Background: Cullin-RING ubiquitin ligases are the largest family of E3 ligases, but their roles in ovarian cancers remain uninvestigated.
Results: Inhibition of CRL4 activity induced ovarian cancer death.
Conclusion: CRL4CDT2 repression and CDT1 accumulation contributed to the genotoxic effects of MLN4924 in ovarian cancer cells.
Significance: Indicating CRL4CDT2 is an effective drug target in ovarian cancers.
Keywords: Apoptosis, Cancer Therapy, DNA Damage, E3 Ubiquitin Ligase, Ovarian Cancer, CDT2, MLN4924
Abstract
Cullin-RING ubiquitin ligases (CRLs) are the largest family of E3 ligases and require cullin neddylation for their activation. The NEDD8-activating enzyme inhibitor MLN4924 reportedly blocked cullin neddylation and inactivated CRLs, which resulted in apoptosis induction and tumor suppression. However, CRL roles in ovarian cancer cell survival and the ovarian tumor repressing effects of MLN4924 are unknown. We show here that CRL4 components are highly expressed in human epithelial ovarian cancer tissues. MLN4924-induced DNA damage, cell cycle arrest, and apoptosis in ovarian cancer cells in a time- and dose-dependent manner. In addition, MLN4924 sensitized ovarian cancer cells to other chemotherapeutic drug treatments. Depletion of CRL4 components Roc1/2, Cul4a, and DDB1 had inhibitory effects on ovarian cancer cells similar to MLN4924 treatment, which suggested that CRL4 inhibition contributed to the chemotherapeutic effect of MLN4924 in ovarian cancers. We also investigated for key CRL4 substrate adaptors required for ovarian cancer cells. Depleting Vprbp/Dcaf1 did not significantly affect ovarian cancer cell growth, even though it was expressed by ovarian cancer tissues. However, depleting Cdt2/Dcaf2 mimicked the pharmacological effects of MLN4924 and caused the accumulation of its substrate, CDT1, both in vitro and in vivo. MLN4924-induced DNA damage and apoptosis were partially rescued by Cdt1 depletion, suggesting that CRL4CDT2 repression and CDT1 accumulation were key biochemical events contributing to the genotoxic effects of MLN4924 in ovarian cancer cells. Taken together, these results indicate that CRL4CDT2 is a potential drug target in ovarian cancers and that MLN4924 may be an effective anticancer agent for targeted ovarian cancer therapy.
Introduction
The core of cullin-RING ubiquitin ligase (CRL)3 ubiquitin ligases is a cullin-RING finger protein complex. In humans and mice, there are seven cullin (CUL 1–3, 4A, 4B, 5, and 7) and two RING (RBX1/RBX2) family members (1). CRL activation requires cullin neddylation, a modification that results from adding ubiquitin-like protein NEDD8 to cullins and catalyzed by NEDD8-activating enzyme E1 (NAE), NEDD8-conjugating enzyme E2 (UBC12), and NEDD8-E3 ligase (2).
An NAE inhibitor, MLN4924, was recently identified by high throughput screening. MLN4924 bound to NAE to create a covalent NEDD8-MLN4924 adduct that blocked NAE enzymatic activity (3). MLN4924 efficiently inhibited cullin neddylation, which inactivated CRL/SCF E3 ligase to cause substrate accumulation. As a result, a DNA damage response was triggered, cellular apoptosis was induced, and remarkable anticancer effects were observed both in vitro and in vivo (4–6). These findings further validated CRL ubiquitin ligases as promising cancer targets and showed MLN4924 to be a novel anticancer agent. Indeed, MLN4924 has advanced to several phase 1 clinical trials for solid tumors and hematological malignancies (3, 7–9). However, MLN4924 actions in ovarian cancer cells are not well defined.
As a founding member of cullin-based E3 ligases, cullin-4 (CUL4A and 4B) differs from other cullins in that it employs the WD40-like repeat-containing protein DDB1 as its adaptor, which has unique structural and biochemical properties (2). DDB1 was initially identified as a damaged DNA-binding protein that recognized UV- or chemical mutagen-induced DNA lesions and recruited the nucleotide excision repair machinery to remove this damage. Subsequently it was revealed that DDB1 participated in a number of fundamental processes, such as transcription, cell cycle progression, cell death, and embryonic development (10, 11). Recent work further identified a family of DDB1 and CUL4-associated factors (DCAFs, which have more than 90 putative members in mammalian genomes) as CUL4-DDB1 substrate receptors, including VPRBP/DCAF1, CDT2/DCAF2, DDB2, and DCAF26 (12–15). This implicated CRL4 in regulating a broad spectrum of cellular processes. It has been reported that DDB1 and DDB2 mutations facilitated liver and skin cancer development (16–19), although the roles of CUL4, DDB1, and their specific substrate adaptors in ovarian cancers remain unknown.
Epithelial ovarian cancer is the most lethal of the gynecologic malignancies and is the fifth most common cause of cancer death for women in the United States (20, 21). Due to the internal localization of the ovaries, the lack of specific symptoms, and a lack of an effective screening method, ovarian cancer usually remains undetected until it has reached an advanced stage. In nearly 70% of patients who present with late-stage disease, it has already spread to other organs in the abdominal cavity and their 5-year survival remains at only 30%. The current standard of care includes surgical resection of the tumor, followed by treatment with genotoxic chemotherapies. However, chemoresistance is a major hurdle to successful cancer therapy (22–24). Therefore, better treatments for ovarian cancer are urgently needed.
Despite the ubiquitous nature of CRL ubiquitin ligase functions and the potential of CRL-targeted chemotherapy for a variety of tumors (25, 26), it remains unknown whether abnormalities in the CRL ubiquitin ligase system and their protein targets are associated with epithelial ovarian cancers. Our findings presented here show that CRL4 components are highly expressed in human ovarian cancer tissues, and that ovarian cancer cell proliferation and survival depend on CRL4CDT2 activity.
In the present study, we also report that MLN4924-mediated apoptosis induction contributes to ovarian cancer growth suppression. As one major target of MLN4924, its inhibition of CRL4CDT2 activity caused the accumulation of its substrate DNA replication licensing factor, CDT1, activation of a DNA damage checkpoint, and cell cycle arrest. By inducing these biochemical effects, MLN4924 could increase the anti-cancer effects of other therapeutic drugs. These results indicate that MLN4924 may be useful as an effective anticancer agent for targeted ovarian cancer therapy.
EXPERIMENTAL PROCEDURES
Reagents and Cell Culture
Human ovarian cell lines ES-2, SKOV3, OV2008, and A2780 were purchased from the ATCC. All cells were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS; Hyclone) and 1% penicillin-streptomycin solution (Invitrogen) at 37 °C in a humidified 5% CO2 incubator.
Mice and Xenograft Models
Mice were housed on a 14:10 h, light:dark schedule and provided food and water ad libitum. All animal protocols were in accordance with the NIH Guide for the Care and Use of Laboratory Animals. To assess proliferation, ovarian cancer cells were injected subcutaneously into 8-week-old female athymic nude mice. Four weeks later, primary tumor masses were excised, fixed in 4% paraformaldehyde, and embedded in paraffin. Sections (5 μm thick) were prepared and stained with hematoxylin and eosin.
Colony Formation Assay
Single-cell suspensions were seeded in 60-mm dishes and treated with MLN4924 or siRNA for 10 days. Colonies were stained and counted. Triplicate cultures were used for each experiment. The plates were placed in a 5% CO2 humidified incubator set at 37 °C. Colonies were counted at 10 days after plating.
Cell Growth Assay
After exposure to various treatments, cells were seeded in 96-well plates at a density of 3 × 103 cells/well in DMEM containing 10% FBS. After they had adhered, cells were assessed for growth using an MTT assay. Briefly, 20 μl of MTT solution (5 mg/ml in PBS) was added into triplicate wells and cells were incubated for 4–6 h in an incubator. Absorbance at 490 nm was read with a microplate reader.
