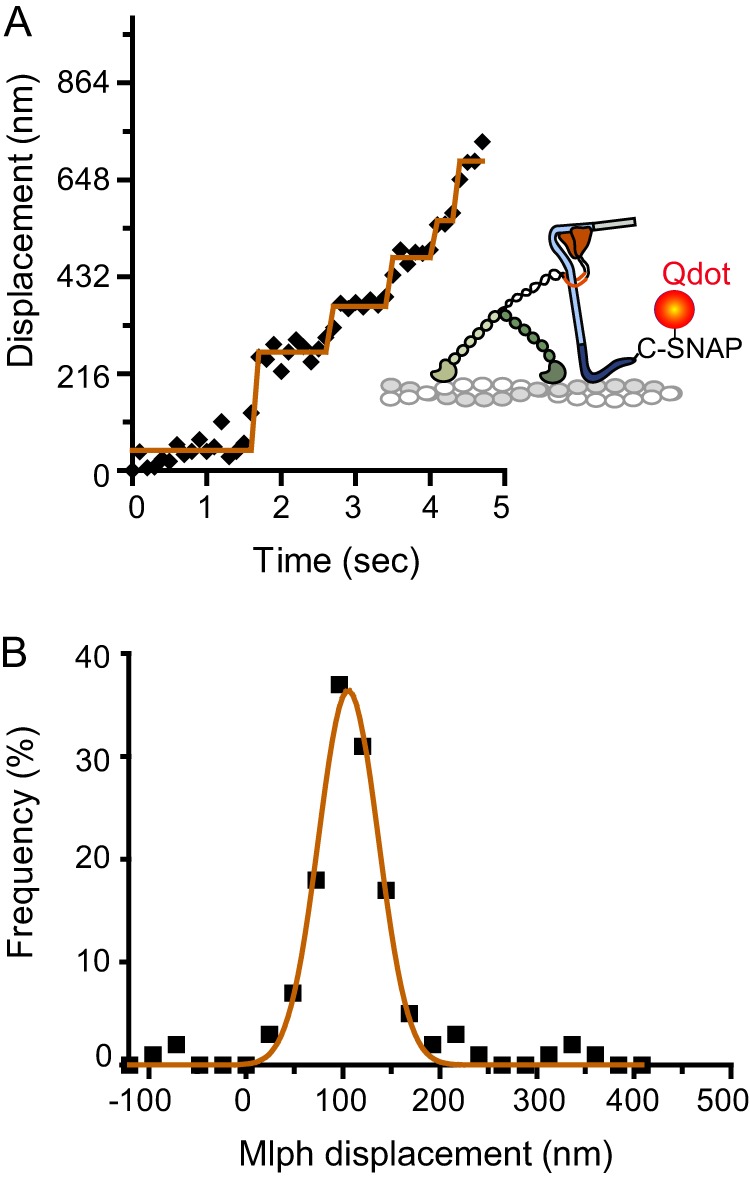
Step-like movement of a Qdot bound to the C terminus of Mlph, near the actin binding site, in complex with unlabeled myoVa. A, Mlph-C-SNAP-biotin was bound to a streptavidin-Qdot. MyoVa did not contain a biotin tag. A representative displacement versus time trace is shown. B, the histogram of Mlph displacement was fit with a single Gaussian (d = 106 ± 31 nm, n = 131).
Mlph Restores a Uniform Stepping Pattern of MyoVa
The stepping behavior of myoVa was compared in the presence or absence of Mlph to gain insight into whether Mlph engages in cargo-dependent activation of myoVa. Because our experiments used a higher salt concentration than is typically used for single-molecule experiments, we first compared the stepping pattern of myoVa-HMM at 25 mm versus 150 mm KCl. It is well established that at low salt, myoVa-HMM has a uniform stepping pattern with an average step size of ∼72 ± 12 nm (data not shown) (35–37). MyoVa-HMM at 150 mm KCl and low ATP concentration (2 μm) also steps uniformly with an average step size of 72 ± 19 nm. A difference from lower salt conditions is that myoVa-HMM shows a lower stepping accuracy at 150 mm KCl, which is reflected by the occasional back step, as well as by a greater spread in the step size data (Fig. 8A). MyoVa at 150 mm KCl showed the expected altered gating: most forward steps are 64 ± 21 nm, but there was a population with a smaller forward step size of 28 ± 7 nm, as evidenced by the front shoulder on the Gaussian fit (Fig. 8B), as well as a small population of back steps.
