Background: Carcinoembryonic antigen-related cellular adhesion molecules (CEACAMs) engage in intercellular binding and influence cellular growth and differentiation.
Results: Calcium-dependent signals cause dissolution of the transmembrane domain-driven basal state CEACAM1 oligomers into monomers that mediate intercellular binding.
Conclusion: Regulated switching in CEACAM1 oligomerization controls cell-cell adhesion and downstream effector recruitment.
Significance: Inside-out signaling effects CEACAM1-dependent cell adhesion by controlling a reversible dimer-to-monomer transition.
Keywords: Bacterial Adhesion, Calcium Signaling, Calmodulin, Cancer, Cell Adhesion, Membrane Proteins, CEACAM1, Carcinoembryonic Antigen-related Cell Adhesion Molecule, Homodimer, Homophilic Binding
Abstract
Cell-cell contacts are fundamental to multicellular organisms and are subject to exquisite levels of control. The carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) can engage in both cis-homophilic (parallel) oligomerization and trans-homophilic (anti-parallel) binding. In this study, we establish that the CEACAM1 transmembrane domain has a propensity to form cis-dimers via the transmembrane-embedded 432GXXXG436 motif and that this basal state is overcome when activated calmodulin binds to the CEACAM1 cytoplasmic domain. Although mutation of the 432GXXXG436 motif reduced CEACAM1 oligomerization, it did not affect surface localization of the receptor or influence CEACAM1-dependent cellular invasion by the pathogenic Neisseria. The mutation did, however, have a striking effect on CEACAM1-dependent cellular aggregation, increasing both the kinetics of cell-cell association and the size of cellular aggregates formed. CEACAM1 association with tyrosine kinase c-Src and tyrosine phosphatases SHP-1 and SHP-2 was not affected by the 432GXXXG436 mutation, consistent with their association with the monomeric form of wild type CEACAM1. Collectively, our results establish that a dynamic oligomer-to-monomer shift in surface-expressed CEACAM1 facilitates trans-homophilic binding and downstream effector signaling.
Introduction
Cell adhesion is a fundamental activity required for the correct functioning of virtually every process in the multicellular organism. Despite this central role, the molecular events that govern intercellular binding and translate this event to intracellular signals with the potential to control cellular growth, differentiation, and development often remain only superficially defined. An emerging appreciation that the misregulation of individual cell adhesion molecules can contribute to pathologies as disparate as cancer, inflammation, pathogenic infections, and autoimmune disease (1) has revealed the complexity of cell adhesion molecule signal integration and prompted an intense effort to understand what governs physical binding and how these molecular events elicit a cellular response.
The carcinoembryonic antigen gene family represents a large but well defined subgroup within the immunoglobulin superfamily, one of the largest and most diverse families of proteins in the body (2). The prototypical family member, CEACAM1,3 consists of one N-terminal IgV-like domain followed by up to three IgC-like domains (3). The largest isoform, CEACAM1–4L, has three IgC-like domains (A1-B-A2) and a 76-amino acid cytoplasmic domain (4, 5). The splice variant CEACAM1–4S shares the extracellular and transmembrane structure of CEACAM1–4L but has a 10-amino acid cytoplasmic domain (5). CEACAM1 undergoes both homophilic (CEACAM1-CEACAM1) and heterophilic (CEACAM1-CEACAM5) intercellular binding between its N-terminal Ig variable-like domains (6, 7) in an antiparallel orientation (8). CEACAM1 homophilic binding has been shown to regulate processes, including cellular proliferation (9), apoptosis (10), angiogenesis (11), tumor growth (12), and immune cell activation (13–22). Remarkably, however, studies by various groups suggest that CEACAM1 may exert either inhibitory or activating functions on the cell (23). One plausible explanation is that different oligomeric states of CEACAM1 may recruit different effectors to elicit opposing signaling responses (24).
In addition to trans-intercellular (antiparallel) binding, CEACAM1 has been shown to self-associate into oligomers of two or more monomers together in cis (parallel) fashion (25). On rat epithelial cells, surface-expressed CEACAM1 has been shown to exist as either a monomer or noncovalently linked dimer (25). Although no experimental evidence yet supports a difference in function of monomeric versus cis-dimeric human CEACAM1, work by Obrink and co-workers (26) suggested that the tyrosine phosphatases tended to associate with dimeric CEACAM1-L, whereas the tyrosine kinase c-Src displayed no obvious preference for dimeric versus monomeric CEACAM1-L. Given the differential function of these effector proteins, the balance between monomeric and dimeric forms of CEACAM1 may result in different, even apparently contradictory, cellular responses.
Early studies with isolated rat liver plasma membrane suggest that CEACAM1 dimerization may be controlled by calcium-loaded calmodulin binding to CEACAM1. This postulate arises from both an observation that the calcium ionophore ionomycin causes a transient increase in the CEACAM1 monomer-dimer ratio (25) and separate biophysical studies revealing a calmodulin-binding site within synthetic peptides based upon the sequence of naturally occurring CEACAM1 splice variants with both the long (L) and short (S) cytoplasmic domains (27). Such observations suggest a means by which CEACAM1 oligomers may be disassociated; yet they do not consider what motivates CEACAM1 dimerization, and there remains no appreciation as to what effect this transition has on the behavior of a cell.
A lack of understanding regarding how cell adhesion molecule oligomerization influences the outcome of cell-cell binding is currently the greatest hurdle to understanding their function in health and disease. In this study, we used in vitro cellular experiments to demonstrate that human CEACAM1 naturally exists as a dimer due to the presence of membrane-buried glycine residues that self-assemble to promote the cis-homodimerization. Our data suggest that, upon cellular activation, calcium-loaded calmodulin binds to the cytoplasmic domain of CEACAM1 and promotes its dissociation into monomers. This effect has direct implications for cell-cell communications, as we establish that monomeric CEACAM1 promotes intercellular binding and that the long cytoplasmic domain then allows intracellular recruitment of cytoplasmic effectors that control the cellular response. These findings provide a molecular level context for recent work demonstrating that forced monomers of CEACAM1–4S reduced anchorage-independent growth in vitro and tumorigenicity in vivo (28), and they provide a new paradigm by which to understand how fluctuations in the oligomeric state of CEACAM1 can control intercellular adhesion and mediate its effect on cell growth and differentiation.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
All reagents were from Sigma unless otherwise indicated. The rabbit CEACAM-specific polyclonal antiserum and normal rabbit serum were from Dako (Mississauga, Ontario, Canada). The CEACAM pan-specific D14HD11 antibody was from Genovac GmbH (Freiburg, Germany). The anti-SHP-1, anti-SHP-2, and anti c-Src antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). HRP- and fluorescent-conjugated secondary antibodies were purchased from Jackson ImmunoResearch (Mississauga, Ontario, Canada).
Cell Culture, Cloning, and Expression Procedures
The stably transfected HeLa cell line expressing defined recombinant CEACAM1 (HeLa-CEACAM1–4L) was described previously (29). HeLa-CEACAM1–4L and the parental HeLa cells were maintained in RPMI 1640 medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Hyclone, Logan, UT) and 4 mm GlutaMAX (Invitrogen). Cells were cultured at 37 °C in humidified air containing 5% CO2.
Plasmids containing CEACAM1 in pRC/CMV were generously provided by Wolfgang Zimmermann (Munich, Germany). cDNAs were amplified via PCR (5′ primers contained a Kozak sequence, GCCACCATG, for protein expression) and subcloned into pMSCV Puro retroviral expression vector (Clontech). The variants of human CEACAM1–4L, CEACAM1–4S (CEACAM1 with short cytoplasmic domain) and truncated CEACAM1 (CEACAM1 lacking the complete cytoplasmic domain) were amplified from CEACAM1–4L originally cloned into the pRC/CMV vector (30) and then subcloned into pMSCV-Puro vector. The R43S/Q44L-CEACAM1–4L variant was generated from the pMSCV-Puro-CEACAM1 by PCR splicing by overlap extension (SOEing) (31). The G432L/G436L-CEACAM1–4L variant was generated using the QuikChange® site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The oligonucleotide primers used to create amino acid substitutions at positions 432 and 436 of the transmembrane domain of CEACAM1–4L are shown in supplemental Table S1. For introducing mutations, CEACAM1–4L was amplified by PCR from pMSCV-Puro-CEACAM1–4L using a forward primer and a reverse primer containing the desired mutations. CEACAM1–4L with appropriate mutation was amplified and cloned into pMSCV-Puro vector using restriction sites XhoI and EcoRI.
The pEYFP-tagged CEACAM1 variants were constructed by cloning CEACAM1 variants into the pEYFP-N1 vector (Clontech). c-Myc-tagged CEACAM1–4L was constructed by PCR addition of the c-Myc protein-derived peptide sequence to the C terminus of CEACAM1–4L using a 3′-edge primer containing the c-Myc sequence and an EcoRI restriction enzyme site and then cloned into pMSCV Puro vector. Primers used for generation of recombinant CEACAM1 alleles are listed in supplemental Table S1. For transient transfection-based assays, the HeLa cell lines were transfected with the indicated CEACAM1 alleles using FuGENE 6 according to manufacturer's instructions (Roche Applied Science).
Bacterial Strains
Neisseria gonorrhoeae strains constitutively expressing either no Opa protein (N302) or the CEACAM-specific Opa52 (N309) were derived from a N. gonorrhoeae strain MS11 mutant that does not express pili (32) and were graciously provided by Prof. T. F. Meyer (Max-Planck-Institut für Infektionsbiologie, Berlin, Germany). N. gonorrhoeae were grown from frozen stocks on 1% (v/v) IsoVitaleXTM (BBLTM, BD Biosciences)-supplemented GC agar (Difco) at 37 °C in a humidified 5% CO2-containing atmosphere. Gonococcal strains were subcultured daily using a binocular microscope to select desired colony opacity phenotype, and Opa protein expression was routinely confirmed by immunoblot analysis.
Chemical Cross-linking and Pulldown
Prior to cross-linking, cells from a low (<50%) confluence culture dish were resuspended using trypsin/EDTA, which does not cleave surface CEACAM1, and then washed twice in PBS supplemented with 0.5 mm MgCl2 and 1 mm CaCl2 (PBS/Mg/Ca) by pelleting and resuspension. Cross-linking was initiated by the addition of dithiobis(succinimidyl) propionate (DSP) at 500 μm or at the indicated concentrations. After gentle agitation for 30 min at room temperature, the reaction was quenched with the addition of Tris/HCl, pH 7.4, at a final concentration of 100 mm. Cells were washed twice with 100 mm Tris/HCl, pH 7.4, before immunoblotting or immunoprecipitation studies. Cells were occasionally visually observed and counted before and after cross-linking to ensure no aggregation of the suspended cells had occurred. In a pulldown experiment, HeLa cells were transiently transfected with a c-Myc- and/or pEYFP-tagged CEACAM1–4L using FuGENE 6 as per the manufacturer's instruction. After cells were cross-linked as described previously, cells were lysed in cold RIPA buffer. c-Myc-containing proteins were recovered from lysates using anti-c-Myc antibody (9E10, Babco, Berkeley, CA). Co-immunoprecipitated proteins were subjected to SDS-PAGE immunoblot analysis with anti-YFP antibody (Santa Cruz Biotechnology). Total cell lysates were probed with mouse CEACAM1-specific monoclonal antibody (D14DH11, Genovac GmbH, Freiburg, Germany) as described previously (29).
For all immunoblot analyses, antigen-specific primary antibodies were detected with horseradish peroxidase-conjugated secondary antibodies and then developed using ECL Plus chemiluminescence substrate (Amersham Biosciences) and x-ray film.
Ionophore and Calmodulin Antagonist Treatment
Unless otherwise noted, HeLa cells transfected with the indicated CEACAM1 mutant constructs were incubated in PBS/Mg/Ca with 1 μm ionomycin (EMD Chemicals Inc., Gibbstown, NJ) or 50 μm N-(6-aminohexyl)-5-chloro-1-naphthalenesulfonamide hydrochloride (W-7) (Sigma). After incubation at 37 °C for various periods of time, the ionophore or W-7 was removed, and the cells were cross-linked as described. For depletion of intracellular calcium, HeLa-CEACAM1–4L cells were incubated with 3 mm EGTA (Sigma) in PBS for 30 min before exposure to ionomycin or other treatments.
