Background: Development of resistance is major problem in TRAIL anti-cancer therapy.
Results: Cryptotanshinone induces death receptor 5 via ROS signaling and therefore enhances TRAIL-induced apoptosis.
Conclusion: Cryptotanshinone restores TRAIL sensitivity in TRAIL-resistant cancer cells.
Significance: We demonstrate a novel TRAIL-enhancing action by natural compound cryptotanshinone.
Keywords: Apoptosis, Cell Death, Melanoma, Reactive Oxygen Species (ROS), Trail
Abstract
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) selectively induces apoptosis and kills cancer cells but not normal cells. However, TRAIL resistance due to low level of TRAIL receptor expression is widely found in cancer cells and hampers its development for cancer treatment. Thus, the agents that can sensitize the tumor cells to TRAIL-mediated apoptosis are urgently needed. We investigated whether tanshinones, the major bioactive compounds of Salvia miltiorrhiza (danshen), can up-regulate TRAIL receptor expression. Among the major tanshinones being tested, cryptotanshinone (CT) showed the best ability to induce TRAIL receptor 2 (DR5) expression. We further showed that CT was capable of promoting TRAIL-induced cell death and apoptosis in A375 melanoma cells. CT-induced DR5 induction was not cell type-specific, as DR5 induction was observed in other cancer cell types. DR5 knockdown abolished the enhancing effect of CT on TRAIL responses. Mechanistically, induction of the DR5 by CT was found to be p53-independent but dependent on the induction of CCAAT/enhancer-binding protein-homologous protein (CHOP). Knockdown of CHOP abolished CT-induced DR5 expression and the associated potentiation of TRAIL-mediated cell death. In addition, CT-induced ROS production preceded up-regulation of CHOP and DR5 and consequent sensitization of cells to TRAIL. Interestingly, CT also converted TRAIL-resistant lung A549 cancer cells into TRAIL-sensitive cells. Taken together, our results indicate that CT can potentiate TRAIL-induced apoptosis through up-regulation of DR5.
Introduction
TNF-related apoptosis inducing ligand (TRAIL)2 is a member of the TNF superfamily of cytokines that represents an attractive therapy targeting on receptor-mediated apoptosis. TRAIL has attracted considerable attention owing to its selective killing of tumor cells but not normal cells (1). TRAIL induces apoptosis through binding to its receptors, namely TRAIL receptor 1 (DR4) and TRAIL receptor 2 (DR5). Upon binding to DR4 or DR5, TRAIL induces receptor trimerization to form the death-inducing signaling complex, which in turn activates caspase-8, leading directly to the activation of caspase-3 and subsequent apoptosis (2).
Various cancer cell types were previously reported to be susceptible to TRAIL-induced apoptosis (3). However, intrinsic resistance to TRAIL, due to the low TRAIL receptor expression levels, always appears as a major barrier for the development of efficient TRAIL-based cancer therapy (4). Therefore, identification of sensitizing agents capable of increasing TRAIL receptor levels is important to facilitate TRAIL-mediated therapy. Recently, the involvement of CCAAT/enhancer binding protein homologous protein (CHOP) and reactive oxygen species (ROS) signaling pathways in the sensitization of cancer cells to TRAIL-mediated apoptosis has been documented in several studies (5, 6). It has been suggested that ROS act as upstream signaling molecules to initiate endoplasmic reticulum (ER) stress, which in turn triggers CHOP-dependent DR5 expression and finally sensitizes cells to TRAIL-induced apoptosis (5, 6). Thus, identification of small molecules compounds capable of up-regulating TRAIL receptor via ROS-CHOP signaling cascades provides a fresh and novel therapeutic approach to target highly TRAIL-resistant cancers.
Tanshinones are diterpenes isolated from Salvia miltiorrhiza (danshen), a well known herb traditionally used for the treatment of cardiovascular diseases (7). Among the major tanshinones isolated, cryptotanshinone, tanshinone I, tanshinone IIA, and dihydrotanshinone, tanshinone IIA have been shown to exert multiple anti-cancer activities including induction of apoptosis and inhibition of angiogenesis and metastasis (8). Furthermore, tanshinones hold promise as sensitizing agents for chemotherapy and radiotherapy to enhance the cytotoxic effects of anti-cancer agents such as TNF-α (9), 5-fluorouracil (10), and γ-irradiation (11). Recently, tanshinones have been found to induce ROS production in various types of cancer cells (12–14). In addition, tanshinones have been reported to exert anti-cancer effects via ROS-mediated ER stress. For example, cryptotanshinone (CT) has been shown to induce ROS and contribute to ER stress-mediated apoptosis in human hepatoma and breast cancer cells (15). Although a large number of studies showed that tanshinones induce ROS and ER stress, the effect of tanshinones on TRAIL receptor and TRAIL-induced apoptosis is not clear.
In the present study, we examined whether tanshinones can induce DR5 expression in TRAIL-resistant cancer cells and, if it does, whether tanshinones can also enhance TRAIL responses. We describe here, for the first time, that cryptotanshinone can potentiate TRAIL-induced apoptosis in TRAIL-resistant cancer cells through up-regulating DR5 via ROS-mediated CHOP activation.
EXPERIMENTAL PROCEDURES
Reagents and Cell Culture
Antibodies against PARP, caspase-9, caspase-8, caspase-3, DR5, CHOP, p53, survivin, X-linked inhibitor of apoptosis protein (XIAP), Mcl-1, cFLIP, cIAP-1, and cIAP-2 were purchased from Cell Signaling (Beverly, MA). Antibodies against bcl-2, bcl-xL, actinin, and actin were purchased from Santa Cruz Biotechnology, Inc. Antibody against FLAG (M2) was purchased from Sigma. Antibody against DR4 was purchased from Millipore. Recombinant human TRAIL/Apo2 ligand (amino acids 114–281), caspase colorimetric QuantiPak, and calcein AM were purchased from Enzo Life Sciences. Crystal violet, propidium iodide, DAPI, N-acetyl-l-cysteine (NAC), cycloheximide, actinomycin, cryptotanshinone, tanshinone I, tanshinone IIA, dihydrotanshinone, tanshinone IIA, dihydroethidium (DHE), and 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) were purchased from Sigma. 6-Carboxy-2′,7′-dichlorofluorescein diacetate (DCF) was purchased from Molecular Probes.
A375, HCT116, HT-29, OVCAR3, MRC-9, and A459 were obtained from the American Type Culture Collection. HCT116 variant with deletion of p53 was kindly supplied by Dr. Bert Vogelstein (The Johns Hopkins University, Baltimore, MD). All cell lines were cultured in DMEM supplemented with 5% fetal calf serum, 100 units/ml penicillin, and 100 mg/ml streptomycin.
Western Blot Analysis
Whole-cell protein was prepared and analyzed by Western blotting as described previously (16).
Annexin V/PI Assay
The early indicator of apoptosis was detected by using annexin V/PI binding kit (Abcam) and then analyzed with a flow cytometer.
Analysis of Cell Surface DR5
Cells were detached with 0.5 mm EDTA and washed three times with PBS supplemented with 0.5% BSA. Cells were then resuspended in 200 μl of PBS, stained with phycoerythrin-conjugated mouse monoclonal anti-human DR5 or isotype control, and incubated for 60 min at 4 °C. Unreacted antibody was removed by washing the cells twice with PBS buffer. Cells were resuspended in 500 μl of PBS. Surface expression of DR5 was determined by flow cytometry. Fluorescence intensity of the cells was directly proportional to the density of receptors. Phycoerythrin-conjugated mouse anti-human DR5 (DJR2–4) and phycoerythrin-mouse IgG1 isotype control (MOPC-21) were purchased from BioLegend.
