Background: ABC transporter OpuA controls cell volume.
Results: Anionic lipids are required for gating and efficient energy coupling of transport.
Conclusion: Ionic strength and excluded volume effects act synergistically in the gating of transport.
Significance: Tight coupling between substrate binding and ATP hydrolysis in ABC transporters allows the study of transmembrane signaling in nanodiscs.
Keywords: ABC Transporter, Energetics, Gating, Membrane, Membrane Reconstitution, Membrane Transport, BC Transporter, Cell Volume Regulation, Gating Mechanism, Ionic Strength, Macromolecular Crowding
Abstract
Cells control their volume through the accumulation of compatible solutes. The bacterial ATP-binding cassette transporter OpuA couples compatible solute uptake to ATP hydrolysis. Here, we study the gating mechanism and energy coupling of OpuA reconstituted in lipid nanodiscs. We show that anionic lipids are essential both for the gating and the energy coupling. The tight coupling between substrate binding on extracellular domains and ATP hydrolysis by cytoplasmic nucleotide-binding domains allows the study of transmembrane signaling in nanodiscs. From the tight coupling between processes at opposite sides of the membrane, we infer that the ATPase activity of OpuA in nanodiscs reflects solute translocation. Intriguingly, the substrate-dependent, ionic strength-gated ATPase activity of OpuA in nanodiscs is at least an order of magnitude higher than in lipid vesicles (i.e. with identical membrane lipid composition, ionic strength, and nucleotide and substrate concentrations). Even with the chemical components the same, the lateral pressure (profile) of the nanodiscs will differ from that of the vesicles. We thus propose that membrane tension limits translocation in vesicular systems. Increased macromolecular crowding does not activate OpuA but acts synergistically with ionic strength, presumably by favoring gating interactions of like-charged surfaces via excluded volume effects.
Introduction
Cells respond to osmotic stress by adjusting the intracellular components to keep the cell volume, crowding, ionic strength, and other (physico)chemical parameters within limits. Under hyperosmotic conditions, bacteria accumulate or synthesize compatible solutes to rehydrate the cytoplasm, to reverse cell shrinkage, and to prevent plasmolysis (1). Osmoregulatory transporters respond to hyperosmotic stress by taking up compatible solutes, such as glycine betaine, carnitine, trimethylamine N-oxide, or proline (2). Three osmoregulatory transporters have been studied in great detail: ProP from Escherichia coli (3–6), BetP from Corynebacterium glutamicum (7–10), and OpuA from Lactococcus lactis (11–13). ProP and BetP are secondary transporters driven by the proton and sodium electrochemical gradient across the membrane, respectively, whereas OpuA is a primary transporter driven by ATP. Although high resolution structures are available for BetP, and a wealth of functional and structural data are available for all three proteins, the gating mechanism of osmoregulatory transporters is poorly understood (14).
The ATP-binding cassette (ABC)3 transporter OpuA couples the hydrolysis of ATP to the uptake of glycine betaine with a stoichiometry of two ATP molecules per substrate molecule (15). The activity of OpuA increases with the osmolality of the medium, which is signaled to the protein as an increase in the cytoplasmic electrolyte concentration (12). We have recently shown that the CBS2 domain of the CBS module of OpuA is critical for electrolyte sensing, whereas the CBS1 domain merely serves as linker between the nucleotide-binding domain and CBS2 domain (13). For OpuA, we have proposed that the interaction of the CBS module with the membrane surface may lock the transporter in an inactive conformation. Increasing the intracellular ionic strength beyond threshold levels would screen the electrostatic interactions of oppositely charged surfaces and activate the transporter (12, 13). Alternatively, the ionic gating of OpuA could involve two like-charged surfaces, such as the anionic membrane and anionic protein residues, in which case a high ionic strength would promote their interaction (16). It is evident that screened electrostatic forces and an anionic membrane surface are intrinsic to the gating mechanism of OpuA and BetP. Less clear is the role of hydration and/or excluded volume (“crowding”) effects on the activation of these transporters. These physicochemical parameters have been shown to influence the activity of ProP, either directly or via interactions of protein residues with the membrane (17, 18).
Because of the difficulty to manipulate (macro)molecular crowding and (bivalent) cation concentrations in vivo or in proteoliposomal systems, we reconstituted OpuA in phospholipid bilayer nanodiscs (Fig. 1) and determined the (ionic) osmolyte and lipid requirements and the crowding effects on the ATPase activity of the transporter. In the last decade, the lipid bilayer nanodisc system developed by the Sligar laboratory has become an important tool to characterize membrane proteins (19–20). Many proteins have been reconstituted in nanodiscs, including human tissue factor (21), bacteriorhodopsin (22–23), G-protein-coupled receptors (24–26), chemoreceptor Tar (27–28), translocon SecYEG (29–31), and F0F1-ATPase (32). ABC transporters, such as the maltose system MalEFGK (33–34), the lipid A exporter MsbA (35), and P-glycoprotein (19, 36), have also been reconstituted in nanodiscs, albeit generally with a low coupling efficiency. Substrate-independent ATPase activity is also observed with ABC transporters in the detergent-solubilized state and is often not meaningful for functional characterization of the proteins. We thus focused on a protocol that ensures tight coupling between substrate binding and ATP hydrolysis. Although the translocation reaction cannot be probed directly, the ionic strength-gated, substrate-dependent ATPase activity of OpuA indicates that we are probing transmembrane transport. The OpuA nanodiscs thus combine full transporter functionality with a non-compartmentalized membrane system, which is ideally suited for biochemical and spectroscopic measurements both in ensemble and at the single molecule level. We used the OpuA nanodiscs to optimize the energy coupling and to determine the gating mechanism of the transporter.
FIGURE 1.
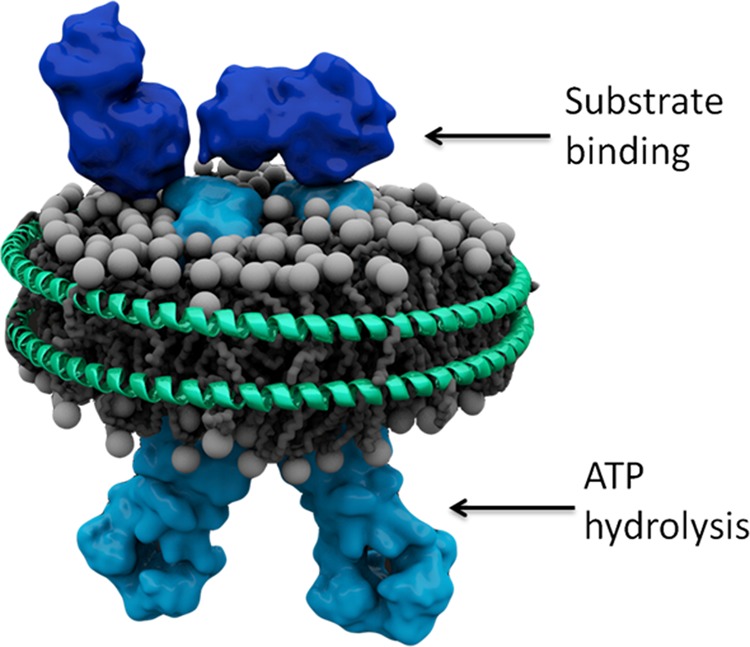
Schematic of OpuA reconstituted in a phospholipid bilayer nanodisc. The schematic is based on the structures of the OpuAC domains (Protein Data Bank codes 3L6G and 3L6H; dark blue (49)) of OpuA, the structure of the membrane domain of the molybdate transporter (Protein Data Bank code 2ONK; light blue (50)), and the structural analysis of the CBS domains of OpuA (15). Arrows indicate the localization of the substrate-binding (extracellular) and nucleotide-binding (cytoplasmic) domains.