Flow Cytometry
Stably transfected HeLa-CEACAM1–4L cells and transiently transfected HeLa cells (5 × 105) with pEYFP-tagged CEACAM1 mutants were collected in PBS containing 2% FBS (v/v) and subsequently fixed in 4% paraformaldehyde. Surface CEACAM1 was stained with the CEACAM-specific monoclonal antibody (mAb) D14HD11 (20 μg/ml) diluted in 3% FCS/PBS for 1 h on ice, washed with ice-cold PBS, and incubated with FITC-conjugated anti-mouse F(ab′)2. Background fluorescence was determined using isotype-matched Ig.
For the intracellular staining of CEACAM1, cells were treated with Cytofix/Cytoperm solution (BD Biosciences) for 20 min at 4 °C, washed with Perm/Wash buffer (BD Biosciences), and stained with 20 μg/ml CEACAM-specific mAb D14HD11 followed by phycoerythrin-conjugated anti-mouse F(ab′)2 antibody (Jackson ImmunoResearch), all diluted in Perm/Wash buffer. A minimum of 1 × 104 gated cells from each sample was analyzed using FACSCalibur flow cytometer with CellQuest software (BD Biosciences).
CEACAM1 Surface Expression
The membrane-impermeable EZ-Link Sulfo-NHS-LC-biotin (Pierce) was added to cross-linked samples at a final concentration of 1 mg/ml. After gentle agitation for 30 min at room temperature, the reaction was quenched with the addition of Tris/HCl, pH 7.4, at a final concentration of 100 mm. Cells were washed twice with 100 mm Tris/HCl, pH 7.4. Following washing, cells were lysed in cold radioimmunoprecipitation assay (RIPA) buffer (1% Triton X-100, 50 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, 1 mm PMSF, 1 μg/ml each aprotinin, leupeptin, and pepstatin A, 1 mm NaF, 100 μm Na3VO4, and 10 mm H2O2). Biotinylated surface proteins were pulled down using streptavidin-agarose (Sigma), and SDS-PAGE immunoblots of nonreduced or reduced pellets were probed to detect CEACAM1 dimers and monomers using mAb against CEACAM1 (D14HD11; Abcam, Cambridge, MA).
Association of Calmodulin
HeLa-CEACAM1–4L cells were treated with the indicated amounts of ionomycin or W-7 for various periods of time. After cells were cross-linked as described, cells were lysed in cold RIPA buffer. Calmodulin was immunoprecipitated from lysates with a mAb antibody specific for calmodulin (EP799Y; Abcam). Recovered proteins were subjected to SDS-PAGE immunoblot analysis with antibodies specific for CEACAM1 (D14HD11).
Computational Searches of CEACAM1 Transmembrane Domain Dimers (TMD) Using CHI
Potential sites of CEACAM1 TMD self-interaction were identified via simulation with the crystallography and NMR system (CNS) searching of helix interactions (CHI) software suite, as described previously (33–35). Briefly, two identical α-helices were generated from the primary sequence of the CEACAM1 TMD (residues 429–451, inclusive), and dimers with either a right-handed or left-handed crossing angle was created. Helix-helix interactions were searched in a parallel orientation. Rotation angles from 0 to 360° were searched in steps of 45°, and molecular dynamics simulation, simulated annealing, and energy minimization were carried out for each step. Energy-minimized dimers were grouped into clusters of at least 10 structures with a cutoff of mean square deviation of 1 Å. The average structure of each cluster was calculated and refined by energy minimization. All simulations were conducted in vacuum (dielectric constant Z = 1) to mimic the low dielectric constant of the membrane core.
Homo-FRET Imaging
Fluorescence anisotropy microscopy (FAIM) for the detection of FRET between like fluorescent proteins (homo-FRET) was performed on an LSM710 NLO confocal microscope (Carl Zeiss Microimaging) equipped with a Chameleon Ti:Sapphire laser (Coherent). Cells were imaged in RPMI at room temperature using a 1.4 NA Plan-Apochromat 63× oil immersion objective lens and pixel dwell time of 12.61 μs. These images were collected from the bottom of the cell at the glass coverslip because this position provides a relatively flat field of the plasma membrane. Aside from focal adhesions, this ventral cell surface was not generally attached to the glass because most of the membrane surface was freely accessible to soluble fluorescent dyes administered to the culture medium (data not shown), and these cells were not polarized, so CEACAM1 and other receptors were distributed along both dorsal and ventral cell surfaces. mVenus and Yellow (YFP) fluorescent protein excitation was achieved with the femtosecond laser set to 950 nm (36). Fluorescence emission was collected using a two-channel LSM BiG GaASP detector (Carl Zeiss Microimaging) equipped with an IR-blocked 500–550-nm emission bandpass filter (Chroma), and a polarizing beam splitter (Edmund Optics) set to collect parallel (channel 1) and perpendicular (channel 2) emission (Chroma). The calculation of anisotropy (r) for homo-FRET analysis was done using custom plug-ins written for ImageJ, National Institutes of Health (37) as shown in Equation 1,
 |
These plug-ins applied corrections for instrumental bias (g) and skewing of parallel (Ix) and perpendicular (Iy) intensities due to a large NA collection of fluorescence, as described previously (36, 38–40). Fluorescent proteins have a long rotational correlation time compared with fluorescence lifetime resulting in an intrinsically high steady-state anisotropy (41). In the case of FRET, donor fluorescent proteins transfer energy to an acceptor through a dipole-dipole interaction. As a result, the emission becomes more randomized leading to a lower anisotropy. This drop in anisotropy can be used to measure both hetero-FRET and homo-FRET. We tested the ability of FAIM to measure homo-FRET in cells expressing monomeric Venus fluorescent protein (mVenus) and a tandem dimer of Venus fluorescent protein (Venus dimer) (supplemental Fig. S1). Cells expressing the Venus tandem dimer consistently exhibited a lower anisotropy value than cells expressing monomeric Venus.
Analysis of Bacterial Binding and Invasion
A gentamycin-based assay to quantify viable total associated and intracellular bacteria was performed as described previously (29). Briefly, transiently transfected HeLa cells were infected with 40 bacteria/cell (multiplicity of infection = 40) for 1 h. Unbound bacteria were washed, and cells were incubated with RPMI containing 50 μg/ml gentamycin to kill extracellularly associated bacteria. After 45 min of incubation with the gentamycin-containing medium, cells were lysed by the addition of 1% saponin in PBS for 15 min. Suitable dilutions were plated in triplicate on GC agar plates to determine the number of recovered viable bacteria. A Student's t test analysis was performed on the data to determine whether statistically significant differences exist in OpaCEA-mediated adhesion and uptake by HeLa cells transfected with CEACAM1–4L compared with the CEACAM1 mutants.
Intercellular Aggregation Assays
For cellular aggregation assays, HeLa cells were transiently transfected with CEACAM1-4L or CEACAM1 mutants. Puromycin (3 μg/ml, Sigma) was added to cells 24 h following transfection to kill the untransfected cells, and the cells were then cultured for 2 more days. Cells were washed and trypsinized from tissue culture dishes and aggressively mixed to produce a single cell suspension. 1 × 106 cells of each transfected cell line were incubated in 1 ml of RPMI 1640 medium containing 0.8% FBS, 10 mg/ml DNase, and incubated at 37 °C with constant gentle rocking. Samples were retrieved at the indicated time periods and evaluated using a hemocytometer to quantify single cells. Viability of cells was also determined by trypan blue dye exclusion. Images were taken on a Leica MZ16F stereomicroscope. The numbers of cells per aggregate was calculated from 10 arbitrarily selected fields of view (∼500 cells per field) for each cell line. For ease of interpretation and to facilitate the creation of a visual image of the aggregates observed, aggregate sizes were expressed in values reflecting ranges of cells and represented in Fig. 8 pie charts.
FIGURE 8.
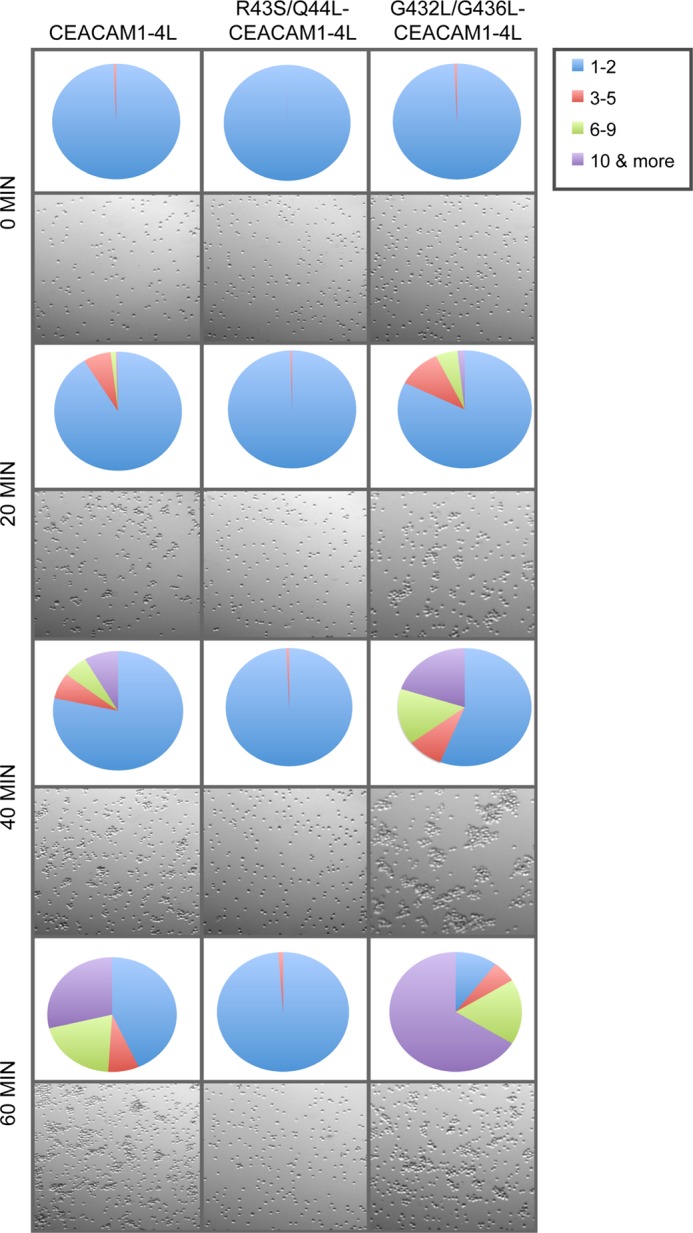
Effects of CEACAM1 dimerization on trans-homophilic cell-cell adhesion. HeLa cells transfected with CEACAM1–4L, R43S/Q44L-CEACAM1–4L, or G432L/G436L-CEACAM1–4L were prepared as single cell suspensions and then allowed to aggregate. Images of cell cultures taken at indicated time points are shown. Quantitative analysis of number and size of aggregates at each time point are represented as a pie chart. Each value is a mean of triplicate samples and are representative of three independent experiments.