FIGURE 8.
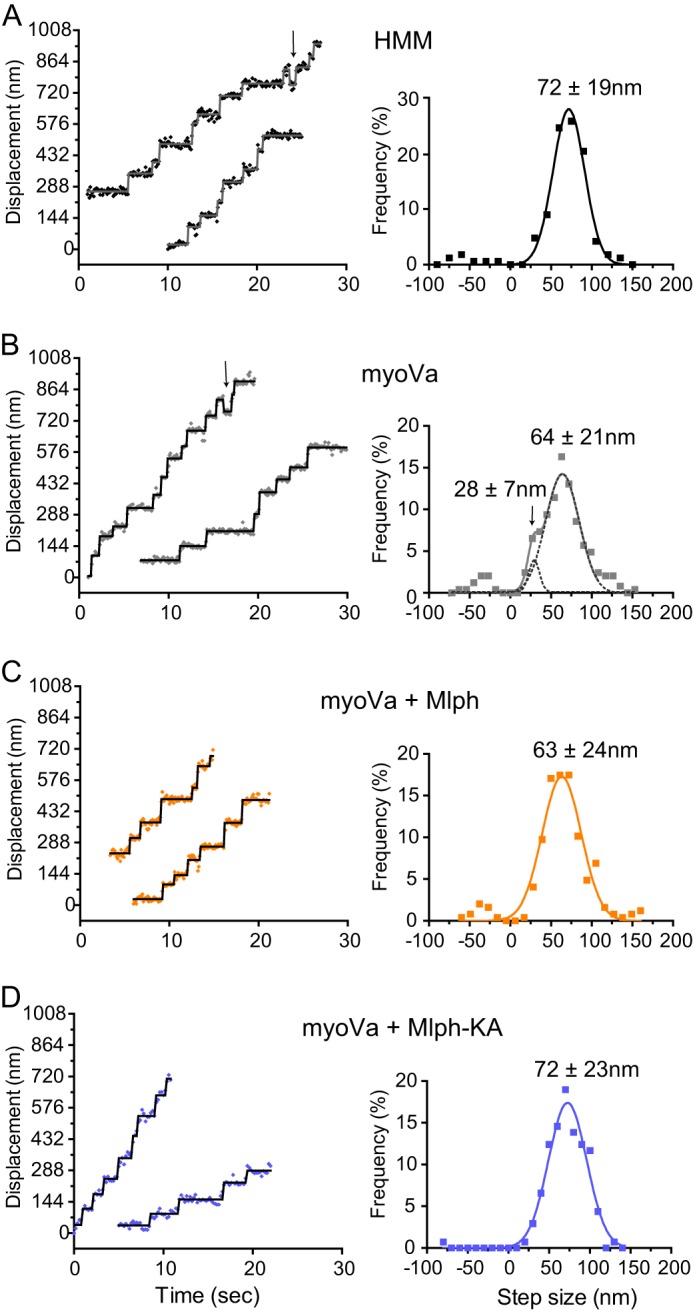
Stepping pattern of myoVa in the presence or absence of Mlph. Representative displacement versus time traces and histograms of step sizes fit with Gaussian distributions are shown. A, myoVa-HMM has a mean step size of 71.9 ± 19.4 nm (n = 161) and shows occasional back steps (arrow). B, myoVa shows two step populations with average sizes of 27.8 ± 6.5 nm and 64.0 ± 20.5 nm (n = 245). Dashed line shows the individual Gaussian fits to the slower shoulder and main peak. Arrow in the left panel points to a back step. C, in the presence of Mlph the distribution becomes uniform with an average step size of 63.0 ± 24.0 nm (n = 246). D, in the presence of Mlph-KA step sizes are uniform and show the same size as myoVa-HMM (71.7 ± 22.4 nm, n = 137). Conditions: 150 mm KCl, pH 7.4, 2–4 μm MgATP.