EXPERIMENTAL PROCEDURES
Materials
The pMSP1D1 plasmid (Addgene plasmid 20061) was purchased from Addgene, and 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phosphatidylethanolamine (DOPE), and 1,2-dioleoyl-sn-glycero-3-phosphatidylglycerol (DOPG) were purchased from Avanti Polar Lipids. Radiolabeled [N-methyl-14C]choline chloride (55 mCi/mmol) was obtained from Amersham Biosciences and converted to [N-methyl-14C]glycine betaine.
Expression and Purification of MSP1D1 and MSP1E3D1
The membrane scaffold proteins (MSPs) MSP1D1 and MSP1E3D1 are based on high density lipoprotein, which physiologically enables the transport of lipids around cells. Expression and purification of MSP1D1 and MSP1E3D1 were performed as described previously with some modifications (19). Briefly, E. coli strain BL21(DE3) carrying the pMSP1D1 or pMSP1E3D1 plasmid was cultivated aerobically at 37 °C in 2 liters of Terrific Broth medium supplemented with 10 μg/ml kanamycin. At A600 of 2, the culture was induced by adding 1 mm isopropyl 1-thio-β-d-galactopyranoside, and after 3 h, the cells were harvested by centrifugation (8,000 × g, 15 min, 4 °C), washed, and resuspended in 50 mm potassium Pi, pH 7.8, and stored at −80 °C. For purification, the cells were resuspended in 50 mm potassium Pi, pH 7.8, supplemented with 1 mm PMSF and 100 μg/ml DNase, and lysed by 2-fold passage through a manual French pressure cell (10,000 p.s.i.). To the cell lysate, 1% Triton X-100 (Sigma) was added, and after centrifugation (30,000 × g, 30 min, 4 °C), 20 mm imidazole was added to the supernatant. MSP1D1 (or MSP1E3D1) was purified from the supernatant by Ni2+-Sepharose (GE Healthcare) affinity chromatography (column volume = 7.5 ml), using 40 ml of each of the three buffers: wash buffer I (40 mm Tris/HCl, pH 8.0, 0.3 m NaCl, 1% Triton X-100); wash buffer II (40 mm Tris/HCl, pH 8.0, 0.3 m NaCl, 50 mm sodium cholate, 20 mm imidazole); and wash buffer III (40 mm Tris/HCl, pH 8.0, 0.3 m NaCl, 50 mm imidazole). MSP1D1 (or MSP1E3D1) was eluted in 20 ml of elution buffer (40 mm Tris/HCl, pH 8.0, 0.3 m NaCl, 500 mm imidazole). Subsequently, the protein sample was dialyzed overnight against 1 liter of 20 mm Tris/HCl, pH 7.4, 0.1 m NaCl, 0.5 mm EDTA to remove the imidazole. After filtering of the MSP1D1 (or MSP1E3D1) sample using a 0.22-μm syringe filter, the protein was diluted to 8 mg/ml, and 0.01% NaN3 was added. Subsequently, aliquots of the purified protein were stored at −80 °C.
Expression of OpuA and Membrane Vesicle Preparation
L. lactis strain Opu401 carrying the pNZopuAHis plasmid was cultivated semianaerobically at 30 °C in a medium containing 2% (w/v) Gistex LS (Strik BV, Eemnes, The Netherlands) and 65 mm sodium phosphate (NaPi), pH 6.5, supplemented with 1.0% (w/v) glucose and 5 μg/ml chloramphenicol (12). The culture was grown in a 2-liter pH-regulated bioreactor to an A600 of 2, after which transcription from the nisA promoter was switched on by the addition of 0.1% (v/v) culture supernatant of the nisin A-producing strain NZ9700. After 2 h, the cells were harvested by centrifugation (7,446 × g, 15 min, 4 °C); washed and resuspended in 50 mm potassium Pi, pH 7.0, 20% (v/v) glycerol; and stored at −80 °C. Before lysing the cells, 5 mm MgSO4, 100 μg/ml DNase, and 1 mm PMSF were added to the cells resuspended in 50 mm potassium Pi, pH 7.0, 20% glycerol. The cells were lysed by high pressure disruption (Constant Systems Ltd.; two passes at 39,000 p.s.i.). Unbroken cells and cell debris were removed by centrifugation (267,000 × g, 30 min, 4 °C), after which the membrane vesicles were collected by ultracentrifugation (280,000 × g, 60 min, 4 °C) and resuspended in 50 mm potassium Pi, pH 7.0, 20% (v/v) glycerol. The protein concentration was determined using the DC Protein Assay (Bio-Rad), and aliquots of the membrane vesicles were stored at −80 °C.
Purification of OpuA and Reconstitution in Lipid Bilayer Nanodiscs
10 mg of total protein in membrane vesicles were washed with Buffer A (50 mm potassium Pi, pH 7.0, 200 mm KCl, 20% glycerol) and solubilized with 0.5% (w/v) DDM. Subsequently, OpuA was allowed to bind to nickel-Sepharose (0.5-ml bed volume; GE Healthcare) and purified by affinity chromatography, using 20 column volumes of wash buffer (50 mm potassium Pi, pH 7.0, 200 mm KCl, 20% (v/v) glycerol, 50 mm imidazole plus 0.04% (w/v) DDM). The protein was eluted in column volumes of elution buffer (50 mm potassium Pi, pH 7.0, 200 mm KCl, 20% (w/v) glycerol, 200 mm imidazole plus 0.04% (w/v) DDM). The reconstitution of purified OpuA in lipid bilayer nanodiscs was performed as described with some modifications (33). The required amount of preformed liposomes, prepared as described previously (37), composed of various mol % of DOPC, DOPE, and DOPG, was thawed and extruded through a polycarbonate filter (400-nm pore size) to generate large unilamelar vesicles. After extrusion, 12 mm DDM (a concentration well above the critical micellar concentration of ∼0.2 mm) was added to the liposomes, and the solution was vortexed until it became optically clear. The solubilized lipids, purified MSP1D1, and purified OpuA were mixed in a final volume of 700 μl, giving rise to the following composition: 50 mm potassium Pi, pH 7.0, 12 mm DDM, 4% (w/v) glycerol, 0.72 μm OpuA, 14.3 μm MSP1D1, and 1.43 mm lipid. This corresponds to a molar ratio of OpuA/MSP1D1/lipid of 1:20:2000 and was obtained after optimization of the procedure (as described under “Results”). The reconstitution mixture was incubated for 60 min at 4 °C with gentle agitation, after which 300 mg of Amberlite XAD-2 polymeric absorbent beads (Supelco, Sigma-Aldrich) were added. The mixture was incubated overnight at 4 °C with gentle agitation. The beads and aggregated protein were removed by transferring the mixture to a clean Eppendorf tube using a syringe and subsequent centrifugation (25,000 × g, 10 min, 4 °C). The supernatant was stored at −80 °C after flash-freezing in liquid nitrogen. The protein and lipid composition of the nanodiscs was determined after fractionation of the reconstitution mixture by size exclusion chromatography, using a Superdex 200 10/300 GL column (GE Healthcare) and 50 mm potassium Pi, pH 7.0, 200 mm NaCl, 4% (w/v) glycerol as buffer. The protein/protein ratios of the OpuA subunits and MSP1D1 of the peak fraction of nanodiscs were analyzed by quantitative SDS-PAGE using 12.5% polyacrylamide gels. Pictures of the gels taken by a Fujifilm LAS 3000 Imager were analyzed using ImageJ. The analysis of the lipids is described in supplemental Fig. 1.