RNA Isolation and Quantitative RT-PCR
Total RNA was isolated from tissues or cultured cells using TRIzol reagent (Invitrogen) following the manufacturer's protocol. Total RNA (5 μg) was reversed transcribed using a cDNA Synthesis Kit (Invitrogen). The following primers were used to amplify target genes: Actin, 5′-GCTCTTTTCCAGCCTTCCTT-3′, 5′-GTACTTGCGCTCAGGAGGAG-3′; Roc1, 5′-CTGGGCCTGGGATATTGTGGTT-3′, 5′-AGCCAGCGAGAGATGCAGTG-3′; Roc2, 5′-ATTGAAAAGCCGACTCCTCCTA-3′, 5′-AAGCCGTTTACCCGACAGTTAC-3′; Cul4a, 5′-GTCCCAAAGGAAAGGAGGTG-3′, 5′-CGATCTGGTACTGTCTATCCTGG-3′; Cul4b, 5′-AGAAACGGTTGAAGAACAAGCAAGC-3′, 5′-TCAGCAGGCTTTACTGGAAATTTCA-3′; Vprbp/Dcaf1, 5′-GTGAAACGGAACATCTTTGACCTG-3′, 5′-CTGGTCCTCCTCTTCATCCTCAT-3′; Cdt2/Dcaf2, 5′-GCCCCCTTCCAGTCATGAAACTT-3′; and 5′-GTTAAGGCTATGGTGTGCTTGGTT-3′.
siRNA-mediated Gene Silencing
Ovarian cancer cells were transfected with siRNA oligonucleotides using Lipofectamine 2000. Briefly, cells were seeded in 6-well plates. siRNA used for transfection and Lipofectamine 2000 were each incubated separately with Opti-MEM for 5 min, mixed together for 10 min at room temperature, after which this mixture was added to the cells plated in 1 ml of medium (final concentration of siRNA: 80 nm). The siRNA sequences were: siCon, 5′-UUCUCCGAACGUGUCACGUTT-3′; siRoc1-1, 5′-GACUUUCCCUGCUGUUACCUAATT-3′; siRoc1-2, 5′-CUGUGCCAUCUGCAGGAACCACAUU-3′; siCul4A, 5′-GAAGCUGGUCAUCAAGAACdTdT-3′; siCul4B, 5′-AAGCCUAAAUUACCAGAAAdTdT-3′; siVprbp1-1, 5′-UCACAGAGUAUCUUAGAGATT-3′; siVprbp1-2, 5′-AAUCACAGAGUAUCUUAGATT-3′; siRoc2-1, 5′-CAAGAGGACUGUGUUGUGGUCUGGUCUGG-3′; siRoc2-2, 5′-UCAGAAAUUCUCUGCGAUUAA-3′; siDdb1, 5′-GGCCAAGAACAUCAGUGUGtt-3′; siCdt1, 5′-GUACCCCCGAGGCCCCAGA-3′; and siCdt2, 5′-GAAUUAUACUGCUUAUCGA-3′. All siRNAs were purchased from GenePharma (Shanghai, China).
Immunohistochemistry
Paraffin-embedded human tissues from ovarian tumors were provided by the Jiaxing Maternity and Child Health Care Hospital, China. The use of archived cancer samples for this study was approved by the Zhejiang University Institutional Review Board.
Sections were cut with a Leica RM2235 microtome at 5-μm thickness and immunohistochemically stained with affinity-purified anti-ROC1, anti-DDB1 (Epitomic), anti-VPRBP (ProteinTech), anti-CDT2 (Bethyl Laboratories), anti-pH2AX, and anti-Cleaved caspase 3 (Cell signaling Technology) antibodies using a Vector ABC kit (Vector Laboratories). Briefly, sections were deparaffinized, rehydrated, and incubated in 0.3% H2O2. After antigen retrieval using 10 mm sodium citrate (pH 6.0), sections were probed with primary and secondary antibodies and washed again with PBS-T prior to incubation with ABC solution. Color was developed using diaminobenzidine substrate (Vector Laboratories). The sections were washed, counterstained, dehydrated, and mounted with Vectamount permanent mounting medium (Vector Laboratories). Sections were observed under a Nikon Eclipse 80i Microscope. Staining intensity for ROC1, DDB1, VPRBP, and CDT2 was scored by a pathologist (L. Guo).
Immunofluorescent Staining
Cultured cells were seeded on coverslips and transfected with siRNA. After 24–48 h, cells were washed with PBS, fixed with 4% paraformaldehyde, permeabilized with PBS containing 0.3% Triton X-100 (PBST), and incubated with blocking buffer (PBST containing 5% bovine serum albumin). Cells were sequentially probed with primary antibodies as indicated under “Results” and Alexa Fluor 594- or 488-conjugated secondary antibodies (Molecular Probes). Slides were mounted using VectaShield with 4′,6-diamidino-2-phenylindole (DAPI, Vector Laboratories). Digital images were acquired using an epifluorescence microscope (Nikon Eclipse 80i) with ×4–100 objectives.
Flow Cytometry
To analyze DNA contents by propidium iodide (PI) staining, OV2008 and A2780 cells were treated with MLN4924 for 24 h or transfected with siRNA for 72–96 h, fixed in 75% ethanol overnight at 4 °C, washed twice with 1× phosphate-buffered saline (PBS), and then permeabilized as previously described. Fixed and permeabilized cells were stained with PI (50 μg/ml) for at least 30 min at room temperature in the dark.
Western Blot Analysis
Cell extracts containing 30 μg of protein were resolved by SDS-PAGE and transferred to PVDF membranes (Millipore Corp., Bedford, MA). After probing with primary antibodies, the membranes were incubated with horseradish peroxidase-linked anti-rabbit antibodies (Cell Signaling Technologies, Danvers, MA) and then washed. Bound antibodies were visualized using an Enhanced Chemiluminescence Detection Kit (Amersham Biosciences). The primary antibodies were: pH2AX (Ser-139), pCHK1 (Ser-345), pCHK2 (Thr-68), Cleaved caspase 3, β-actin (Cell signaling), p27, p21, p53 (Santa Cruz), ROC1, DDB1, VPRBP, CDT2, and CDT1.
Statistical Analysis
All in vitro assays were done in triplicate. Group results were compared by two-tailed t-tests or analysis of variance using GraphPad Prism statistical programs (GraphPad Prism, San Diego, CA). p < 0.05 was considered significant.
RESULTS
CRL4DCAF2 E3 Ligase Complexes Are Highly Expressed in Human Ovarian Cancer Tissues and Cells
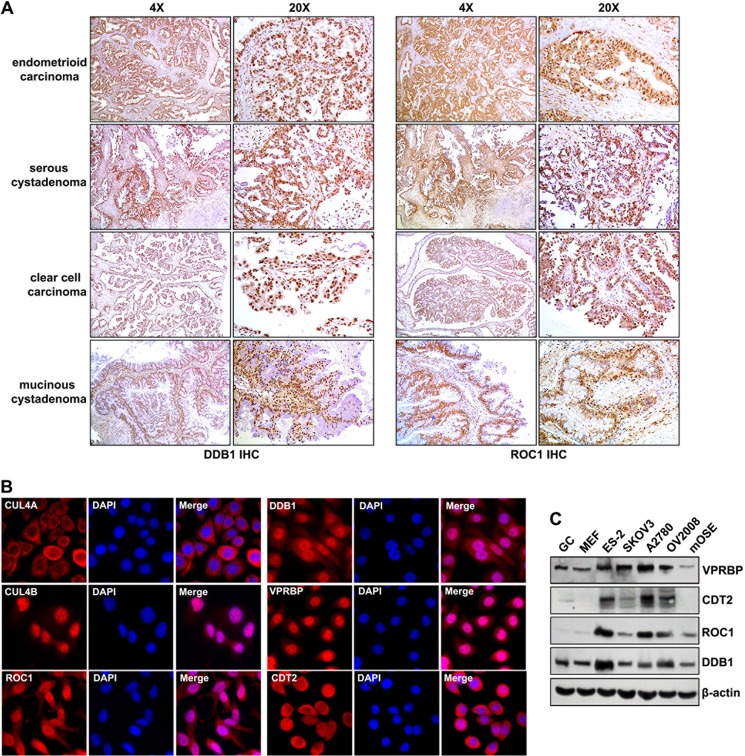
ROC1 and DDB1 expression in human ovarian cancer tissues was assessed by immunohistochemistry. As shown in Fig. 1A, significant ROC1 and DDB1 expression was detected in endometrioid carcinoma, serous cystadenoma, clear cell carcinoma, and mucinous cystadenoma. Furthermore, DDB1 was highly expressed in all serous cyctadenoma samples examined (n = 12), but was weakly expressed in normal human ovarian tissues. The subcellular localization of CRL4 E3 ligase components were also examined in ovarian cancer cell lines. DDB1, VPRBP/DCAF1, CUL4B, and ROC1 were primarily localized in nuclei (Fig. 1B), whereas more CUL4A and CDT2 signals were detected in the cytoplasm of ovarian cancer cells. At the protein level, compared with non-tumor somatic cells, including mouse embryonic fibroblasts (MEFs), granular cells (GCs), and mouse ovarian surface epithelia (mOSE) cells, CRL4 E3 ligase components were highly expressed in ovarian cancer cell lines (Fig. 1C). These results suggested that CRL4 E3 ligase complexes might play an essential role in ovarian cancer cell proliferation and apoptosis.