MTT Cytotoxicity Assay and Crystal Violet Staining
For MTT assay, cells were treated as indicated with TRAIL alone or pretreated with CT for 12 h, washed with PBS, and then treated with TRAIL for an additional 24 h. Then, the effects of CT on TRAIL-induced cytotoxicity were determined by the MTT uptake method (17). For crystal violet staining assay, cells (1 × 105) were seeded in 60-mm dishes and then untreated or pretreated with CT for 12 h, washed with PBS, and then treated with TRAIL. After 5 days, cells were stained with crystal violet.
Live/dead Assay
To determine cell death, cells were stained with Calcein-AM (1 μm) and PI (25 μg/ml) for 30 min at 37 °C, and the fraction of live (Calcein-AM-positive) and dead (PI) cells was determined under fluorescence microscope (Nikon eclipse 80i).
siRNA Treatment
Silencing of DR5 or CHOP was achieved by transfecting DR5, CHOP, or control siRNAs (Dharmacon) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations. The gene silencing effect was evaluated by Western blot analysis.
Plasmids and Transfection
DR5 and CHOP luciferase promoter constructs were synthesized by PCR using A375 genomic DNA (isolated by DNAzol reagent, Invitrogen) as the template, and the PCR products were inspected on agarose gel, gel purified, and cloned into pGL3-basic vector for the luciferase gene expression assay. pDR5-310ΔCHOP luciferase reporter, which bearing mutation of CHOP binding site in the DR5 promoter, was generated with a site-directed mutagenesis kit (Stratagene, La Jolla, CA). Superoxide dismutase 1 (SOD1) and catalase full-length expression constructs were synthesized by PCR using A375 total mRNA and subcloned into the pcDNA3-FLAG-tagged vector. The plasmid transfection and luciferase assay were the same as described previously (18). Firefly and Renilla luciferase activities were assayed according to the manufacturer's instructions (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity in cell lysate and expressed as an average of three independent experiments.
Measurement of Reactive Oxygen Species
Cells were plated at a density of 2 × 104/well in black 96-well plates and allowed to attach for 24 h. Then cells were loaded with fluorescent dyes, DCF or dihydroethidium (DHE), and further stimulated with 20 μm CT with or without the pretreatment of 5 mm NAC for 1 h. DCF and DHE fluorescence was measured using a fluorescence microplate reader (EnVision® Multilabel Reader, PerkinElmer Life Science) at indicated time point at 37 °C.
Asp-Glu-Val-Asp-ase (DEVDase) and Ile-Glu-Thr-Asp-ase (IETDase) Activity Assays
To evaluate DEVDase (caspase-3) or IETDase (caspase-8) activities, cell lysates were prepared after their respective treatment with TRAIL or CT. Assays were performed in 96-well microtiter plates by incubating 20 μg of cell lysates in 100 μl of reaction buffer (50 mm HEPES, pH 7.4, 100 mm NaCl, 0.1% (w/v) CHAPS, 10 mm DTT, 1 mm EDTA, 10% (v/v) glycerol) containing the corresponding caspase substrates at 4 μm. Lysates were incubated at 37 °C for 2 h. Thereafter, the absorbance at 405 nm was measured with a spectrophotometer.
Reverse Transcription-PCR and Real-time PCR Analysis
Total RNA was extracted using the TRIzol reagent (Invitrogen), and RT-PCR was conducted. PCR products were analyzed by agarose gel electrophoresis and visualized by GelRed. Alternatively, real-time PCR on RNA was carried out in an Applied Biosystems ViiA 7 real-time PCR machine using SYBR Green assays. The PCR primers were as follows: 5′-GAATGACCTCCTTTTCTGCTTGC-3′ and 5′-GCTTCCCCACTGTGCTTTGTA-3′ for DR5, and 5′-ATGAGGACCTGCAAGAGGTCC-3′ and 5′-TCCTCCTCAGTCAGCCAAGC-3′ for CHOP.
RESULTS
Tanshinones Induce DR5 Expression in Human Melanoma Cells
We first examined the ability of tanshinones to modulate TRAIL receptors expression in human A375 melanoma cells. As shown in Fig. 1B, all four tanshinones from Salvia miltiorrhiza induced DR5 but not DR4 expression in A375 cells. Among the tanshinones tested, CT showed the best ability to induce DR5 expression; therefore, CT was chosen for further studies.
FIGURE 1.
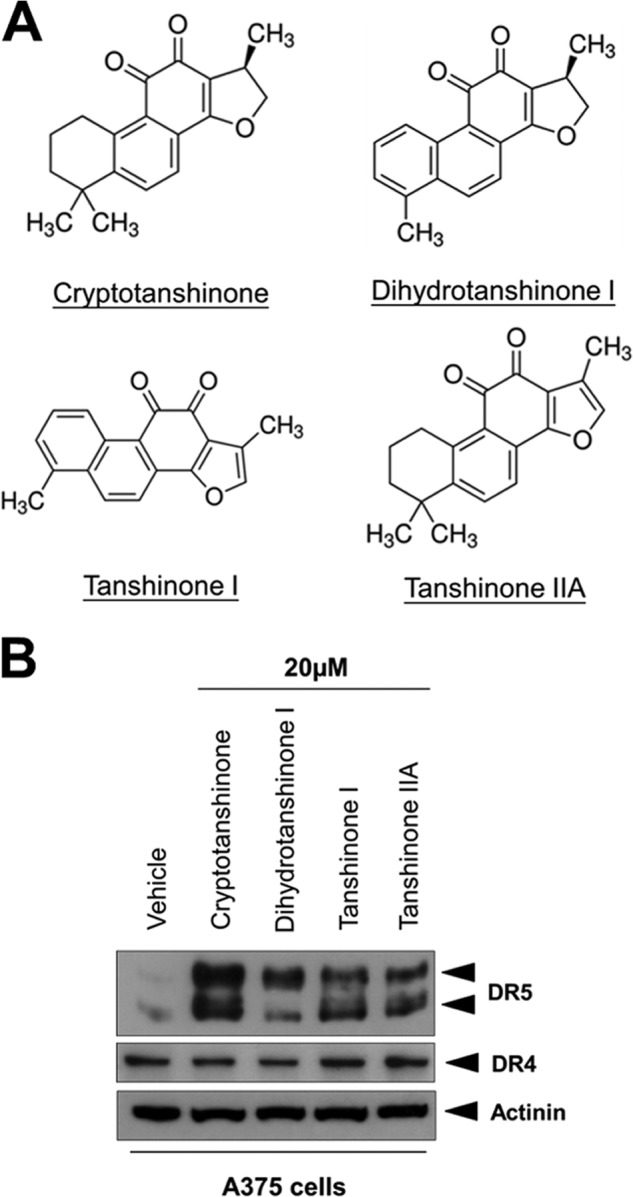
Tanshinones induce DR5 expression in A375 melanoma cells. A, chemical structure of tanshinones. B, Western blotting of A375 lysates from cells treated with 20 μm of indicated tanshinones for 12 h.