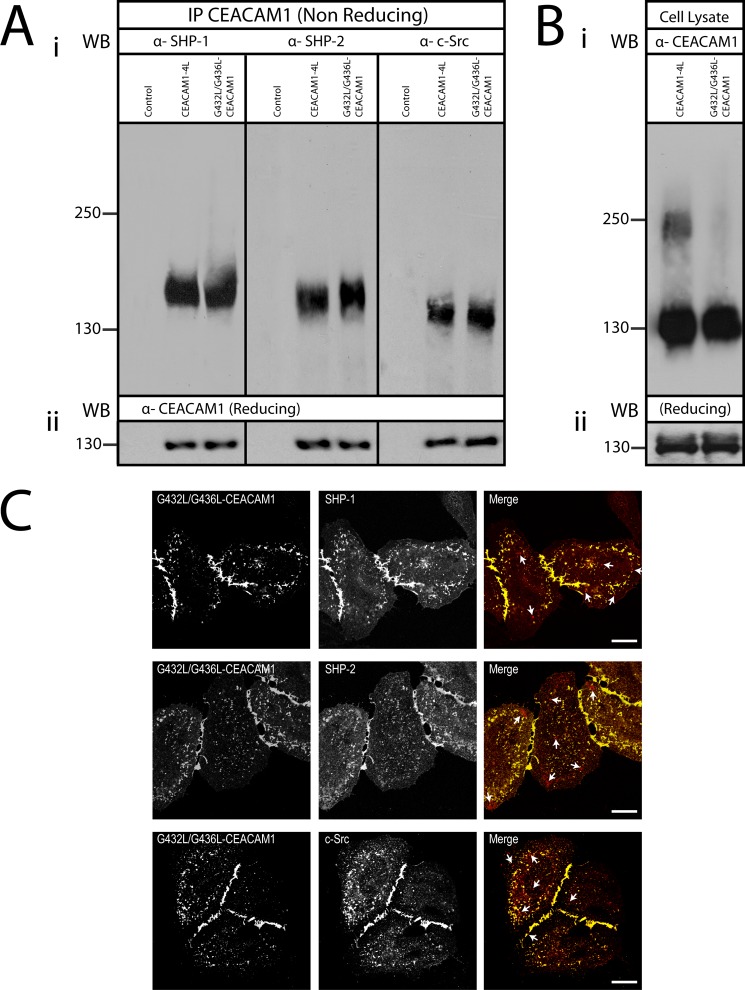
Association of SHP-1, SHP-2 Phosphatase, and c-Src Kinase with CEACAM1
HeLa cells that were transiently transfected with CEACAM1–4L or G432L/G436L-CEACAM1–4L constructs were treated with 100 μm pervanadate for 1 h at 37 °C. Cells were pelleted, and cellular proteins were cross-linked as described above. The cells were lysed in cold RIPA buffer. CEACAM1 was immunoprecipitated using DynabeadsTM magnetic beads covalently coupled with protein A (Invitrogen) according to the manufacturer's instructions. Recovered proteins were subjected to SDS-PAGE immunoblot analysis under nonreducing conditions with mouse monoclonal anti-SH-PTP1 antibody (BD Biosciences), mouse monoclonal anti-SH-PTP2 antibody (Abcam, Cambridge, MA), and mouse monoclonal anti c-Src antibody (Santa Cruz Biotechnology). Proteins were also probed with mouse CEACAM1-specific monoclonal antibody (D14DH11, Genovac GmbH, Freiburg, Germany) under reducing conditions.
Immunofluorescence Staining
HeLa cells seeded on glass coverslips transfected with the pEYFP-CEACAM1–4L or pEYFP-G432L/G436L-CEACAM1–4L were fixed with 4% paraformaldehyde in PBS after treatment with 100 μm pervanadate for 1 h at 37 °C. Cells were washed and permeabilized with 0.4% Triton X-100 for 15 min, prior to incubation in blocking buffer (PBS, 5% FCS) for 1 h. Cells were incubated with primary monoclonal antibodies followed by incubation with Texas Red-conjugated goat anti-mouse secondary antibody. Each coverslip was mounted onto a glass slide and images were taken with a Zeiss LSM 510 laser scanning confocal microscope.
RESULTS
CEACAM1 Exists as a Monomer and Dimer in HeLa Epithelial Cells
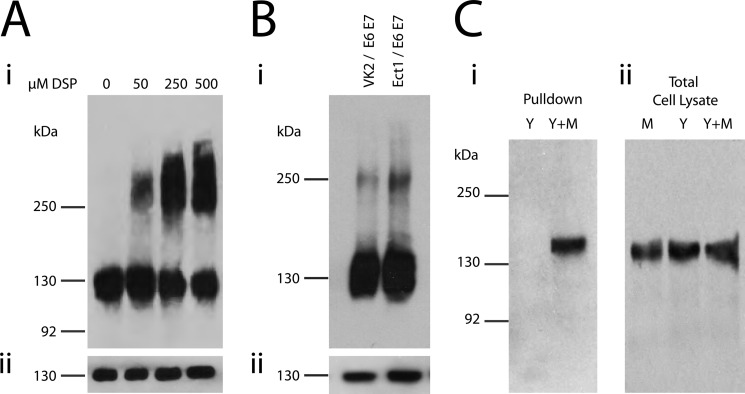
Previous studies have observed that CEACAM1 can exist as dimers in rat hepatocytes, but dimers could not be detected in Chinese hamster ovary (CHO) cells stably transfected with CEACAM1 (25). HeLa cells have routinely been used by ourselves (29, 42, 43) and others (44–50) for CEACAM1 structure-function analyses because these human endocervical epithelium-derived cells do not express endogenous CEACAMs. To determine the molecular organization of human CEACAM1, HeLa cells stably expressing CEACAM1–4L were used for cross-linking analysis with DSP, a membrane-permeable homobifunctional reagent, to detect the interactions between CEACAM1 and associated cellular proteins. Treatment of HeLa-CEACAM1–4L cells with increasing DSP concentrations resulted in an increasing intensity of a large molecular mass species of ∼260 kDa, with a concomitant decrease in intensity of the smaller molecular mass species, signifying a decrease in the amount of monomeric CEACAM1 (Fig. 1A). To confirm that this effect was not the simple result of CEACAM1 overexpression in a transfected cell line, the human ectocervical (Ect1/E6E7) and vaginal (Vk2/E6E7) epithelial cells, which naturally express CEACAM1, were exposed to DSP. In both cell lines, a CEACAM1 complex with a mobility reflecting that seen in the transfected HeLa cells was apparent (Fig. 1B).
FIGURE 1.
CEACAM1 exists as monomers and oligomers in HeLa cells. A, HeLa-CEACAM1 cells were treated with the indicated concentrations of DSP; total cell lysates were resolved under nonreducing (panel i) or reducing (panel ii) conditions by SDS-PAGE and probed with antibodies specific for CEACAM1. B, ectocervical (Ect1/E6E7) and vaginal (Vk2/E6E7) epithelial cells were cross-linked with DSP. Total cell lysates were resolved under nonreducing (panel i) or reducing (panel ii) conditions by SDS-PAGE, and immunoblots were probed with antibodies specific for CEACAM1. C, HeLa cells were transfected with only pEYFP-CEACAM1–4L (Y) or pEYFP-CEACAM1–4L and CEACAM1–4L-c-Myc (Y+M) and treated with DSP. c-Myc-containing proteins were immunoprecipitated and SDS-PAGE immunoblots run in reducing conditions were probed with antibodies specific for YFP (C, panel i). Cell lysates were probed with anti-CEACAM1 antibody to ensure all cells were expressing the transfected proteins (C, panel ii). Blots are representative of three independent experiments.
Although the emergence of a higher molecular weight CEACAM1 species after exposure to DSP is consistent with the cross-linker's stabilization of CEACAM1 homodimers, it could also indicate CEACAM1 association with a different protein. To detect CEACAM1-CEACAM1 oligomers, we performed co-immunoprecipitation assays with two different epitope-tagged CEACAM1–4L constructs. pEYFP-CEACAM1–4L and CEACAM1–4L-myc, which consist of CEACAM1–4L with C-terminal YFP and Myc tags, respectively, were co-transfected into HeLa cells. Immunoprecipitation of the chemically cross-linked lysates with Myc tag-specific antibody led to the recovery of pEYFP-CEACAM1–4L (Fig. 1C), providing strong support for the existence of a homodimer and/or homo-oligomers of CEACAM1–4L.
cis-CEACAM1 Dimerization Is Controlled by Intracellular Calcium Levels
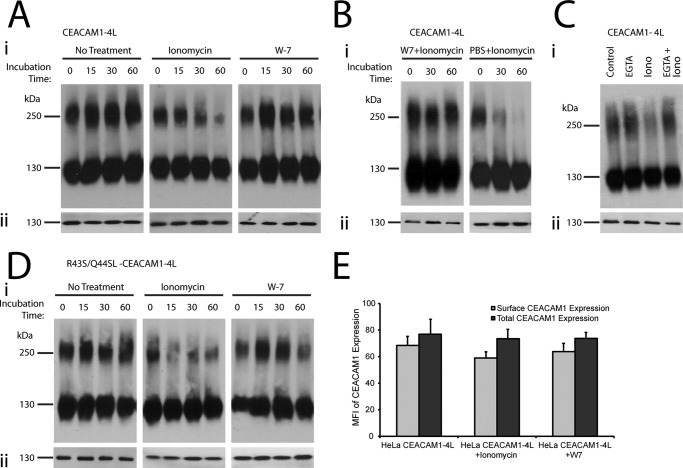
Previous studies have indicated that treatment of cells with a Ca2+ ionophore alters levels of CEACAM1 dimerization in rat hepatocytes (25). To establish that similar effects were apparent in the human-derived cells, HeLa-CEACAM1–4L were treated with the Ca2+ ionophore, ionomycin. A decrease in the levels of CEACAM1–4L dimers was readily apparent (Fig. 2A, panel i). Calmodulin was one of the first cytoplasmic proteins identified to bind to the CEACAM1 cytoplasmic domain (51), and previous studies have demonstrated specific binding of calmodulin to the cytoplasmic domain of CEACAM1–4L, inhibiting its self-association in rat hepatocyte (27, 51, 52). To confirm the effect of calmodulin on human CEACAM1 dimerization, the cells were treated with calmodulin antagonist, W-7. When used alone, W-7 does not have any apparent effect on the level of CEACAM1 dimerization (Fig. 2A). To test whether the effect of ionomycin was dependent upon calmodulin and to confirm the activity of our W-7 inhibitor, the cells were incubated with W-7 and then exposed to ionomycin. As illustrated in Fig. 2B, W-7 effectively blocked the dissociation of dimeric CEACAM1–4L otherwise apparent upon treatment with ionomycin. To ensure that the reduction of CEACAM1–4L dimers upon ionomycin treatment was not from an overall decrease in CEACAM1–4L expression, the HeLa-CEACAM1–4L cross-linked lysates were also treated with reducing agent to detect any changes in total CEACAM1 protein (Fig. 2, A and B, panel ii). No change in total CEACAM1 was observed, indicating that the ionomycin treatment has no effect on total CEACAM1 levels. Combined, these results indicate that CEACAM1–4L dimers are not covalently linked and that ionomycin causes a calmodulin-dependent disruption of these dimers.
FIGURE 2.
CEACAM1 dimers are formed by cis-interactions and regulated by intracellular Ca2+. Transfected HeLa cells expressing CEACAM1–4L (A–C) or CEACAM1–4L containing R43S/Q44L mutations (D) were either untreated or treated with ionomycin or W-7 or W-7 and ionomycin or PBS + ionomycin, EGTA, and EGTA + ionomycin for various time points, and then proteins were cross-linked with DSP. Ionomycin was used at 10 μm for B but used at 1 μm for all other experiments. Total cell lysates were resolved under nonreducing (panel i) or reducing (panel ii) conditions by SDS-PAGE, and immunoblots were probed with antibodies specific for CEACAM1. Depicted blots are representative of three independent experiments. E, quantitative evaluation of surface and total CEACAM1 expression using flow cytometry, plotted as mean fluorescence intensity (MFI). Data represent mean ± S.E. from three separate experiments.
To confirm that the effect of ionomycin depended upon its ability to permit calcium flux into the cell, we tested the effect of the Ca2+-chelating agent EGTA on ionomycin-dependent dissolution of CEACAM1 dimers. In the presence of EGTA, ionomycin had little effect on the ratio of monomer to dimer within the cell (Fig. 2C), consistent with a Ca2+-dependent effect of calmodulin on CEACAM1 dimer dissociation.
While providing the evidence to support the existence of CEACAM1–4L oligomers that can be associated by Ca2+-calmodulin, the aforementioned results cannot distinguish between cis- (parallel) and trans- (anti-parallel) interactions. We therefore took advantage of a mutant CEACAM1 allele containing Arg-Gln to Ser-Leu mutations at positions 43 and 44 (R43S/Q44L), which abrogates trans-homophilic binding (53), to confirm that dimeric CEACAM1 detected in our assays was in the cis-conformation (Fig. 2D, No treatment). As with HeLa cells expressing CEACAM1–4L, treatment of HeLa cells expressing R43S/Q44L-CEACAM1–4L with ionomycin resulted in a decrease in cis-dimers, although W-7 treatment had no effect (Fig. 2D, panel i). Portions of the cross-linked lysates were then treated with a reducing agent to ensure equal expression of total CEACAM1 upon ionophore or calmodulin antagonist treatment (Fig. 2D, panel ii). These results thereby confirm that CEACAM1–4L dimers are formed by cis-interactions and also suggest that inactivity of calmodulin, either due to the absence of free intracellular calcium or the presence of W-7, allows the coalescence of CEACAM1 into basal state cis-dimers.