FIGURE 1.
CRL4DCAF2 E3 ligase complex expression patterns in human ovarian cancer samples and cell lines. A, immunohistochemistry results for DDB1 and ROC1 expression in ovarian cancer tissues. Sections were counterstained with hematoxylin. B, immunofluorescent staining results for the expression of CRL4 components in OV2008 ovarian cancer cells: ROC1, CUL4A, CUL4B, DDB1, VPRBP, and CDT2. Cells were seeded on glass coverslips in 24-well plates overnight before staining. Nuclei were stained with 4′,6′-diamidno-2-phenylindole (DAPI; blue). C, Western blotting results for CRL4 E3 component proteins (ROC1, DDB1, VPRBP, and CDT2) in ovarian cancer cells (ES-2, SKOV3, OV2008, and A2780), and non-tumor cells (mOSEs, MEFs, and GCs).
MLN4924 Induces DNA Damage and Apoptosis in Ovarian Cancer Cells
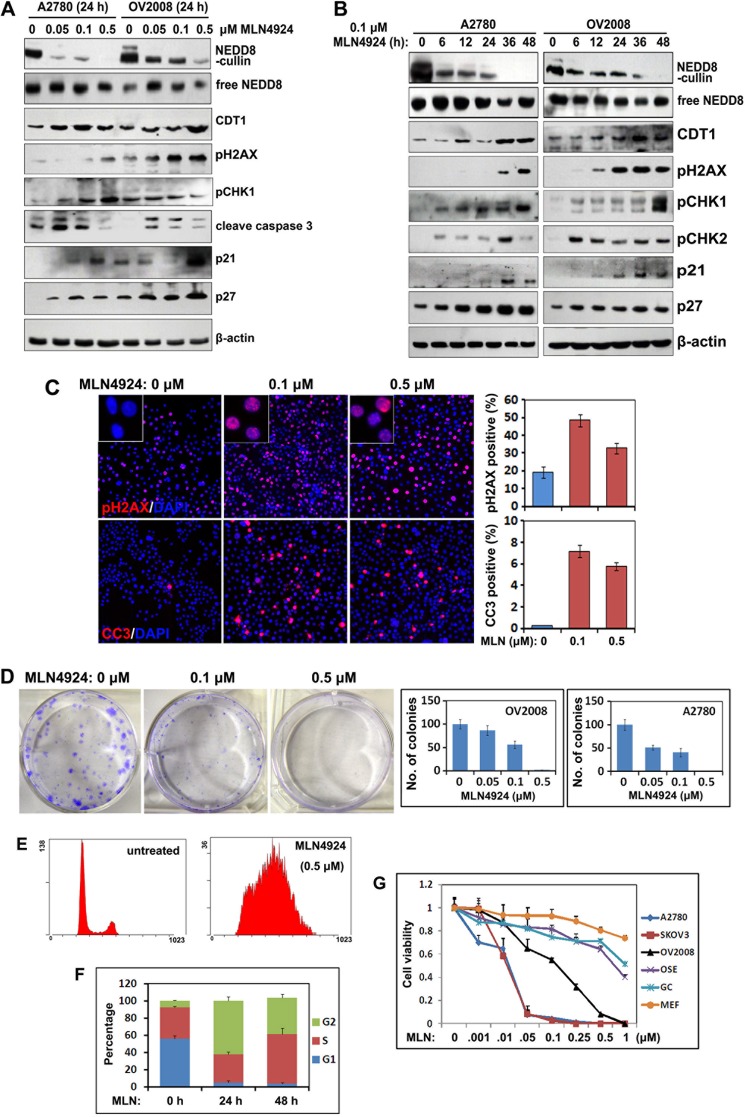
Recent studies showed that MLN4924 was a potent inhibitor of cullin neddylation, and thereby inactivated CRL E3 ligase and caused accumulation of a number of its substrates. MLN4924 treatment resulted in tumor cell growth suppression both in vitro and in vivo by promoting abnormal cell cycle progression and inducing apoptosis. To test the effects of MLN4924 in ovarian cancer cells, we examined for cullin neddylation by Western blotting. At a concentration as low as 0.05 μm, MLN4924 treatment inhibited cullin neddylation within 24 h in both A2780 and OV2008 cells (Fig. 2A). After longer periods of MLN4924 treatment (0.1 μm), cullin neddylation decreased to undetectable levels after 36 h in A2780 cells and after 48 h in OV2008 cells (Fig. 2B). However, free NEDD8 levels were not affected by MLN4924 in both of these cell lines.
FIGURE 2.
Effects of MLN4924 treatment on ovarian cancer cell growth, apoptosis, and DNA damage. A, Western blot results for the changes in the levels of the indicated proteins in ovarian cancer cells (A2780 and OV2008) treated with increased concentrations of MLN4924 for 24 h. B, Western blot results showing the changes in the levels of the indicated proteins in ovarian cancer cells (A2780 and OV2008) treated with 0.1 μm MLN4924 for increased periods of time. C, immunofluorescent staining results showing increased pH2AX and cleaved caspase 3 (CC3) levels at 24 h after treatment with 0.1 or 0.5 μm MLN4924. The percentages of pH2AX and CC3 positive cells were determined by counting the total cells in 10 independent areas. D, colony formation assay for OV2008 and A2780 cells after treatment with 0.1 or 0.5 μm MLN4924. Colony numbers were counted after 10 days in culture. E and F, cell cycle phase determined by flow cytometry. OV2008 cells (1 × 105 cells per 60-mm dish) were cultured overnight, treated with or without MLN4924 (0.5 μm), and then subjected to PI staining and FACS analysis at the indicated time points. The percentages of cells in the G1, S, and G2/M phases are indicated. G, sensitivity to MLN4924 by different cancer cell lines (SKOV3, OV2008, and A2780) and normal cells (mOSEs, MEFs, and GCs). Cells were seeded in 96-well plates in triplicate and treated with increased concentrations of MLN4924 for 72 h, followed by MTT assay.
We next determined the effects of MLN4924 on inhibiting the growth of several human ovarian cancer cell lines, including OV2008, SKOV3, and A2780 cells, and on normal cells (GCs, MEFs, and mOSEs). MLN4924 treatment effectively inhibited ovarian cancer cell proliferation with inhibitory concentrations ranging from 10 to 100 nm, but MLN4924 did not inhibit normal cell proliferation when using the same concentrations as for ovarian cancer cells (Fig. 2G).
It was recently shown that CDT1 was a substrate for CRL4 E3 ligase and that it played a critical role in DNA re-replication (10). Western blot results showed that MLN4924 induced both dose- and time-dependent accumulations of CDT1 (Fig. 2, A and B). In addition, MLN4924 activated DNA damage responses and apoptosis pathways in these cells, as shown by the accumulations of pH2AX, pCHK1, pCHK2, cleaved caspase-3, and the cyclin-dependent kinase inhibitors p27 and p21 (Fig. 2, A and B). The key apoptosis regulator p53 was not detected because it was mutated in these ovarian cancer cell lines (data not shown). MLN4924-induced histone H2AX phosphorylation and caspase 3 cleavage were further confirmed and quantified by immunofluorescent staining (Fig. 2C).