Cryptotanshinone Sensitizes Human Melanoma Cells to TRAIL-mediated Apoptosis
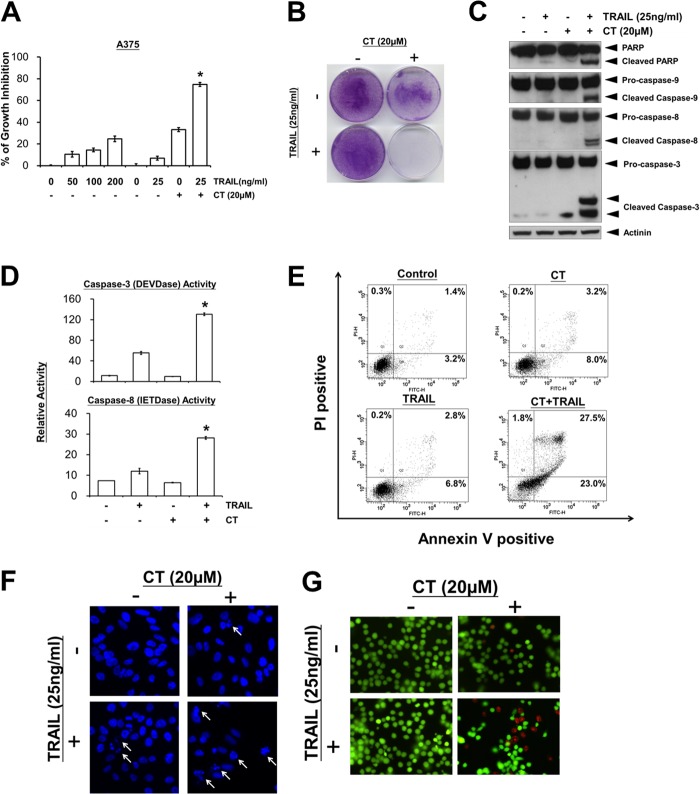
To examine the effect of CT on TRAIL-induced cell death in human melanoma cells, A375 cells were pretreated with CT and then treated with TRAIL, after which cell death was investigated by the MTT method. A375 melanoma cells displayed low rates of TRAIL-induced cell death when TRAIL was used alone at concentration as high as 200 ng/ml (Fig. 2A). However, pretreatment with CT significantly enhanced TRAIL-induced cytotoxicity (Fig. 2A).
FIGURE 2.
Cryptotanshinone sensitizes TRAIL-induced apoptosis in A375 melanoma cells. A, cell viability was assessed by the MTT assay. *, p < 0.05 versus control, CT, and TRAIL. B, crystal violet staining of A375 cells treated with CT and TRAIL as described under “Experimental Procedures.” C, Western blotting of lysates from A375 cells that had been pretreated with CT for 12 h followed by treatment with TRAIL for 3 h using the indicated antibodies. D, caspase activities in A375 cells that had been treated with 20 μm CT for 12 h and then TRAIL for 8 h. *, p < 0.05 versus control, CT, and TRAIL. E, cells were treated as described above and stained with PI/annexin V and then analyzed by FACS. At least two independent experiments revealed largely comparable results. F, DAPI staining. Cells were treated as described above nuclei with apoptotic morphological change (arrow) were visualized using fluorescence microscopy (original magnification, ×400). G, cells were treated as described above. Cell death was determined by the live/dead cell viability assay.
To determine the long term growth inhibitory effects, we pre-incubated A375 cells with vehicle or CT for 12 h, washed the cells, and grew them for 5 additional days in the presence or absence of TRAIL. Under these conditions, cells treated with vehicle, CT alone, or TRAIL alone remained viable and continued to grow (Fig. 2B). In contrast, the combination of CT and TRAIL led to the elimination of all viable cells, indicating that the TRAIL-sensitizing action of CT significantly reduced long term cell survival.
We also investigated whether CT increases TRAIL-induced activation of caspase-3, -8, and -9 and consequent PARP cleavage. We found that CT enhanced TRAIL-induced activation of all three caspases, which in turn led to increased PARP cleavage (Fig. 2C). In addition, combinatory treatment of A375 with CT and TRAIL strongly stimulated caspase-3 and caspase-8 activities (Fig. 2D). Furthermore, pretreatment of A375 cells with CT resulted in a markedly increased apoptotic cells (Fig. 2E), apoptotic nuclei (Fig. 2F), and cytotoxic effect (Fig. 2G) upon TRAIL treatment. Taken together, these results suggest that CT enhances TRAIL-induced apoptosis and cell death.
Up-regulation of DR5 by Cryptotanshinone Is Not Cell-type-specific and Contributes to the Enhancement of TRAIL-induced Cell Death
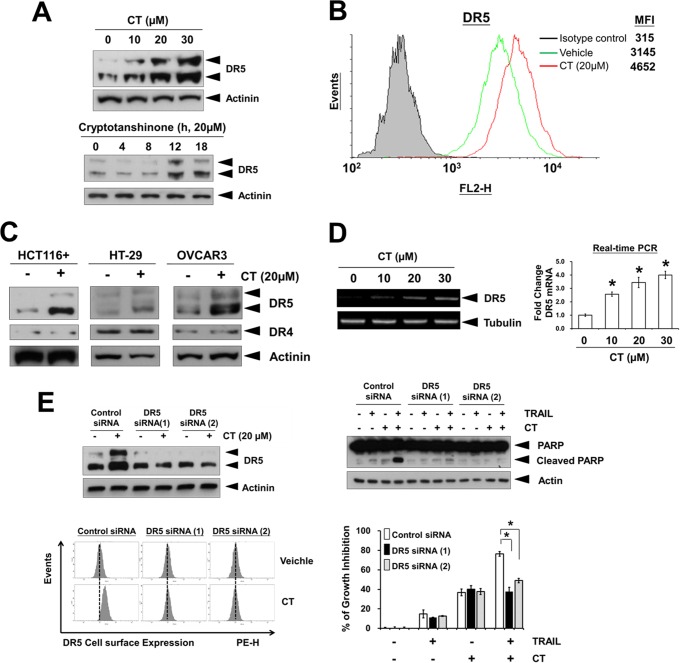
TRAIL mediates its activity through its interaction with death receptors DR4 and DR5 to trigger apoptotic signals (19). Treatment of A375 cells with CT for 12 h resulted in an increased protein expression of DR5 in a dose-dependent manner (Fig. 3A, top panel). CT also induced DR5 expression in a time-dependent manner (Fig. 3A, bottom panel). To determine whether CT induces expression of the DR5 on the cell surface, we analyzed the cell surface expression of DR5 in cells using flow cytometry analysis. After treatment of CT, the level of DR5 on the cell surface increased (Fig. 3B). Collectively, these results indicate that CT up-regulates the expression of DR5 on the cell surface.
FIGURE 3.
Cryptotanshinone-induced DR5 up-regulation is essential for sensitization of TRAIL-mediated apoptosis. A, A375 cells were treated with indicated and whole cell extracts were then prepared and analyzed for DR5 expression by Western blotting. B, the cell surface expression levels of DR5 in A375 cells treated with 20 μm CT for 12 h were measured by flow cytometry analysis using phycoerythrin (PE)-conjugated DR5 and isotype control antibodies. MFI, mean fluorescence intensity. C, CT up-regulated DR5 in various types of cancer cells. Cells were treated with 20 μm CT for 12 h, after which whole-cell extracts were prepared and analyzed by Western blotting. D, CT up-regulates DR5 mRNA expression. Left, ethidium bromide-agarose gels of RT-PCR product from A375 cells treated with the indicated concentrations of CT for 12 h. Right, A375 cells were treated as above, and total RNA was examined by real-time PCR analysis. *, p < 0.05 versus control. E, cells were transfected with siRNAs and subjected to Western blot analysis of DR5 (top left) and PARP (top right) and for DR5 cell surface staining by FACS (bottom left). Right bottom panel, after transfection with siRNA for 24 h, A375 cells were pretreated with 20 μm CT for 12 h, washed with PBS, and then treated with TRAIL (25 ng/ml) for an additional 24 h. Cell viability was assessed by the MTT assay. *, p < 0.05.
We next determined whether up-regulation of DR5 by CT is specific to A375 melanoma cells or also happens in other cell types. As shown in Fig. 3C, CT induced DR5 expression in colon (HCT116, HT-29) and ovarian (OVCAR3) cancer cells. Thus, we concluded that up-regulation of DR5 by CT is probably not cell-type specific.
Next, we asked whether DR5 expression is induced by CT at the transcriptional level. Using RT-PCR, we found that CT markedly up-regulated DR5 mRNA expression in a dose-dependent manner (Fig. 3D), suggesting that CT does modulate DR5 expression at the transcriptional level.