Ca2+-Calmodulin-regulated CEACAM1 Dimers Are at the Cell Surface
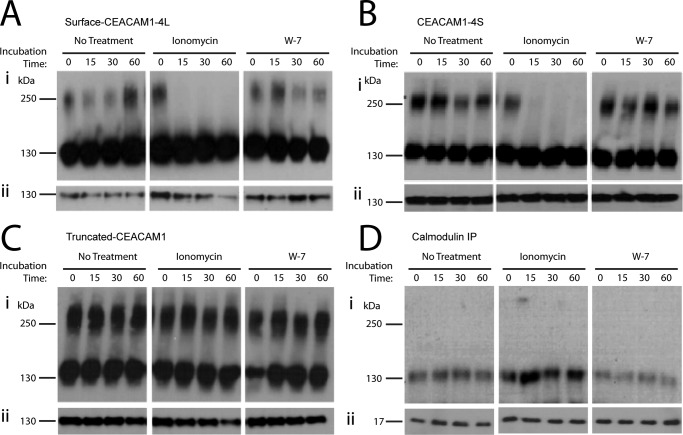
Because CEACAM1 is actively internalized in response to various cues (54), we investigated whether ionomycin or W-7 have an effect on the surface expression of CEACAM1–4L in HeLa cells by flow cytometric analysis. Our results (Fig. 2E) show that surface expression of CEACAM1 was unaffected by treatment of HeLa-CEACAM1–4L cells with ionomycin or W-7. While providing the evidence for the existence of surface CEACAM1 upon ionomycin treatment, these flow cytometry studies with HeLa-CEACAM1–4L cannot distinguish between monomeric or dimeric states of the CEACAM1 at the cell surface. To confirm that CEACAM1 dimers were present on the cell surface and that these were affected by ionomycin, surface proteins were labeled by exposing HeLa-CEACAM1–4L cells to membrane-impermeant Sulfo-NHS-LC-biotin along with the DSP cross-linker and then isolating the biotinylated surface protein with streptavidin-agarose. Similar to what was seen when monitoring total cellular CEACAM1, ionomycin treatment caused a rapid decrease in CEACAM1 dimers at the cell surface (Fig. 3A, panel i). Although the sample-to-sample consistency of isolating biotinylated samples was a persistent problem in this assay (note variability in CEACAM1 in reduced samples (Fig. 3A, panel ii) and between the level of dimers seen in the unreduced samples of untreated and W-7-treated samples (Fig. 3A, panel i), surface dimers were apparent under both of these conditions. Based upon these results, we conclude that the ionomycin and W-7 did not affect surface CEACAM1 uptake or degradation, but it did affect the oligomeric state of surface-expressed CEACAM1 structural organization. Considering that the intercellular adhesion function occurs at the cell surface, the intracellular Ca2+- and calmodulin-dependent change in monomer-dimer equilibrium might have implications in trans-intercellular binding and/or cytoplasmic signaling if downstream effector proteins preferentially bind to either the monomeric or dimeric CEACAM1.
FIGURE 3.
Cell surface CEACAM1 dimers are dissociated by Ca2+-calmodulin binding. Transfected HeLa cells expressing full-length CEACAM1 (CEACAM1–4L) were either untreated or treated with ionomycin or W-7, and then proteins were cross-linked with DSP. Cell surface-exposed CEACAM1 was labeled by conjugation to biotin and recovered using streptavidin-agarose beads; biotinylated lysates were resolved under nonreducing (A, panel i) or reducing (A, panel ii) conditions by SDS-PAGE, and immunoblots were probed with antibodies specific for CEACAM1. Transfected HeLa cells expressing a natural splice variant of CEACAM1 containing a short cytoplasmic domain, 10 amino acids (CEACAM1–4S) (B), or a recombinant form completely lacking a cytoplasmic domain (truncated CEACAM1) (C) were either untreated or treated with ionomycin or W-7, and then proteins were cross-linked with DSP. D, co-immunoprecipitation (IP) of HeLa cells expressing full-length CEACAM1–4L using calmodulin-specific antibody after the treatment with or without ionomycin or W-7 and DSP. Calmodulin-associated proteins were resolved under nonreducing conditions, and immunoblots were probed with antibodies specific for CEACAM1 (panel i) or separated under reducing conditions, and immunoblots were probed to detect total levels of calmodulin (panel ii). Blots are representative of three independent experiments.
Calmodulin-dependent Changes in CEACAM1 Dimerization Require Calmodulin-binding Motifs in the Cytoplasmic Domain
The cytoplasmic domain of CEACAM1 is believed to directly participate in control of CEACAM1 dimerization because studies have demonstrated that calmodulin binds to the cytoplasmic domains of both CEACAM1–4L and the naturally occurring splice variant CEACAM1–4S (27, 51). In rat hepatocyte studies, the levels of CEACAM1 dimers decrease with the addition of activated calmodulin (25). As well, changes in intracellular Ca2+ levels, which could activate calmodulin, also affect the levels of CEACAM1 monomers and dimers. Calmodulin is capable of binding to peptides synthesized to reflect the 10 most membrane-proximal amino acid residues in the cytoplasmic domain of CEACAM1 (55). Therefore, we decided to verify the role that the CEACAM1 cytoplasmic domain plays in control of CEACAM1 dimerization levels in vivo using HeLa cells transfected with a construct expressing CEACAM1–4S, because the 10-residue-long cytoplasmic domain of this splice variant encodes a single calmodulin-binding site. We also expressed a recombinant (non-natural) truncated form of CEACAM1 that completely lacks the cytoplasmic domain. As predicted, because CEACAM1–4S contains a calmodulin-binding site, there was a decrease in the level of CEACAM1–4S dimers when the cells were treated with ionomycin (Fig. 3B, panel i). Interestingly, treatment of HeLa cells expressing truncated CEACAM1 with ionomycin caused no effect on the dimers (Fig. 3C, panel i). Similar to previous observations when using CEACAM1–4L (Fig. 2), W-7 did not alter the level of dimers consisting of CEACAM1–4S or truncated CEACAM1. Once again, cross-linked lysates were treated with a reducing agent to ensure equal expression of total CEACAM1-S and truncated CEACAM1 upon ionophore or calmodulin antagonist treatment (Fig. 3, B and C, panel ii). The sustained level of CEACAM1 dimers upon ionomycin treatment verifies that the cytoplasmic membrane-proximal 10 amino acids are required for the calmodulin-dependent disruption of cell surface dimers.
Calmodulin Binding to CEACAM1 Regulates CEACAM1 Dimerization
Previous studies have suggested that the binding of activated calmodulin to the calmodulin-binding site located in the CEACAM1 cytoplasmic domain disrupts CEACAM1 dimers (25). Using a co-immunoprecipitation assay, we observed a physical association between calmodulin and CEACAM1 monomer but not with the CEACAM1 dimeric form (Fig. 3D, panel i). Strikingly, we also observed an increased association between calmodulin and the CEACAM1 monomer upon treatment of HeLa-CEACAM1–4L cells with ionomycin (Fig. 3D). Together with our previous results, this suggests that calcium-activated calmodulin binds to CEACAM1–4L in a manner that promotes its dissociation from the dimeric to the monomeric form.
Mapping of the Region Involved in Dimerization of CEACAM1
Our observations regarding the behavior of truncated CEACAM1 indicate that the cytoplasmic domain does not play a significant role in promoting CEACAM1 dimerization. In addition, we showed that a CEACAM1 allele containing an R43S/Q44L mutation in the extracellular domain of CEACAM1–4L that abrogates trans-homophilic binding does not affect the cis-homophilic interactions of CEACAM1. We therefore sought to determine whether the transmembrane domain might contribute to dimerization. The CEACAM1 TMD sequence, as defined by the program TMpred (56), encompasses residues 429–452 and contains two glycines that are separated by three intervening residues (Gly-432, Gly-436; see Fig. 4A, panel i). Two established dimerization motifs in transmembrane α-helices are consistent with the patterning of these glycine residues; they could mediate right-handed helix packing via a GXXXG/tetrad motif (57, 58), or participate in a small residue heptad repeat pattern of left-handed packing (59).
FIGURE 4.
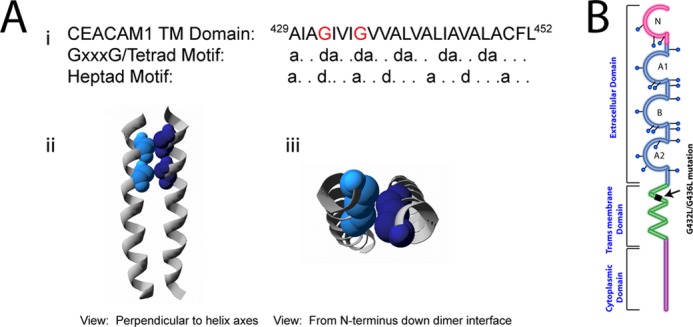
Computational model of the CEACAM1 transmembrane domain dimers. A, sequence of the human CEACAM1 transmembrane domain is shown. The position of two glycine residues (red text) is consistent with the GXXXG/tetrad motif repeat patterning (at position d) and with heptad repeat motif patterning (at positions d and a) (panel i). A view perpendicular to the helix axes and from the N terminus down the interface of a representative left-handed TMD dimer model is shown. Individual helices are represented as gray ribbons, and the glycine residues at the interface are depicted as space-filling spheres in shades of blue (panels ii and iii). Note the close packing of Gly-432 and Gly-436 in the contact surface between the two helices. B, schematic of CEACAM1 to illustrate location of transmembrane domain mutations of glycine 432 and glycine 436 to Leu residues.
To determine whether the CEACAM1 TMD sequence has the potential to exist as a dimer, computational modeling of a CEACAM1–4L TMD dimer was accomplished using the crystallography and NMR system-searching (CNS) of helix interactions (CHI) software suite (33, 34, 60) to examine whether any of the resulting structures were compatible with CEACAM1 self-association with this motif. Modeling of the CEACAM1–4L TMD dimer produced multiple structures, one of which had the Gly-432 and Gly-436 residues in contact at the helix-helix interface, as shown in Fig. 4A, panels ii and iii). This dimer exhibited a parallel, left-handed orientation consistent with packing via the small residue heptad motif, where the “small” Gly-432 and Gly-436 residues localized to the same surface of the transmembrane helix permitted close approach of the CEACAM1–4L TMDs in a parallel fashion.
To subsequently test whether the glycine residues positioned at 432 and 436 of the TMD are critical for dimerization of CEACAM1, we replaced them with leucine, a large hydrophobic amino acid that maintained the hydrophobic character typical of most transmembrane domains (Fig. 4B). Transiently transfected HeLa cells with CEACAM1–4L or G432L/G436L-CEACAM1–4L (carrying the two glycine-to-leucine mutations within the TMD GXXXG) were evaluated for their ability to form dimers based on cross-linking. CEACAM1–4L formed dimers upon treatment with increasing concentrations of DSP, as shown in Fig. 5A, whereas the G432L/G436L-CEACAM1–4L mutant failed to form dimers even in the presence of the highest concentration of DSP (Fig. 5B). Mutations introduced into the transmembrane domain of the receptors have been shown to cause misfolding or inappropriate membrane expression (61). We analyzed CEACAM1 surface expression by flow cytometry using the CEACAM1-specific D14HD11 mouse monoclonal antibody. As shown in Fig. 5C, CEACAM1–4L and G432L/G436L-CEACAM1–4L mutants displayed similar levels of both surface and (after permeabilization of the cell membrane) total CEACAM1 expression. These results showed that the mutated CEACAM1 expressed on the outer cell surface, indicating that protein transport to and/or from the plasma membrane was not impaired by the transmembrane domain mutations. When combined, these results indicate that these glycine residues are essential for the oligomerization of CEACAM1.
FIGURE 5.
