Background: We previously identified GAREM1 as a downstream adaptor of the EGF receptor.
Results: GAREM2 is a brain-specific GAREM subtype that is also tyrosine-phosphorylated and binds Grb2.
Conclusion: GAREM2 is a regulator of neurite outgrowth of neuroblastoma cells in the presence of IGF-1.
Significance: This study demonstrates the biological function of the GAREM family proteins, including their expression and subcellular localization.
Keywords: Adaptor Proteins, Epidermal Growth Factor Receptor (EGFR), ERK, MAP Kinases (MAPKs), Neurite Outgrowth, Nuclear Translocation
Abstract
Grb2-associated regulator of Erk/MAPK1 (GAREM) is an adaptor molecule in the EGF-mediated signaling pathway. GAREM is expressed ubiquitously in human organs and cultured cells. Two GAREM homologues are encoded by the human genome. Therefore, previously identified GAREM is named GAREM1. Here we characterized a new subtype of GAREM, GAREM2, that is specifically expressed in the mouse, rat, and human brain. Three GAREM2 tyrosines (Tyr-102, Tyr-429, and Tyr-551) are phosphorylated upon EGF stimulation and are necessary for binding to Grb2. Furthermore, GAREM2 and Shp2 regulate Erk activity in EGF-stimulated cells. These characteristics are similar to those of GAREM1. GAREM2 is expressed in some neuroblastoma cell lines and is also tyrosine-phosphorylated and bound to Grb2 after treatment with EGF. Eventually, GAREM2 regulates Erk activation in the presence of EGF or insulin like growth factor 1. GAREM2 also regulates insulin-like growth factor 1-induced neuronal differentiation of the SH-SY5Y neuroblastoma cell line. Although the structure and function of both GAREM subtypes are similar, GAREM1 is recruited into the nucleus and GAREM2 is not. Nuclear localization of GAREM1 might be controlled by a GAREM1-specific nuclear localization sequence and 14-3-3ϵ binding. The N-terminal 20 amino acids of GAREM1 make up its nuclear localization sequence that is also a 14-3-3ϵ binding site. The GAREM family is a new class of adaptor molecules with subtype-specific biological functions.
Introduction
Receptor tyrosine kinases mediate cell growth and differentiation and have been associated with various human diseases (1). The EGF receptor is linked to the generation and development of human cancer, and EGF antagonists have been used to develop effective cancer therapeutics (2, 3). The interactions between receptor tyrosine kinases and adaptor proteins are crucial for the transduction of intracellular growth signals from the plasma membrane to the nucleus (4, 5). Grb2-associated binder (Gab) and insulin receptor substrate (IRS)2 family proteins transduce physiological cell growth and differentiation signals and are candidate intracellular therapeutic targets for some diseases (5–7). These adaptor proteins contain functional regions such as SH2, SH3, pleckstrin homology, and phosphotyrosine binding domains that mediate interactions with the growth factor receptor, other adaptor proteins, and phospholipids at the plasma membrane (8). Complex formation of these molecules is regulated by tyrosine phosphorylation of the growth factor receptors or adaptor proteins after growth factor stimulation. Tyrosine phosphorylation triggers a conformational change or protein-protein interactions. The Gab and IRS families in mammals include subtypes with functional differences that depend on their expression in various organs (8–10). The Gab subtypes Gab1, Gab2, and Gab3 are widely expressed in various mammalian tissues. Their expression levels differ in lymphoid tissues, and individual Gab knockout mice yielded independent phenotypes (11–14), suggesting independent functions in the human body.
GAREM was originally identified as FLJ21610 (or FAM59A) in phosphoproteomics studies (15–19). We characterized it as an adaptor protein in the EGF signaling pathway. GAREM has two tyrosine phosphorylation sites (Tyr-105 and Tyr-453) that are activated by EGF stimulation (20). The amino acid sequence surrounding Tyr-453 is similar to the ITIM motif that is a consensus binding region for Shp2 (21). Shp2 is an Erk/MAPK-specific phosphatase. Thus, GAREM may regulate Erk activation in EGF-stimulated cells. Tumor cell malignancy is also regulated by GAREM expression (20). We have also identified a GAREM homologue (also named as GAREM-L(like) or FAM59B) encoded on several mammalian genomes. The first identified GAREM was renamed GAREM1, and the homologue is GAREM2. The human GAREM1 and GAREM2 genes are located on chromosomes 18q12.1 and 2p23.3. GAREM1 and GAREM2 are composed of 875 and 874 amino acid residues and share 62% sequence homology. We have shown that GAREM1 is expressed ubiquitously throughout human tissues and a wide variety of cultured cells (20). Therefore, GAREM1 may be a general mediator of growth factor signaling in various mammalian organs. In contrast, GAREM2 is specifically expressed in the mouse and rat brain and cultured neuronal cell lines. In this study, we compared GAREM1 and GAREM2 and identified their common and unique characteristics.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
COS-7, SH-SY5Y, HEK293T, and HeLa cells were maintained in DMEM supplemented with 10% FBS, 100 μg/ml streptomycin, and 100 units/ml penicillin. Plasmid transfection into COS-7 and 293T cells was carried out by electroporation by using a Gene-Pulser (Bio-Rad). Prior to EGF or IGF-1 stimulation, the cells were serum-starved for 16 h, and 100 ng/ml of EGF (Sigma) or 10 nm IGF-1 (Peprotech) dissolved in a serum-free medium was added.
cDNA Cloning and Vector Construction
GAREM2 cDNA of the Flexi ORF clone purchased from Promega was subcloned into pFLAG-CMV6a to be expressed as an N-terminal FLAG-tagged protein. Point mutations or internal deletions were introduced by using the QuikChange kit (Stratagene) according to the protocol of the manufacturer. The expression plasmid for myc-tagged Shp2 has been described previously (20). All nucleotide sequences were determined and verified using an ABI Prism dye terminator cycle sequencing kit (PerkinElmer Life Sciences) and an ABI Prism 310 genetic analyzer.
Antibodies
An anti-GAREM2 rabbit polyclonal antibody was raised against a GAREM2 fragment containing residues 535–679 that was bacterially produced as a GST fusion protein by using the pGEX4T vector (GE Healthcare). This antibody was purified by using HiTrap N-hydroxysuccinimide-activated Sepharose columns (GE Healthcare) coupled with an immunizing antigen. An anti-EGF receptor rabbit polyclonal antibody has been described previously (20). The following other antibodies were obtained commercially from various companies: anti-FLAG M2 and anti-β-actin (Sigma), anti-phosphotyrosine (catalog no. 4G10, Upstate Biotech Inc.), anti-Grb2 and anti-cytochrome c, (Santa Cruz Biotechnology), anti-Erk1/2 and anti-phospho-Erk1/2 (Cell Signaling Technology), anti-Lamin B and anti-Hsp90 (BD Transduction Laboratories), and anti-myc (catalog no. 9E10, Roche).