To determine the functional role of DR5 in the enhancement of TRAIL-induced apoptosis by CT, we used DR5-specific siRNA to knock down the expression of this receptor. Transfection of A375 cells with DR5 siRNAs prevented CT-induced DR5 expression (Fig. 3E, top left and bottom left panels). PARP cleavage induced by CT plus TRAIL was significantly inhibited in cells transfected with DR5 siRNAs, compared with control siRNA-transfected cells (Fig. 3E, top right panel). In addition, the effect of CT on TRAIL-induced cell growth inhibition was significantly reduced in cells transfected with DR5 siRNAs compared with control siRNA (Fig. 3E, bottom right panel). Taken together, these results suggest that CT-induced DR5 induction is critical for TRAIL enhancing effects in A375 cells.
Cryptotanshinone Activates DR5 Transcription in a CHOP-dependent Manner
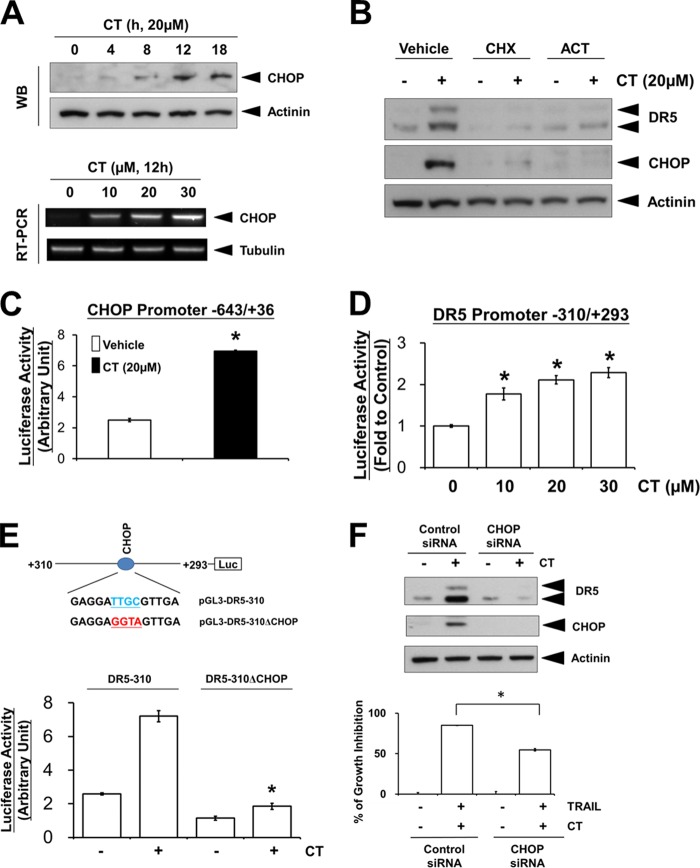
Recently, it has been shown that the induction of DR5 can be mediated through the activation of CHOP (20) and contributes to the sensitization of TRAIL-mediated apoptosis (5, 6). Therefore, we next investigated whether CHOP is involved in CT-induced DR5 up-regulation. We found that the protein expression of CHOP was significantly increased by CT treatment in a time-dependent manner (Fig. 4A, top panel). We further found that CT induced CHOP mRNA expression in a dose-dependent manner (Fig. 4A, bottom panel). Pre-treatment of cells with transcriptional (actinomycin D) and translational (cycloheximide) inhibitors blocked CT-induced DR5 and CHOP expression (Fig. 4B), indicating that DR5 and CHOP induction occurred at both transcriptional and post-transcriptional levels. Furthermore, treatment with CT was able to induce luciferase activity of a reporter plasmid containing 643 bp of the upstream portion of CHOP promoter (Fig. 4C). Together, these results suggest that CT regulates the transcription of CHOP.
FIGURE 4.
Induction of CHOP is required for cryptotanshinone-induced DR5 expression. A (top), Western blotting of A375 lysates from cells treated with 20 μm CT for indicated time periods; bottom, RT-PCR analysis of CHOP. B, A375 cells were pretreated with 1.5 μg/ml cycloheximide (CHX) or 2 μg/ml actinomycin D (ACT) for 30 min before incubation with 20 μm CT for 12 h. The whole-cell extracts were subjected to Western blot analysis using the anti-DR5 and anti-CHOP antibodies. C, A375 cells were transfected with pGL3-CHOP-643/+36 and then treated with 20 μm CT. After 12 h, cells were lysed and assayed for luciferase activity. *, p < 0.05 compared with vehicle-treated cells. D, luciferase assay with reporter pDR5-310/+293 was performed as described above. E, cells were transfected with pDR5-310 or pDR5-310ΔCHOP and then treated with 20 μm CT for 12 h and then assayed for luciferase activity. *, p < 0.05 compared with CT-treated pDR5-310-transfected cells. F (top), cells were transfected with siRNAs and cell extracts were prepared for Western blot analysis of DR5 and CHOP; bottom, after transfection with siRNA for 24 h, A375 cells were pretreated with 20 μm CT for 12 h, washed with PBS, and then treated with TRAIL (25 ng/ml) for an additional 24 h. Cell viability was assessed by the MTT assay. *, p < 0.05.
To determine whether CHOP induction by CT is involved in DR5 transcription, we began our investigation by examining the effects of CT on the transactivation of DR5 promoter. We observed that CT significantly induced the luciferase activity of DR5 promoter (−310/+293) in a dose-dependent manner (Fig. 4D), indicating CT-induced DR5 promoter transactivation. Given that a CHOP binding site is located in this region (20), we further examined the effects of CT on the transactivation of DR5 reporter constructs carrying wild-type or mutated CHOP binding sites. Whereas the promoter activity of pDR5-310 was significantly increased by CT treatment, the promoter activity of pDR5-310ΔCHOP was not enhanced by CT (Fig. 4E). Moreover, knockdown of CHOP expression by siRNA transfection significantly inhibited CT-induced DR5 up-regulation (Fig. 4F, top panel), indicating again that CHOP induction is essential in CT-induced DR5 up-regulation. In line with this, knockdown of CHOP significantly attenuated the cells from death induced by the combined treatment of CT and TRAIL (Fig. 4F, bottom panel). Taken together, these results suggest that CHOP induction plays an essential role in both CT-induced DR5 up-regulation and CT-mediated TRAIL-induced cell death enhancement.