We then investigated the effects of MLN4924 on ovarian cancer cell growth. Colony formation by OV2008 and A2780 cells was remarkably inhibited by MLN4924 treatment (Fig. 2D). FACS analysis revealed that these ovarian cancer cells were arrested in the G2 phase of the cell cycle at 24 h after MLN4924 treatment (Fig. 2, E and F). This was consistent with the observation that a DNA damage checkpoint had been activated by MLN4924. A cell viability assay showed that MLN4924 caused significantly increased ovarian cancer cell death at increased concentrations (A2780, SKOV3, and OV2008 cells). However, the non-oncogenic somatic cells, including immortalized mouse OSEs, primary mouse GCs, and MEFs, were resistant to MLN4924 treatment, even at a very high concentration (1 μm). Collectively, these results showed that MLN4924 effectively inhibited cullin neddylation, promoted the accumulations of CDT1, triggered DNA damage responses, and promoted apoptosis in ovarian cancer cells.
MLN4924 Sensitizes Ovarian Cancer Cells to Chemotherapeutic Drug Treatment
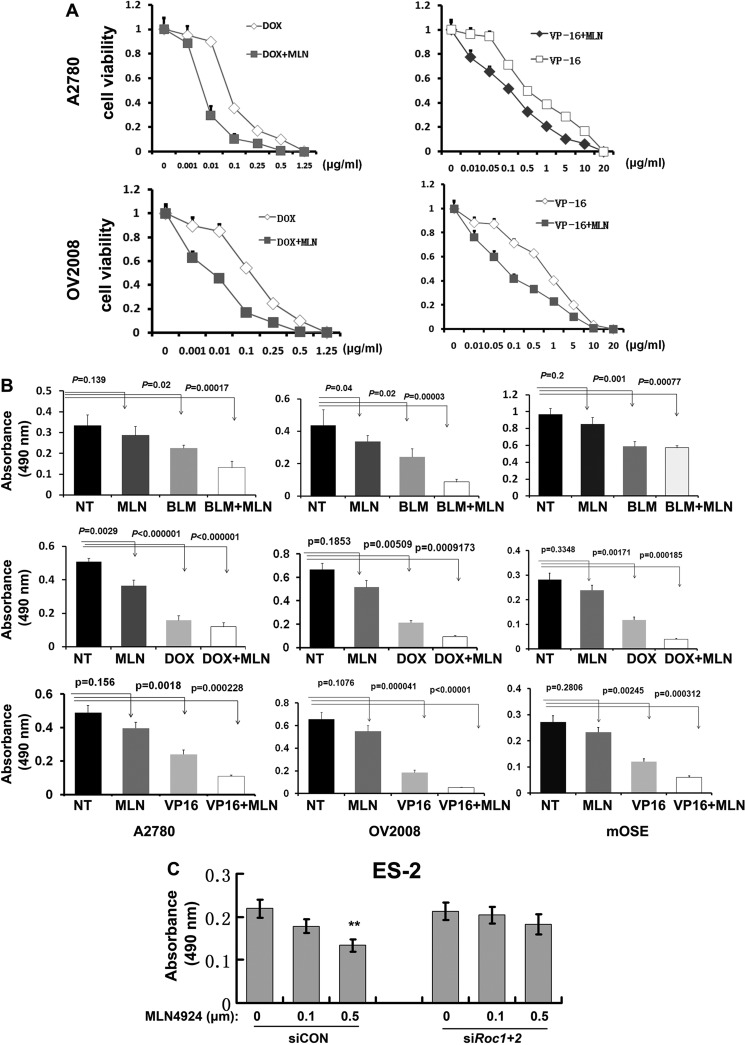
We next investigated possible chemosensitizing effects of MLN4924 in ovarian cancer cells. We first confirmed that bleomycin, doxorubicin, and etoposide (VP-16), which are first-line anticancer agents for human ovarian cancer, inhibited the proliferation of ovarian cancer cells (A2780 and OV2008 cells) and non-cancer cells (mOSEs and GCs) (Fig. 3, A and B). Then we fixed the concentration of MLN4924 at low-toxic dose (0.05 μm) and then determined whether low-toxic MLN4924 sensitizes ovarian cancer cells to other anticancer drugs.
FIGURE 3.
MLN4924 sensitizes ovarian cancer cells to chemotherapeutic drug treatment. A, cell viability after treatment with varying concentrations of chemotherapy drugs. B, MLN4924 (0.05 μm) treatment was combined with agents used for the clinical management of ovarian cancer. A2780, OV2008, and mOSE cells were seeded in 96-well plates in triplicate and treated with MLN4924 and the indicated therapeutic drugs for 96 h, followed by MTT assay. For A2780 and OV2008 cells, drug concentrations were: bleomycin (BLM), 20 μg/ml; doxorubicin (DOX), 0.5 μg/ml; and etoposide (VP-16), 5 μg/ml. For mOSE cells, drug concentrations were: bleomycin (20 μg/ml), doxorubicin (0.1 μg/ml), and etoposide (1 μg/ml). C, cell viability of MLN4924-treated ES-2 ovarian cancer cells, with or without Roc1/2 RNAi depletion.
For OV2008 cells, MLN4924 showed significant additive effects with all three of these drugs (Fig. 3, A and B). Similarly, MLN4924 also increased the proliferation-inhibiting effects of bleomycin and VP-16 on A2780 cells (Fig. 3, A and B). However, for untransformed mOSE cells, MLN4924 had little effect by itself for inhibiting proliferation and did not enhance the effect of bleomycin when they were added together in the cell culture medium (Fig. 3B). However, MLN4924 had an additive effect when mOSE cells were treated with doxorubicin and VP-16. These results showed that MLN4924 increased the effects of other chemotherapeutic drugs in ovarian cancer cells.
Furthermore, in ES-2 ovarian cancer cells, RNAi depletion of Roc1/2 did not significantly affect cell viability. However, the Roc1/2-depleted ES-2 cells were more resistant to MLN4924 treatment than the Roc1/2-intact cells (Fig. 3C). This result indicated that MLN4924 relied on ROC1/2 to exert its growth inhibition effects.
Roc1/2 Depletion in Ovarian Cancer Cells Mimics the Pharmacological Effects of MLN4924
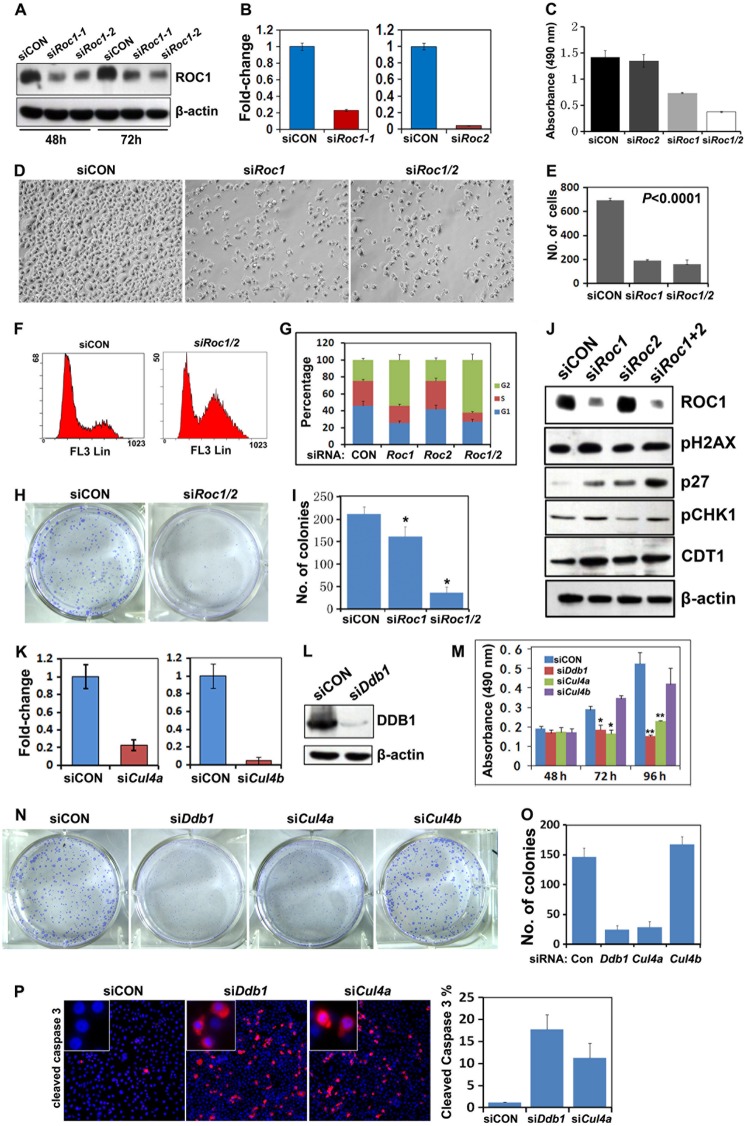
MLN4924 treatment had inhibited cullin neddylation and caused DNA damage accumulation in ovarian cancer cells. To confirm that the pharmacological effects of MLN4924 were due to disrupting CRLs, we depleted Roc1/2, the ring finger proteins shared by all CRL complexes, in ovarian cancer cells. Roc1 and Roc2 were efficiently knocked down in OV2008 cells by using siRNA oligos, as shown by quantitative real-time PCR and Western blot results (Fig. 4, A and B). Roc1 and Roc1/2 double depletion significantly inhibited the proliferation of the OV2008 cells, as assessed by MTT proliferation assay (Fig. 4C) and by counting cell numbers (Fig. 4, D and E). FACS analysis also showed that ovarian cancer cells were arrested in the G2-M phase of the cell cycle with either Roc1 depletion or Roc1/2 double silencing (Fig. 4, F and G). By itself, Roc2 depletion did not affect cell growth, but it did enhance the effect of Roc1 depletion (Fig. 4, C and G). Roc1/2 double silencing also suppressed colony formation by OV2008 cells (Fig. 4, H and I). Western blot results showed that CDT1, p27, pH2AX, and pCHK1 expressions were significantly increased after Roc1 and Roc1/2 double silencing (Fig. 4J). These results showed that Roc1/2 depletion mimicked the pharmacological effects of MLN4924, which suggested that CRLs were the major pharmacological targets of MLN4924 in ovarian cancer cells.
