Background: The carboxyl-terminal tail domain of the paramyxovirus nucleoprotein is considered instrumental for polymerase recruitment to the template.
Results: Truncated nucleoproteins reveal that the tail domain is dispensable for viral polymerase loading and activity.
Conclusion: The viral polymerase complex is capable of productively docking directly to the nucleocapsid core.
Significance: This finding alters the current paradigm of paramyxovirus polymerase recruitment.
Keywords: Animal Viruses, Microbiology, Viral Polymerase, Viral Replication, Virus
Abstract
Paramyxovirus genomes are ribonucleoprotein (RNP) complexes consisting of nucleoprotein (N)-encapsidated viral RNA. Measles virus (MeV) N features an amino-terminal RNA-binding core and a 125-residue tail domain, of which only the last 75 residues are considered fully mobile on the nucleocapsid surface. A molecular recognition element (MoRE) domain mediates binding of the viral phosphoprotein (P). This P N-tail interaction is considered instrumental for recruiting the polymerase complex to the template. We have engineered MeV N variants with tail truncations progressively eliminating the MoRE domain and upstream tail sections. Confirming previous reports, RNPs with N truncations lacking the carboxyl-terminal 43-residues harboring the MoRE domain cannot serve as polymerase template. Remarkably, further removal of all tail residues predicted to be surface-exposed significantly restores RNP bioactivity. Insertion of structurally dominant tags into the central N-tail section reduces bioactivity, but the negative regulatory effect of exposed N-tail stems is sequence-independent. Bioactive nucleocapsids lacking exposed N-tail sections are unable to sustain virus replication, because of weakened interaction of the advancing polymerase complex with the template. Deletion of the N-MoRE-binding domain in P abrogates polymerase recruitment to standard nucleocapsids, but polymerase activity is partially restored when N-tail truncated RNPs serve as template. Revising central elements of the current replication model, these data reveal that MeV polymerase is capable of productively docking directly to the nucleocapsid core. Dispensable for polymerase recruitment, N-MoRE binding to P-tail stabilizes the advancing polymerase-RNP complex and may rearrange unstructured central tail sections to facilitate polymerase access to the template.
Introduction
The order of nonsegmented negative strand RNA viruses comprises viral families of major clinical significance such as Ebola virus of the Filoviridae, rabies virus of the Rhabdoviridae, and MeV,2 mumps virus, and respiratory syncytial virus of the Paramyxoviridae. For all nonsegmented negative strand RNA virus, only assembled RNPs can serve as a template for transcription and replication, which are both mediated by the viral RNA-dependent RNA-polymerase (RdRp) complex (1). Despite the health impact of these pathogens, our current understanding of central aspects of RdRp recruitment onto the template and progression along the nucleocapsid remains limited.
Paramyxovirus RdRps are hetero-oligomers composed of the polymerase (L) protein and an essential co-factor, the P protein, which is required for nucleocapsid binding. Important functional insight has come from the MeV and related Sendai virus (SeV) systems (2–5). MeV RNPs assume a helical organization characteristic for the Paramyxoviridae and other nonsegmented negative strand RNA virus (1). The amino-terminal 400 residues of the viral N protein form the RNA-binding N-core, which determines the spatial organization of the nucleocapsid (6, 7). The carboxyl-terminal 125-residue N-tail domain is intrinsically disordered but considered essential for RNA transcription and replication (3, 7–10). In addition, the tail domain modulates RNP structure, because EM studies have shown that tail removal decreases diameter and pitch of the nucleocapsids, resulting in a rigid, rodlike organization (1, 7, 11, 12). Docking of respiratory syncytial virus nucleoprotein-RNA crystal structures (13) into EM density maps of MeV RNPs posited the beginning of the MeV N-tail domain at the interior of the RNP helix (7). In situ structural analysis of viral nucleocapsids then suggested that N-tails protrude through the interstitial spaces between adjacent RNP helical turns, freely exposing only the carboxyl-terminal half of the tail, approximately MeV N residues 450–525, on the surface of assembled RNPs (8). Supporting the validity of this respiratory syncytial virus-based MeV nucleocapsid model, removal of the interstitial tail residues should result in direct contact between adjacent RNP turns, rigidifying the helical structure as observed experimentally (7).