Cryptotanshinone Potentiates TRAIL-induced Apoptosis through ROS Generation
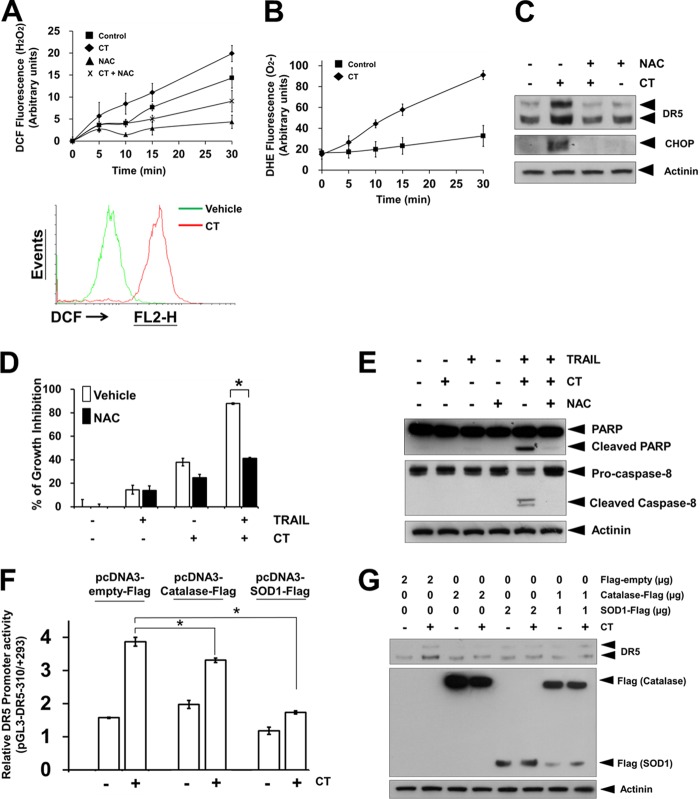
Recently, ROS has been reported to play a critical role in both DR5 up-regulation and TRAIL-induced apoptosis (5, 6). Given that tanshinones have been found to induce ROS production (12–14), we examined whether the generation of ROS could be involved in CT-induced TRAIL sensitization. The intracellular hydrogen peroxide (H2O2) levels were detected using fluorescent probe H2DCFDA. CT strongly induced the production of H2O2 in A375 cells (Fig. 5A). Pretreatment with ROS scavenger NAC effectively blocked H2O2 production induced by CT (Fig. 5A). We also found that CT increased superoxide (O2⨪) production using the fluorescent probe DHE (Fig. 5B). Recently reports have revealed that ROS induces up-regulation of CHOP and therefore the DR5 induction (21–23). Therefore, we next determined whether ROS regulates CT-induced expression of CHOP and TRAIL receptor. As shown in Fig. 5C, pretreatment of A375 cells with the antioxidant NAC reduced the CT-induced CHOP and DR5 up-regulation. Under these conditions, NAC also suppressed the enhancement effect of CT on TRAIL-induced cell death (Fig. 5D) and PARP/caspase-8 cleavage (Fig. 5E). To further confirm the critical role of H2O2 and O2⨪ in CT-induced DR5 promoter activity, we performed DR5 promoter studies with the overexpression of either empty, catalase, or SOD1 expression vectors. As shown in Fig. 5F, the CT-mediated DR5 promoter activity was significantly blocked by the overexpression of catalase or SOD1 in A375 cells, suggesting the essential role of H2O2 and O2⨪ induction in CT-mediated DR5 up-regulation. In addition to this, overexpression of catalase and/or SOD1 suppressed CT-induced DR5 protein up-regulation as determined by Western blotting (Fig. 5G). Taken together, these data clearly indicate that ROS generation by CT is critical for the up-regulation of CHOP and DR5 and contributing to CT-stimulated TRAIL-induced cell death and apoptosis.
FIGURE 5.
ROS mediates CT-induced DR5 and CHOP up-regulation. A (top panel), A375 cells were loaded with DCF (H2O2 probe) and further stimulated with 20 μm CT with or without pretreatment of 5 mm NAC for 1 h. Fluorescence was measured using a fluorescence microplate reader; bottom panel, A375 cells were loaded with DCF, and fluorescence was measured by flow cytometry. B, A375 cells were loaded with DHE (O2⨪ probe) and further stimulated with 20 μm CT. Fluorescence was measured using a fluorescence microplate reader. C, A375 cells were treated with 20 μm CT for 12 h with or without pretreatment of 5 mm NAC for 1 h. Western blotting analysis was performed using anti-DR5 and anti-CHOP antibodies. D, A375 cells were pretreated with vehicle or 5 mm NAC for 1 h, and then cells were further treated with 20 μm CT for 12 h, washed with PBS, and then treated with TRAIL (25 ng/ml) for additional 24 h. Cell viability was assessed by the MTT assay. *, p < 0.05. E, A375 cells were pretreated with vehicle or 5 mm NAC for 1 h, and then cells were further treated with 20 μm CT for 12 h and then incubated with 25 ng/ml TRAIL for 3 h. Then, Western blotting analysis was performed using anti-PARP and anti-caspase-8 antibodies. F, A375 cells were transfected with pDR5-310 together with either empty, catalase, or SOD1 expression vectors and then treated with 20 μm CT. After 12 h, cells were lysed and assayed for luciferase activity. *, p < 0.05 compared with CT-treated empty vector transfected cells. G, A375 cells were transfected with either empty, catalase, or SOD1 expression vectors for 48 h and then treated with 20 μm CT. The expression levels of DR5, catalase, and SOD1 were detected by Western blotting.
Cryptotanshinone-induced Up-regulation of DR5 Is p53-independent
Recent studies have revealed that p53 regulates DR5 expression and have identified the p53-responsive element as being in the first intron region of DR5 (24, 25). To determine whether p53 is involved in CT-induced up-regulation of DR5, we treated p53 wild-type and p53 knock-out HCT-116 cells with CT and tested their DR5 expression with Western blot analysis. As shown in Fig. 6A, CT was able to induce DR5 expression in both p53 wild-type and knock-out cells. These results indicate that CT-induced up-regulation of DR5 is independent of p53.
FIGURE 6.
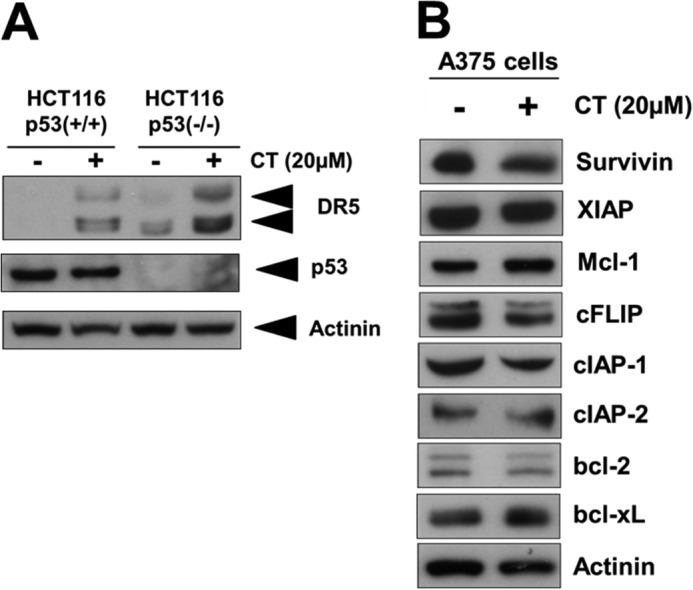
Cryptotanshinone-induced up-regulation of DR5 up-regulation is p53-independent. A, Western blotting of DR5 and p53 in wild-type and p53 knock-out HCT116 cells treated with 20 μm CT for 12 h. B, A375 cells were treated with 20 μm CT for 12 h. Whole-cell extracts were prepared and analyzed by Western blotting with the indicated antibodies.
A number of anti-apoptotic proteins such as survivin, X-linked inhibitor of apoptosis protein (XIAP), Mcl-1, cIAP-1, cIAP-2, cFLIP, Bcl-2, Bcl-xL have been shown to be responsible for the TRAIL resistance (4). Therefore, we examined whether CT sensitized TRAIL-induced cell death through modulation of these cell survival proteins. We found that CT had no apparent effect on the expression of anti-apoptotic proteins being tested in A375 cells (Fig. 6B).