Strikingly, addition of Mlph to myoVa restored a uniform stepping pattern, with steps centered at 63 ± 24 nm (Fig. 8C). A small population of back steps, as seen with myoVa-HMM, remained. The stepping pattern of myoVa in the presence of Mlph-KA was also uniform and fit a single Gaussian centered at 72 ± 23 nm, with no evidence of short forward steps or back steps (Fig. 8D). t test analysis showed that the difference between the step size of both myoVa with Mlph, and the major peak of myoVa, are significantly different from the step sizes of both myoVa-HMM and myoVa in the presence of Mlph-KA (p < 0.001).
DISCUSSION
Here we show that the adapter protein Mlph, which links the melanocyte isoform of myoVa to Rab27a-melanosomes, is an active player in cargo transport and causes significant changes to the single-molecule properties of myoVa. First, the number of full-length myoVa motors recruited to actin in the presence of Mlph is much higher than can be accounted for by simply assuming that Mlph causes the motor to assume the active conformation. Second, there is a significant reduction in the average speed of movement of myoVa in the presence of Mlph. Similar and even slower speeds (<100 nm/s) have been observed for melanosome movement in vivo (3, 38) and in vitro (39). Our in vitro data can explain why melanosome movement in the cell does not occur at the characteristic speed of myoVa (∼500 nm/s) and show that intracellular cargo transport cannot be elucidated by considering only the properties of the isolated motor. Within the cell, it is possible that other features, such as drag from other motors bound to the melanosome or the complex architecture of the actin meshwork, may also contribute to the reduced rates of motility. Third, the median run length more than doubled in the presence of Mlph. Mlph thus causes a large increase (∼8-fold) in the time that myoVa remains actively engaged with actin, a feature that is likely to facilitate the transfer of melanosomes to the adjacent keratinocyte.
We propose that Mlph acts as a tether between the motor and the actin track. This would lead to an effective increased affinity for actin and decreased off-rate of the myoVa-Mlph complex and could account for the increased run length and event frequency. A sufficiently strong tether could also impose a drag force on myoVa, which would explain the substantial decrease in speed. The strength of the electrostatic interaction between Mlph and actin must be quite strong, as it persists near physiological ionic strength (150 mm KCl). A mutant Mlph with less positive charge in the actin binding domain showed a diminished ability to act as a tether.
Single-molecule techniques showed that Mlph dwells at discrete sites on actin. Given that the position of Mlph is determined by the steps of myoVa, the 106 nm between binding sites corresponds to the center of mass of the motor moving 3 × 36 nm before Mlph detaches and reattaches further along the filament. The unstructured central region of Mlph (22) could provide the necessary flexibility to accommodate the stretch in the molecule as it moves with the globular tail of myoVa on its N terminus and dwells on actin with its C terminus, for several steps taken by myoVa.
Evidence for Cargo Activation by Mlph
In addition to acting as a tether, does binding of Mlph favor the active form of myoVa, a mechanism known as cargo-dependent activation? Previous actin-activated ATPase assays with Mlph showed only partial activation at 0.2 m KCl, conditions where myoVa is essentially unfolded and active even in the absence of cargo (40). We performed ATPase assays at 150 mm NaCl and also showed only partial activation by Mlph relative to that obtained in calcium (data not shown). Our single-molecule data provide a potential explanation. Tethering by Mlph slows the speed of movement and ATP turnover, which would also depress the ATPase activity and thus result in an apparent partial activation.