Semi-quantitative RT-PCR
To assess the relative expression levels of the GAREM family transcripts, RT-PCR was performed on each mouse tissue or human tissue cDNA (human tissue MTC panel, Clontech) by using the following primers: 5′-AAGCCCCACCCTGTCTTACT-3′ and 5′-GGACTTCCAAATGGGGACTT-3′ for mouse GAREM1, 5′-ACATCCTGCTGATCCACTCC-3′ and 5′-ATCACGAAGATTCGGTCAGG-3′ for mouse GAREM2, and 5′-TTCCCCAAGCTGCAGCCGGTA-3′ and 5′-GAATTCGGGAGCAGCTGAATAGGCCTG-3′ for human GAREM2. Total RNA was prepared by using an RNeasy kit (Qiagen). cDNA was synthesized from total RNA by using ReverTra-Ace (TOYOBO).
siRNA
Synthetic siRNA duplexes were used for the knockdown of GAREM2 expression in SH-SY5Y cells. The targeted human GAREM2 sequences were 5′-CTGTGTCTGTGAGATGCCTTT-3′ (GAREM-siRNA1) and 5′-GGATCTATGTCGAGACTGATT-3′ (GARM-siRNA2), both of which are derived from the 3′ non-coding region of human GAREM2. The siRNA for GFP was used as a control.
Immunoprecipitation and Immunoblot Analysis
The following procedures were carried out at 0–4 °C. The transfected cells were lysed in a lysis buffer containing 20 mm Tris-HCl (pH 7.5), 1 mm EDTA, 10 mm DTT, 1% Triton X-100, 150 mm NaCl, 10 mm NaF, 1 mm Na3VO4, and a complete protease inhibitor mixture (Roche) to produce a total cell lysate. For the immunoprecipitation experiments, the total cell lysate was centrifuged, and the supernatant was incubated for 2 h with either the primary antibody or an anti-FLAG affinity gel (Sigma). Protein G-Sepharose (GE Healthcare) was added, and the resulting mixture was rotated at 4 °C for 1 h. The beads were subsequently washed three times with the lysis buffer. The processed samples were performed as described previously (21).
Purification of FLAG-GAREM1
HEK293T cells (1 × 108 cells) transiently expressing FLAG-tagged GAREM1 were lysed with the lysis buffer. The lysate was centrifuged, and the supernatant was incubated with a FLAG affinity gel (bed volume, 50 μl) for 2 h. The gel was applied to an empty minicolumn (Bio-Rad) and washed several times with the lysis buffer. FLAG-GAREM1 was eluted with the FLAG peptide. All bands that were visualized by Coomassie Brilliant Blue staining were subjected to mass spectrometric analysis following in-gel digestion with trypsin. The in-gel digestion and mass spectrometric analysis were performed as described previously (21).
Analysis of Neurite Outgrowth
SH-SY5Y cells transfected with either control siRNA or GAREM2-siRNA1 were grown in DMEM supplemented with 10% FBS for 24 h and serum-starved for 16 h. Then, serum-starved cells were treated with or without 10 nm IGF-I for 48 h for neurite outgrowth experiments.
Cell Fractionation
The subcellular fractions of HeLa or SH-SY5Y cells were prepared by using an appropriate kit from Pierce Biotech, Inc.
Immunohistochemistry and Fluorescence Microscopy Analysis
Transfected and nontransfected cells were fixed with 5% formaldehyde in PBS or cold methanol for 10 min, washed with PBS, permeabilized with 0.1% Triton X-100 in PBS for 10 min, and washed with PBS once again. Following a blocking step with 3% bovine serum albumin in PBS for 30 min, the primary antibodies (anti-FLAG, anti-GAREM1, and anti-GAREM2) were applied for 1 h. After washing with PBS, the cells were incubated with the appropriate secondary antibodies conjugated with Alexa fluorescent dyes (Molecular Probes) for 45 min. If necessary, the cells were treated with rhodamine phalloidin (Molecular Probes), and the nuclei were simultaneously stained with 2 μm Hoechst 33342 (Molecular Probes). Finally, the cells were rinsed three times with PBS and mounted onto microscope slides with ProLong antifade reagents (Molecular Probes).
Adult Wistar Rats (Charles River Laboratories) were deep-anesthetized with halothane and perfusion-fixed with 4% paraformaldehyde in 0.1 m phosphate buffer (pH 7.4). Each brain was removed and post-fixed in the same fixative overnight at 4 °C, followed by cryoprotection with 30% sucrose solution. We prepared 14-μm-thick sections using a cryostat, thaw-mounted them on MAS-coated glass slides (Matsunami), and stored them at −80 °C until use (22). Each section was air-dried and washed with PBS (pH 7.4). For fluorescent immunostaining, each section was immunostained with anti-FAM59B rabbit polyclonal antibody (1:400), followed by Alexa Fluor 488-conjugated anti-rabbit IgG antibodies (1:1000). Fluorescent images were obtained using a confocal microscope (LSM510META, Carl Zeiss) and a fluorescent microscope (BZ-9000, Keyence). Neurite images were captured using a BX51 fluorescence microscope (Olympus) and ORCA-ER (Hamamatsu photonics). The figures were prepared using Adobe Photoshop.
RESULTS
Comparison of GAREM1 and GAREM2
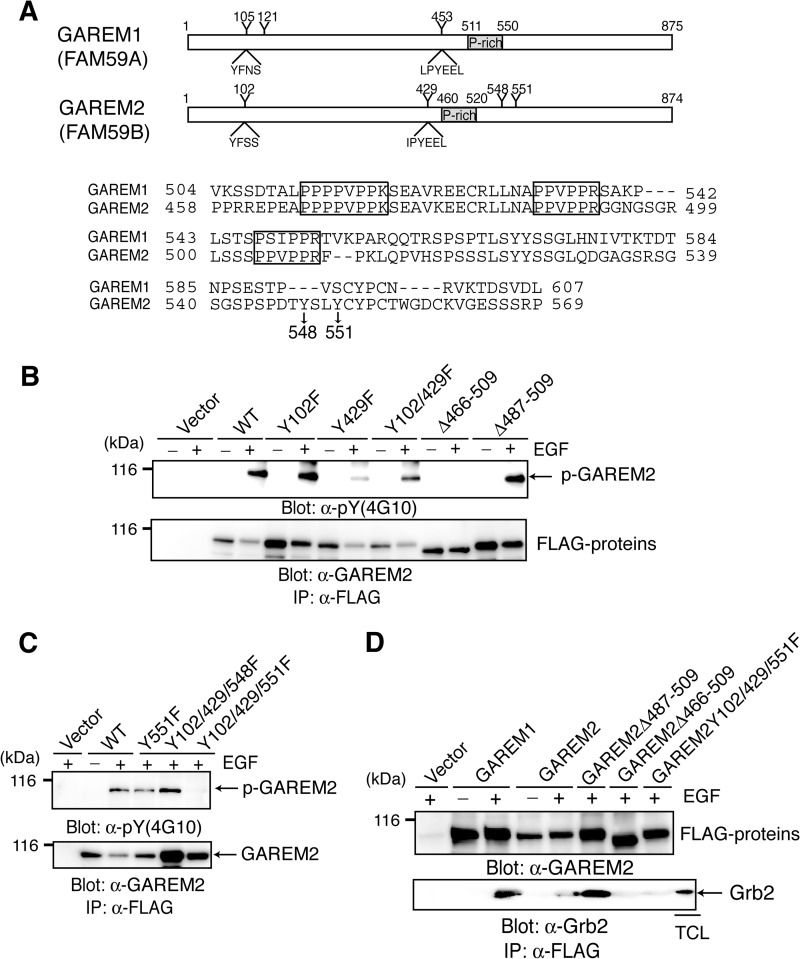
GAREM1 Tyr-105 and Tyr-453 were phosphorylated in EGF-stimulated cells (20). Alignment of the GAREM1 and GAREM2 phosphorylation sites suggested that Tyr-102 and Tyr-429 are the GAREM2 phosphorylation sites (Fig. 1A). However, substitution of these residues with Phe did not eliminate phosphorylation in EGF-stimulated COS-7 cells (Fig. 1B), indicating that GAREM2 contains a unique tyrosine phosphorylation site. Focusing on the position of tyrosine residues of GAREM1 and GAREM2 that are not conserved with each other, we estimated two tyrosine residues (Tyr-548 and Tyr-551) as candidates for the additional phosphorylation site in GAREM2 (Fig. 1A). A mutation analysis revealed that Tyr-551 is the tyrosine phosphorylation site of GAREM2 in addition to Tyr-102 and Tyr-429. It is not conserved in GAREM1 (Fig. 1C). Furthermore, the proline-rich motif in GAREM2 is necessary for Grb2 binding (Fig. 1D). As shown in Fig. 1B, tyrosine phosphorylation was abolished in a GAREM2 mutant in which the proline-rich region (amino acids 466–509) is deleted. This region is the Grb2-binding domain and is indispensable for tyrosine phosphorylation of GAREM2. Therefore, GAREM2 may bind to Grb2 in EGF-stimulated cells as GAREM1 does, but it is dependent on the N-terminal proline-rich motif (amino acids 466–474) and phosphorylation of three tyrosine residues. GAREM family tyrosine phosphorylation and Grb2 binding are similar in EGF-stimulated cells.