ATPase Activity Assay with OpuA Reconstituted in Nanodiscs
The activity of OpuA was determined by measuring the ATPase activity of the transporter using a coupled enzyme assay. The measurements were performed at 30 °C in a 96-well plate using a Synergy MX 96-well plate reader (BioTek Instruments, Inc.). A standard measurement solution of 200 μl/well was composed of 50 mm potassium Pi, pH 7.0, 57 nm OpuA incorporated in nanodiscs, 4 mm sodium phosphoenolpyruvate, 0.3 mm NADH, and 3.5 μl of pyruvate kinase/lactic dehydrogenase enzyme mixture from rabbit muscle (Sigma-Aldrich) in 50% glycerol, with or without 62 μm glycine betaine and/or other compounds as indicated in the figure legends. After incubation of the assay mixture for 3 min at 30 °C, 10 mm MgATP, pH 7.0, was added to each well, and the absorbance of NADH at 340 nm was followed for 7 min. Because the oxidation of NADH is stoichiometric with the amount of ATP consumed, the ATPase activity of OpuA could be expressed as the μmol of ATP hydrolyzed/(min × mg of OpuA). For analysis, the background activity measured in the presence of empty nanodiscs was subtracted from the activity measured in the presence of OpuA nanodiscs. Using SigmaPlot version 11.0 software, the data were fitted to the appropriate biophysical models.
Because ionic strength or crowding could affect the activity of pyruvate kinase/lactic dehydrogenase, we tested the effect of these additives on the enzyme system (see supplemental Fig. 2). The standard assay had a pyruvate kinase/lactic dehydrogenase activity at least 4 times higher than the maximal OpuA activity.
Cryoelectron Microscopy
After self-assembly of the nanodiscs at an OpuA/MSP1D1/lipid ratio of 1:20:2000, 14 ml of the solution was concentrated to 600 μl, using an Amicon Ultra-15 centrifugal filter device (Merck Millipore) with a molecular weight cut-off of 50,000. This sample was applied to a Superdex 200 10/300 GL column (GE Healthcare) equilibrated with 50 mm potassium Pi, pH 7.0, 200 mm NaCl, 4% (w/v) glycerol. The elution fractions containing nanodiscs, including OpuA, empty nanodiscs, and larger, aggregate-like particles were used for cryoelectron microscopy. A drop of these elution fractions was placed on a glow-discharged holey carbon-coated copper grid (Quantifoil 3.5/1). After blotting, the sample was rapidly frozen in liquid ethane (Vitrobot, FEI, Eindhoven, The Netherlands) and analyzed in a CM120 electron microscope operating at 120 keV. Images were recorded under low dose conditions on a slow scan CCD camera.
Reconstitution of OpuA in Liposomes and Transport Assay
Purified OpuA was reconstituted in liposomes composed of synthetic lipids as described previously (37). The final protein/lipid ratio was 1:100 (w/w), which corresponds to an OpuA/lipid molar ratio of about 1:30,000. The ATP-regenerating system was enclosed in the proteoliposomes in two cycles of flash-freezing in liquid N2 and thawing at room temperature (37), and uptake activity of glycine betaine under different KCl concentrations was measured as described (13).
RESULTS
Reconstitution and Characterization of OpuA Nanodiscs
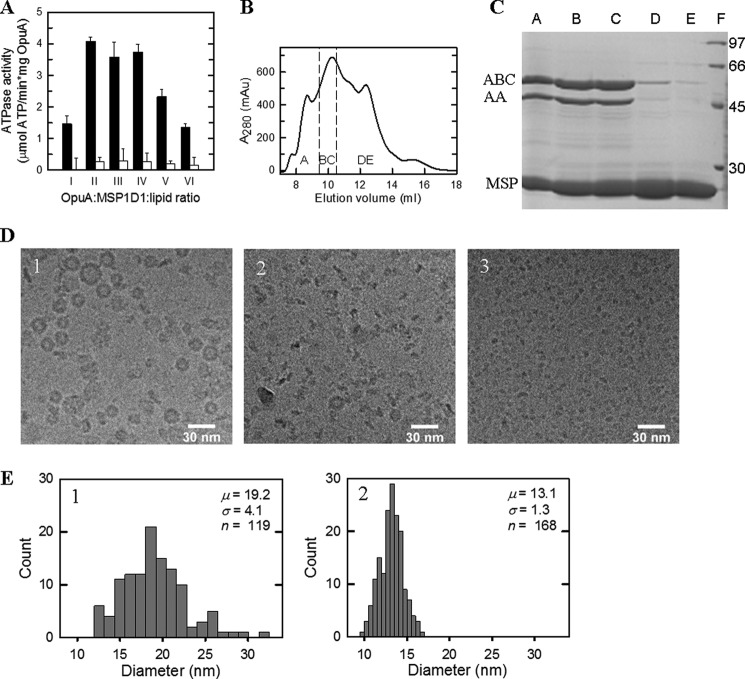
Nanodiscs are formed by mixing purified membrane protein, phospholipids, and MSP in the presence of detergent, followed by the removal of detergent by adsorption onto polystyrene beads or dialysis (19). The molar ratio of protein, lipid, and MSP is critically important for obtaining monodisperse nanodiscs. We optimized this ratio for two different MSPs: MSP1D1 (diameter of 9.5 nm when wrapped around the proteolipid complex) and MSP1E3D1 (diameter of 12.1 nm). The ATPase activity of OpuA reconstituted in nanodiscs formed at different molar ratios of OpuA, MSPD1, and lipids (38 mol % DOPG, 12 mol % DOPC, and 50 mol % DOPE) is shown in Fig. 2A. The OpuA/MSP1D1/lipid molar ratio of 1:20:2000 showed the highest ATPase activity. We also tested MSP1D1E3 under identical conditions, but the ATPase activity was on average 20% lower than with MSP1D1. We thus optimized the nanodisc formation further with MSP1D1 as scaffold protein and substrate-activated ATPase activity as readout. The rate of detergent removal was optimal with 300 mg of polystyrene beads from Sigma, but we have noticed batch-to-batch variation. Another critical parameter is the co-solvent concentration; glycerol inhibits nanodisc formation but is necessary to keep the OpuA complex (two OpuAA and two OpuABC subunits) together in the presence of detergent. Lowering of the glycerol concentration from 20 to 4% (w/v) prior to nanodisc formation proved optimal for OpuA activity.
FIGURE 2.