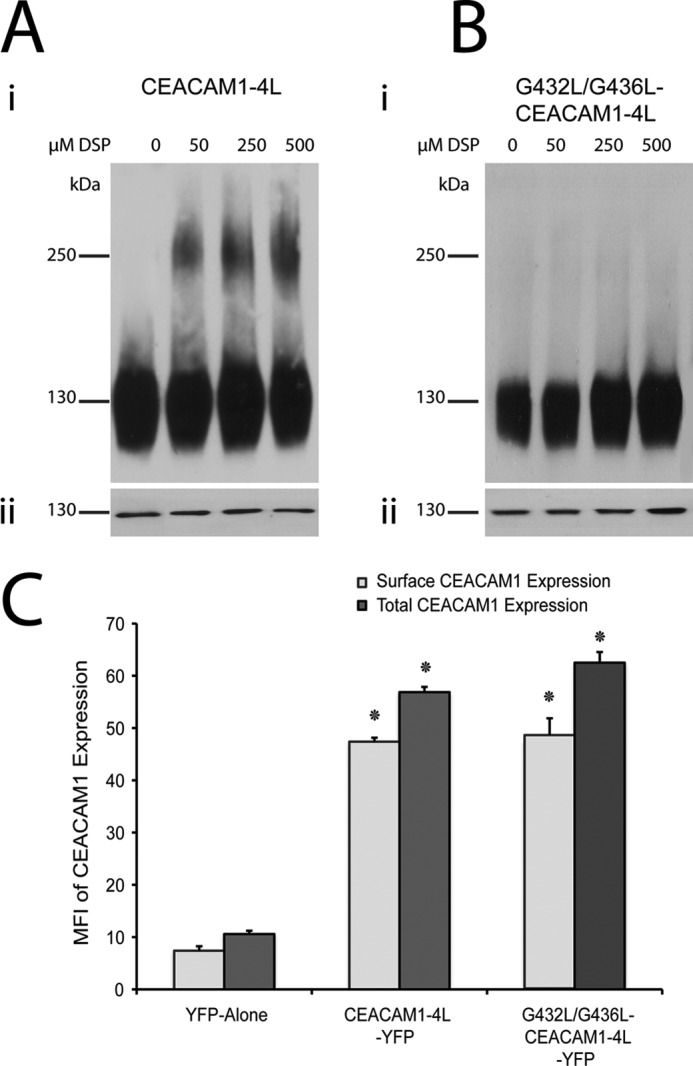
Oligomerization of wild type CEACAM1 and its G432L/G436L transmembrane mutant. Transiently transfected HeLa cells expressing CEACAM1–4L (A) or CEACAM1–4L containing G432L/G436L mutations (B) were treated with various concentrations of the homobifunctional chemical cross-linker DSP; total cell lysates were resolved under nonreducing conditions (panel i) or reducing conditions (panel ii) by SDS-PAGE and probed with antibodies specific for CEACAM1. Blots are representative of three independent experiments. B, quantitative evaluation of CEACAM1 surface as well as total expression by flow cytometry, plotted as mean fluorescence intensity (MFI). The x axis indicates transfected allele. Data represent the mean ± S.E. from three separate experiments. *, p value < 0.05 compared with pEYFP alone control.
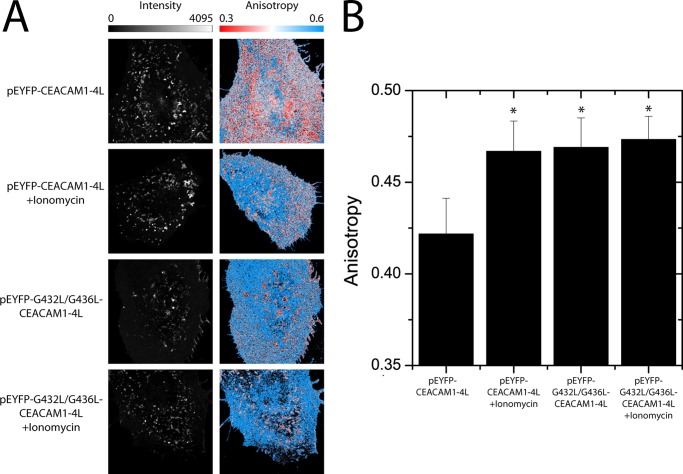
To provide an alternative method by which to compare the oligomeric state of CEACAM1, and to observe the distribution of these oligomers on the plasma membrane of living cells, FAIM was performed on HeLa cells expressing CEACAM1–4L-YFP or G432L/G436L-CEACAM1–4L-YFP. FAIM is a Förster resonance energy transfer (FRET)-based assay that can be used to measure protein self-oligomerization in live cells. As applied here, this technique detects fluorescence depolarization due to energy transfer between identical fluorophores (homo-FRET). The various YFP-tagged CEACAM1–4L variants were indistinguishable based on intensity alone, showing fluorescence consistent with plasma membrane association (Fig. 6A, Intensity). In contrast, these same cells showed significantly different calculated anisotropies (Fig. 6A, Anisotropy). CEACAM1–4L-YFP-expressing cells showed a significantly higher anisotropy when treated with ionomycin (Fig. 6, A and B). This increase in anisotropy was similar to the difference observed in anisotropy between the monomeric and tandem-dimer Venus constructs (supplemental Fig. S1) supporting the biochemical data suggesting that CEACAM1–4L-YFP becomes more monomeric upon ionomycin treatment. Importantly, cells expressing G432L/G436L-CEACAM1–4L-YFP showed a high (monomeric) anisotropy value reminiscent of that seen with the ionomycin-treated wild type form (Fig. 6, A and B). The lack of an effect of ionomycin on the G432L/G436L-CEACAM1–4L anisotropy also suggests that this mutant exists exclusively as a monomer. Together, these data indicate that CEACAM1 tends to exist in dimeric form on the plasma membrane of living cells due to the GXXXG transmembrane domain motif and becomes more monomeric upon association with calcium-loaded calmodulin.
FIGURE 6.
FAIM reveals varied oligomerization of CEACAM1–4L and G432L/G436L-CEACAM1–4L. Fluorescence anisotropy measurements were made on HeLa cells expressing enhanced YFP-CEACAM1–4L and enhanced YFP-G432L/G436L-CEACAM1–4L with and without ionomycin treatment. A, intensity images (left column) show similar expression and distribution of wild type and mutant CEACAM1 on the plasma membrane. A comparison of the anisotropy images (right column) show that ionomycin treatment increases the anisotropy of CEACAM1–4L-YFP, indicating that there is an increase in the proportion of CEACAM1 that is in monomeric form. Anisotropy images for the untreated G432L/G436L mutant reflect that seen with ionomycin-treated CEACAM1, and this is not affected by exposure to ionomycin. B, summarized anisotropy values for CEACAM1–4L-YFP and G432L/G436L-CEACAM1–4L-YFP with and without ionomycin. The data represent the mean ± S.E. from measurements on 30 or more cells collected from six independent experiments. Asterisk indicates a p value of <0.05 by a Student's t test compared with the CEACAM1–4L-YFP.
Monomeric CEACAM Can Mediate Bacterial Binding and Engulfment
A remarkable number of protein classes form reversible homodimers, and such an association has been observed to regulate biological functions ranging from protein folding and trafficking to ligand binding and signaling (62). Because the G432L/G436L mutations did not seem to affect the trafficking of CEACAM1, we considered whether the mutant's inability to form dimers might affect its various known functions. The human pathogenic bacteria N. gonorrhoeae express Opa protein adhesins that bind CEACAM1 and trigger effective bacterial engulfment into these otherwise nonphagocytic epithelial cells (43, 45, 47, 63, 64). To assess whether CEACAM1 mutations affect bacterial adhesion and/or engulfment, we analyzed HeLa cells transiently transfected with CEACAM1–4L, G432L/G436L-CEACAM1–4L (which are impaired in cis-dimerization), or R43S/Q44L-CEACAM1–4L (which are impaired in cell-cell adhesion). There was a significant decrease in the number of bound and internalized OpaCEA-expressing gonococci when the cells were incubated with HeLa cells expressing R43S/Q44L-CEACAM1–4L compared with CEACAM1–4L (Fig. 7, A and B). Residues Arg-43 and Gln-44 are located in the exposed loops of the GFCC′C″ face of the CEACAM1 N domain (8) and are required for effective binding by some Opa variants, including the one used in this study (48, 65, 66). No significant differences were detected when considering bacterial binding or internalization by cells expressing wild type versus the G432L/G436L mutant of CEACAM1–4L (Fig. 7, A and B). This suggests that the bacterial adhesin does not have a preference for monomeric versus dimeric CEACAM1 and/or that the preferred monomeric receptor form is not in limiting abundance on the cells expressing CEACAM1–4L.
FIGURE 7.
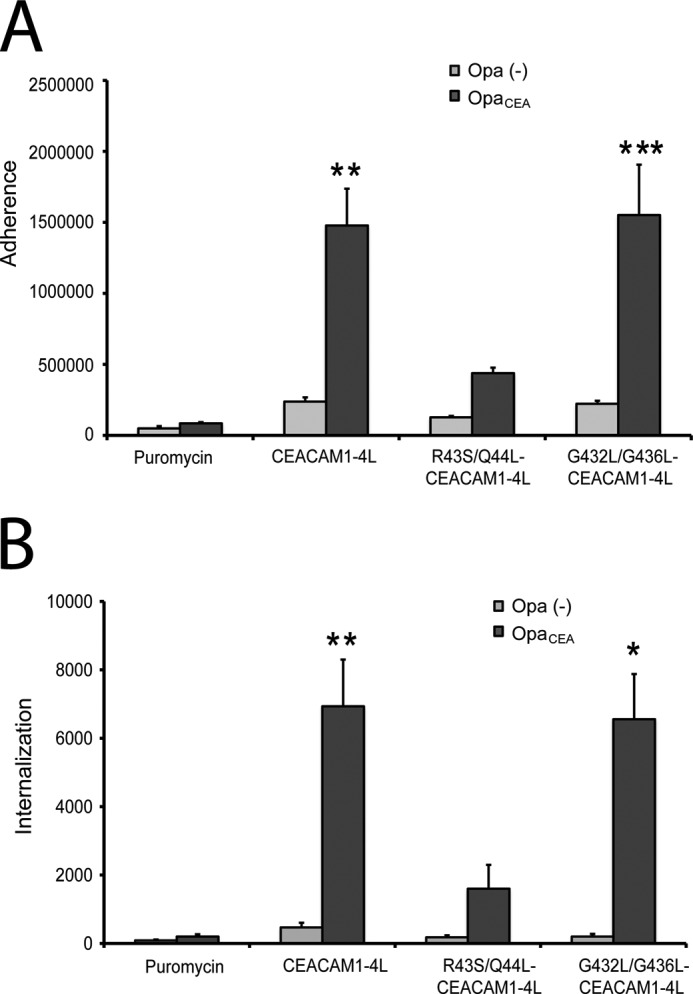
Effects of CEACAM1 dimerization on adhesion and engulfment of N. gonorrhoeae. HeLa cells transfected with vector control, CEACAM1–4L, CEACAM1–4L containing R43S/Q44L mutations, or CEACAM1–4L containing G432L/G436L mutations were infected with N. gonorrhoeae expressing either no Opa proteins (gray bars) or a CEACAM1-specific OpaCEA protein (black bars). Means ± S.E. of triplicate samples were graphed to illustrate bacterial attachment (A) and internalization (B) by these cells. Asterisk denotes p < 0.05; double asterisk, p < 0.01; and triple asterisk, p < 0.005 compared with the untreated samples. The results are representative of three independent experiments.
CEACAM1 trans-Homophilic Interactions Are Mediated by cis-Monomers of CEACAM1
CEACAM1 is capable of binding to itself in a trans-homophilic manner and has been demonstrated to mediate aggregation of a variety of cell types (67–70). Moreover, the ability of CEACAM1 to control growth and differentiation relies on this antiparallel binding (23). However, critically, it remains unknown whether intercellular adhesion is mediated by interactions between monomeric or dimeric CEACAM1. Because the transmembrane G432L/G436L mutant of CEACAM1–4L does not dimerize, we sought to assess whether replacement of these residues affected cell-cell binding. As expected, cells expressing CEACAM1–4L proceeded to aggregate following resuspension into single cells, with ∼30% of cells in aggregates of 10 or more and ∼49% of cells in aggregates of 6 or more by 60 min (Fig. 8). This aggregation depends on CEACAM1 because cells expressing the R43S/Q44L-containing mutant, which is unable to engage in trans-homophilic binding (53), remained largely unaggregated throughout the experiment (Fig. 8). Strikingly, expression of transmembrane domain mutant G432L/G436L-CEACAM1–4L caused a substantial increase in cellular aggregation, with the end result being ∼90% of cells found within aggregates of three or more cells by 60 min (Fig. 8). Taken together, these results suggest that CEACAM1–4L mediates intercellular adhesion through homophilic interactions involving cis-monomeric CEACAM1 binding to cis-monomeric CEACAM1 on the adjacent cells, forming the trans-homophilic dimers where the residues Arg-43 and Gln-44 at the N-terminal domain of the CEACAM1 are critical in cell-cell adhesion.