So how does one establish whether binding of Mlph activates the motor? One cannot simply ask whether the myoVa-Mlph complex behaves like the constitutively active truncated myoVa-HMM in terms of speed, run length, and run frequency, because the tethering ability of Mlph alters these properties of myoVa. Single-molecule stepping dynamics were therefore used as an indicator of whether myoVa was activated by Mlph.
Hydrodynamic measurements and a comparison of single-molecule run frequencies were first used to establish that under our ionic conditions cargo-free myoVa is predominantly in the inhibited conformation. The 3.3-fold lower processive run frequency of full-length myoVa compared with myoVa-HMM suggests that ∼70% of full-length myoVa is in the inactive conformation. However, those full-length motors that move at 150 mm KCl have the same speed and run length as myoVa-HMM. The altered gating between the heads of full-length myoVa is present but less prominent at 150 mm KCl than previously observed at lower salt concentrations (18).
The single-molecule signature of an active myoVa is that it steps in a uniform hand-over-hand fashion, which results in a single Gaussian peak centered at ∼70 nm. Multiple peaks in the step size histogram, which include short forward steps and back steps, in addition to the normal ∼70-nm forward steps, are suggestive of an equilibrium between inhibited and active motors (18). As expected, the step size distribution for myoVa alone showed these three populations of step sizes. In contrast, the stepping data of myoVa in the presence of WT-Mlph and Mlph-KA showed a uniform Gaussian distribution of steps. These observations suggest that Mlph stabilizes the extended, active conformation of myoVa independent of its tethering ability and support the idea of cargo-dependent activation of myoVa. The stepping data also show that the slower speed of movement in the presence of Mlph cannot be accounted for by more short forward steps, consistent with the idea that tethering by Mlph slows the stepping rate.
The mean of the step size distribution for myoVa with WT-Mlph was 9 nm less than in the presence of Mlph-KA. The attachment of Mlph to actin is the most likely reason for the reduced average step size. This effect is probably not the result of a load imposed by the tethering because the step size of myoVa was shown to be insensitive to load (41). A similar effect of a slightly shorter step size with a broader distribution was seen with a myoVa-HMM mutant that had two alanine residues inserted between the third and fourth IQ motif of the neck linker, which increased lever arm flexibility (36). Interactions of Mlph with the lever arm are possible from a geometrical perspective.
Of note, even myoVa-HMM showed some back steps at 150 mm KCl. Back steps are rare under unloaded conditions at low salt, but their probability increases with load (42, 43). When they occur, back steps are triggered by the motor taking a shorter forward step, likely one actin monomer before its preferred binding site (42). The motor generally corrects its mistake after a back step and continues forward processive motion. The lower affinity of the head for actin at higher salt concentrations may cause more forward missteps that in turn trigger back steps. The greater stepping inaccuracy at higher salt concentrations is also reflected in the greater standard deviation for these step size distributions.