Characterization of OpuA nanodiscs. A, ATPase activity of OpuA in nanodiscs formed from reconstitution mixtures consisting of OpuA/MSP1D1/lipid ratios of 1:20:1000 (I), 1:20:1500 (II), 1:20:2000 (III), 1:20:2500 (IV), 1:20:3000 (V), or 1:20:4000 (VI), measured in 50 mm potassium Pi, pH 7.0, 300 mm KCl and in the presence (black bars) or absence (white bars) of 62 μm glycine betaine. B, size exclusion chromatography profile of nanodiscs formed under the 1:20:2000 ratio. The peak fractions indicated by A–E were used for further analysis. C, the peak fractions A–E from B were analyzed by 12.5% polyacrylamide SDS-PAGE and showed the presence of OpuABC (62.8 kDa), OpuAA (45.8 kDa), and MSP1D1 (24.7 kDa). Lane F, molecular size markers, the sizes of which are given in kDa. D, cryoelectron microscopy images of the peak fractions A (1), C (2), and D (3) of B. E, histograms showing the diameter distributions of the particles as observed in the cryo-EM images; the numbers are the same as those in D. The mean diameter (μ), S.D. (σ), and sample size (n) of the statistical analysis are given. Error bars, S.D.
Fig. 2B shows the size exclusion chromatography profile of OpuA reconstituted in nanodiscs. We analyzed the fractions by SDS-PAGE (Fig. 2C), cryoelectron microscopy (Fig. 2D), and histograms of size distributions (Fig. 2E). Peak A (∼9 ml) contains non-symmetric donut-like structures, peak BC (∼10 ml) contains the OpuA-containing nanodiscs, and peak DE (∼12.5 ml) contains empty nanodiscs. The electron microscopy shows that the donut-shaped particles that peak after the void volume have a diameter of about 20 nm (top and side views are visible) (Fig. 2, D (1) and E (1)). This diameter is bigger than expected for nanodiscs (19). The peak fractions BC show disc-shaped particles with a diameter of about 13 nm (Fig. 2, D (2) and E (2)), which is somewhat bigger than expected for MSP1D1 as scaffolding protein. A larger diameter of nanodiscs formed by MSP1D1 has been seen for the reconstitution of the maltose ABC transporter MalFGK2 as well (34). The empty nanodiscs have a diameter of about 8 nm (Fig. 2D, 3), which is a little smaller than observed previously (19).
We examined the lipid composition of the nanodiscs after reconstitution and purification, using different ratios of DOPE, DOPG, and DOPC. The molar ratio of lipids in the nanodiscs and the starting liposome mixture were identical (supplemental Fig. 1), indicating that specific lipids were not enriched or depleted upon nanodisc formation. We did not detect natural lipids from L. lactis that could have been co-purified with the OpuA protein. We then determined the ratio of OpuA and MSP1D1 using quantitative protein detection. Due to the reduction of glycerol to 4% (w/v), we lose a fraction of the OpuAA subunit in the reconstitution (OpuABC/OpuAA ratio of 1.9:1); the nanodiscs devoid of OpuAA are inactive. Furthermore, the OpuABC/MSP1D1 and OpuAA/MSP1D1 ratios were 1:1.2 and 1:2.2, respectively. We conclude that the active 13-nm nanodiscs are composed of one OpuA molecule (two OpuABC plus two OpuAA subunits) and two MSP1D1 molecules.
Benchmarking of the OpuA Nanodiscs
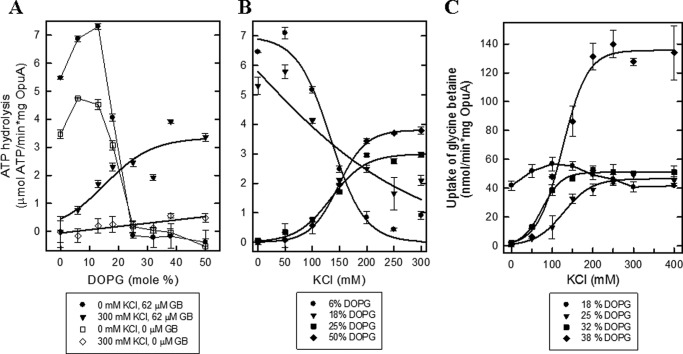
Fig. 3, A and B, compares the ATPase activity of OpuA in nanodiscs with the transport activity of OpuA reconstituted in proteoliposomes, both with identical lipid composition. Importantly, the ATPase activity in the nanodiscs is dependent of glycine betaine and a high salt concentration, which mimics the tight coupling and regulation of ATP hydrolysis and glycine betaine transport in cells and in proteoliposomes (11, 38). Surprisingly, the ATPase activity in the nanodiscs is more than an order of magnitude higher than the transport activity in proteoliposomes (Fig. 3, A (I) versus B (VII)). We noted that in proteoliposomes, two molecules of ATP are hydrolyzed per molecule of glycine betaine translocated (15). Even if we assume that due to a random orientation of the transporter molecules in the proteoliposomes (11, 15), half of the OpuA molecules do not participate in uptake, then still the ATPase activity is 10-fold higher. This suggests that either OpuA is more efficiently reconstituted in nanodiscs than in proteoliposomes or the activation energy barrier for translocation is lowered in the nanodiscs (e.g. due to an increased lipid chain order and thus changes in the lateral pressure profile (39)). We emphasize that the high ATPase activity requires glycine betaine binding to the extracytoplasmic SBDs plus threshold levels of salt, acting at the cytoplasmic face of OpuA, and thus both events signal each other. Uncoupled ATPase activity (i.e. ATP hydrolysis) in the absence of glycine betaine was observed only when the fraction of anionic lipids was below 18 mol % and in the absence of salt (Fig. 4A).
FIGURE 3.
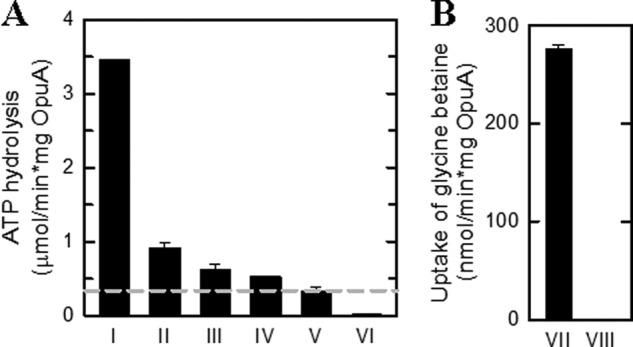
Activity of OpuA in nanodiscs and proteoliposomes. A, ATPase activity of OpuA nanodiscs at an OpuA/MSP1D1/lipid ratio of 1:20:2000 in the presence of 300 mm KCl plus 62 μm glycine betaine (I), 62 μm glycine betaine (II), and 300 mm KCl (III) and without additions (IV). The ATPase activity of empty nanodiscs (V) and DDM-solubilized OpuA (VI) were done in the presence of 300 mm KCl plus 62 μm glycine betaine. The residual ATPase activity of empty nanodiscs was used as the zero value in all subsequent experiments (dashed line in A). B, the uptake of [14C]glycine betaine by OpuA reconstituted in liposomes (VII) and empty liposomes (VIII) in 90 mm potassium Pi, pH 7.0, supplemented with 300 mm KCl plus 51 μm glycine betaine. The lipid composition used for the reconstitutions was in all cases 50 mol % DOPE, 12 mol % DOPC, and 38 mol % DOPG. Error bars, S.D.