Src Tyrosine Kinases and SHP Tyrosine Phosphatases Associate with CEACAM1 Monomers
Previous studies have demonstrated an interaction between the CEACAM1 long cytoplasmic domain and Src family protein-tyrosine kinases (71) and with the protein-tyrosine phosphatases SHP-1 and SHP-2 (72, 73). Recently, Obrink and co-workers (26) suggested that the molecular organization of rat CEACAM1 might be a factor that regulates the cytoplasmic interaction. Because the G432L/G436L mutations occur in the transmembrane domain, these residues are unlikely to interact with these downstream effector proteins. As such, we sought to determine whether the forced monomers of CEACAM1 that are involved in cell-cell binding would recruit Src kinases, SHP-1 and/or SHP-2. To detect the interactions, untransfected or transfected HeLa cells expressing CEACAM1–4L or G432L/G436L-CEACAM1–4L were subjected to DSP and pervanadate treatment, the latter of which has been shown to stabilize phosphotyrosine-dependent recruitment of these effector proteins by CEACAM1 (72, 74). The cells were lysed with nonionic detergent-containing buffers, and CEACAM1 was immunoprecipitated using a CEACAM1-specific antibody. Association of recovered proteins was then detected using immunoblot analysis. We found that SHP-1, SHP-2, and c-Src were all associated with CEACAM1–4L monomers but not with CEACAM1–4L dimers or with untransfected control (Fig. 9A). This preferential co-immunoprecipitation of downstream effectors with monomeric CEACAM1 suggests that the mode of molecular signaling may be attributed to a structural organization of the CEACAM1. It also suggests that the ratio of CEACAM1 dimers and monomers expressed on the cell surface has implications for the effect of CEACAM1, with the basal (dimeric) state of CEACAM1 present in resting cells not engaging these effector proteins.
FIGURE 9.
Association of downstream effector proteins with CEACAM1 monomers. A, HeLa cells untransfected (control) or transfected with CEACAM1–4L or G432L/G436L-CEACAM1–4L were pretreated with pervanadate and then cross-linked with DSP. CEACAM1 and associated proteins were co-immunoprecipitated (IP) with CEACAM1-specific antibodies. Proteins were resolved under nonreducing conditions, and immunoblots were probed with antibodies specific for SHP-1, SHP-2, and c-Src (panel i) or separated under reducing conditions and immunoblots probed to detect total levels of CEACAM1 (panel ii). WB, Western blot. B, cell lysates were probed with anti-CEACAM1 antibody to ensure the cells were expressing the transfected proteins under nonreducing (panel i) or reducing conditions (panel ii). Note that CEACAM1-effector complexes migrate at sizes greater than CEACAM1 alone (compare A with B). C, immunostaining of SHP-1, SHP-2 phosphatases, and c-Src kinase in HeLa cells expressing pEYFP-G432L/G436L-CEACAM1–4L. The cellular localization of pEYFP-G432L/G436L-CEACAM1–4L (green false-color YFP) with SHP-1 (red), with SHP-2 (red), or with c-Src (red), as indicated, were analyzed by confocal microscopy. Arrows indicate an area devoid of CEACAM1. Bars, 10 μm. Data are representative of three independent experiments.
Furthermore, to consider whether the interaction between CEACAM and the downstream effectors occurred at sites relevant for intercellular adhesion, transfected cells expressing CEACAM1–4L or the G432L/G436L-CEACAM1–4L mutant were pretreated with pervanadate to preserve phosphotyrosines and then immunostained with CEACAM1 and effector-specific antibodies. Cell-cell contact areas displayed an intense co-localization of SHP-1, SHP-2, and c-Src associated with both the CEACAM1–4L (data not shown) and G432L/G436L-CEACAM1–4L mutant cell lines (Fig. 9, B and C). Altogether, our data indicate that CEACAM1 molecular organization, phosphorylation, and recruitment of downstream effectors are interdependent events that regulate the cell adhesion function of CEACAM1 and the cellular response that results from its trans-homophilic binding.
DISCUSSION
Despite a general assumption that the oligomeric state of CEACAM1 must affect its function (23, 24), the functional implications of altering the CEACAM1 monomer-dimer equilibrium have only been indirectly explored. Previous studies have observed that CEACAM1 exists as either monomers or cis-homodimers on the surface of rat epithelial cells, although the dimers are absent in CHO cells (25). This is indicative of CEACAM1 dimerization being controlled by a cell type-specific process, perhaps due to basal state calcium signaling. Our chemical cross-linking and high resolution anisotropy-based homo-FRET analyses both indicate that CEACAM1 exists as an equilibrium of cis-monomers and cis-homodimers on the surface of human epithelium-derived cells. Although the low molecular weight form of CEACAM1 tends to be the predominant form detected by our cross-linking experiments, we are not inclined to conclude that the monomer comprises the majority of CEACAM1 on the surface of cells. This could, instead, reflect the efficiency of cross-linking at the DSP concentrations and cell density used in our studies and would presumably be regulated by growth factors and/or cellular localization within the tissues in vivo.
Calmodulin has been shown to bind synthetic peptides corresponding to the cytoplasmic domain of both CEACAM1–4L and CEACAM1–4S splice variant sequences in a calcium-dependent manner (55), and purified Ca2+-calmodulin was seen to suppress the assembly of CEACAM1 dimers in protein suspensions (25). These studies suggested that calmodulin was an important regulator of CEACAM1 monomer-dimer shifts (24). Our studies both support and extend this premise by showing that ionomycin-induced calcium fluxes dissociate dimers of both CEACAM1–4L and CEACAM1–4S but do not affect a truncated form of CEACAM1 that completely lacks the calmodulin-binding site. This effect requires active calmodulin because the calmodulin-specific inhibitor W-7 completely abrogates the dissolution of CEACAM1 dimers.
In view of our observation that CEACAM1 lacking the entire cytoplasmic sequence persisted as a dimer and previous work indicating that the soluble extracellular domain of CEACAM1 is itself monomeric (75), we considered that the transmembrane domain of CEACAM1 might be a driving force for the establishment of the basal state dimers. Diverse motifs that may promote association between transmembrane helices have been identified (76). Of particular interest here are the small residue GXXXG/tetrad and heptad motifs (where small represents Gly, Ala, or Ser). These two sequence patterns each place the small side chains on the same helix surface to allow helix-helix association, but the former mediates right-handed and the latter left-handed helix packing angles. Inspection of the CEACAM1–4L transmembrane domain sequence indicated that pairs of residues were positioned appropriately for participation in either small residue interaction motif. We explored the potential for helix-helix association in CEACAM1–4L using a computational tool that predicts possible conformations for membrane-embedded helical pairs based upon an exhaustive search for structures that exist as tightly packed helices. This analysis predicted that the CEACAM1–4L transmembrane domain has the potential to undergo dimerization via a small residue heptad repeat motif requiring the two Gly residues at positions 432 and 436. Consistent with this, our biochemical and biophysical analyses revealed that replacement of these glycine residues with leucine resulted in mutants of human CEACAM1–4L that were strictly monomeric, showing no evidence of the dimeric form normally apparent with the CEACAM1–4L.
The monomeric CEACAM1–4L mutant was expressed at the cell surface, a fact that was critical for our ability to compare the function of monomeric versus wild type CEACAM1–4L. Indeed, the monomeric G432L/G436L mutant retains the capacity to function as a host cellular receptor for N. gonorrhoeae expressing the CEACAM1-specific Opa protein adhesins, with no apparent effect on either bacterial binding or engulfment by cells expressing the wild type or mutant alleles. Although this result does not exclude the possibility that dimers of CEACAM1 may also facilitate infection by these pathogenic bacteria, it nonetheless proves that the monomeric form has the capacity to do so.
Recently, Lawson et al. (28) reported that rat CEACAM1–4S encodes two different GXXXG sequences that must be simultaneously mutated to prevent the formation of dimers in rat hepatocellular carcinoma cell line. These mutations had a profound effect on rCEACAM1–4S function, abrogating the anchorage-independent growth potential normally observed with rCEACAM1–4S-expressing cells in their system. The mechanistic reason for the effect of these mutations was not further explored (28). Human CEACAM1 dimers dissociated when we mutated a single GXXXG sequence within the wild type CEACAM1–4L transmembrane domain to G432L/G436L-CEACAM1–4L. Moreover, our results provide the first mechanistic insight as to how monomer-dimer shifts may affect the cell; monomeric CEACAM1 at the cell surface promoted intercellular aggregation by virtue of its ability to engage in trans-homophilic binding and recruit cytoplasmic effector proteins to the site of cell-cell contact. In considering that the Src family tyrosine kinases and SHP-1 and SHP-2 tyrosine phosphatases each appear to associate exclusively with the monomeric form of CEACAM1, evidenced both by co-immunoprecipitation and immunofluorescence microscopy-based analysis of cell-cell junctions, it is enticing to propose that the immunoreceptor tyrosine-based inhibitory motifs, which recruit the cytoplasmic effectors, are sterically occluded within the CEACAM1 dimer.
Our findings seem to contradict the previous suggestion that SHP-2 preferentially binds to dimeric forms of CEACAM1–4S (26); however, that study relied on the effects of a two monoclonal antibodies that seem to either promote or prevent CEACAM1 oligomerization, whereas ours tests the effect of a mutant that cannot oligomerize. It seems reasonable that the conformation and/or localization of CEACAM1 may not be equivalent under these very different conditions. Although the effectors responsible for the CEACAM1–4S-driven anchorage-independent growth have yet to be defined, it seems reasonable to consider that masking/unmasking of cytoplasmically localized binding motifs may also occur in this context. Indeed, such a model fits nicely with observations described in the rCEACAM1–4S/rat hepatocellular carcinoma study (28).
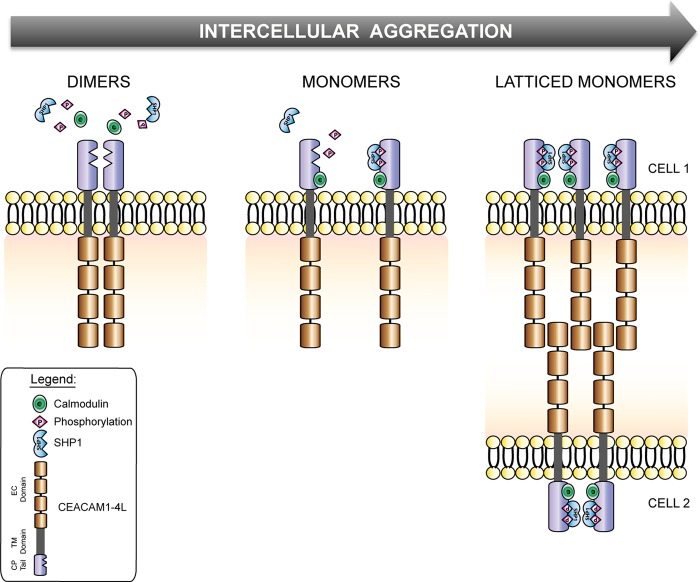
Because the immunoreceptor tyrosine-based inhibitory motif-dependent recruitment of tyrosine phosphatases has been shown to be required for the tumor inhibitory function (77) and for CEACAM1-L-dependent inhibition of T lymphocyte activation and proliferation (78), our results predict that the basal state dimers on the cell surface would be silent, and monomeric CEACAM1 at cell-cell junctions would liberate growth inhibitory signals. The increased CEACAM1-dependent cell aggregation kinetics and increased size of aggregates formed by the G432L/G436L mutant clearly suggest that monomeric CEACAM1 facilitates intercellular binding. However, it is important to consider the studies of Muller et al. (26) that suggest that trans-homophilic CEACAM1 binding triggers increased cis-oligomerization. The most satisfying means by which to integrate the data from our study with this and other studies mentioned above is to consider that basal state CEACAM1 dimers must dissociate to engage in intercellular binding, which would then promote the progressive recruitment of antiparallel trans-dimers that would allow the accumulation of downstream effector proteins at the cell-cell interaction plane (Fig. 10). In addition to providing a new paradigm for cell adhesion by the immunoglobulin superfamily members, this scheme provides an exciting opportunity to affect cancer progression through the pharmacologic control of receptor oligomerization.