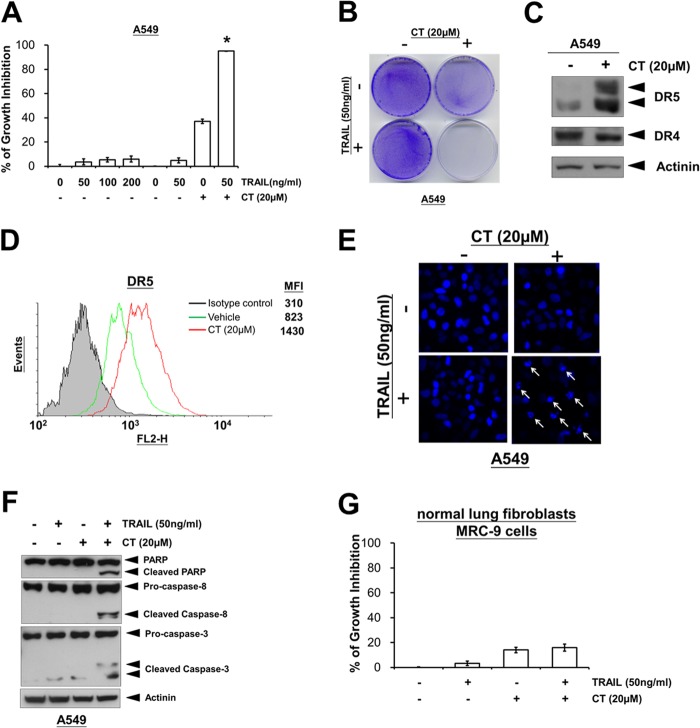
Cryptotanshinone Sensitizes TRAIL-resistant Cancer Cells
It has been shown that some cancer cells such as lung A549 cancer cells are completely resistant to TRAIL (26, 27). We therefore investigated whether CT could also sensitize A549 cells to TRAIL-induced apoptosis. We found that A549 cells were completely resistant to TRAIL treatment alone (Fig. 7A). However, pretreatment with CT significantly enhanced TRAIL-induced cell death, as determined by MTT (Fig. 7A) and crystal violet staining analysis (Fig. 7B). To determine how CT sensitizes A549 cells to TRAIL-induced cell death, we determined its effect on TRAIL receptors DR4 and DR5. We found that CT induced up-regulation of DR5 but not DR4 (Fig. 7C). Results of flow cytometry analysis also revealed the increase in DR5 cell surface expression in A549 cells (Fig. 7D). Furthermore, we found that pretreatment of cells with CT resulted in a markedly increased accumulation of apoptotic nuclei (Fig. 7E) and PARP, caspase-8, and caspase-3 cleavages (Fig. 7F) under TRAIL treatment. Interestingly, no enhancement effect was observed in normal lung MRC-9 fibroblast cells (Fig. 7G). Overall, we found that CT can also enhance TRAIL-induced apoptosis in TRAIL-resistant lung cancer cells associated with the up-regulation of DR5 expression.
FIGURE 7.
Cryptotanshinone sensitizes TRAIL-resistant lung cancer cells. A, TRAIL-resistant A549 lung cancer cells were treated as indicated with TRAIL alone or pretreated with 20 μm CT for 12 h, washed with PBS, and then treated with TRAIL for an additional 24 h. Cell viability was assessed by the MTT assay. *, p < 0.05 versus control, CT, and TRAIL. B, crystal violet staining of A549 cells treated with CT and TRAIL as described under “Experimental Procedures.” C, A549 cells were treated with 20 μm CT for 12 h. Whole-cell extracts were analyzed for expression of DR5 and DR4 by Western blotting. D, the cell surface expression levels of DR5 in A549 cells treated with 20 μm CT for 12 h were measured by flow cytometry analysis using phycoerythrin (PE)-conjugated DR5 and isotype control antibodies. MFI, mean fluorescence intensity. E, A549 cells were treated as described above. Cells were fixed and stained with DAPI. Nuclei morphological change was visualized using fluorescence microscopy (×400). F, Western blotting of lysates from A549 cells pretreated with 20 μm CT for 12 h followed by treatment with TRAIL for 3 h using the indicated antibodies. G, normal lung MRC-9 fibroblast cells were treated as indicated with TRAIL alone or pretreated with 20 μm CT for 12 h, washed with PBS, and then treated with TRAIL for an additional 24 h. Cell viability was assessed by the MTT assay.
DISCUSSION
TRAIL is considered as a highly promising anticancer agent because it has remarkable specificity for inducing apoptosis in tumor cell lines but not in normal cells (1). However, a considerable number of cancer cell types are highly resistant to TRAIL treatment due to low expression levels of TRAIL receptors (4). Therefore, novel therapeutic strategies to restore TRAIL receptor levels are urgently needed to overcome TRAIL resistance cancer cells to TRAIL. In this study, we demonstrate for the first time that CT effectively sensitizes human melanoma cells to TRAIL-induced apoptosis through up-regulation of DR5 via ROS signaling pathways. CT-induced ROS mediates the expression of CHOP and further up-regulation of DR5 via the CHOP binding element in the DR5 promoter. We also found that CT can also enhance TRAIL-induced apoptosis in TRAIL-resistant lung cancer cells (summarized in Fig. 8).
FIGURE 8.
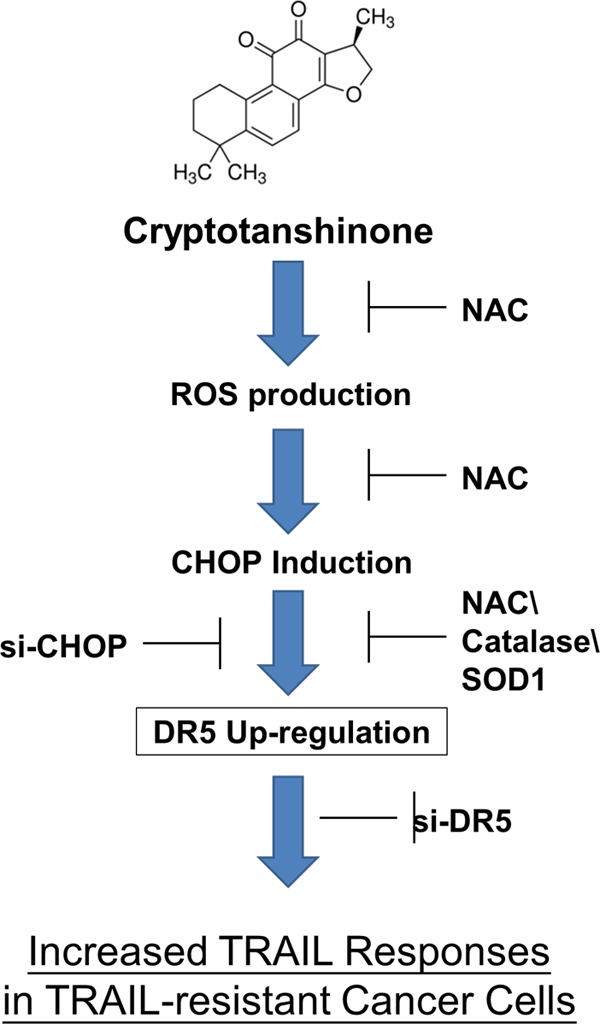
Schematic diagram of the mechanism by which cryptotanshinone potentiates TRAIL-induced apoptosis.
In the present study, we showed that cryptotanshinone, a component derived from Salvia miltiorrhiza (danshen), can enhance the apoptotic effects of TRAIL against human melanoma cells. Primary melanoma without any evidence of metastases is mostly treated by surgery. The alkylating agents dacarbazine and temozolomide are the first line therapy for metastatic melanoma; however, these treatments fail to help most patients (80–87%) (28, 29). Two therapeutic regimens, vemurafenib (targeting the BRAF V600E mutation) and combination therapy utilizing ipilimumab (targeting CTLA-4) and dacarbazine (DTIC), have improved overall survival compared with dacarbazine monotherapy (29); however, they are not suitable for many patients due to toxicity, lack of the BRAF V600E mutation and/or development of resistance. Therefore, other treatment strategies are still required (29). Melanoma cells were previously reported to be susceptible to different degrees of TRAIL-induced apoptosis (30, 31). However, only low levels of death receptors for TRAIL were observed in fresh melanoma isolates and melanoma tissue sections (31). Therefore, melanoma may not respond to TRAIL treatment unless increasing the expression of TRAIL death receptors by other agents. In this study, we found that CT is able to enhance TRAIL response in TRAIL-resistant melanoma cells by up-regulating DR5. Thus, the combination of TRAIL and CT may be an effective cancer therapy that warrants additional study in vivo.