Implications for Melanosome Transport
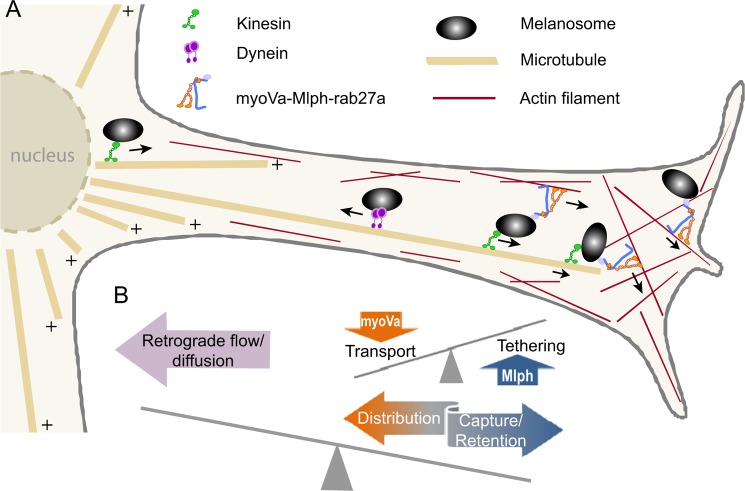
A capture model was proposed to explain melanosome localization in melanocytes (3). In this model, melanosomes are transported along microtubules by kinesin to the cell periphery where they are then transferred to the actin cytoskeletal mesh that forms a layer next to the cell membrane (Fig. 9). Redistribution within this peripheral actin mesh depends on myoVa. For this model to work two conditions have to be fulfilled. (i) Retention: The affinity of the melanosome for the actin mesh has to be high to prevent retrograde transport back to the cell center along microtubules and to prevent dissociation and diffusion of melanosomes away from the cell periphery where they are positioned for transfer to adjacent keratinocytes. Our data provide evidence that Mlph acts as a strong tether which would ensure sufficient retention. Similar functions were attributed to MyRIP (Slac-2c), which is involved in myoVa dependent exocytosis of secretory granules in enterochromaffin cells (44) and is also involved in myoVIIa-dependent transport of melanosomes in retinal epithelial cells (45). MyRIP shares homology with Mlph in the actin binding region (26). (ii) Distribution: Melanocytes have elaborate dendritic networks to allow the transfer of melanosomes to several dozen keratinocytes. Reasonable transport rates have to be accomplished to reach all destinations. Our data again support this idea because myoVa can walk while being tethered to actin via Mlph. A motor that can move under high load, like myoVa, is needed to counteract a tether that is sufficiently strong to oppose microtubule directed retrograde transport. The slowness of the speed is probably irrelevant in the cellular context because long range transport has already occurred on microtubule tracks. Only a short range movement within the cell cortex is required for melanosomes to become distributed near the cell membrane.
FIGURE 9.
Model of strategies for correct melanosome localization in the cell. A, schematic diagram of a melanosome dendritic protrusion. Mature melanosomes can undergo bidirectional transport along microtubules by using either kinesin for anterograde movement or dynein for retrograde transport. At the interface with the cortical actin network at the cell periphery melanosomes are transferred and tethered to actin by the action of myoVa and Mlph. Transport through the cortical actin mesh to the plasma membrane requires both the tethering action of Mlph as well as the motor properties of myoVa. B, tethering to actin has to both outweigh the propensity of the microtubule-based transporters to carry melanosomes back to the cell body and prevent dissociation and diffusion away from the cell cortex. MyoVa-directed transport has to counterbalance tethering forces to allow for adequate transport and distribution of melanosomes within the cell cortex.
Roles for Adapter Proteins
Our data show that near physiological ionic strength Mlph acts as a tether as well as a cargo adapter to myoVa. What is not clear at present is whether this is a general phenomenon or a feature unique to Mlph. Given that MyRIP also contains an actin binding domain, we predict that it would have a similar effect on motor driven transport. It has not been demonstrated if adapter proteins like rabphilin and granuphilin, which couple the brain isoform of myoVa to secretory granules in pancreatic beta cells, have actin binding regions that could act as tethers (46, 47). One might speculate, however, that all myoVa-based transport would benefit from having an actin tether which would help retain the cargo in the actin periphery. In a more general sense, our results suggest that adapter proteins that link motors to cargo are not passive players in cargo transport, but can modify motor properties in ways favorable for their biological role.
Acknowledgments
We thank Paul Selvin for the anti-HIS Qdots and Guy Kennedy from the University of Vermont Instrumentation and Model Facility for expert microscopy design and assistance.
This work was supported by National Institutes of Health Grants GM078097 (to K. M. T.) and GM094229 (to D. M. W.), and Training Grant T32HL07647 (to M. S.).

This article contains supplemental Movies S1–S3.