FIGURE 4.
The effect of DOPG lipids on the activity of OpuA. A, ATPase activity of OpuA reconstituted in nanodiscs at an OpuA/MSP1D1/lipid ratio of 1:20:2000. The lipid composition of all samples consisted of 50 mol % DOPE. The molar fraction of DOPG was varied from 0 to 50 mol %, whereas the molar fraction of DOPC was adjusted reciprocally. The ATPase activity assay was performed in 50 mm potassium Pi, pH 7.0, supplemented with different concentrations of KCl and glycine betaine, as indicated in the key. B, ATPase activity of OpuA reconstituted as in A. The molar fraction of DOPG was varied as indicated in the key, whereas the molar fraction of DOPC was adjusted reciprocally. The ATPase activity assay was performed in 50 mm potassium Pi, pH 7.0, 62 μm glycine betaine plus various concentrations of KCl. C, the uptake of [14C]glycine betaine by OpuA reconstituted in liposomes. The lipid composition of all samples consisted of 50 mol % DOPE. The molar fraction of DOPG was varied as indicated in the key, whereas the molar fraction of DOPC was adjusted reciprocally. The uptake activity was assayed in 90 mm potassium Pi, pH 7.0, 51 μm glycine betaine plus various concentrations of KCl. Error bars, S.D.
Effect of Anionic Lipids
Because anionic lipids are critical for the gating of OpuA, we reconstituted the transporter in nanodiscs or liposomes composed of different lipid composition. Specifically, we varied the fraction of DOPG and DOPC reciprocally and kept DOPE at 50 mol %. Fig. 4 shows that a high concentration of DOPG is needed for maximal activity of OpuA, both in the ATPase assay in the nanodiscs (Fig. 4, A and B) and in the transport assay in the proteoliposomes (Fig. 4C). A high concentration of DOPG also favored nanodisc formation (supplemental Fig. 3). Remarkably, at 18 mol % DOPG (Fig. 4B) or below, the ATPase activity increases with decreasing salt concentration, whereas uptake of glycine betaine in the proteoliposomes decreases (Fig. 4C). These data indicate that at low concentrations of anionic lipids, the ATPase activity and substrate translocation are no longer coupled. To test this postulate, we analyzed the ATPase activity in the presence and absence of glycine betaine. Indeed, below 18 mol % DOPG, the ATPase activity is largely independent of glycine betaine (Fig. 4A), which is indicative of a loss of transmembrane signaling to the nucleotide-binding domains upon binding of the substrate to the extracytoplasmic SBDs.
Effect of Non-bilayer Lipids
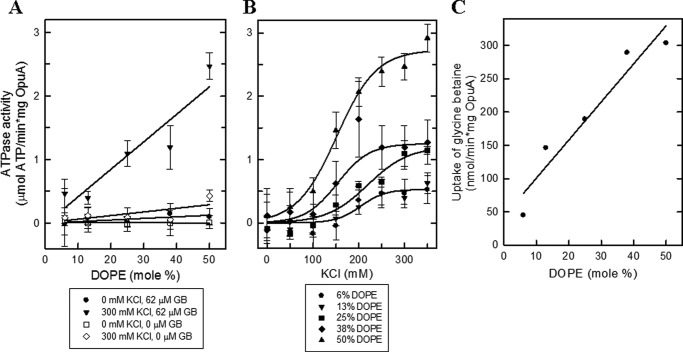
The non-bilayer-forming lipid DOPE exhibits a more negative spontaneous curvature than, for instance, DOPC. Changes in DOPE/DOPC ratio alter the attractive and repulsive forces along the bilayer normal, which is generally described as change in the lateral pressure profile (40, 41). We used this property to optimize the activity of OpuA nanodiscs. We find that the activity of OpuA increases linearly with the fraction of DOPE both in nanodiscs (Fig. 5A) and in proteoliposomes (Fig. 5C); the fraction of DOPE did not affect the ionic activation profile of OpuA (Fig. 5B). Thus, DOPE is essential for a high substrate-dependent ATPase activity of OpuA but does not impact the ionic gating mechanism of the transporter.
FIGURE 5.
The effect of DOPE lipids on the activity of OpuA. A, ATPase activity of OpuA reconstituted in nanodiscs at an OpuA/MSP1D1/lipid ratio of 1:20:2000. The lipid composition of all samples consisted of 38 mol % DOPG. The molar fraction of DOPE was varied from 6 mol % to 50 mol %, whereas the molar fraction of DOPC was adjusted reciprocally. The ATPase activity assay was performed in 50 mm potassium Pi, pH 7.0, supplemented with different concentrations of KCl and glycine betaine as indicated in the key. B, ATPase activity of OpuA reconstituted as in A. The molar fraction of DOPE was varied as indicated in the key, whereas the molar fraction of DOPC was adjusted reciprocally. The ATPase activity assay was performed in 50 mm potassium Pi, pH 7.0, 62 μm glycine betaine plus various concentration of KCl. C, the uptake of [14C]glycine betaine by OpuA reconstituted in liposomes was taken from Ref. 12. The lipid composition of all samples consisted of 38 mol % DOPG. The molar fraction of DOPE was varied from 6 to 50 mol %, whereas the molar fraction of DOPC was adjusted reciprocally. The maximal activities were plotted as a function of the molar fraction of DOPE. Error bars, S.D.
Specificity of Ionic Activation
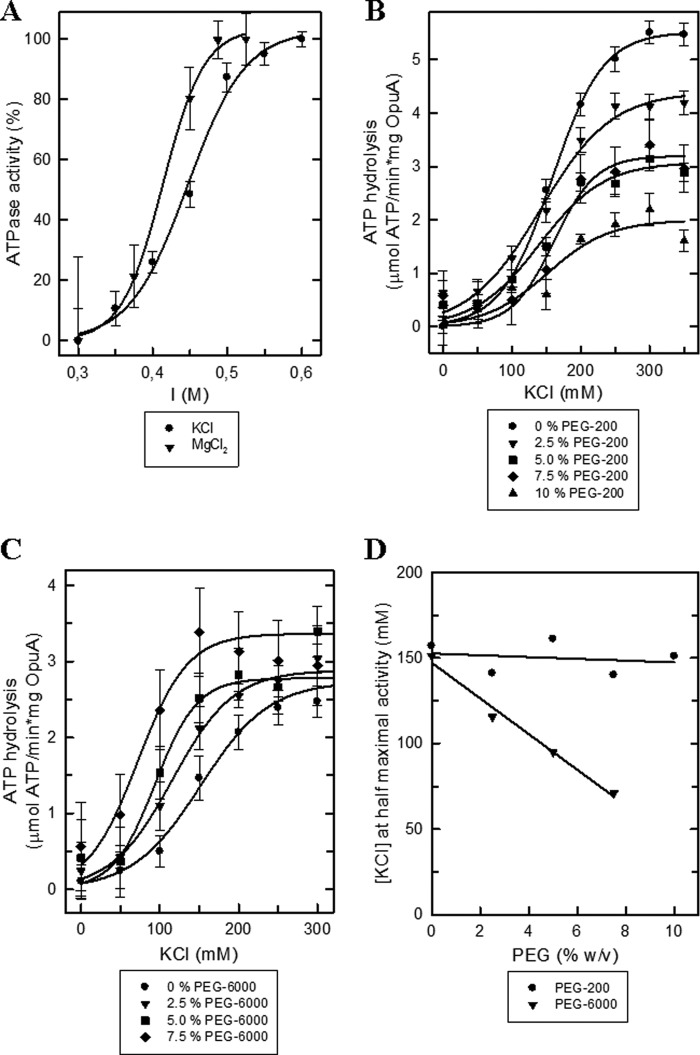
We have previously shown that Cs+, Rb+, K+, Na+, and Li+ (with chloride or phosphate as counter ion) activate OpuA (38), which indicates that the system is activated by electrolytes rather than a specific ion. However, in proteoliposomes, the concentration range of bivalent cations that can be tested is limited because, for instance, Mg2+ induces leaky membrane fusion. The OpuA-containing nanodiscs proved to be much more stable, and Mg2+ could be tested up to a concentration of at least 75 mm, which in the used buffer system corresponds to an ionic strength of 0.525 m (Fig. 6A). As a function of the concentration, MgCl2 proved 4–5-fold more effective than KCl; similar results were obtained when sulfate salts were used. We could not test Ca2+ salts because calcium ions compete with Mg2+ for binding to ATP and inhibit the ATPase activity of the nucleotide-binding domains.