When examined for the role of ROS in the induction of death receptors, we found that ROS plays a critical role in the expression of DR5 induced by CT. First, we showed that CT induced the production of ROS including H2O2 and O2⨪ (Fig. 5, A and B). Second, pretreatment of cells with the antioxidant NAC abolished the effect of CT on the induction of DR5 (Fig. 5C). Third, NAC treatment also abolished the enhancing effect of CT on cell death and apoptosis induced by TRAIL (Fig. 5, D and E). Taken together, this evidence indicates that ROS plays an essential role in the action of CT. Moreover, induction of ROS by CT treatment was involved in CHOP induction, which is critical for CT-mediated DR5 induction (Fig. 5C). CHOP is a typical ER stress-regulated protein that is involved in ER stress-induced apoptosis. Previous studies reported that CT-induced ER stress is involved in its apoptotic activities (15). Our data also suggest that CT can induce CHOP expression in melanoma cells. The detailed mechanism underlying CT-induced CHOP up-regulation via the ROS pathway during ER stress needs further investigation. Interestingly, overexpression of catalase gave a lesser suppressive function for DR5 promoter activity compared with SOD1 overexpression (Fig. 5F). This phenomenon can be explained by (i) overexpression of catalase alone induced slightly up-regulation of DR5 promoter activity (Fig. 5F) and (ii) superoxide generation may be more critical for CHOP/DR5 induction by CT. Further investigation is needed to clarify the individual role of superoxide and hydrogen peroxide in CT-induced ER stress and CHOP up-regulation.
In previous studies, it has been suggested that induction of DR5 can be mediated through the p53 intronic binding site in DR5 promoter (24, 25). However, we found that DR5 was induced by CT through a p53-independent mechanism, as determined using HCT116 p53 knock-out cells (Fig. 6A). Moreover, CT was able to induce DR5 expression in p53-mutated HT-29 and OVCAR3 cells (Fig. 3C) (32–34), further suggesting that p53 is not involved in CT-induced DR5 up-regulation. Interestingly, we found that CT induced higher expression of DR5 in HCT116 p53 knock-out cells (Fig. 6A). Previous studies have reported that therapeutic agents such as glucocorticoids, interferon-γ, UV irradiation, and TNF-α can induce p53-independent up-regulation of DR5 (35, 36), indicating that DR5 can be regulated in a cell type-specific and p53-independent manner. Further studies are needed to investigate the possible inhibitory role of p53 in CT-induced DR5 induction.
S. miltiorrhiza (danshen) has been employed for use as traditional Chinese medicine for cardiovascular and cerebrovascular disease. Compound danshen dripping pills, consisting of Radix salviae miltiorrhizae, Radix notoginseng, and Borneolum, is an herbal medicine recognized in the official Chinese Pharmacopoeia (37). Recently, compound danshen dripping pills has been approved by the U. S. Food and Drug Administration for clinical trials in the United States (www.clinicaltrials.gov), with four clinical trials on hypertension, coronary heart problems, and polycystic ovary disease are being conducted or have been completed. Given that cryptotanshinone, dihydrotanshinone I, tanshinone I, and tanshinone IIA are the main tanshinones contained in root of danshen (38) and all four tanshinones can increase DR5 levels (Fig. 1B), its worthy to test whether compound danshen dripping pills can up-regulate DR5 and enhance TRAIL-induced apoptosis in the future.
In conclusion, we show here for the first time that cryptotanshinone can effectively restore TRAIL sensitivity in cancer cells by up-regulation of DR5. Furthermore, ROS generated by CT play a critical role in CHOP-mediated DR5 up-regulation. Because the CT-related product is being tested clinically, animal studies are warranted to explore the combined effects of CT and TRAIL, with the goal of providing new, safe, and effective TRAIL treatment regimens for cancer.
This work was supported by Hong Kong Baptist University Grants HKBU 38-40-076, FRG1/11-12/053, FRG2/11-12/057, and FRG1/13-14/002 and the University Grants Committee of Hong Kong Grant HKBU 262512.
- TRAIL
- TNF-related apoptosis inducing ligand
- CT
- cryptotanshinone
- DR5
- death receptor 5
- ROS
- reactive oxygen species
- CHOP
- CAAT/enhancer-binding protein homologous protein
- DCF
- 6-carboxy-2′,7′-dichlorofluorescein diacetate
- NAC
- N-acetyl-l-cysteine
- ER
- endoplasmic reticulum
- MTT
- 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide
- DHE
- dihydroethidium
- SOD1
- superoxide dismutase 1
- PI
- propidium iodide.
REFERENCES
- 1. Pan G., Ni J., Wei Y. F., Yu G., Gentz R., Dixit V. M. (1997) An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science 277, 815–818 [DOI] [PubMed] [Google Scholar]
- 2. Pennarun B., Meijer A., de Vries E. G., Kleibeuker J. H., Kruyt F., de Jong S. (2010) Playing the DISC: turning on TRAIL death receptor-mediated apoptosis in cancer. Biochim. Biophys. Acta 1805, 123–140 [DOI] [PubMed] [Google Scholar]
- 3. Russo M., Mupo A., Spagnuolo C., Russo G. L. (2010) Exploring death receptor pathways as selective targets in cancer therapy. Biochem. Pharmacol. 80, 674–682 [DOI] [PubMed] [Google Scholar]
- 4. Dimberg L. Y., Anderson C. K., Camidge R., Behbakht K., Thorburn A., Ford H. L. (2013) On the TRAIL to successful cancer therapy? Predicting and counteracting resistance against TRAIL-based therapeutics. Oncogene 32, 1341–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mellier G., Pervaiz S. (2012) The three Rs along the TRAIL: resistance, re-sensitization and reactive oxygen species (ROS). Free Radic. Res. 46, 996–1003 [DOI] [PubMed] [Google Scholar]
- 6. Siegelin M. D. (2012) Utilization of the cellular stress response to sensitize cancer cells to TRAIL-mediated apoptosis. Expert Opin. Ther. Targets 16, 801–817 [DOI] [PubMed] [Google Scholar]
- 7. Zhou L., Zuo Z., Chow M. S. (2005) Danshen: an overview of its chemistry, pharmacology, pharmacokinetics, and clinical use. J. Clin. Pharmacol. 45, 1345–1359 [DOI] [PubMed] [Google Scholar]
- 8. Zhang Y., Jiang P., Ye M., Kim S. H., Jiang C., Lü J. (2012) Tanshinones: sources, pharmacokinetics and anti-cancer activities. Int. J. Mol. Sci. 13, 13621–13666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim J. H., Jeong S. J., Kwon T. R., Yun S. M., Jung J. H., Kim M., Lee H. J., Lee M. H., Ko S. G., Chen C. Y., Kim S. H. (2011) Cryptotanshinone enhances TNF-α-induced apoptosis in chronic myeloid leukemia KBM-5 cells. Apoptosis 16, 696–707 [DOI] [PubMed] [Google Scholar]
- 10. Su C. C. (2012) Tanshinone IIA potentiates the efficacy of 5-FU in Colo205 colon cancer cells in vivo through downregulation of P-gp and LC3-II. Exp. Ther. Med. 3, 555–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ye Y., Xu W., Zhong W., Li Y., Wang C. (2012) Combination treatment with dihydrotanshinone I and irradiation enhances apoptotic effects in human cervical cancer by HPV E6 down-regulation and caspases activation. Mol. Cell. Biochem. 363, 191–202 [DOI] [PubMed] [Google Scholar]