FIGURE 4.
Growth inhibiting effects of Roc1, Ddb1, and Cul4a silencing on ovarian cancer cell in vitro. A and B, Western blot (A) and quantitative RT-PCR (B) results for Roc1 and Roc2 RNAi depletion efficiency. OV2008 cells were transfected with control siRNA (siCON), siRoc1, or siRoc2. Total RNAs and proteins were isolated 48 h after transfection. C, effects of Roc1/2 RNAi depletion on ovarian cancer cell growth. OV2008 cells were seeded in 96-well plates (3,000 cells/well) at 48 h after transfection, and then assessed by MTT assay. D and E, Roc1 or Roc1/2 double silencing inhibited ovarian cancer cell proliferation. Cells were counted at 72 h after transfection. F and G, Roc1/2 depletion induced G2-M cell cycle arrest in ovarian cancer cells. Cells were transfected with siCON, siRoc1, siRoc2, or siRoc1/2 for 72 h and then subjected to PI staining and FACS analysis to determine cell cycle profiles. H and I, Roc1/2 depletion inhibited colony formation of OV2008 cells. *, p < 0.01; **, p < 0.001. J, Western blot results for the levels of pH2AX, pCHK1, p27, and CDT1 after RNAi depletion of Roc1, Roc2, and Roc1/2 in OV2008 cells. K, quantitative RT-PCR results for Cul4a and Cul4b RNAi depletion efficiency. L, Western blot results for Ddb1 RNAi depletion efficiency. M, effects of Ddb1, Cul4a, and Cul4b depletion on ovarian cancer cell proliferation as assessed by MTT assay. N and O, effects of Ddb1, Cul4a, and Cul4b depletion on colony formation by OV2008 cells. P, immunofluorescent staining results for increased caspase 3 cleavage after RNAi depletion of Ddb1 and Cul4a.
CRL4 Inhibition Contributes to the Chemotherapeutic Effects of MLN4924
There are 5 major CRL complexes in mammalian cells: CRL1–5. Because CRL4 components were highly expressed in human ovarian cancer tissues, we further investigated if CRL4 inhibition contributed to the chemotherapeutic effects of MLN4924. To address this, we knocked down Ddb1, Cul4a, and Cul4b expression by using siRNA oligos. Cul4a, Cul4b, and Ddb1 were efficiently depleted in OV2008 cells, as shown by quantitative RT-PCR and Western blotting results (Fig. 4, K and L). Ddb1 and Cul4a silencing significantly inhibited cell proliferation, as assessed by MTT assay, but Cul4b depletion did not affect the proliferation of OV2008 cells (Fig. 4M). Ddb1 and Cul4a depletion also notably inhibited colony formation by these cells, whereas Cul4b depletion did not (Fig. 4, N and O). Similar to MLN4924 treatment, Ddb1 or Cul4a depletion induced OV2008 cell apoptosis, as shown by anti-cleaved caspase 3 immunofluorescence (Fig. 4P). These results demonstrated that the CRL4 core components ROC1/2, DDB1, and CUL4A were required for ovarian cancer cell proliferation and survival.
CRL4 Substrate Adaptor VPRBP/DCAF1 Is Dispensable for Ovarian Cancer Cell Survival and Proliferation
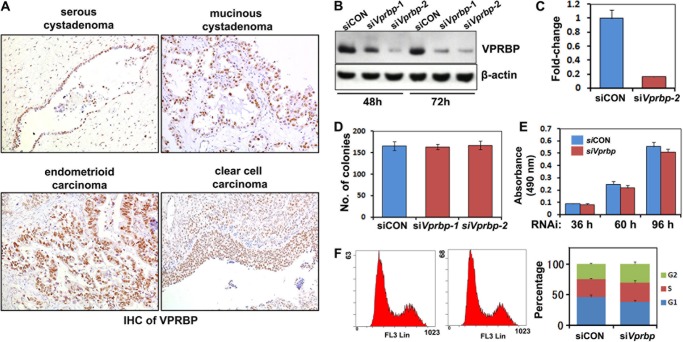
We hypothesized that those substrate receptor proteins with a WD40 domain might be required for substrate recognition by ubiquitin-dependent degradation machinery. VPRBP is one of the common components of a CUL4-containing complex. First, we examined for VPRBP protein expression in human tissues by immunohistochemistry. As shown in Fig. 5A, VPRBP expression was found in human ovarian cancer tissues as were other CRL4 components. Therefore, we knocked down Vprbp using siRNA oligos. Efficient Vprbp depletion in OV2008 cells was shown by quantitative RT-PCR and Western blotting results (Fig. 5, B and C). However, Vprbp depletion did not affect the proliferation of OV2008 cells, colony formation, or cell cycle progression (Fig. 5, D–F). This was also the case for several other ovarian cancer cell lines (data not shown). Taken together, these results demonstrated that although it was expressed in ovarian cancer cells, VPRBP was dispensable for the survival and proliferation of these cells. Therefore, VPRBP/DCAF1 was not an essential CRL4 substrate adaptor for ovarian cancer cell survival and proliferation.
FIGURE 5.
Vprbp silencing does not affect the proliferation of ovarian cancer cells. A, immunohistochemistry results for VPRBP expression in ovarian cancer tissues. Sections were counterstained with hematoxylin and eosin. B and C, Western blot (B) and quantitative RT-PCR (C) results for Vprbp RNAi depletion efficiency. D, Vprbp knockdown did not affect colony formation by OV2008 cells. E, Vprbp knockdown did not affect ovarian cancer cell proliferation. Cells were seeded in 96-well plates (3,000 cells/well) at 48 h after transfection and then assessed by MTT assay. F, Vprbp depletion did not induce cell cycle arrest in ovarian cancer cells. Cells were transfected with siCON or siVprbp for 72 h and then subjected to PI staining and FACS analysis to determine cell cycle profiles.