FIGURE 6.
The effect of ionic strength and molecular crowders on OpuA activity. A, relative ATPase activity of OpuA reconstituted in nanodiscs at a OpuA/MSP1D1/lipid ratio of 1:20:2000 and lipid composition of 50 mol % DOPE, 12 mol % DOPC, and 38 mol % DOPG. The ATPase activity assay was performed in 50 mm potassium Pi, pH 7.0, 62 μm glycine betaine supplemented with various concentrations of KCl (circles) or MgCl2 (inverted triangles). The maximal activity was 4.0 and 1.6 μmol of ATP/(min × mg of OpuA) for KCl and MgCl2, respectively. B, the ATPase activity assay was performed as described in A in the presence of various w/v percentages of PEG 200 as indicated in the key. C, same as for B, with PEG 6000 instead of PEG 200. D, the concentration of KCl required for half-maximal activity as a function of PEG 200 (circles) and PEG 6000 (inverted triangles), recalculated from B and C. Error bars, S.D.
Dehydration and Excluded Volume Effects
Next, we addressed the effect of a high concentration of polyethylene glycol (PEG) on the ATPase activity of OpuA in nanodiscs. High concentrations of PEG dehydrate macromolecules, and depending on their size, they may destabilize or stabilize proteins (42, 43). We thus tested low and high molecular weight PEG molecules. We ensured that the activity of pyruvate kinase/lactic dehydrogenase was never limiting in the enzyme assay (see “Experimental Procedures”).
Fig. 6, A and C, shows that over a wide range of concentrations, PEG molecules do not activate OpuA when the ionic strength is below threshold values. Increasing concentrations of PEG 200 inhibit KCl-activated OpuA (Fig. 6B), presumably because the low molecular weight PEG molecules enter protein cavities and bind and interfere with conformational transitions. The binding of PEG 200 may even lead to protein denaturation (43). Importantly, PEG 6000 did not inhibit OpuA but shifted the ionic activation profile to lower salt concentrations (Fig. 6C). PEG 6000 is large enough to exert crowding-induced protein stabilization, and, besides stabilizing the overall protein structure, it may act by favoring the interaction of like-charged surfaces that form the gating switch of OpuA. In Fig. 6D, we plot the concentration at half-maximal activity as a function of the PEG concentration. Clearly, PEG 6000 and ionic strength act synergistically, whereas PEG 200 does not affect the activation profile.
DISCUSSION
We show strict coupling between glycine betaine binding and ATPase activity in OpuA nanodiscs, provided the chemical composition of the bilayer membrane and ionic strength of the medium are taken care of. A caveat of the nanodisc system is that vectorial transport cannot be measured directly, and enzyme activity does not necessarily reflect translocation activity. In fact, in vitro, the number of ATP molecules hydrolyzed per substrate translocated of many ABC transporters is very high and does not reflect the mechanistic stoichiometry and thus does not reflect translocation (44–45). For OpuA, we do not have this drawback. We observe low ATPase activity in the absence of glycine betaine and very high activity in its presence when the fraction of DOPG is well above 25 mol %. At intermediate levels of anionic lipids (18 mol %), the transporter is partially coupled. We note that the relatively high fraction of anionic lipids required for glycine betaine-dependent ATPase activity is physiologically relevant (46). In addition to glycine betaine, threshold levels of salt are required for ATP hydrolysis of OpuA nanodiscs high in anionic lipids. With 25 mol % of DOPG or more, the ATPase activity is inhibited at low but not at high ionic strength. Transmembrane signaling (from the SBDs to the nucleotide-binding domains) and full ionic regulation are retained in the nanodiscs with a high fraction of anionic lipids. Thus, the osmoregulatory ABC transporter OpuA incorporated into phospholipid bilayer nanodiscs displays full functionality and very high and coupled activity.
The main advantages of the nanodiscs are the relatively high stability, the accessibility of the cis and trans side of the protein, the low scattering of light compared with the proteoliposomes, and the control of protein number per particle. Using the nanodisc system, we now provide support for ionic strength-based gating of transport: (i) Mg2+ ions are 4–5-fold more effective in stimulating OpuA than monovalent cations; (ii) molecular crowders in the form of low or high molecular weight PEG molecules do not activate OpuA; (iii) high molecular weight PEG molecules exert excluded volume effects and shift the activation profile to lower ionic strength.
PEG molecules have been used in numerous studies to probe osmolality (hydration state) and molecular crowding as parameters of enzyme and transporter activity (47, 48), and the molecules can have destabilizing and stabilizing effects on proteins. Destabilizing effects of PEG molecules originate from the Lifshitz electrodynamic interaction of the co-solvent molecules with proteins, and these are attractive regardless of the chain length of the PEG molecules (43). The stabilization of protein structure by PEG molecules is attributed to the steric excluded volume effects, which are caused by entropic forces of the PEG molecules. The larger the PEG molecule, the more the attractive forces are overcome by the repulsive forces (i.e. the protein destabilization by binding of PEG molecules is overcome by the excluded volume effects). We found that PEG molecules themselves do not activate OpuA; rather, the low molecular weight PEG 200 inhibited the ATPase activity of the transporter. On the contrary, the high molecular weight PEG 6000 shifted the activation profile of OpuA to a lower salt concentration. If we assume two like-charged surfaces to form the gating switch of OpuA, then the ionic strength and macromolecular crowding (excluded volume) effects would act synergistically. The requirement for anionic lipids strongly suggests that the membrane lipids and protein residues form the like-charged surfaces.