- 12. Lee W. Y., Liu K. W., Yeung J. H. (2009) Reactive oxygen species-mediated kinase activation by dihydrotanshinone in tanshinones-induced apoptosis in HepG2 cells. Cancer Lett. 285, 46–57 [DOI] [PubMed] [Google Scholar]
- 13. Cheng C. Y., Su C. C. (2010) Tanshinone IIA may inhibit the growth of small cell lung cancer H146 cells by up-regulating the Bax/Bcl-2 ratio and decreasing mitochondrial membrane potential. Mol. Med. Rep. 3, 645–650 [DOI] [PubMed] [Google Scholar]
- 14. Chiu T. L., Su CC. (2010) Tanshinone IIA induces apoptosis in human lung cancer A549 cells through the induction of reactive oxygen species and decreasing the mitochondrial membrane potential. Int. J. Mol. Med. 25, 231–236 [PubMed] [Google Scholar]
- 15. Park I. J., Kim M. J., Park O. J., Choe W., Kang I., Kim S. S., Ha J. (2012) Cryptotanshinone induces ER stress-mediated apoptosis in HepG2 and MCF7 cells. Apoptosis 17, 248–257 [DOI] [PubMed] [Google Scholar]
- 16. Tse A. K., Wan C. K., Shen X. L., Yang M., Fong W. F. (2005) Honokiol inhibits TNF-α-stimulated NF-κB activation and NF-κB-regulated gene expression through suppression of IKK activation. Biochem. Pharmacol. 70, 1443–1457 [DOI] [PubMed] [Google Scholar]
- 17. Yang Y., Tse A. K., Li P., Ma Q., Xiang S., Nicosia S. V., Seto E., Zhang X., Bai W. (2011) Inhibition of androgen receptor activity by histone deacetylase 4 through receptor SUMOylation. Oncogene 30, 2207–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tse A. K., Zhu G. Y., Wan C. K., Shen X. L., Yu Z. L., Fong W. F. (2010) 1α,25-Dihydroxyvitamin D3 inhibits transcriptional potential of nuclear factor κB in breast cancer cells. Mol. Immunol. 47, 1728–1738 [DOI] [PubMed] [Google Scholar]
- 19. Abdulghani J., El-Deiry W. S. (2010) TRAIL receptor signaling and therapeutics. Expert Opin. Ther. Targets 14, 1091–1108 [DOI] [PubMed] [Google Scholar]
- 20. Yamaguchi H., Wang H. G. (2004) CHOP is involved in endoplasmic reticulum stress-induced apoptosis by enhancing DR5 expression in human carcinoma cells. J. Biol. Chem. 279, 45495–45502 [DOI] [PubMed] [Google Scholar]
- 21. Kim Y. H., Jung E. M., Lee T. J., Kim S. H., Choi Y. H., Park J. W., Park J. W., Choi K. S., Kwon T. K. (2008) Rosiglitazone promotes tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by reactive oxygen species-mediated up-regulation of death receptor 5 and down-regulation of c-FLIP. Free Radic. Biol. Med. 44, 1055–1068 [DOI] [PubMed] [Google Scholar]
- 22. Yokouchi M., Hiramatsu N., Hayakawa K., Okamura M., Du S., Kasai A., Takano Y., Shitamura A., Shimada T., Yao J., Kitamura M. (2008) Involvement of selective reactive oxygen species upstream of proapoptotic branches of unfolded protein response. J. Biol. Chem. 283, 4252–4260 [DOI] [PubMed] [Google Scholar]
- 23. Moon D. O., Park S. Y., Choi Y. H., Ahn J. S., Kim G. Y. (2011) Guggulsterone sensitizes hepatoma cells to TRAIL-induced apoptosis through the induction of CHOP-dependent DR5: involvement of ROS-dependent ER-stress. Biochem. Pharmacol. 82, 1641–1650 [DOI] [PubMed] [Google Scholar]
- 24. Wu G. S., Burns T. F., McDonald E. R., 3rd., Jiang W., Meng R., Krantz I. D., Kao G., Gan D. D., Zhou J. Y., Muschel R., Hamilton S. R., Spinner N. B., Markowitz S., Wu G., el-Deiry W. S. (1997) KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat. Genet. 17, 141–143 [DOI] [PubMed] [Google Scholar]
- 25. Takimoto R., El-Deiry W. S. (2000) Wild-type p53 transactivates the KILLER/DR5 gene through an intronic sequence-specific DNA-binding site. Oncogene 19, 1735–1743 [DOI] [PubMed] [Google Scholar]
- 26. Seol J. Y., Park K. H., Hwang C. I., Park W. Y., Yoo C. G., Kim Y. W., Han S. K., Shim Y. S., Lee C. T. (2003) Adenovirus-TRAIL can overcome TRAIL resistance and induce a bystander effect. Cancer Gene Ther. 10, 540–548 [DOI] [PubMed] [Google Scholar]
- 27. Aydin C., Sanlioglu A. D., Bisgin A., Yoldas B., Dertsiz L., Karacay B., Griffith T. S., Sanlioglu S. (2010) NF-κB targeting by way of IKK inhibition sensitizes lung cancer cells to adenovirus delivery of TRAIL. BMC Cancer 10, 584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eigentler T. K., Caroli U. M., Radny P., Garbe C. (2003) Palliative therapy of disseminated malignant melanoma: a systematic review of 41 randomised clinical trials. Lancet Oncol. 4, 748–759 [DOI] [PubMed] [Google Scholar]
- 29. Julia F., Thomas L., Dalle S. (2012) New therapeutical strategies in the treatment of metastatic disease. Dermatol. Ther. 25, 452–457 [DOI] [PubMed] [Google Scholar]
- 30. Griffith T. S., Chin W. A., Jackson G. C., Lynch D. H., Kubin M. Z. (1998) Intracellular regulation of TRAIL-induced apoptosis in human melanoma cells. J. Immunol. 161, 2833–2840 [PubMed] [Google Scholar]
- 31. Nguyen T., Zhang X. D., Hersey P. (2001) Relative resistance of fresh isolates of melanoma to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis. Clin. Cancer Res. 7, 966s–973s [PubMed] [Google Scholar]
- 32. Yaginuma Y., Westphal H. (1992) Abnormal structure and expression of the p53 gene in human ovarian carcinoma cell lines. Cancer Res. 52, 4196–4199 [PubMed] [Google Scholar]
- 33. Kastrinakis W. V., Ramchurren N., Rieger K. M., Hess D. T., Loda M., Steele G., Summerhayes I. C. (1995) Increased incidence of p53 mutations is associated with hepatic metastasis in colorectal neoplastic progression. Oncogene 11, 647–652 [PubMed] [Google Scholar]
- 34. O'Connor P. M., Jackman J., Bae I., Myers T. G., Fan S., Mutoh M., Scudiero D. A., Monks A., Sausville E. A., Weinstein J. N., Friend S., Fornace A. J., Jr., Kohn K. W. (1997) Characterization of the p53 tumor suppressor pathway in cell lines of the National Cancer Institute anticancer drug screen and correlations with the growth-inhibitory potency of 123 anticancer agents. Cancer Res. 57, 4285–4300 [PubMed] [Google Scholar]
- 35. Meng R. D., El-Deiry W. S. (2001) p53-independent upregulation of KILLER/DR5 TRAIL receptor expression by glucocorticoids and interferon-gamma. Exp. Cell Res. 262, 154–169 [DOI] [PubMed] [Google Scholar]
- 36. Sheikh M. S., Burns T. F., Huang Y., Wu G. S., Amundson S., Brooks K. S., Fornace A. J., Jr., el-Deiry W. S. (1998) p53-dependent and -independent regulation of the death receptor KILLER/DR5 gene expression in response to genotoxic stress and tumor necrosis factor alpha. Cancer Res. 58, 1593–1598 [PubMed] [Google Scholar]
- 37. The State Pharmacopoeia Commission of China (2005) Chinese Pharmacopoeia, 528 [Google Scholar]
- 38. Liu A. H., Lin Y. H., Yang M., Sun J. H., Guo H., Guo D. A. (2006) High-performance liquid chromatographic determination of tanshinones in the roots of Salvia miltiorrhiza and related traditional Chinese medicinal preparations. J. Pharm. Pharm. Sci. 9, 1–9 [PubMed] [Google Scholar]