CDT2/DCAF2 Is a Major CRL4 Substrate Adaptor That Maintains Ovarian Cancer Cell Survival and Proliferation
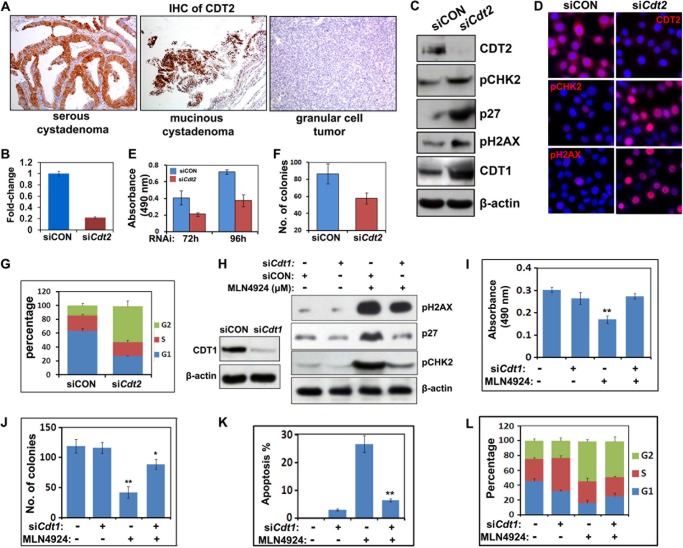
It was previously reported that CDT2/DCAF2 was an important CRL4 substrate adaptor that controlled the protein levels of the DNA replication licensing factor CDT1. As shown in Fig. 6A, significant CDT2 expression was found by immunohistochemistry in clear cell carcinoma and mucinous cystadenoma, but not in granulosa cell tumors and other types of ovarian epithelial tumors. To investigate the function of CDT2 in ovarian cancer cells, we deleted Cdt2 using siRNA oligos. As shown by quantitative RT-PCR (Fig. 6B), Western blotting (Fig. 6C), and immunofluorescence (Fig. 6D) results, Cdt2 was efficiently knocked down in OV2008 cells. Cdt2 depletion inhibited the proliferation and colony formation of OV2008 cells and caused their cell cycle arrest in the G2 phase (Fig. 6, E–G). Western blot results showed remarkable CDT1 and p27 expression in Cdt2-depleted cells (Fig. 6C). Cdt2 depletion also caused DNA damage checkpoint activation, as shown by increases in pH2AX and pCHK2 (Fig. 6, C and D). These results demonstrated that CDT2 was an essential CRL4 substrate adaptor that maintained ovarian cancer cell survival and proliferation.
FIGURE 6.
Effects of Cdt1/2 depletion on the proliferation and survival of ovarian cancer cells. A, immunohistochemistry results for CDT2 expression in ovarian cancer tissues. Sections were counterstained with hematoxylin and eosin. B, quantitative RT-PCR results for Cdt2 RNAi depletion efficiency. C, Western blot results for the levels of the indicated proteins in OV2008 cells transfected with control or Cdt2 siRNAs. D, immunofluorescent staining results for increase of pH2AX and pCHK2 after Cdt2 RNAi depletion. E, effect of Cdt2 depletion on OV2008 cell growth as assessed by MTT assay. F, effect of Cdt2 depletion on colony formation by OV2008 cells. G, Cdt2 depletion induced G2-M cell cycle arrest in ovarian cancer cells. Cells were transfected with siCON or siCdt2 for 72 h and then subjected to PI staining and FACS analysis to determine cell cycle profiles. H, Western blot results for MLN4924-induced accumulation of pH2AX, pCHK2, and p27 in OV2008 cells that were transfected with control or Cdt1 siRNAs. Cells were harvested at 48 h after MLN4924 treatment (0.1 μm). I and J, effects of MLN4924 (0.1 μm) on growth (I) and colony formation (J) by OV2008 cells with or without Cdt1 RNAi depletion. *, p < 0.01; **, p < 0.001. K and L, flow cytometry results for MLN4924 (0.5 μm)-induced apoptosis (K) and cell cycle arrest (L) of OV2008 cells with or without Cdt1 RNAi depletion. **, p < 0.001.
Because both MLN4924 treatment and Cdt2 depletion could enhance DNA damage and cause G2/M arrest, we hypothesized that accumulation of the CRL4CDT2 substrate CDT1 might be a key biochemical event that contributed to the pharmacological effects of MLN4924. Western blotting results showed that CDT1 knockdown using siRNA oligos decreased MLN4924-induced pCHK2, p27, and pH2AX accumulation (Fig. 6H).
By itself, Cdt1 depletion did not affect the survival or colony formation of OV2008 cells, but it did significantly rescue MLN4924-induced cell death in both of these experiments (Fig. 6, I and J). In MLN4924-treated cells, CDT1 knockdown reduced apoptosis from ∼26.7 to ∼6.46%, as quantified by flow cytometry (Fig. 6K). However, MLN4924-induced cell cycle arrest at the G2/M phase was not rescued by Cdt1 depletion, which indicated that this MLN4924 effect was mediated by some other factor(s) (Fig. 6L). These results clearly suggested a causal effect of CDT1 accumulation on MLN4924-treated ovarian cancer cells. However, the incomplete rescue of MLN4924-treated cells by CDT1 knockdown suggested that other downstream targets, such as CRL1 complexes, might also mediate the pharmacological effects of MLN4924.
Disrupting CRL4CDT2 E3 Ligase Complexes Blocks Ovarian Cancer Cell Proliferation in Vivo
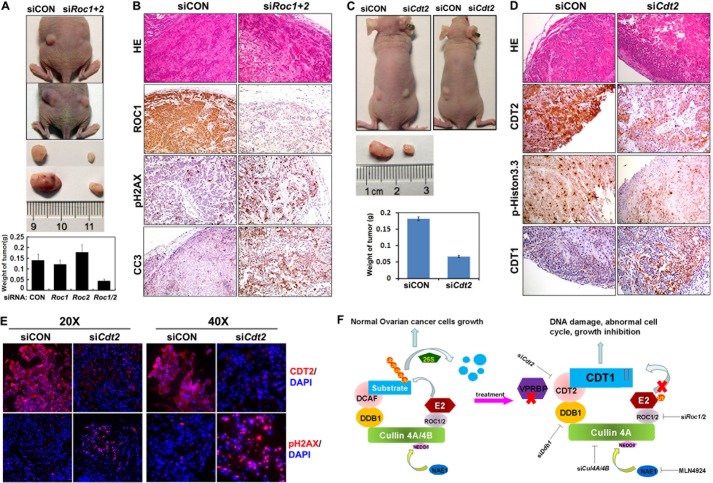
Based on our in vitro findings, we next investigated if ROC1/2, the core components of the CRL complex, were also required for ovarian cancer cell survival and growth in vivo. OV2008 cells transfected with control and Roc1/2 siRNAs were implanted subcutaneously into the left and right flanks of female nude mice, respectively. As shown in Fig. 7A, implanted control cells formed significantly larger tumors than did siRoc1/2-treated cells. Immunohistochemical examination confirmed that ROC1 protein levels were remarkably down-regulated in tumor tissues derived from siRoc1/2-treated cells. Significant signals for phospho-H2AX and cleaved caspase 3 were also detected in these tumor tissues (Fig. 7B), which indicated extensive DNA damage and apoptosis in the ROC1/2-depleted cells in vivo.
FIGURE 7.
RNAi depletion of Roc1/2 and Cdt2 inhibits ovarian cancer cell growth and survival in vivo. A, OV2008 ovarian cancer cells were transfected with siCON or siRoc1/2 (106 cells for each) and then implanted subcutaneously into the left and right flanks of nude mice, respectively. After 30 days, tumors were excised and weighed (n = 6). B, immunohistochemistry results for ROC1, cleaved caspase 3 (CC3), and pH2AX in tumor tissues without or with Roc1/2 depletion. C, OV2008 ovarian cancer cells were transfected with siCON or siCdt2 (106 cells for each) and then implanted subcutaneously into the left and right flanks of nude mice, respectively. After 30 days, tumors were excised and weighed (n = 6). D, immunohistochemistry results for CDT2, CDT1, and pH2AX in tumor tissues without or with Cdt2 depletion. E, immunofluorescent signals for CDT2 and pH2AX in tumor tissues without or with Cdt2 depletion. F, schematic diagram showing CRL4 function in ovarian cancer cells. Under normal circumstances, a CRL4 E3 complex catalyzes ubiquitin transfer from E2 to protein substrates, which results in their polyubiquitination and targeted degradation by the 26 S proteasome. Thus, CRL4 activity maintains ovarian cancer cell growth. When a CRL4DCAF2 E3 complex is inactivated by RNAi depletion of its essential components, including Roc1/2, Cul4a/4b, Ddb1, or Cdt2, or by pharmaceutical inhibition of cullin neddylation with MLN4924, its substrates CDT1 and others accumulate, which suppresses cancer cell growth and triggers ovarian cancer cell killing pathways, including apoptosis, DNA damage, and G2-M arrest.