FIGURE 1.
GAREM2, like GAREM1, is a Grb2-binding protein. A, schematics of the primary structure of GAREM1 and GAREM2 (upper panel). Representative tyrosine residues and the surrounding amino acid sequence in GAREM1 and GAREM2 are indicated. Numbers indicate amino acid residues. Also shown are amino acid sequences of the proline-rich region of GAREM1 and GAREM2 (lower panel). Proline-rich (P-rich) regions that may bind the SH3 domain are indicated in the box. Tyrosines 548 and 551 in GAREM2, which are not conserved in GAREM1, are indicated by arrows. B and C, the unique tyrosine phosphorylation site (Tyr-551) in GAREM2. COS7 cells were transfected with plasmids encoding the FLAG-tagged construct of GAREM2 derivatives in which tyrosine residues (Y) were substituted with phenylalanine (F) and deleted (Δ) in the proline-rich motifs. The number of substituted or deleted amino acid residues is indicated. Immunoprecipitation (IP) studies were performed with lysates from COS-7 cells transfected with the empty vector or indicated plasmids. Cells were treated with (+) or without (−) EGF stimulation for 10 min, and each FLAG-tagged molecule was immunoprecipitated with the corresponding anti-FLAG antibody. Immunoblotting was performed using anti-phosphotyrosine (top panel) and anti-FLAG antibodies (bottom panel). D, the proline-rich motifs in GAREM2 enable binding to Grb2. This binding is dependent on tyrosine phosphorylation of GAREM upon EGF stimulation. Coimmunoprecipitation studies were performed with the anti-FLAG antibody by using the lysates of COS-7 cells transfected with plasmids carrying FLAG-GAREM2 derivatives and treated with EGF. Immunoblotting was performed with anti-FLAG (top panel) or anti-Grb2 antibodies (bottom panel).
GAREM2 Is Specifically Expressed in the Brain
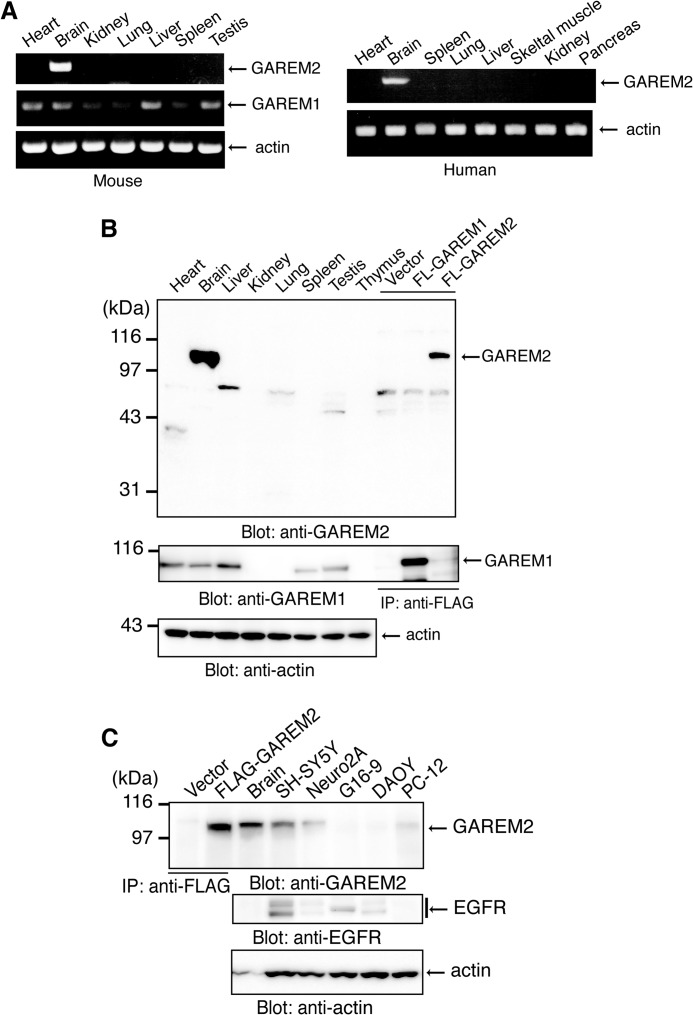
The GAREM1 transcript is observed in a wide variety of human tissues (20). RT-PCR showed a similar pattern for mouse GAREM1 in this study (Fig. 2A, right panel). In contrast, GAREM2 was mainly expressed in the mouse brain, and a faint band was observed in the lung (Fig. 2A, left panel). We developed a specific antibody to GAREM2 that does not react to GAREM1. We used this antibody to detect GAREM2 protein expression. Consistent with the RT-PCR results, GAREM2 protein expression was detected only in the mouse brain (Fig. 2B, upper panel). GAREM1 protein is expressed in various tissues (Fig. 2B, center panel).
FIGURE 2.