FIGURE 10.
Dynamic reassortment of CEACAM1 oligomers to facilitate trans-homophilic binding and downstream effector recruitment. A schematic diagram depicts the progression of CEACAM1 oligomeric states in a basal state (dimers), as monomers bound to Ca2+-calmodulin, and in a lattice-like arrangement at cell-cell contacts.
Acknowledgment
We thank Dr. Bjorn Obrink (Karolinska Institute, Sweden) for our engaging and insightful discussions during the course of this work.
This work was supported in part by the Canadian Institutes of Health Research Training Program in Protein Folding and Interaction Dynamics (to P. C. P.), Canadian Institutes of Health Research Grant FRN-5810 (to C. M. D.), the Canada Research Chairs Program (to C. M. Y.), Natural Sciences and Engineering Research Council of Canada/Canadian Institutes of Health Research Collaborative Health Research Program Grant 365861-09 (to C. M. Y. and S. D. G.), and Canadian Foundation for Innovation Grant JVR-NMD106997.

This article contains supplemental Fig. S1 and Table S1.
- CEACAM1
- carcinoembryonic antigen-related cellular adhesion molecule 1
- DSP
- dithiobis(succinimidyl)propionate
- TMD
- transmembrane domain dimer
- ITIM
- immunoreceptor tyrosine-based inhibitory motif
- SHP-1
- SH2-domain-containing protein-tyrosine phosphatase-1
- SHP-2
- SH2-domain-containing protein-tyrosine phosphatase-2
- SH
- Src homology
- FAIM
- fluorescence anisotropy imaging microscopy
- L
- long
- S
- short
- W-7
- N-(6-aminohexyl)-5-chloro-1-naphthalenesulfonamide hydrochloride.
REFERENCES
- 1. Okegawa T., Pong R. C., Li Y., Hsieh J. T. (2004) The role of cell adhesion molecule in cancer progression and its application in cancer therapy. Acta Biochim. Pol. 51, 445–457 [PubMed] [Google Scholar]
- 2. Beauchemin N., Draber P., Dveksler G., Gold P., Gray-Owen S., Grunert F., Hammarström S., Holmes K. V., Karlsson A., Kuroki M., Lin S. H., Lucka L., Najjar S. M., Neumaier M., Obrink B., Shively J. E., Skubitz K. M., Stanners C. P., Thomas P., Thompson J. A., Virji M., von Kleist S., Wagener C., Watt S., Zimmermann W. (1999) Redefined nomenclature for members of the carcinoembryonic antigen family. Exp. Cell Res. 252, 243–249 [DOI] [PubMed] [Google Scholar]
- 3. Barnett T. R., Drake L., Pickle W., 2nd (1993) Human biliary glycoprotein gene: characterization of a family of novel alternatively spliced RNAs and their expressed proteins. Mol. Cell Biol. 13, 1273–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hinoda Y., Neumaier M., Hefta S. A., Drzeniek Z., Wagener C., Shively L., Hefta L. J., Shively J. E., Paxton R. J. (1988) Molecular cloning of a cDNA coding biliary glycoprotein I: primary structure of a glycoprotein immunologically cross-reactive with carcinoembryonic antigen. Proc. Natl. Acad. Sci. U.S.A. 85, 6959–6963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barnett T. R., Kretschmer A., Austen D. A., Goebel S. J., Hart J. T., Elting J. J., Kamarck M. E. (1989) Carcinoembryonic antigens: alternative splicing accounts for the multiple mRNAs that code for novel members of the carcinoembryonic antigen family. J. Cell Biol. 108, 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watt S. M., Teixeira A. M., Zhou G. Q., Doyonnas R., Zhang Y., Grunert F., Blumberg R. S., Kuroki M., Skubitz K. M., Bates P. A. (2001) Homophilic adhesion of human CEACAM1 involves N-terminal domain interactions: structural analysis of the binding site. Blood 98, 1469–1479 [DOI] [PubMed] [Google Scholar]
- 7. Wikström K., Kjellström G., Obrink B. (1996) Homophilic intercellular adhesion mediated by C-CAM is due to a domain 1-domain 1 reciprocal binding. Exp. Cell Res. 227, 360–366 [DOI] [PubMed] [Google Scholar]
- 8. Tan K., Zelus B. D., Meijers R., Liu J. H., Bergelson J. M., Duke N., Zhang R., Joachimiak A., Holmes K. V., Wang J. H. (2002) Crystal structure of murine sCEACAM1a[1,4]: a coronavirus receptor in the CEA family. EMBO J. 21, 2076–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Singer B. B., Scheffrahn I., Obrink B. (2000) The tumor growth-inhibiting cell adhesion molecule CEACAM1 (C-CAM) is differently expressed in proliferating and quiescent epithelial cells and regulates cell proliferation. Cancer Res. 60, 1236–1244 [PubMed] [Google Scholar]
- 10. Nittka S., Günther J., Ebisch C., Erbersdobler A., Neumaier M. (2004) The human tumor suppressor CEACAM1 modulates apoptosis and is implicated in early colorectal tumorigenesis. Oncogene 23, 9306–9313 [DOI] [PubMed] [Google Scholar]
- 11. Wagener C., Ergün S. (2000) Angiogenic properties of the carcinoembryonic antigen-related cell adhesion molecule 1. Exp. Cell Res. 261, 19–24 [DOI] [PubMed] [Google Scholar]
- 12. Luo W., Wood C. G., Earley K., Hung M. C., Lin S. H. (1997) Suppression of tumorigenicity of breast cancer cells by an epithelial cell adhesion molecule (C-CAM1): the adhesion and growth suppression are mediated by different domains. Oncogene 14, 1697–1704 [DOI] [PubMed] [Google Scholar]
- 13. Boulton I. C., Gray-Owen S. D. (2002) Neisserial binding to CEACAM1 arrests the activation and proliferation of CD4+ T lymphocytes. Nat. Immunol. 3, 229–236 [DOI] [PubMed] [Google Scholar]
- 14. Chen D., Iijima H., Nagaishi T., Nakajima A., Russell S., Raychowdhury R., Morales V., Rudd C. E., Utku N., Blumberg R. S. (2004) Carcinoembryonic antigen-related cellular adhesion molecule 1 isoforms alternatively inhibit and costimulate human T cell function. J. Immunol. 172, 3535–3543 [DOI] [PubMed] [Google Scholar]
- 15. Kammerer R., Stober D., Singer B. B., Obrink B., Reimann J. (2001) Carcinoembryonic antigen-related cell adhesion molecule 1 on murine dendritic cells is a potent regulator of T cell stimulation. J. Immunol. 166, 6537–6544 [DOI] [PubMed] [Google Scholar]
- 16. Lee H. S., Ostrowski M. A., Gray-Owen S. D. (2008) CEACAM1 dynamics during Neisseria gonorrhoeae suppression of CD4+ T lymphocyte activation. J. Immunol. 180, 6827–6835 [DOI] [PubMed] [Google Scholar]
- 17. Markel G., Lieberman N., Katz G., Arnon T. I., Lotem M., Drize O., Blumberg R. S., Bar-Haim E., Mader R., Eisenbach L., Mandelboim O. (2002) CD66a interactions between human melanoma and NK cells: a novel class I MHC-independent inhibitory mechanism of cytotoxicity. J. Immunol. 168, 2803–2810 [DOI] [PubMed] [Google Scholar]
- 18. Markel G., Wolf D., Hanna J., Gazit R., Goldman-Wohl D., Lavy Y., Yagel S., Mandelboim O. (2002) Pivotal role of CEACAM1 protein in the inhibition of activated decidual lymphocyte functions. J. Clin. Invest. 110, 943–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morales V. M., Christ A., Watt S. M., Kim H. S., Johnson K. W., Utku N., Texieira A. M., Mizoguchi A., Mizoguchi E., Russell G. J., Russell S. E., Bhan A. K., Freeman G. J., Blumberg R. S. (1999) Regulation of human intestinal intraepithelial lymphocyte cytolytic function by biliary glycoprotein (CD66a). J. Immunol. 163, 1363–1370 [PubMed] [Google Scholar]
- 20. Skubitz K. M., Campbell K. D., Skubitz A. P. (1996) CD66a, CD66b, CD66c, and CD66d each independently stimulate neutrophils. J. Leukoc. Biol. 60, 106–117 [DOI] [PubMed] [Google Scholar]
- 21. Chen C. J., Shively J. E. (2004) The cell-cell adhesion molecule carcinoembryonic antigen-related cellular adhesion molecule 1 inhibits IL-2 production and proliferation in human T cells by association with Src homology protein-1 and down-regulates IL-2 receptor. J. Immunol. 172, 3544–3552 [DOI] [PubMed] [Google Scholar]
- 22. Stern N., Markel G., Arnon T. I., Gruda R., Wong H., Gray-Owen S. D., Mandelboim O. (2005) Carcinoembryonic antigen (CEA) inhibits NK killing via interaction with CEA-related cell adhesion molecule 1. J. Immunol. 174, 6692–6701 [DOI] [PubMed] [Google Scholar]
- 23. Gray-Owen S. D., Blumberg R. S. (2006) CEACAM1: contact-dependent control of immunity. Nat. Rev. Immunol. 6, 433–446 [DOI] [PubMed] [Google Scholar]
- 24. Obrink B., Sawa H., Scheffrahn I., Singer B. B., Sigmundsson K., Sundberg U., Heymann R., Beauchemin N., Weng G., Ram P., Iyengar R. (2002) Computational analysis of isoform-specific signal regulation by CEACAM1-A cell adhesion molecule expressed in PC12 cells. Ann. N.Y. Acad. Sci. 971, 597–607 [DOI] [PubMed] [Google Scholar]
- 25. Hunter I., Sawa H., Edlund M., Obrink B. (1996) Evidence for regulated dimerization of cell-cell adhesion molecule (C-CAM) in epithelial cells. Biochem. J. 320, 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Müller M. M., Klaile E., Vorontsova O., Singer B. B., Obrink B. (2009) Homophilic adhesion and CEACAM1-S regulate dimerization of CEACAM1-L and recruitment of SHP-2 and c-Src. J. Cell Biol. 187, 569–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Edlund M., Obrink B. (1993) Evidence for calmodulin binding to the cytoplasmic domains of two C-CAM isoforms. FEBS Lett. 327, 90–94 [DOI] [PubMed] [Google Scholar]
- 28. Lawson E. L., Mills D. R., Brilliant K. E., Hixson D. C. (2012) The transmembrane domain of CEACAM1–4S is a determinant of anchorage independent growth and tumorigenicity. PLoS One 7, e29606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gray-Owen S. D., Dehio C., Haude A., Grunert F., Meyer T. F. (1997) CD66 carcinoembryonic antigens mediate interactions between Opa-expressing Neisseria gonorrhoeae and human polymorphonuclear phagocytes. EMBO J. 16, 3435–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuroki M., Arakawa F., Matsuo Y., Oikawa S., Misumi Y., Nakazato H., Matsuoka Y. (1991) Molecular cloning of nonspecific cross-reacting antigens in human granulocytes. J. Biol. Chem. 266, 11810–11817 [PubMed] [Google Scholar]
- 31. Horton R. M., Cai Z. L., Ho S. N., Pease L. R. (1990) Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8, 528–535 [PubMed] [Google Scholar]
- 32. Kupsch E. M., Knepper B., Kuroki T., Heuer I., Meyer T. F. (1993) Variable opacity (Opa) outer membrane proteins account for the cell tropisms displayed by Neisseria gonorrhoeae for human leukocytes and epithelial cells. EMBO J. 12, 641–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Adams P. D., Arkin I. T., Engelman D. M., Brünger A. T. (1995) Computational searching and mutagenesis suggest a structure for the pentameric transmembrane domain of phospholamban. Nat. Struct. Biol. 2, 154–162 [DOI] [PubMed] [Google Scholar]
- 34. Adams P. D., Engelman D. M., Brünger A. T. (1996) Improved prediction for the structure of the dimeric transmembrane domain of glycophorin A obtained through global searching. Proteins 26, 257–261 [DOI] [PubMed] [Google Scholar]