To assess the effect of CDT2 on tumor cell survival and growth, we also implanted siCON or siCdt2-transfected OV2008 cells subcutaneously into the left and right flanks of nude mice, respectively. As shown in Fig. 7C, tumors that formed due to siCON-treated cells were significantly larger than those formed due to siCdt2-treated cells. Immunohistochemical and immunofluorescence examinations confirmed that CDT2 protein levels were down-regulated in tumor tissues derived from siCdt2-treated cells (Fig. 7, D and E). Immunohistochemistry staining for phospho-histone H3 indicated that there were more proliferating cells in control tumors than in Cdt2-depleted tumors (Fig. 7D). CDT1 accumulation was also confirmed by immunohistochemistry staining in Cdt2-depleted tumors by immunohistochemistry. Similar to Roc1/2-depleted cells, Cdt2-depleted tumor tissues contained notably increased numbers of pH2AX positive cells than did control tissues (Fig. 7E). Taken together, these results demonstrated that CRL4CDT2 complexes were required for ovarian cancer cell growth in vivo.
DISCUSSION
In this study, we carefully investigated the pharmacological effects of the newly developed anti-cancer molecule MLN4924 in ovarian cancer cells and the biochemical mechanisms underlying its effects. It appeared that ovarian cancer cells were more sensitive to MLN4924 treatment than normal cells, such as cultured ovarian granulosa cells and immortalized ovarian surface epithelial cells. This may have been because the growth and survival of ovarian cancer cells were more dependent on CRL4 activity.
In support of this speculation, our results showed that CRL4 components were highly expressed in human ovarian cancer tissues and ovarian cancer cell lines. Furthermore, depleting these cells of CRL4 components using RNAi mimicked the growth inhibition effects of MLN4924. Thus, MLN4924 may be useful as a new therapeutic drug for ovarian cancer patients with the advantages of high target selectivity in tumor cells and low toxicity for normal cells. Furthermore, overexpression of CRL4 components may become useful biomarkers for ovarian tumor malignancy and for determining MLN4924 efficacy in cancer treatment.
Our results also indicated that different CRL4 components played functionally overlapping or specific roles in ovarian cancer cells. For example, previous studies showed that ROC1 silencing triggered multiple death and growth arrest pathways and effectively suppressed tumor cell growth (27, 28). We confirmed the growth inhibiting effects of ROC1 depletion in ovarian cancer cells. We also found that simultaneous knockdown of ROC1/2 had a stronger effect than depletion of either ROC1 or ROC2 alone. This suggested that ROC2 could partially substitute for ROC1 function for maintaining ovarian cancer cell survival.
Recent gene knock-out experiments showed that Cul4a and Cul4b were functionally redundant during most developmental events in mouse embryos (29). However, each of them was indispensable in certain tissues; as examples, Cul4a in testis and Cul4b in placenta (30–32). Interestingly, RNAi depletion of Cul4a rather than Cul4b mimicked the growth inhibiting effects of MLN4924, which suggested that CUL4A, but not CUL4B, was the major functional CUL4 subtype in ovarian cancer cells.
The CRL4 substrate adaptors DCAFs comprise more than 90 proteins with WD40 repeats (12, 33). Although the diverse biological functions of CRL4 have been extensively studied in multiple organisms and cell types, the contributions of specific DCAFs in most of these scenarios were uncertain. The best studied CRL4 substrate adaptors, VPRBP/DCAF1 and CDT2/DCAF2, were both expressed in human ovarian cancer samples. Thus, we investigated their roles in ovarian cancer cells and their contributions to the pharmacological effects of MLN4924. Gene knock-out experiments indicated that, similar to Roc1 and Ddb1, Vprbp and Cdt2 were essential for mouse early embryo development. Other studies also identified katanin p60 and merlin as selective substrates for CRL4VPRBP E3 complexes (34, 35).
Interestingly, our results suggested that CRL4CDT2, rather than CRL4VPRBP, was one of the major targets of MLM4924 in ovarian cancer cells. This is the first report of distinct contributions by specific DCAFs in mediating the pharmacological effects of MLN4924. This observation helped to explain why MLN4924 had weaker growth suppression effects in non-cancer cells, such as granulosa cells, mouse MEFs, and normal OSE cells. As shown in Fig. 1C, CDT2 protein levels were high in all four of the ovarian cancer cell lines examined in this experiment, but CDT2 levels were very low in non-cancer cells. Thus, these non-cancer cells were more resistant to MLN4924 treatment because one of its major targets was missing. Clinically, evaluating CDT2 levels might be a practical approach to predict the effectiveness of MLN4924 for a specific cancer type or an individual patient.
Although our results suggested that DNA damage caused by CDT1 accumulation significantly contributed to the genotoxic effects of MLN4924, other MLN4924 targets must be involved to cause cell cycle arrest, as CDT1 depletion rescued MLN4924-induced DNA damage and apoptosis, but not G2/M arrest of the cells. A recent report indicated that, in addition to its genotoxic effects, MLN4924 also induced senescence in cancer cells, presumably by inhibiting CRL1 activity and causing p21/p27 accumulation (5). Our results confirmed that MLN4924 also induced p21/p27 accumulation (Fig. 3, A and B) and senescence (data not shown) in ovarian cancer cells.
In many cases, the well known tumor suppressor p53 promotes p21/p27 expression in response to DNA damage signals. However, p53 mutations are found in 50% of ovarian cancer samples (36, 37). In particular, the OV2008 cell line we used in this study did not express any p53. MLN4924, as well as CRL4 depletion, appeared to induce ovarian cancer cell apoptosis and cell cycle arrest independently of p53, and increased cancer cell sensitivities to traditional chemotherapeutic drugs, including bleomycin, etoposide, and doxorubicin. Therefore, our findings suggest that CRL4 inhibition may become an effective novel approach for treating ovarian cancer cases that are p53 negative and chemoresistant.
This work was supported by National Basic Research Program of China Grants 2011CB944504 and 2012CB944403, National Natural Science Foundation of China Grant 81172473, Basic Scientific Research Funding of Zhejiang University Grant 2011QN81001 (to H. Y. F.), and Natural Science Foundation of Zhejiang Province Grant LQ13H160020 (to W. W. P.). This project was also supported by the Open Research Fund of State Key Laboratory of Cellular Stress Biology, Xiamen University Grant SKLCSB2013KF001.
- CRL
- cullin-RING ubiquitin ligase
- DCAF
- DDB1 and CUL4-associated factor
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PI
- propidium iodide
- MEF
- mouse embryonic fibroblasts
- GC
- granular cells
- mOSE
- mouse ovarian surface epithelia.