Brain-specific expression of mouse and human GAREM2. A, expression of GAREM2 mRNA in various mouse (left panel) and human (right panel) tissues. Shown is RT-PCR of GAREM1 (center row) and GAREM2 (upper rows) with β-actin as the loading control (bottom rows). B, protein expression of GAREM2 (top panel), GAREM1 (center panel), and β-actin (bottom panel) in various mouse tissues. The total cell lysate (20 μg) was separated by SDS-PAGE and analyzed by immunoblotting with each antibody. Immunoprecipitated (IP) FLAG-GAREM proteins as the positive control were prepared from lysates from COS-7 cells that were transfected with empty vector (Vector) or expression vectors for overexpressing the recombinant proteins of FLAG-GAREM1 (FL-GAREM1) and FLAG-GAREM2 (FL-GAREM2), with β-actin as the loading control (bottom panel). C, GAREM2 (top panel), EGFR (center panel), and β-actin (bottom panel) in neuronal cell lines or cultured cells derived from brain. Immunoprecipitated FLAG-GAREM proteins as the positive control were prepared from lysates from COS-7 cells that were transfected with empty vector or expression vectors for FLAG-GAREM1 (FL-GAREM1). Total cell lysates (20 μg of protein of cell lysate and 5 μg of protein of mouse brain lysate) were separated by SDS-PAGE and immunoblotted with each antibody with β-actin loading control (bottom panel). Neuro2A, mouse neuroblastoma cells; SH-SY5Y, human neuroblastoma cells; DAOY, human medulloblastoma cells; G16-9, human glioma cells.
To characterize the basic information of GAREM2, we screened neuronal cultured cells. GAREM2 was expressed in the neuroblastoma cell lines SH-SY5Y (human) and Neuro2A (mouse) and in the rat adrenal pheochromocytoma line PC12, which also differentiate neuronal cells (Fig. 2C).
Regulation of Erk Activity in Response to EGF Stimulation by GAREM2 Expression in SH-SY5Y Cells
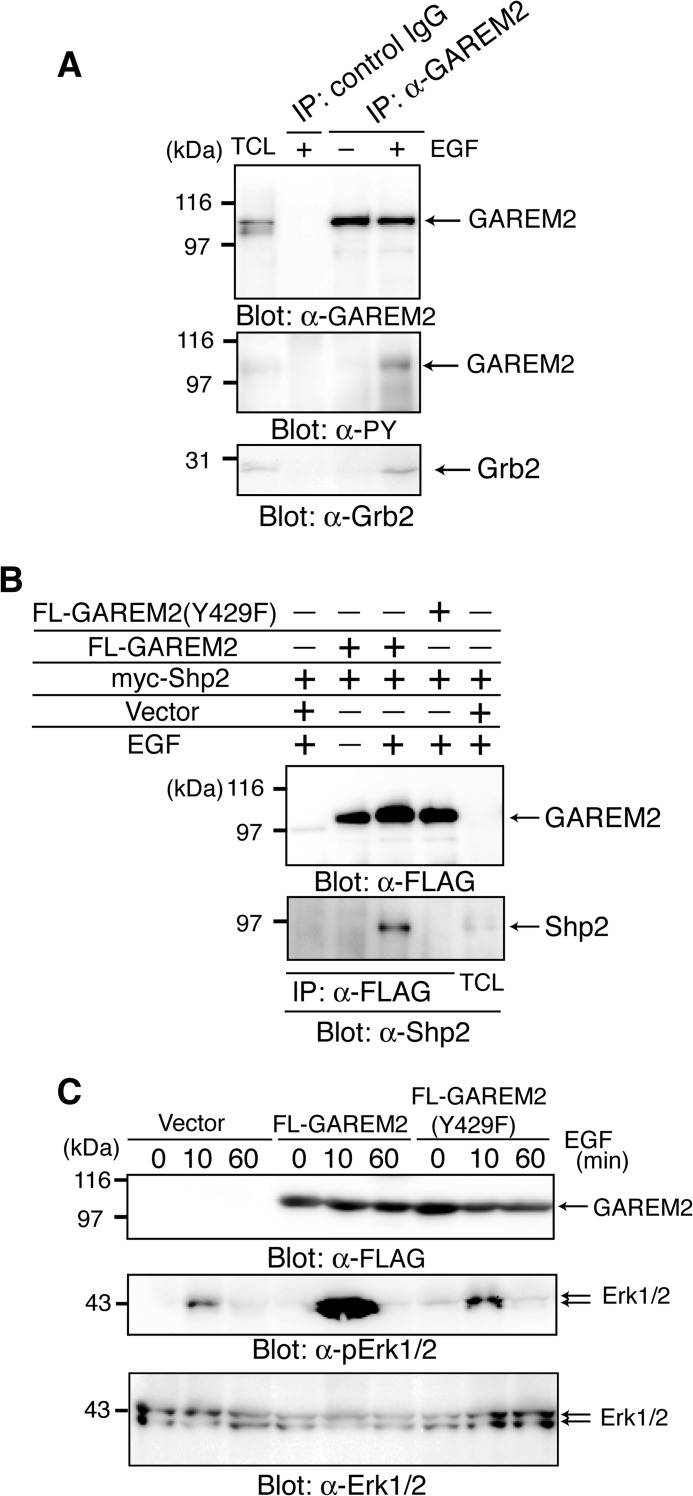
First, we confirmed the tyrosine phosphorylation of endogenous GAREM2 in SH-SY5Y cells. Immunoprecipitated GAREM2, with its specific antibody from EGF-stimulated SH-SY5Y cells, was tyrosine-phosphorylated, and immunoblotting was confirmed with Grb2 binding to GAREM2 in SH-SY5Y cells in response to EGF stimulation (Fig. 3A).
FIGURE 3.
Characterization of GAREM2 in SH-SY5Y cells. A, tyrosine phosphorylation of endogenous GAREM2 in EGF-stimulated SH-SY5Y cells, which associate with Grb2. Immunoprecipitation (IP) studies were performed with lysates from SH-SY5Y cells treated with (+) or without (−) EGF for 10 min, and GAREM2 was immunoprecipitated with anti-GAREM2 antibody. Immunoblotting was performed using anti-GAREM2 (top panel), anti-phosphotyrosine antibody (center panel), and anti-Grb2 (bottom panel). B, binding of Shp2 to GAREM2 upon EGF stimulation. COS-7 cells were transfected with a plasmid expressing myc-Shp2 and each molecule of FLAG-GAREM2 and FLAG-GAREM2(Y429F). Coimmunoprecipitation experiments and EGF stimulation were performed as described in Fig. 1. Immunoblotting was performed using anti-FLAG (top panel) and anti-Shp2 (bottom panel) antibodies. C, effect of GAREM2 expression on Erk activation in response to EGF stimulation in COS-7 cells. The amount of each protein was compared by immunoblotting with anti-GAREM2 (top panel), anti-phospho-Erk1/2 (center panel), and anti-Erk1/2 antibodies (bottom panel). Total cell lysates were prepared from COS-7 cells transfected with the empty vector (left three lanes) and the expression plasmid for FLAG-GAREM2 (center three lanes) and FLAG-GAREM2(Y429F) (right three lanes) and were stimulated with EGF for the indicated times. 20 μg of each lysate was run in each lane.
We demonstrated previously that GAREM1 up-regulates Erk activity by binding to Shp2 downstream of the EGF receptor (20). The association between FLAG-GAREM2 and myc-Shp2 is dependent on EGF stimulation (Fig. 3B), and Erk activation is enhanced after EGF stimulation in COS-7 cells that overexpress FLAG-GAREM2 (C). A point mutant of GAREM2 (Y429F) did not bind to Shp2 and did not induce the elevation of Erk activation. The region surrounding the phosphorylation site, Tyr-429:IPphosphoYEEL, is a good match for the consensus binding site of Shp1 and Shp2, (I/V/L)XphosphoYXX(I/V/L) (where X is any amino acid residue). These data suggest that GAREM1 and GAREM2 have a positive effect on Erk activation via Shp2 binding, which is dependent on the phosphorylation of Tyr-429 in GAREM2.