- myoVa
- myosin Va
- AEBSF
- 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride
- CI
- confidence interval
- EB1
- microtubule plus-end tip tracking protein end-binding protein 1
- Mlph
- melanophilin
- myoVa-HMM
- truncated, constitutively active heavy meromyosin subfragment of myosin Va
- Qdot
- quantum dot
- TIRF
- total internal reflection fluorescence
- TLCK
- Nα-p-tosyl-l-lysine chloromethyl ketone.
REFERENCES
- 1. Arnold D. B. (2009) Actin and microtubule-based cytoskeletal cues direct polarized targeting of proteins in neurons. Sci. Signal. 2, pe49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hume A. N., Seabra M. C. (2011) Melanosomes on the move: a model to understand organelle dynamics. Biochem. Soc. Trans. 39, 1191–1196 [DOI] [PubMed] [Google Scholar]
- 3. Wu X., Bowers B., Rao K., Wei Q., Hammer J. A. (1998) Visualization of melanosome dynamics within wild-type and dilute melanocytes suggests a paradigm for myosin V function in vivo. J. Cell Biol. 143, 1899–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu X. S., Tsan G. L., Hammer J. A. (2005) Melanophilin and myosin Va track the microtubule plus end on EB1. J. Cell Biol. 171, 201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marks M. S., Seabra M. C. (2001) The melanosome: membrane dynamics in black and white. Nat. Rev. Mol. Cell Biol. 2, 738–748 [DOI] [PubMed] [Google Scholar]
- 6. Wu X., Rao K., Bowers M. B., Copeland N. G., Jenkins N. A., Hammer J. A. (2001) Rab27a enables myosin Va-dependent melanosome capture by recruiting the myosin to the organelle. J. Cell Sci. 114, 1091–1100 [DOI] [PubMed] [Google Scholar]
- 7. Wu X. S., Rao K., Zhang H., Wang F., Sellers J. R., Matesic L. E., Copeland N. G., Jenkins N. A., Hammer J. A., 3rd (2002) Identification of an organelle receptor for myosin-Va. Nat. Cell Biol. 4, 271–278 [DOI] [PubMed] [Google Scholar]
- 8. Trybus K. M. (2008) Myosin V from head to tail. Cell. Mol. Life Sci. 65, 1378–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang J. D., Mermall V., Strobel M. C., Russell L. B., Mooseker M. S., Copeland N. G., Jenkins N. A. (1998) Molecular genetic dissection of mouse unconventional myosin-VA: tail region mutations. Genetics 148, 1963–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lambert J., Naeyaert J. M., Callens T., De Paepe A., Messiaen L. (1998) Human myosin V gene produces different transcripts in a cell type-specific manner. Biochem. Biophys. Res. Commun. 252, 329–333 [DOI] [PubMed] [Google Scholar]
- 11. Mercer J. A., Seperack P. K., Strobel M. C., Copeland N. G., Jenkins N. A. (1991) Novel myosin heavy-chain encoded by murine dilute coat color locus. Nature 349, 709–713 [DOI] [PubMed] [Google Scholar]
- 12. Pashkova N., Jin Y., Ramaswamy S., Weisman L. S. (2006) Structural basis for myosin V discrimination between distinct cargoes. EMBO J. 25, 693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu X., Wang F., Rao K., Sellers J. R., Hammer J. A., 3rd (2002) Rab27a is an essential component of melanosome receptor for myosin Va. Mol. Biol. Cell 13, 1735–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thirumurugan K., Sakamoto T., Hammer J. A., 3rd, Sellers J. R., Knight P. J. (2006) The cargo binding domain regulates structure and activity of myosin 5. Nature 442, 212–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu J., Taylor D. W., Krementsova E. B., Trybus K. M., Taylor K. A. (2006) Three-dimensional structure of the myosin V-inhibited state by cryoelectron tomography. Nature 442, 208–211 [DOI] [PubMed] [Google Scholar]