Our data on OpuA differ from those of the Wood laboratory (17, 18) on the osmoregulatory transporter ProP. Here, lumenal PEG molecules activate the transporter, and the extent of the activation correlates with PEG molecule size (17, 18). Because high concentrations of PEG molecules expel water from protein surfaces by steric effects (47), the protein is dehydrated. Wood and colleagues (18) propose that ProP activity is influenced by the hydration state of the protein, preferential interaction of PEG molecules, and macromolecular crowding on the membrane or protein itself. A large impact of hydration state on protein activity has been observed for hexokinase (48). Here, a high concentration of PEG molecules lowered the availability of water in the active site of the enzyme and thereby increased the affinity for glucose. Our work and these examples illustrate that the gating mechanisms of osmotically regulated systems can differ significantly, but with the appropriate model systems, the individual physicochemical factors can be delineated.
In line with previous observations (12, 38), OpuA in nanodiscs requires relatively high levels of anionic lipids for its ionic gating. In proteoliposomes, hardly any uptake of glycine betaine is observed below 12 mol % of DOPG. We now show that under these conditions, ATP is hydrolyzed at a high rate, indicating that anionic lipids are required not only for gating of the transporter but also for tight coupling between ATP hydrolysis and glycine betaine uptake. Remarkably, the coupled activity of OpuA in the nanodiscs is about an order of magnitude higher than in the proteoliposomes. It has been argued that the lateral tension exerted by the membrane lipids onto a membrane protein is different in nanodiscs and lamellar vesicular membrane systems. In fact, there is evidence that the lipid acyl chains are stretched (and as a corollary, the bilayer thickness is increased) in the nanodiscs as compared with vesicular membranes (39). We propose that conformational transitions associated with OpuA-mediated transport are facilitated in the nanodiscs as compared with the proteoliposomes, which may relate to a different lipid order and consequently a different lateral pressure (profile). Support for this notion comes from the observation that OpuA activity is linearly dependent on the fraction of DOPE in the bilayer. Non-bilayer lipids like DOPE also influence the lateral pressure profile of the membrane (40).
In conclusion, we show that the ABC transporter OpuA can be functionally reconstituted into bilayer nanodiscs, provided great care is taken in the optimization of the lipid composition of the membrane. Under optimal conditions, the ATPase activity of the transporter is fully linked to the presence of substrate (glycine betaine), and the activity is gated by ionic strength. We show that above threshold levels of electrolytes, excluded volume effects act synergistically with ionic strength.
This work was supported by Netherlands Organization for Scientific Research (NWO) Top-Subsidy Grant 700.56.302 and ChemThem “Chemical Biology” Grant 728.011.104.

This article contains supplemental Figs. 1–3.
- ABC
- ATP-binding cassette
- CBS
- cystathionine β-synthase
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine
- DOPE
- 1,2-dioleoyl-sn-glycero-3-phosphatidylethanolamine
- DOPG
- 1,2-dioleoyl-sn-glycero-3-phosphatidylglycerol
- MSP
- membrane scaffold protein.
REFERENCES
- 1. Wood J. M., Bremer E., Csonka L. N., Kraemer R., Poolman B., van der Heide T., Smith L. T. (2001) Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 130, 437–460 [DOI] [PubMed] [Google Scholar]
- 2. Bolen D. W. (2001) Protein stabilization by naturally occurring osmolytes. Methods Mol. Biol. 168, 17–36 [DOI] [PubMed] [Google Scholar]
- 3. Racher K. I., Culham D. E., Wood J. M. (2001) Requirements for osmosensing and osmotic activation of transporter ProP from Escherichia coli. Biochemistry 40, 7324–7333 [DOI] [PubMed] [Google Scholar]
- 4. Tsatskis Y., Khambati J., Dobson M., Bogdanov M., Dowhan W., Wood J. M. (2005) The osmotic activation of transporter ProP is tuned by both its C-terminal coiled-coil and osmotically induced changes in phospholipid composition. J. Biol. Chem. 280, 41387–41394 [DOI] [PubMed] [Google Scholar]
- 5. Culham D. E., Romantsov T., Wood J. M. (2008) Roles of K+, H+, H2O, and ΔΨ in solute transport mediated by major facilitator superfamily members ProP and LacY. Biochemistry 47, 8176–8185 [DOI] [PubMed] [Google Scholar]
- 6. Keates R. A., Culham D. E., Vernikovska Y. I., Zuiani A. J., Boggs J. M., Wood J. M. (2010) Transmembrane helix I and periplasmic loop 1 of Escherichia coli ProP are involved in osmosensing and osmoprotectant transport. Biochemistry 49, 8847–8856 [DOI] [PubMed] [Google Scholar]
- 7. Rübenhagen R., Morbach S., Krämer R. (2001) The osmoreactive betaine carrier BetP from Corynebacterium glutamicum is a sensor for cytoplasmic K+. EMBO J. 20, 5412–5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ziegler C., Bremer E., Krämer R. (2010) The BCCT family of carriers. From physiology to crystal structure. Mol. Microbiol. 78, 13–34 [DOI] [PubMed] [Google Scholar]
- 9. Perez C., Koshy C., Ressl S., Nicklisch S., Krämer R., Ziegler C. (2011) Substrate specificity and ion coupling in the Na+/betaine symporter BetP. EMBO J. 30, 1221–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nicklisch S.C., Wunnicke D., Borovykh I. V., Morbach S., Klare J. P., Steinhoff H. J., Krämer R. (2012) Conformational changes of the betaine transporter BetP from Corynebacterium glutamicum studied by pulse EPR spectroscopy. Biochim. Biophys. Acta 1818, 359–366 [DOI] [PubMed] [Google Scholar]
- 11. van der Heide T., Stuart M. C., Poolman B. (2001) On the osmotic signal and osmosensing mechanism of an ABC transport system for glycine betaine. EMBO J. 20, 7022–7032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Biemans-Oldehinkel E., Mahmood N. A., Poolman B. (2006) A sensor for intracellular ionic strength. Proc. Natl. Acad. Sci. U.S.A. 103, 10624–10629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karasawa A., Erkens G. B., Berntsson R. P., Otten R., Schuurman-Wolters G. K., Mulder F. A., Poolman B. (2011) Cystathionine β-synthase (CBS) domains 1 and 2 fulfill different roles in ionic strength sensing of the ATP-binding cassette (ABC) transporter OpuA. J. Biol. Chem. 286, 37280–37291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Perez C., Koshy C., Yildiz O., Ziegler C. (2012) Alternating-access mechanism in conformationally asymmetric trimers of the betaine transporter BetP. Nature 490, 126–130 [DOI] [PubMed] [Google Scholar]
- 15. Patzlaff J. S., van der Heide T., Poolman B. (2003) The ATP/substrate stoichiometry of the ATP-binding cassette (ABC) transporter OpuA. J. Biol. Chem. 278, 29546–29551 [DOI] [PubMed] [Google Scholar]
- 16. Spitzer J. J., Poolman B. (2005) Electrochemical structure of the crowded cytoplasm. Clues from bacterial osmosensors. Trends Biochem. Sci. 30, 536–541 [DOI] [PubMed] [Google Scholar]
- 17. Culham D. E., Henderson J., Crane R. A., Wood J. M. (2003) Osmosensor ProP of Escherichia coli responds to the concentration, chemistry, and molecular size of osmolytes in the proteoliposome lumen. Biochemistry 42, 410–420 [DOI] [PubMed] [Google Scholar]