If overexpression of GAREM2 enhances Erk activation, knockdown of the protein should yield the opposite effect. We designed siRNA to knock down GAREM2 expression in SH-SY5Y cells (Fig. 4A). GAREM2 knockdown suppressed Erk activation in EGF- or IGF-I-stimulated SH-SY5Y cells (Fig. 4B). Therefore, GAREM2 regulates Erk activation after growth factor stimulation in SH-SY5Y cells.
FIGURE 4.
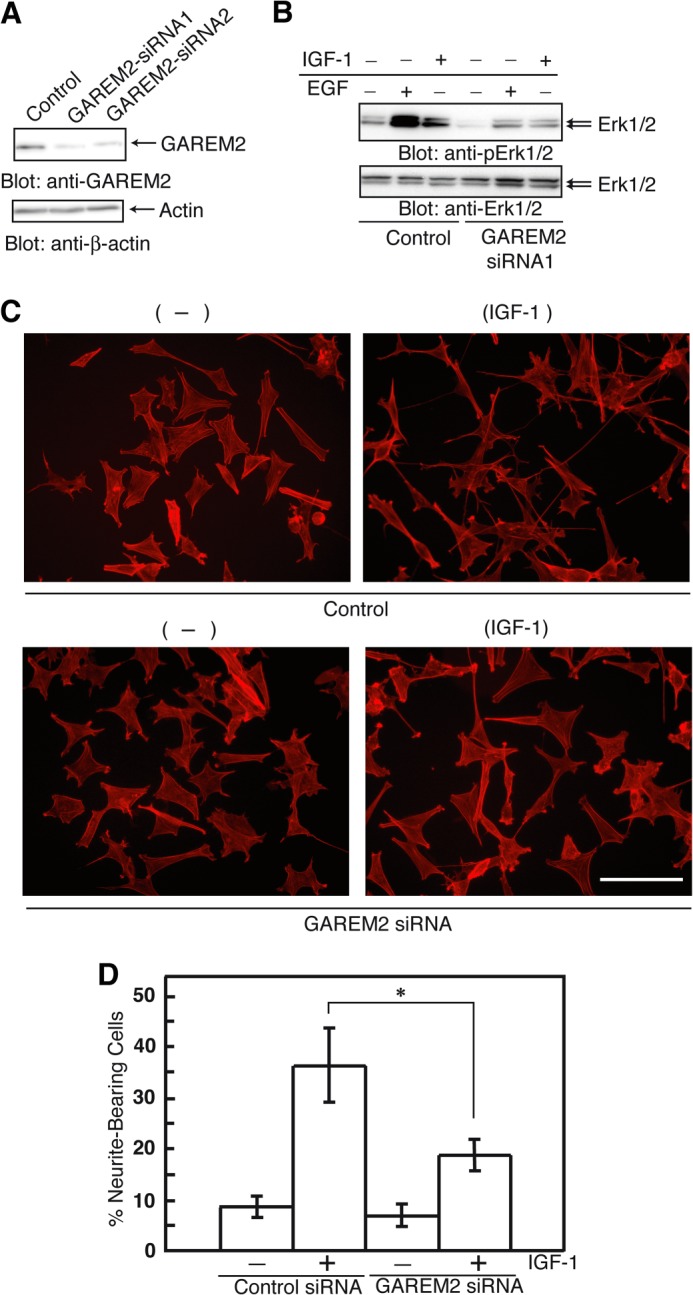
Effect of GAREM2 expression on neurite outgrowth in neuroblastoma cells. A, GAREM2 knockdown in SH-SY5Y cells. B, effect of siRNA knockdown of GAREM2 on Erk activation in response to EGF or IGF-1. The amounts of phospho-Erk1/2 (top panel) and Erk1/2 (center panel) were compared. Total cell lysates were prepared using control siRNA-transfected (left three lanes) and GAREM2-siRNA-transfected (right three lanes) SH-SY5Y cells stimulated with EGF or IGF-1. 20 μg of each lysate was applied in each lane. C, effects of GAREM knockdown on neurite outgrowth in IGF-1 stimulated SH-SY5Y cells. Control siRNAs (bottom panel) or GAREM2-siRNAs were transfected into SH-SY5Y cells for 12 h. Then, cells were treated with IGF-1 for 48 h and fixed and stained with actin with rhodamine-phalloidin. Scale bars = 50 μm. D, the percentage of neurite outgrowth cells. This value was calculated by measuring the neurite-bearing cells in C. Data were obtained from 300 randomly selected cells in a single experiment. Three independent sets of experiments were performed. Error bars show S.E. *, p < 0.05 compared with control siRNA-treated cells.
GAREM2 Expression and IGF-I induced Neurite Outgrowth of SH-SY5Y Cells
Various stimuli, including IGF-I treatment, trigger neurite outgrowth of SH-SY5Y cells (23–27). Activation of Erk with IGF-I stimulation is necessary for this phenomenon (28). To test whether GAREM2 expression is involved in the IGF-I-induced promotion of neurite outgrowth of SH-SY5Y cells, we knocked down GAREM2 with siRNA. Neurite outgrowth of SH-SY5Y cells after IGF-I treatment was clearly inhibited by the loss of GAREM2 expression (Fig. 4, C and D), suggesting that GAREM2 is an important adaptor for transduction of IGF-I signaling.
Subcellular Localization of GAREM1 and GAREM2
Although the protein structure and function of both GAREM subtypes are similar, biochemical studies revealed that the subcellular localization of each molecule was slightly different. GAREM1, but not GAREM2, localized to the nucleus (Fig. 5A). In quiescent COS-7 cells, endogenous GAREM1 is expressed in the cytosol, plasma membrane, and nucleus, as observed by immunofluorescent microscopic studies (Fig. 5B). Furthermore, GAREM1 and the EGF receptor colocalized as vesicular forms in EGF-stimulated cells (Fig. 5B, bottom panel). In SH-SY5Y cells, GAREM2 was present in the cytosol but not in the nucleus. GAREM2 expression in the rat brain cortex was confirmed by immunofluorescence staining to be localized outside of the nucleus (Fig. 5C). Therefore, the immunofluorescent microscopic data support the results of immunoblotting by using the samples of subcellular fractionation with each GAREM-specific antibody.
FIGURE 5.