- 16. Li X. D., Jung H. S., Wang Q., Ikebe R., Craig R., Ikebe M. (2008) The globular tail domain puts on the brake to stop the ATPase cycle of myosin Va. Proc. Natl. Acad. Sci. U.S.A. 105, 1140–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krementsov D. N., Krementsova E. B., Trybus K. M. (2004) Myosin V: regulation by calcium, calmodulin, and the tail domain. J. Cell Biol. 164, 877–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Armstrong J. M., Krementsova E., Michalek A. J., Heaslip A. T., Nelson S. R., Trybus K. M., Warshaw D. M. (2012) Full-length myosin Va exhibits altered gating during processive movement on actin. Proc. Natl. Acad. Sci. U.S.A. 109, E218–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Strom M., Hume A. N., Tarafder A. K., Barkagianni E., Seabra M. C. (2002) A family of Rab27-binding proteins. J. Biol. Chem. 277, 25423–25430 [DOI] [PubMed] [Google Scholar]
- 20. Nagashima K., Torii S., Yi Z., Igarashi M., Okamoto K., Takeuchi T., Izumi T. (2002) Melanophilin directly links Rab27a and myosin Va through its distinct coiled-coil regions. FEBS Lett. 517, 233–238 [DOI] [PubMed] [Google Scholar]
- 21. Kukimoto-Niino M., Sakamoto A., Kanno E., Hanawa-Suetsugu K., Terada T., Shirouzu M., Fukuda M., Yokoyama S. (2008) Structural basis for the exclusive specificity of Slac2-a/melanophilin for the Rab27 GTPases. Structure 16, 1478–1490 [DOI] [PubMed] [Google Scholar]
- 22. Geething N. C., Spudich J. A. (2007) Identification of a minimal myosin Va binding site within an intrinsically unstructured domain of melanophilin. J. Biol. Chem. 282, 21518–21528 [DOI] [PubMed] [Google Scholar]
- 23. Fukuda M., Kuroda T. S. (2004) Missense mutations in the globular tail of myosin-Va in dilute mice partially impair binding of Slac2-a/melanophilin. J. Cell Sci. 117, 583–591 [DOI] [PubMed] [Google Scholar]
- 24. Hume A. N., Tarafder A. K., Ramalho J. S., Sviderskaya E. V., Seabra M. C. (2006) A coiled-coil domain of melanophilin is essential for myosin Va recruitment and melanosome transport in melanocytes. Mol. Biol. Cell 17, 4720–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fukuda M., Kuroda T. S. (2002) Slac2-c (synaptotagmin-like protein homologue lacking C2 domains-c), a novel linker protein that interacts with Rab27, myosin Va/VIIa, and actin. J. Biol. Chem. 277, 43096–43103 [DOI] [PubMed] [Google Scholar]
- 26. Kuroda T. S., Ariga H., Fukuda M. (2003) The actin binding domain of Slac2-a/melanophilin is required for melanosome distribution in melanocytes. Mol. Cell. Biol. 23, 5245–5255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pylypenko O., Goud B. (2012) Posttranslational modifications of Rab GTPases help their insertion into membranes. Proc. Natl. Acad. Sci. U.S.A. 109, 5555–5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu X., Sakamoto T., Zhang F., Sellers J. R., Hammer J. A., 3rd (2006) In vitro reconstitution of a transport complex containing Rab27a, melanophilin and myosin Va. FEBS Lett. 580, 5863–5868 [DOI] [PubMed] [Google Scholar]
- 29. Philo J. S. (2000) A method for directly fitting the time derivative of sedimentation velocity data and an alternative algorithm for calculating sedimentation coefficient distribution functions. Anal. Biochem. 279, 151–163 [DOI] [PubMed] [Google Scholar]
- 30. Sage D., Neumann F. R., Hediger F., Gasser S. M., Unser M. (2005) Automatic tracking of individual fluorescence particles: application to the study of chromosome dynamics. IEEE Trans. Image Process. 14, 1372–1383 [DOI] [PubMed] [Google Scholar]
- 31. Kaplan E. L., Meier P. (1958) Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 53, 457–481 [Google Scholar]