- 18. Culham D. E., Meinecke M., Wood J. M. (2012) Impacts of the osmolality and the lumenal ionic strength on osmosensory transporter ProP in proteoliposomes. J. Biol. Chem. 287, 27813–27822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ritchie T. K., Grinkova Y. V., Bayburt T. H., Denisov I. G., Zolnerciks J. K., Atkins W. M., Sligar S. G. (2009) Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bayburt T. H., Sligar S. G. (2010) Membrane protein assembly into nanodiscs. FEBS Lett. 584, 1721–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shaw A. W., Pureza V. S., Sligar S. G., Morrissey J. H. (2007) The local phospholipid environment modulates the activation of blood clotting. J. Biol. Chem. 282, 6556–6563 [DOI] [PubMed] [Google Scholar]
- 22. Bayburt T. H., Grinkova Y. V., Sligar S. G. (2006) Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch. Biochem. Biophys. 450, 215–222 [DOI] [PubMed] [Google Scholar]
- 23. Ranaghan M. J., Schwall C. T., Alder N. N., Birge R. R. (2011) Green proteorhodopsin reconstituted into nanoscale phospholipid bilayers (nanodiscs) as photoactive monomers. J. Am. Chem. Soc. 133, 18318–18327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Whorton M. R., Bokoch M. P., Rasmussen S. G., Huang B., Zare R. N., Kobilka B., Sunahara R. K. (2007) A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc. Natl. Acad. Sci. U.S.A. 104, 7682–7687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bayburt T. H., Vishnivetskiy S. A., McLean M. A., Morizumi T., Huang C. C., Tesmer J. J., Ernst O. P., Sligar S. G., Gurevich V. V. (2011) Monomeric rhodopsin is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J. Biol. Chem. 286, 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Inagaki S., Ghirlando R., White J. F., Gvozdenovic-Jeremic J., Northup J. K., Grisshammer R. (2012) Modulation of the interaction between neurotensin receptor NTS1 and Gq protein by lipid. J. Mol. Biol. 417, 95–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boldog T., Grimme S., Li M., Sligar S. G., Hazelbauer G. L. (2006) Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc. Natl. Acad. Sci. U.S.A. 103, 11509–11514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li M., Hazelbauer G. L. (2011) Core unit of chemotaxis signaling complexes. Proc. Natl. Acad. Sci. U.S.A. 108, 9390–9395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alami M., Dalal K., Lelj-Garolla B., Sligar S. G., Duong F. (2007) Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. EMBO J. 26, 1995–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dalal K., Nguyen N., Alami M., Tan J., Moraes T. F., Lee W. C., Maurus R., Sligar S. S., Brayer G. D., Duong F. (2009) Structure, binding, and activity of Syd, a SecY-interacting protein. J. Biol. Chem. 284, 7897–7902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dalal K., Chan C. S., Sligar S. G., Duong F. (2012) Two copies of the SecY channel and acidic lipids are necessary to activate the SecA translocation ATPase. Proc. Natl. Acad. Sci. U.S.A. 109, 4104–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ishmukhametov R., Hornung T., Spetzler D., Frasch W. D. (2010) Direct observation of stepped proteolipid ring rotation in E. coli F0F1-ATP synthase. EMBO J. 29, 3911–3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alvarez F. J., Orelle C., Davidson A. L. (2010) Functional reconstitution of an ABC transporter in nanodiscs for use in electron paramagnetic resonance spectroscopy. J. Am. Chem. Soc. 132, 9513–9515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bao H., Duong F. (2012) Discovery of an auto-regulation mechanism for the maltose ABC transporter MalFGK2. PLoS ONE 7, e34836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kawai T., Caaveiro J. M., Abe R., Katagiri T., Tsumoto K. (2011) Catalytic activity of MsbA reconstituted in nanodisc particles is modulated by remote interactions with the bilayer. FEBS Lett. 585, 3533–3537 [DOI] [PubMed] [Google Scholar]
- 36. Ritchie T. K., Kwon H., Atkins W. M. (2011) Conformational analysis of human ATP-binding cassette transporter ABCB1 in lipid nanodiscs and inhibition by the antibodies MRK16 and UIC2. J. Biol. Chem. 286, 39489–39496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Geertsma E. R., Nik Mahmood N. A., Schuurman-Wolters G. K., Poolman B. (2008) Membrane reconstitution of ABC transporters and assays of translocator function. Nat. Protoc. 3, 256–266 [DOI] [PubMed] [Google Scholar]
- 38. Mahmood N. A., Biemans-Oldehinkel E., Patzlaff J. S., Schuurman-Wolters G. K., Poolman B. (2006) Ion specificity and ionic strength dependence of the osmoregulatory ABC transporter OpuA. J. Biol. Chem. 281, 29830–29839 [DOI] [PubMed] [Google Scholar]
- 39. Mörs K., Roos C., Scholz F., Wachtveitl J., Dötsch V., Bernhard F., Glaubitz C. (2013) Modified lipid and protein dynamics in nanodiscs. Biochim. Biophys. Acta 1828, 1222–1229 [DOI] [PubMed] [Google Scholar]
- 40. Marsh D. (2007) Lateral pressure profile, spontaneous curvature frustration, and the incorporation and conformation of proteins in membranes. Biophys. J. 93, 3884–3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brown M. F. (2012) Curvature forces in membrane lipid-protein interactions. Biochemistry 51, 9782–9795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tokuriki N., Kinjo M., Negi S., Hoshino M., Goto Y., Urabe I., Yomo T. (2004) Protein folding by the effects of macromolecular crowding. Protein Sci. 13, 125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Damodaran S. (2012) On the molecular mechanism of stabilization of proteins by cosolvents. Role of Lifshitz electrodynamic forces. Langmuir 28, 9475–9486 [DOI] [PubMed] [Google Scholar]
- 44. Shapiro A. B., Ling V. (1995) Reconstitution of drug transport by purified P-glycoprotein. J. Biol. Chem. 270, 16167–16175 [DOI] [PubMed] [Google Scholar]
- 45. Borths E. L., Poolman B., Hvorup R. N., Locher K. P., Rees D. C. (2005) In vitro functional characterization of BtuCD-F, the Escherichia coli ABC transporter for vitamin B12 uptake. Biochemistry 44, 16301–16309 [DOI] [PubMed] [Google Scholar]
- 46. Driessen A. J., Zheng T., In't Veld G., Op den Kamp J. A., Konings W. N. (1988) Lipid requirement of the branched-chain amino acid transport system of Streptococcus cremoris. Biochemistry 27, 865–872 [DOI] [PubMed] [Google Scholar]
- 47. Parsegian V. A., Rand R. P., Rau D. C. (2000) Osmotic stress, crowding, preferential hydration, and binding. A comparison of perspectives. Proc. Natl. Acad. Sci. U.S.A. 97, 3987–3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reid C., Rand R. P. (1997) Probing protein hydration and conformational states in solution. Biophys. J. 72, 1022–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wolters J. C., Berntsson R. P., Gul N., Karasawa A., Thunnissen A. M., Slotboom D. J., Poolman B. (2010) Ligand binding and crystal structures of the substrate-binding domain of the ABC transporter OpuA. PLoS ONE 5, e10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hollenstein K., Frei D. C., Locher K. P. (2007) Structure of an ABC transporter in complex with its binding proteins. Nature 446, 213–216 [DOI] [PubMed] [Google Scholar]