REFERENCES
- 1. Jia L., Sun Y. (2011) SCF E3 ubiquitin ligases as anticancer targets. Curr. Cancer Drug. Targets 11, 347–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jackson S., Xiong Y. (2009) CRL4s. The CUL4-RING E3 ubiquitin ligases. Trends Biochem. Sci. 34, 562–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Soucy T. A., Smith P. G., Milhollen M. A., Berger A. J., Gavin J. M., Adhikari S., Brownell J. E., Burke K. E., Cardin D. P., Critchley S., Cullis C. A., Doucette A., Garnsey J. J., Gaulin J. L., Gershman R. E., Lublinsky A. R., McDonald A., Mizutani H., Narayanan U., Olhava E. J., Peluso S., Rezaei M., Sintchak M. D., Talreja T., Thomas M. P., Traore T., Vyskocil S., Weatherhead G. S., Yu J., Zhang J., Dick L. R., Claiborne C. F., Rolfe M., Bolen J. B., Langston S. P. (2009) An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 [DOI] [PubMed] [Google Scholar]
- 4. Jia L., Li H., Sun Y. (2011) Induction of p21-dependent senescence by an NAE inhibitor, MLN4924, as a mechanism of growth suppression. Neoplasia 13, 561–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Luo Z., Pan Y., Jeong L. S., Liu J., Jia L. (2012) Inactivation of the Cullin (CUL)-RING E3 ligase by the NEDD8-activating enzyme inhibitor MLN4924 triggers protective autophagy in cancer cells. Autophagy 8, 1677–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luo Z., Yu G., Lee H. W., Li L., Wang L., Yang D., Pan Y., Ding C., Qian J., Wu L., Chu Y., Yi J., Wang X., Sun Y., Jeong L. S., Liu J., Jia L. (2012) The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res. 72, 3360–3371 [DOI] [PubMed] [Google Scholar]
- 7. Yang D., Tan M., Wang G., Sun Y. (2012) The p21-dependent radiosensitization of human breast cancer cells by MLN4924, an investigational inhibitor of NEDD8 activating enzyme. PLoS One 7, e34079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao Y., Xiong X., Jia L., Sun Y. (2012) Targeting Cullin-RING ligases by MLN4924 induces autophagy via modulating the HIF1-REDD1-TSC1-mTORC1-DEPTOR axis. Cell Death Dis. 3, e386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Soucy T. A., Dick L. R., Smith P. G., Milhollen M. A., Brownell J. E. (2010) The NEDD8 conjugation pathway and its relevance in cancer biology and therapy. Genes Cancer 1, 708–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cang Y., Zhang J., Nicholas S. A., Bastien J., Li B., Zhou P., Goff S. P. (2006) Deletion of DDB1 in mouse brain and lens leads to p53-dependent elimination of proliferating cells. Cell 127, 929–940 [DOI] [PubMed] [Google Scholar]
- 11. Cang Y., Zhang J., Nicholas S. A., Kim A. L., Zhou P., Goff S. P. (2007) DDB1 is essential for genomic stability in developing epidermis. Proc. Natl. Acad. Sci. U.S.A. 104, 2733–2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee J., Zhou P. (2007) DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol. Cell 26, 775–780 [DOI] [PubMed] [Google Scholar]
- 13. Xu H., Wang J., Hu Q., Quan Y., Chen H., Cao Y., Li C., Wang Y., He Q. (2010) PLoS Genet. 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McCall C. M., Miliani de Marval P. L., Chastain P. D., 2nd, Jackson S. C., He Y. J., Kotake Y., Cook J. G., Xiong Y. (2008) Human immunodeficiency virus type 1 Vpr-binding protein VprBP, a WD40 protein associated with the DDB1-CUL4 E3 ubiquitin ligase, is essential for DNA replication and embryonic development. Mol. Cell. Biol. 28, 5621–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim Y., Kipreos E. T. (2007) The Caenorhabditis elegans replication licensing factor CDT-1 is targeted for degradation by the CUL-4/DDB-1 complex. Mol. Cell. Biol. 27, 1394–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Endo Y., Zhang M., Yamaji S., Cang Y. (2012) Genetic abolishment of hepatocyte proliferation activates hepatic stem cells. PLoS One 7, e31846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yamaji S., Zhang M., Zhang J., Endo Y., Bibikova E., Goff S. P., Cang Y. (2010) Hepatocyte-specific deletion of DDB1 induces liver regeneration and tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 22237–22242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yoon T., Chakrabortty A., Franks R., Valli T., Kiyokawa H., Raychaudhuri P. (2005) Tumor-prone phenotype of the DDB2-deficient mice. Oncogene 24, 469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pines A., Backendorf C., Alekseev S., Jansen J. G., de Gruijl F. R., Vrieling H., Mullenders L. H. (2009) Differential activity of UV-DDB in mouse keratinocytes and fibroblasts. Impact on DNA repair and UV-induced skin cancer. DNA Repair 8, 153–161 [DOI] [PubMed] [Google Scholar]
- 20. Mullany L. K., Fan H. Y., Liu Z., White L. D., Marshall A., Gunaratne P., Anderson M. L., Creighton C. J., Xin L., Deavers M., Wong K. K., Richards J. S. (2011) Molecular and functional characteristics of ovarian surface epithelial cells transformed by KrasG12D and loss of Pten in a mouse model in vivo. Oncogene 30, 3522–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Richards J. S., Fan H. Y., Liu Z., Tsoi M., Laguë M. N., Boyer A., Boerboom D. (2012) Either Kras activation or Pten loss similarly enhance the dominant-stable CTNNB1-induced genetic program to promote granulosa cell tumor development in the ovary and testis. Oncogene 31, 1504–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abedini M. R., Muller E. J., Bergeron R., Gray D. A., Tsang B. K. (2010) Akt promotes chemoresistance in human ovarian cancer cells by modulating cisplatin-induced, p53-dependent ubiquitination of FLICE-like inhibitory protein. Oncogene 29, 11–25 [DOI] [PubMed] [Google Scholar]
- 23. Abedini M. R., Qiu Q., Yan X., Tsang B. K. (2004) Possible role of FLICE-like inhibitory protein (FLIP) in chemoresistant ovarian cancer cells in vitro. Oncogene 23, 6997–7004 [DOI] [PubMed] [Google Scholar]
- 24. Ali A. Y., Abedini M. R., Tsang B. K. (2012) The oncogenic phosphatase PPM1D confers cisplatin resistance in ovarian carcinoma cells by attenuating checkpoint kinase 1 and p53 activation. Oncogene 31, 2175–2186 [DOI] [PubMed] [Google Scholar]
- 25. Wei D., Morgan M. A., Sun Y. (2012) Radiosensitization of cancer cells by inactivation of Cullin-RING E3 ubiquitin ligases. Transl. Oncol. 5, 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao Y., Sun Y. (2013) Cullin-RING ligases as attractive anti-cancer targets. Curr. Pharm. Des. 19, 3215–3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jia L., Sun Y. (2009) RBX1/ROC1-SCF E3 ubiquitin ligase is required for mouse embryogenesis and cancer cell survival. Cell Div. 4, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jia L., Soengas M. S., Sun Y. (2009) ROC1/RBX1 E3 ubiquitin ligase silencing suppresses tumor cell growth via sequential induction of G2-M arrest, apoptosis, and senescence. Cancer Res. 69, 4974–4982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu L., Lee S., Zhang J., Peters S. B., Hannah J., Zhang Y., Yin Y., Koff A., Ma L., Zhou P. (2009) CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol. Cell 34, 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu L., Yin Y., Li Y., Prevedel L., Lacy E. H., Ma L., Zhou P. (2012) Essential role of the CUL4B ubiquitin ligase in extra-embryonic tissue development during mouse embryogenesis. Cell Res. 22, 1258–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kopanja D., Roy N., Stoyanova T., Hess R. A., Bagchi S., Raychaudhuri P. (2011) Cul4A is essential for spermatogenesis and male fertility. Dev. Biol. 352, 278–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yin Y., Lin C., Kim S. T., Roig I., Chen H., Liu L., Veith G. M., Jin R. U., Keeney S., Jasin M., Moley K., Zhou P., Ma L. (2011) The E3 ubiquitin ligase Cullin 4A regulates meiotic progression in mouse spermatogenesis. Dev. Biol. 356, 51–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Angers S., Li T., Yi X., MacCoss M. J., Moon R. T., Zheng N. (2006) Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443, 590–593 [DOI] [PubMed] [Google Scholar]
- 34. Huang J., Chen J. (2008) VprBP targets Merlin to the Roc1-Cul4A-DDB1 E3 ligase complex for degradation. Oncogene 27, 4056–4064 [DOI] [PubMed] [Google Scholar]
- 35. Maddika S., Chen J. (2009) Protein kinase DYRK2 is a scaffold that facilitates assembly of an E3 ligase. Nat. Cell Biol. 11, 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mullany L. K., Liu Z., King E. R., Wong K. K., Richards J. S. (2012) Wild-type tumor repressor protein 53 (Trp53) promotes ovarian cancer cell survival. Endocrinology 153, 1638–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mullany L. K., Richards J. S. (2012) Minireview. Animal models and mechanisms of ovarian cancer development. Endocrinology 153, 1585–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]