- 32. Kerssemakers J. W., Munteanu E. L., Laan L., Noetzel T. L., Janson M. E., Dogterom M. (2006) Assembly dynamics of microtubules at molecular resolution. Nature 442, 709–712 [DOI] [PubMed] [Google Scholar]
- 33. Pardee J. D., Spudich J. A. (1982) Purification of muscle actin. Methods Enzymol. 85, 164–181 [DOI] [PubMed] [Google Scholar]
- 34. Wang F., Thirumurugan K., Stafford W. F., Hammer J. A., 3rd, Knight P. J., Sellers J. R. (2004) Regulated conformation of myosin V. J. Biol. Chem. 279, 2333–2336 [DOI] [PubMed] [Google Scholar]
- 35. Snyder G. E., Sakamoto T., Hammer J. A., 3rd, Sellers J. R., Selvin P. R. (2004) Nanometer localization of single green fluorescent proteins: evidence that myosin V walks hand-over-hand via telemark configuration. Biophys. J. 87, 1776–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sakamoto T., Yildez A., Selvin P. R., Sellers J. R. (2005) Step-size is determined by neck length in myosin V. Biochemistry 44, 16203–16210 [DOI] [PubMed] [Google Scholar]
- 37. Warshaw D. M., Kennedy G. G., Work S. S., Krementsova E. B., Beck S., Trybus K. M. (2005) Differential labeling of myosin V heads with quantum dots allows direct visualization of hand-over-hand processivity. Biophys. J. 88, L30–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brunstein M., Bruno L., Desposito M., Levi V. (2009) Anomalous dynamics of melanosomes driven by myosin-V in Xenopus laevis melanophores. Biophys. J. 97, 1548–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rogers S. L., Gelfand V. I. (1998) Myosin cooperates with microtubule motors during organelle transport in melanophores. Curr. Biol. 8, 161–164 [DOI] [PubMed] [Google Scholar]
- 40. Li X. D., Ikebe R., Ikebe M. (2005) Activation of myosin Va function by melanophilin, a specific docking partner of myosin Va. J. Biol. Chem. 280, 17815–17822 [DOI] [PubMed] [Google Scholar]
- 41. Clemen A. E., Vilfan M., Jaud J., Zhang J., Bärmann M., Rief M. (2005) Force-dependent stepping kinetics of myosin-V. Biophys. J. 88, 4402–4410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lu H., Kennedy G. G., Warshaw D. M., Trybus K. M. (2010) Simultaneous observation of tail and head movements of myosin V during processive motion. J. Biol. Chem. 285, 42068–42074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kad N. M., Trybus K. M., Warshaw D. M. (2008) Load and Pi control flux through the branched kinetic cycle of myosin V. J. Biol. Chem. 283, 17477–17484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huet S., Fanget I., Jouannot O., Meireles P., Zeiske T., Larochette N., Darchen F., Desnos C. (2012) Myrip couples the capture of secretory granules by the actin-rich cell cortex and their attachment to the plasma membrane. J. Neurosci. 32, 2564–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lopes V. S., Ramalho J. S., Owen D. M., Karl M. O., Strauss O., Futter C. E., Seabra M. C. (2007) The ternary Rab27a-myrip-myosin VIIa complex regulates melanosome motility in the retinal pigment epithelium. Traffic 8, 486–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang J., Takeuchi T., Yokota H., Izumi T. (1999) Novel rabphilin-3-like protein associates with insulin-containing granules in pancreatic beta cells. J. Biol. Chem. 274, 28542–28548 [DOI] [PubMed] [Google Scholar]
- 47. Brozzi F., Diraison F., Lajus S., Rajatileka S., Philips T., Regazzi R., Fukuda M., Verkade P., Molnár E., Váradi A. (2012) Molecular mechanism of myosin Va recruitment to dense core secretory granules. Traffic 13, 54–69 [DOI] [PubMed] [Google Scholar]