- 35. Brunger A. T. (2007) Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 36. Rizzo M. A., Springer G., Segawa K., Zipfel W. R., Piston D. W. (2006) Optimization of pairings and detection conditions for measurement of FRET between cyan and yellow fluorescent proteins. Microsc. Microanal. 12, 238–254 [DOI] [PubMed] [Google Scholar]
- 37. Abramoff M. D., Magelhaes P. J., Ram S. J. (2004) Image processing with ImageJ. Biophotonics Int. 11, 36–42 [Google Scholar]
- 38. Axelrod D. (1979) Carbocyanine dye orientation in red cell membrane studied by microscopic fluorescence polarization. Biophys. J. 26, 557–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Axelrod D. (1989) Fluorescence polarization microscopy. Methods Cell Biol. 30, 333–352 [PubMed] [Google Scholar]
- 40. Rocheleau J. V., Edidin M., Piston D. W. (2003) Intrasequence GFP in class I MHC molecules, a rigid probe for fluorescence anisotropy measurements of the membrane environment. Biophys. J. 84, 4078–4086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Volkmer A., Subramaniam V., Birch D. J., Jovin T. M. (2000) One- and two-photon excited fluorescence lifetimes and anisotropy decays of green fluorescent proteins. Biophys. J. 78, 1589–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Booth J. W., Telio D., Liao E. H., McCaw S. E., Matsuo T., Grinstein S., Gray-Owen S. D. (2003) Phosphatidylinositol 3-kinases in carcinoembryonic antigen-related cellular adhesion molecule-mediated internalization of Neisseria gonorrhoeae. J. Biol. Chem. 278, 14037–14045 [DOI] [PubMed] [Google Scholar]
- 43. McCaw S. E., Liao E. H., Gray-Owen S. D. (2004) Engulfment of Neisseria gonorrhoeae: revealing distinct processes of bacterial entry by individual carcinoembryonic antigen-related cellular adhesion molecule family receptors. Infect. Immun. 72, 2742–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Watt S. M., Sala-Newby G., Hoang T., Gilmore D. J., Grunert F., Nagel G., Murdoch S. J., Tchilian E., Lennox E. S., Waldmann H. (1991) CD66 identifies a neutrophil-specific epitope within the hematopoietic system that is expressed by members of the carcinoembryonic antigen family of adhesion molecules. Blood 78, 63–74 [PubMed] [Google Scholar]
- 45. Chen T., Grunert F., Medina-Marino A., Gotschlich E. C. (1997) Several carcinoembryonic antigens (CD66) serve as receptors for gonococcal opacity proteins. J. Exp. Med. 185, 1557–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rowe H. A., Griffiths N. J., Hill D. J., Virji M. (2007) Co-ordinate action of bacterial adhesins and human carcinoembryonic antigen receptors in enhanced cellular invasion by capsulate serum resistant Neisseria meningitidis. Cell Microbiol. 9, 154–168 [DOI] [PubMed] [Google Scholar]
- 47. Bos M. P., Grunert F., Belland R. J. (1997) Differential recognition of members of the carcinoembryonic antigen family by Opa variants of Neisseria gonorrhoeae. Infect. Immun. 65, 2353–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Virji M., Evans D., Hadfield A., Grunert F., Teixeira A. M., Watt S. M. (1999) Critical determinants of host receptor targeting by Neisseria meningitidis and Neisseria gonorrhoeae: identification of Opa adhesiotopes on the N-domain of CD66 molecules. Mol. Microbiol. 34, 538–551 [DOI] [PubMed] [Google Scholar]
- 49. Rougeaux C., Berger C. N., Servin A. L. (2008) hCEACAM1–4L down-regulates hDAF-associated signalling after being recognized by the Dr adhesin of diffusely adhering Escherichia coli. Cell Microbiol. 10, 632–654 [DOI] [PubMed] [Google Scholar]
- 50. N′Guessan P. D., Vigelahn M., Bachmann S., Zabel S., Opitz B., Schmeck B., Hippenstiel S., Zweigner J., Riesbeck K., Singer B. B., Suttorp N., Slevogt H. (2007) The UspA1 protein of Moraxella catarrhalis induces CEACAM-1-dependent apoptosis in alveolar epithelial cells. J. Infect. Dis. 195, 1651–1660 [DOI] [PubMed] [Google Scholar]
- 51. Blikstad I., Wikström T., Aurivillius M., Obrink B. (1992) C-CAM (Cell-CAM 105) is a calmodulin binding protein. FEBS Lett. 302, 26–30 [DOI] [PubMed] [Google Scholar]
- 52. Edlund M., Gaardsvoll H., Bock E., Obrink B. (1993) Different isoforms and stock-specific variants of the cell adhesion molecule C-CAM (cell-CAM 105) in rat liver. Eur. J. Biochem. 213, 1109–1116 [DOI] [PubMed] [Google Scholar]
- 53. Markel G., Gruda R., Achdout H., Katz G., Nechama M., Blumberg R. S., Kammerer R., Zimmermann W., Mandelboim O. (2004) The critical role of residues 43R and 44Q of carcinoembryonic antigen cell adhesion molecules-1 in the protection from killing by human NK cells. J. Immunol. 173, 3732–3739 [DOI] [PubMed] [Google Scholar]
- 54. Sundberg U., Beauchemin N., Obrink B. (2004) The cytoplasmic domain of CEACAM1-L controls its lateral localization and the organization of desmosomes in polarized epithelial cells. J. Cell Sci. 117, 1091–1104 [DOI] [PubMed] [Google Scholar]
- 55. Edlund M., Blikstad I., Obrink B. (1996) Calmodulin binds to specific sequences in the cytoplasmic domain of C-CAM and down-regulates C-CAM self-association. J. Biol. Chem. 271, 1393–1399 [DOI] [PubMed] [Google Scholar]
- 56. Hofmann K., Stoffel W. (1993) A database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 347, 166 [Google Scholar]
- 57. Russ W. P., Engelman D. M. (2000) The GxxxG motif: a framework for transmembrane helix-helix association. J. Mol. Biol. 296, 911–919 [DOI] [PubMed] [Google Scholar]
- 58. Langosch D., Arkin I. T. (2009) Interaction and conformational dynamics of membrane-spanning protein helices. Protein Sci. 18, 1343–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lear J. D., Stouffer A. L., Gratkowski H., Nanda V., Degrado W. F. (2004) Association of a model transmembrane peptide containing gly in a heptad sequence motif. Biophys. J. 87, 3421–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 61. Tamamizu S., Lee Y., Hung B., McNamee M. G., Lasalde-Dominicci J. A. (1999) Alteration in ion channel function of mouse nicotinic acetylcholine receptor by mutations in the M4 transmembrane domain. J. Membr. Biol. 170, 157–164 [DOI] [PubMed] [Google Scholar]
- 62. Paavola C. D., Hemmerich S., Grunberger D., Polsky I., Bloom A., Freedman R., Mulkins M., Bhakta S., McCarley D., Wiesent L., Wong B., Jarnagin K., Handel T. M. (1998) Monomeric monocyte chemoattractant protein-1 (MCP-1) binds and activates the MCP-1 receptor CCR2B. J. Biol. Chem. 273, 33157–33165 [DOI] [PubMed] [Google Scholar]
- 63. Gray-Owen S. D., Lorenzen D. R., Haude A., Meyer T. F., Dehio C. (1997) Differential Opa specificities for CD66 receptors influence tissue interactions and cellular response to Neisseria gonorrhoeae. Mol. Microbiol. 26, 971–980 [DOI] [PubMed] [Google Scholar]
- 64. Virji M., Makepeace K., Ferguson D. J., Watt S. M. (1996) Carcinoembryonic antigens (CD66) on epithelial cells and neutrophils are receptors for Opa proteins of pathogenic neisseriae. Mol. Microbiol. 22, 941–950 [DOI] [PubMed] [Google Scholar]
- 65. Popp A., Dehio C., Grunert F., Meyer T. F., Gray-Owen S. D. (1999) Molecular analysis of neisserial Opa protein interactions with the CEA family of receptors: identification of determinants contributing to the differential specificities of binding. Cell Microbiol. 1, 169–181 [DOI] [PubMed] [Google Scholar]
- 66. Billker O., Popp A., Gray-Owen S. D., Meyer T. F. (2000) The structural basis of CEACAM-receptor targeting by neisserial Opa proteins. Trends Microbiol. 8, 258–260; discussion 260–251 [DOI] [PubMed] [Google Scholar]
- 67. Tingstrom A., Blikstad I., Aurivillius M., Obrink B. (1990) C-CAM (cell-CAM 105) is an adhesive cell surface glycoprotein with homophilic binding properties. J. Cell Sci. 96, 17–25 [DOI] [PubMed] [Google Scholar]
- 68. Rojas M., Fuks A., Stanners C. P. (1990) Biliary glycoprotein, a member of the immunoglobulin supergene family, functions in vitro as a Ca2+-dependent intercellular adhesion molecule. Cell Growth & Differ. 1, 527–533 [PubMed] [Google Scholar]
- 69. Oikawa S., Kuroki M., Matsuoka Y., Kosaki G., Nakazato H. (1992) Homotypic and heterotypic Ca++-independent cell adhesion activities of biliary glycoprotein, a member of carcinoembryonic antigen family, expressed on CHO cell surface. Biochem. Biophys. Res. Commun. 186, 881–887 [DOI] [PubMed] [Google Scholar]
- 70. Lin S. H., Luo W., Earley K., Cheung P., Hixson D. C. (1995) Structure and function of C-Cam1: effects of the cytoplasmic domain on cell aggregation. Biochem. J. 311 (Pt. 1), 239–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Brümmer J., Neumaier M., Göpfert C., Wagener C. (1995) Association of pp60c-src with biliary glycoprotein (CD66a), an adhesion molecule of the carcinoembryonic antigen family down-regulated in colorectal carcinomas. Oncogene 11, 1649–1655 [PubMed] [Google Scholar]
- 72. Beauchemin N., Kunath T., Robitaille J., Chow B., Turbide C., Daniels E., Veillette A. (1997) Association of biliary glycoprotein with protein tyrosine phosphatase SHP-1 in malignant colon epithelial cells. Oncogene 14, 783–790 [DOI] [PubMed] [Google Scholar]
- 73. Huber M., Izzi L., Grondin P., Houde C., Kunath T., Veillette A., Beauchemin N. (1999) The carboxyl-terminal region of biliary glycoprotein controls its tyrosine phosphorylation and association with protein-tyrosine phosphatases SHP-1 and SHP-2 in epithelial cells. J. Biol. Chem. 274, 335–344 [DOI] [PubMed] [Google Scholar]
- 74. Skubitz K. M., Campbell K. D., Ahmed K., Skubitz A. P. (1995) CD66 family members are associated with tyrosine kinase activity in human neutrophils. J. Immunol. 155, 5382–5390 [PubMed] [Google Scholar]
- 75. Schumann D., Huang J., Clarke P. E., Kirshner J., Tsai S. W., Schumaker V. N., Shively J. E. (2004) Characterization of recombinant soluble carcinoembryonic antigen cell adhesion molecule 1. Biochem. Biophys. Res. Commun. 318, 227–233 [DOI] [PubMed] [Google Scholar]
- 76. Ng D. P., Poulsen B. E., Deber C. M. (2012) Membrane protein misassembly in disease. Biochim. Biophys. Acta 1818, 1115–1122 [DOI] [PubMed] [Google Scholar]
- 77. Izzi L., Turbide C., Houde C., Kunath T., Beauchemin N. (1999) cis-Determinants in the cytoplasmic domain of CEACAM1 responsible for its tumor inhibitory function. Oncogene 18, 5563–5572 [DOI] [PubMed] [Google Scholar]
- 78. Nagaishi T., Pao L., Lin S. H., Iijima H., Kaser A., Qiao S. W., Chen Z., Glickman J., Najjar S. M., Nakajima A., Neel B. G., Blumberg R. S. (2006) SHP1 phosphatase-dependent T cell inhibition by CEACAM1 adhesion molecule isoforms. Immunity 25, 769–781 [DOI] [PubMed] [Google Scholar]