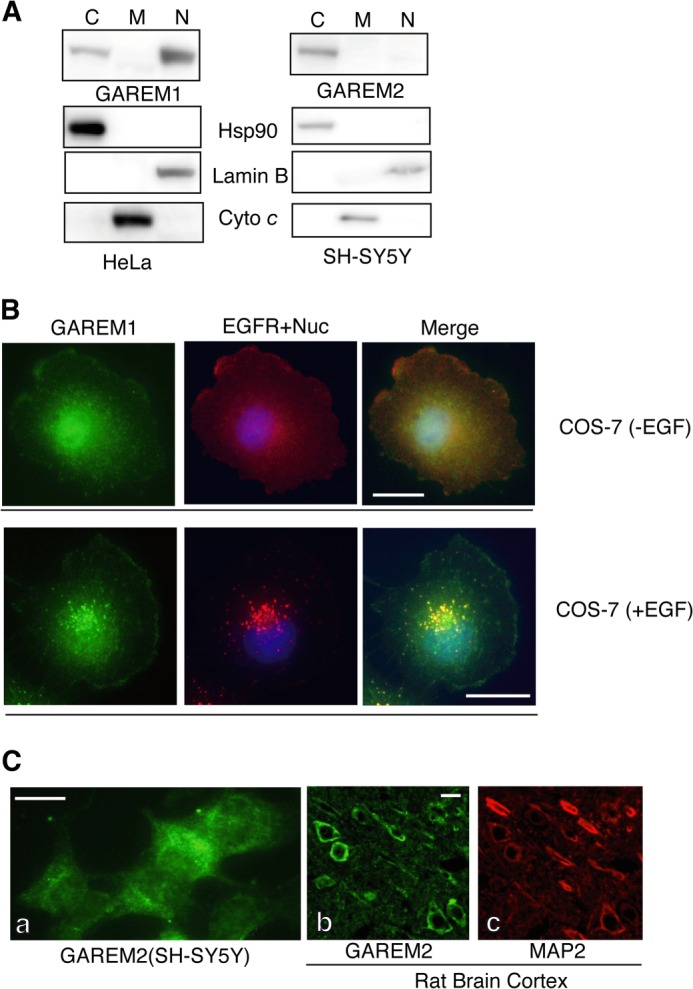
Subcellular localization of the GAREM family. A, biochemical analysis of the subcellular distribution of endogenous GAREM1 and GAREM2. Proteins from the cytosolic (C), nuclear (N), and mitochondrial (M) fractions of HeLa (GAREM1) and SH-SY5Y (GAREM2) cells (20 μg each) were used as indicated for immunoblotting. Cyto c, cytochrome c. B, subcellular localization of endogenous GAREM1 and GAREM2 was characterized by immunofluorescence staining. COS-7 cells with (top panel) or without (bottom panel) EGF stimulation were processed for immunofluorescence staining using anti-GAREM (green) and anti-EGF receptor (red) antibodies. The merged image is shown on the right. C, subcellular localization of GAREM2 (green) in SH-SY5Y cells (a). The rat brain cortex was characterized by immunofluorescence staining with anti-GAREM2 antibody (b). MAP2 (red) was used as a neuronal cell marker (c). Scale bars = 10 μm.
Identification of the GAREM1 Nuclear Localization Sequence (NLS)
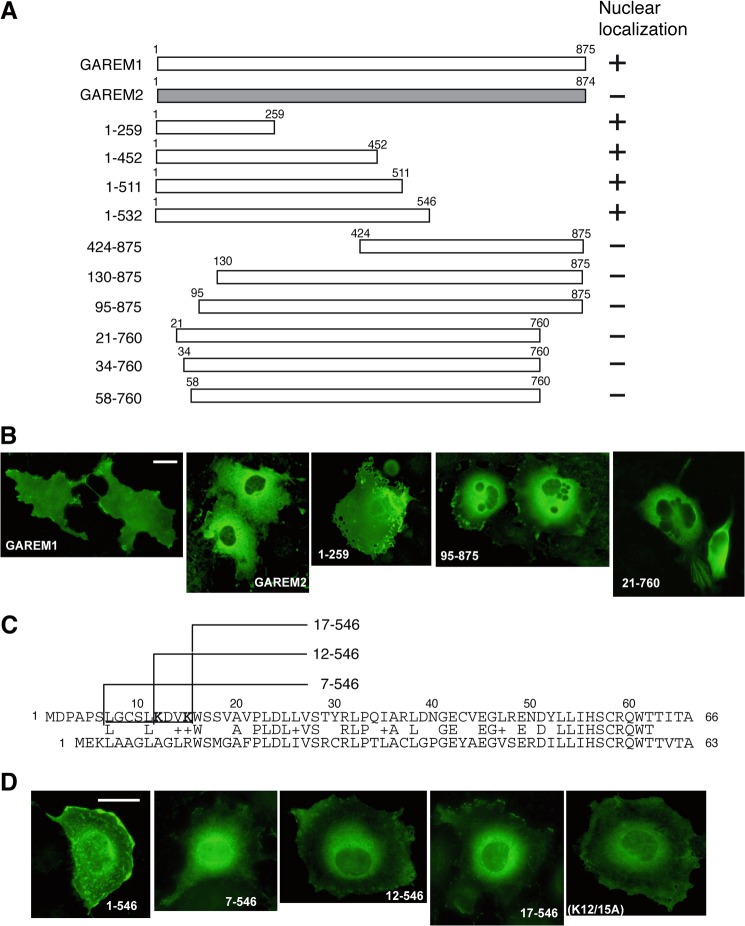
As with the endogenous GAREM family, subcellular localization of N-terminal FLAG-tagged GAREM1 and GAREM2 in COS-7 cells clearly differed. GAREM2 localized to the cytosol, and GAREM1 localized to the cytosol and nucleus (Fig. 6B, two left panels). To identify the GAREM1 NLS, several N-terminal FLAG-tagged deletion mutants were constructed and expressed in COS-7 cells (Fig. 6A). As shown in Fig. 6B, the N-terminal 20 amino acids were important for nuclear localization of GAREM1. Comparing the 60 N-terminal amino acids of both GAREMs reveals a poor similarity, especially in the most N-terminal 20 amino acids (Fig. 6C). Additional truncation mutants revealed that residues 7–17 might be necessary for GAREM1 nuclear localization (Fig. 6D, four left panels). In this region, we focused on two lysine residues (Lys-12 and Lys-15) because typical NLSs such as c-myc and NF-κB have two Lys residues (31, 32). A double Ala substitution partially inhibited nuclear localization (Fig. 6D, rightmost panel). These data suggest that the 10 N-terminal amino acids, including two lysines, might be the NLS of GAREM1. GAREM2 does not possess this motif and is not translocated into the nucleus.
FIGURE 6.
The nuclear localization sequence of GAREM1. A, schematics of the full-length and deletion mutant constructs of GAREM1 and full-length GAREM2. The numbers indicate amino acid residues. B, all recombinant proteins were expressed as the N-terminal FLAG-tagged form. Each construct was transfected into COS-7 cells, and then we analyzed their localization by immunofluorescence staining using anti-FLAG antibody. Representative results are shown. The result of nuclear localization of GAREM derivatives are indicated as (+) or (−) at the right side of each schematic in A. Scale bars = 10 μm. C, amino acid sequences of the N-terminal region of GAREM1 (top) and GAREM2 (bottom). The numbers indicate amino acid residues. Lys-12 and Lys-15 in the putative nuclear localization sequence of GAREM1 (underlined) are shown in bold. D, all recombinant proteins were expressed as the N-terminal FLAG-tagged form. The results of the immunofluorescence staining using anti-FLAG antibody are indicated. The numbers indicate amino acid residues. K12/15A, double point mutant of full-length GAREM1 with Lys-12 and Lys-15 replaced with Ala.
DISCUSSION
GAREM family proteins are categorized as adaptor proteins for the transduction of growth factor receptor signaling. Like the Gab and IRS families that have many important biological roles (8–10), there are subtypes in the GAREM family that give them a wide variety of functions. We have shown that GAREM1 and GAREM2 are structurally and functionally similar. Although their tyrosine phosphorylation sites differ slightly, the binding of Grb2 and Shp2 to GAREMs may be regulated by phosphorylation induced by growth factor stimulation. The role of EGF-stimulated Tyr-551 phosphorylation in GAREM2 is not fully understood. It is possible that Tyr-551 phosphorylation is necessary for Grb2 binding. It is also possible that this phosphorylation site is a target of other GAREM2-binding proteins in neuronal cells because GAREM2 is specifically expressed in the brain. The promoter regions of each mouse GAREM subtype are over 10 kb long, and their transcriptional regulation is not yet understood. Further study is needed to define the mechanism of brain-specific GAREM2 expression and the GAREM2-specific binding partners in neuronal cells. GAREM2 is a key mediator of neuronal differentiation in IGF-1 stimulated SH-SY5Y cells and possibly functions through Erk regulation. The GAREM1 transcript is expressed in the mouse brain (Fig. 2) and in SH-SY5Y cells (data not shown). However, protein expression of GAREM1 was quite low and could not be detected by immunofluorescence staining of the rat brain cortex or immunoblotting of SH-SY5Y cell lysates. Therefore, the effect of GAREM2 siRNA on neuronal differentiation of SH-SY5Y was detected without GAREM1 knockdown.
Characterizing the differences in subcellular localization may be important for identifying the specific binding partner of each GAREM subtype. Nuclear localization of GAREM1 may depend on its N-terminal NLS, and we identified 14-3-3ϵ as a GAREM1-specific binding protein.3 The 14-3-3 family binds phosphoserine-containing domains in its target proteins and retains some phosphorylated transcription factors in the cytosol. After dephosphorylation, the transcription factors enter the nucleus, where they regulate gene expression (29, 30). The NLS of GAREM1 contains four serine residues that may be phosphorylated and serve as the binding motif for 14-3-3 binding. Although the precise phosphorylation site in this region has not been identified, 14-3-3ϵ that is bound to the N terminus of GAREM1 may mask its NLS and inhibit its entry into the nucleus. In contrast, GAREM2 does not contain an NLS and is mainly localized in the cytosol. It is unclear whether GAREM1 binds to a nuclear-specific partner or whether GAREM2 recognizes its cytosol-specific partner. Answering these questions may reveal the functional relevance of each GAREM subtype. Another interesting aspect of the differential subcellular localization of GAREM is its ability to colocalize with EGF receptors in EGF-stimulated cells. These data suggest that GAREM may enter the endosomal transport pathway and regulate the stability of the EGF receptor and GAREM itself. The EGF receptor family is also capable of nuclear translocation (5, 33–35). Thus, it is possible that GAREM1 is a nuclear ligand of the EGF receptor.
Recently, GAREM2 (FAM59B) was identified as an Intersectin-binding protein (36). A cDNA clone containing the proline-rich region of GAREM2 was derived from a human brain cDNA library by yeast two-hybrid screening with the most N-terminal SH3 domain of Intersectin-1 as bait. The Intersectin family is a scaffold protein containing multiple modular domains such as the Eps15 homology, coiled-coil, SH, Dbl homology, and pleckstrin homology domains. Numerous Intersectin-binding proteins have been identified and linked to various biological functions, such as neurodegeneration via the JNK/MAPK pathway (37, 38). The GAREM2-Intersectin 1 complex may mediate neuronal function in the brain.
In contrast, GAREM1 is involved in the interactome of the scaffold protein Shc1 (39), and the tyrosine phosphorylation of GAREM1 was confirmed as being tightly involved in the ErbB family signaling cascade (40). Recent data suggest that the GAREM family may participate in the various signaling pathways as an important downstream molecule of receptor tyrosine kinases.
In conclusion, we identified GAREM2, a new GAREM subtype that may regulate Erk activity in growth factor-stimulated neuronal cells. Diversity of expression of GAREM1 and GAREM2 in each organ and their subcellular localization lead to functional differences in mammals. To analyze the physiological function of GAREM1 and GAREM2, we are developing knockout mice.
This work was supported by grants from the Satake Technical Foundation.
H. Konishi, unpublished observations.
- IRS
- insulin receptor substrate
- IGF
- insulin-like growth factor
- NLS
- nuclear localization sequence
- SH
- Src homology.
REFERENCES
- 1. Lemmon M. A., Schlessinger J. (2010) Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Citri A., Yarden Y. (2006) EGF-ERBB signalling. Towards the systems level. Nat. Rev. Mol. Cell Biol. 7, 505–516 [DOI] [PubMed] [Google Scholar]
- 3. Riese D. J., 2nd, Gallo R. M., Settleman J. (2007) Mutational activation of ErbB family receptor tyrosine kinases. Insights into mechanisms of signal transduction and tumorigenesis. BioEssays 29, 558–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kolch W. (2005) Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 6, 827–837 [DOI] [PubMed] [Google Scholar]
- 5. Gibson S. L., Ma Z., Shaw L. M. (2007) Divergent roles for IRS-1 and IRS-2 in breast cancer metastasis. Cell Cycle 6, 631–637 [DOI] [PubMed] [Google Scholar]
- 6. Shaw L. M. (2011) The insulin receptor substrate (IRS) proteins at the intersection of metabolism and cancer. Cell Cycle 10, 1750–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Verma S., Vaughan T., Bunting K. D. (2012) Gab adapter proteins as therapeutic targets for hematologic disease. Adv. Hematol. 2012, 380635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu Y., Rohrschneider L. R. (2002) The gift of Gab. FEBS Lett. 515, 1–7 [DOI] [PubMed] [Google Scholar]
- 9. White M. F. (2002) IRS proteins and the common path to diabetes. Am. J. Physiol. Endocrinol. Metab. 283, E413–E422 [DOI] [PubMed] [Google Scholar]
- 10. Gu H., Neel B. G. (2003) The “Gab” in signal transduction. Trends Cell Biol. 13, 122–130 [DOI] [PubMed] [Google Scholar]
- 11. Itoh M., Yoshida Y., Nishida K., Narimatsu M., Hibi M., Hirano T. (2000) Role of Gabl in heart, placenta, and skin development and growth factor and cytokine-induced extracellular signal-regulated kinase mitogen-activated protein kinase activation. Mol. Cell Biol. 20, 3695–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sachs M., Brohmann H., Zechner D., Müller T., Hülsken J., Walther I., Schaeper U., Birchmeier C., Birchmeier W. (2000) Essential role of Gab1 for signaling by the c-Met receptor in vivo. J. Cell Biol. 150, 1375–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nishida K., Wang L., Morii E., Park S. J., Narimatsu M., Itoh S., Yamasaki S., Fujishima M., Ishihara K., Hibi M., Kitamura Y., Hirano T. (2002) Requirement of Gab2 for mast cell development and KitL/c-Kit signaling. Blood 99, 1866–1869 [DOI] [PubMed] [Google Scholar]
- 14. Seiffert M., Custodio J. M., Wolf I., Harkey M., Liu Y., Blattman J. N., Greenberg P. D., Rohrschneider L. R. (2003) Gab3-deficient mice exhibit normal development and hematopoiesis and are immunocompetent. Mol. Cell Biol. 23, 2415–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blagoev B., Ong S. E., Kratchmarova I., Mann M. (2004) Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat. Biotechnol. 22, 1139–1145 [DOI] [PubMed] [Google Scholar]
- 16. Kratchmarova I., Blagoev B., Haack-Sorensen M., Kassem M., Mann M. (2005) Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science 308, 1472–1477 [DOI] [PubMed] [Google Scholar]
- 17. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 18. Rush J., Moritz A., Lee K. A., Guo A., Goss V. L., Spek E. J., Zhang H., Zha X. M., Polakiewicz R. D., Comb M. J. (2005) Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 23, 94–101 [DOI] [PubMed] [Google Scholar]
- 19. Thelemann A., Petti F., Griffin G., Iwata K., Hunt T., Settinari T., Fenyo D., Gibson N., Haley J. D. (2005) Phosphotyrosine signaling networks in epidermal growth factor receptor overexpressing squamous carcinoma cells. Mol. Cell Proteomics 4, 356–376 [DOI] [PubMed] [Google Scholar]
- 20. Tashiro K., Tsunematsu T., Okubo H., Ohta T., Sano E., Yamauchi E., Taniguchi H., Konishi H. (2009) GAREM, a novel adaptor protein for growth factor receptor-bound protein 2, contributes to cellular transformation through the activation of extracellular signal-regulated kinase signaling. J. Biol. Chem. 284, 20206–20214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Konishi H., Tashiro K., Murata Y., Nabeshi H., Yamauchi E., Taniguchi H. (2006) CFBP is a novel tyrosine-phosphorylated protein that might function as a regulator of CIN85/CD2AP. J. Biol. Chem. 281, 28919–28931 [DOI] [PubMed] [Google Scholar]
- 22. Dohi E., Tanaka S., Seki T., Miyagi T., Hide I., Takahashi T., Matsumoto M., Sakai N. (2012) Hypoxic stress activates chaperone-mediated autophagy and modulates neuronal cell survival. Neurochem. Int. 60, 431–442 [DOI] [PubMed] [Google Scholar]
- 23. Spinelli W., Sonnenfeld K. H., Ishii D. N. (1982) Effects of phorbol ester tumor promoters and nerve growth factor on neurite outgrowth in cultured human neuroblastoma cells. Cancer Res. 42, 5067–5073 [PubMed] [Google Scholar]
- 24. Recio-Pinto E., Ishii D. N. (1984) Effects of insulin, insulin-like growth factor-II and nerve growth factor on neurite outgrowth in cultured human neuroblastoma cells. Brain. Res. 302, 323–334 [DOI] [PubMed] [Google Scholar]
- 25. Jalava A., Heikkilä J., Lintunen M., Akerman K., Påhlman S. (1992) Staurosporine induces a neuronal phenotype in SH-SY5Y human neuroblastoma cells that resembles that induced by the phorbol ester 12-O-tetradecanoyl phorbol-13 acetate (TPA). FEBS Lett. 300, 114–118 [DOI] [PubMed] [Google Scholar]
- 26. Rogers M. V., Buensuceso C., Montague F., Mahadevan L. (1994) Vanadate stimulates differentiation and neurite outgrowth in rat pheochromocytoma PC12 cells and neurite extension in human neuroblastoma SH-SY5Y cells. Neuroscience 60, 479–494 [DOI] [PubMed] [Google Scholar]
- 27. Grynspan F., Griffin W. B., Mohan P. S., Shea T. B., Nixon R. A. (1997) Calpains and calpastatin in SH-SY5Y neuroblastoma cells during retinoic acid-induced differentiation and neurite outgrowth. Comparison with the human brain calpain system. J. Neurosci. Res. 48, 181–191 [PubMed] [Google Scholar]
- 28. Kim B., Leventhal P. S., Saltiel A. R., Feldman E. L. (1997) Insulin-like growth factor-I-mediated neurite outgrowth in vitro requires mitogen-activated protein kinase activation. J. Biol. Chem. 272, 21268–21273 [DOI] [PubMed] [Google Scholar]
- 29. Makkerh J. P., Dingwall C., Laskey R. A. (1996) Comparative mutagenesis of nuclear localization signals reveals the importance of neutral and acidic amino acids. Curr. Biol. 6, 1025–1027 [DOI] [PubMed] [Google Scholar]
- 30. Beg A. A., Ruben S. M., Scheinman R. I., Haskill S., Rosen C. A., Baldwin A. S., Jr. (1992) IκB interacts with the nuclear localization sequences of the subunits of NF-κB. A mechanism for cytoplasmic retention. Genes Dev. 6, 1899–1913 [DOI] [PubMed] [Google Scholar]
- 31. Morrison D. K. (2009) The 14-3-3 proteins. Integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 19, 16–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang X., Grammatikakis N., Siganou A., Calderwood S. K. (2003) Regulation of molecular chaperone gene transcription involves the serine phosphorylation, 14-3-3ϵ binding, and cytoplasmic sequestration of heat shock factor 1. Mol. Cell Biol. 23, 6013–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin S. Y., Makino K., Xia W., Matin A., Wen Y., Kwong K. Y., Bourguignon L., Hung M. C. (2001) Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 3, 802–808 [DOI] [PubMed] [Google Scholar]
- 34. Carpenter G. (2003) Nuclear localization and possible functions of receptor tyrosine kinases. Curr. Opin. Cell Biol. 15, 143–148 [DOI] [PubMed] [Google Scholar]
- 35. Wang S. C., Hung M. C. (2009) Nuclear translocation of the epidermal growth factor receptor family membrane tyrosine kinase receptors. Clin. Cancer Res. 15, 6484–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wong K. A., Wilson J., Russo A., Wang L., Okur M. N., Wang X., Martin N. P., Scappini E., Carnegie G. K., O'Bryan J. P. (2012) Intersectin (ITSN) family of scaffolds function as molecular hubs in protein interaction networks. PLoS ONE 7, e36023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. O'Bryan J. P. (2010) Intersecting pathways in cell biology. Sci. Signal 3, re10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsyba L., Nikolaienko O., Dergai O., Dergai M., Novokhatska O., Skrypkina I., Rynditch A. (2011) Intersectin multidomain adaptor proteins. Regulation of functional diversity. Gene 473, 67–75 [DOI] [PubMed] [Google Scholar]
- 39. Zheng Y., Zhang C., Croucher D. R., Soliman M. A., St-Denis N., Pasculescu A., Taylor L., Tate S. A., Hardy W. R., Colwill K., Dai A. Y., Bagshaw R., Dennis J. W., Gingras A. C., Daly R. J., Pawson T. (2013) Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature 499, 166–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tzouros M., Golling S., Avila D., Lamerz J., Berrera M., Ebeling M., Langen H., Augustin A. (2013) Development of a 5-plex SILAC method tuned for the quantitation of tyrosine phosphorylation dynamics. Mol. Cell Proteomics 10.1074/mcp.O113.027342 [DOI] [PMC free article] [PubMed] [Google Scholar]