According to the current paradigm of paramyxovirus RNP replication, these exposed N-tail sections are thought to serve as essential anchor points for recruitment of the polymerase complex (6, 14, 15). In the case of MeV N, the MoRE domain (amino acids 488–499), which is located within a conserved box 2 region (amino acids 489–506), and flanking tail residues 486–502 assume an α-helical configuration when binding to the carboxyl-terminal X-domain of the P protein (6, 10, 16). Exposed tail residues 450–487 are thought to provide flexibility for the MoRE domain to recruit soluble polymerase complexes from the cytosol to the RNP through a casting mechanism (17) and allow close proximity of the MoRE-P-L complex with N-core (8). Once RdRp is loaded onto the template, the X domain interactions of tetrameric P (18) with the N-tails may allow progress of the polymerase along the nucleocapsid through iterative cycles of XD to N-tail binding and release (19–21).
Consistent with this view, previous functional studies with carboxyl-terminally truncated SeV and MeV N lacking the P binding domains suggested an inability of N-tail truncated nucleocapsids to serve as template for RdRp activity (9, 14). Biochemical binding studies with truncated MeV N and functional assays combining purified, standard SeV RNPs with soluble truncated SeV N demonstrated that the N-tails are not required for the formation of proper P-L complexes itself or the interaction of P with free N (3, 14). Somewhat unexpectedly, a recent study found that individual point mutations located in the MeV N box 2 region and flanking the N-MoRE domain measurably reduced P-XD affinity to N-tail but did not abolish polymerase activity (22). However, this may be due to the high avidity of tetrameric P interaction with nucleocapsid, because measurable affinity of the mutated MoRE domains for P-XD was maintained in these N variants.
Building on the structural reconstructions of MeV nucleocapsids, we test in this study central elements of the current paramyxovirus replication model in the context of transient replicon systems and virus replication. Specifically, we examine the importance of the carboxyl-terminal P-binding region in the MeV N-tail for polymerase recruitment to the nucleocapsid and explore the mechanistic contribution of the unstructured central N-tail section to RNP template function. Our results redefine the role of the exposed N-tail sections in RdRp loading onto, and movement along, the viral nucleocapsid.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, and Virus Stocks
Baby hamster kidney cells (C-13; ATCC) stably expressing T7 polymerase (BSR-T7/5 (23)) and African green monkey kidney epithelial cells (CCL-81; ATCC) stably expressing human signaling lymphocytic activation molecule (Vero/hSLAM (24)) were maintained at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Both cell lines were incubated in the presence of G-418 (100 μg/ml) every fifth passage. The cells were transfected using either Lipofectamine 2000 (Invitrogen) or, for virus recovery transfections, calcium phosphate precipitation (Promega). Virus stocks were prepared by infecting Vero/hSLAM cells at a multiplicity of infection of 0.001, followed by incubation at 37 °C. When microscopically observed virus-induced cytopathicity reached ∼90%, cell-associated progeny particles were released through freeze/thaw, and titers were determined by 50% tissue culture infectious dose (TCID50) as described (25).
Recombinant MeV
RecMeV particles were generated using a modified recovery protocol as described (26). Emerging infectious particles were transferred onto Vero/hSLAM cells for generation of passage two virus stocks. To confirm integrity of recombinant viruses, RNA was extracted from infected cells using the RNeasy mini kit (Qiagen), and cDNAs were created using random hexamer primers and Superscript III reverse transcriptase (Invitrogen). Modified genome regions were amplified using appropriate primers and sequenced.
Molecular Biology
Plasmids encoding the MeV (−) replicon (27); MeV Edm N, P, or L (28); or MeV IC-B N or P or MeV 9301B L (29) were previously described. For N-tail truncation screening, tandem stop codons (TAGTGA) were introduced into the N-tail ORF through site-directed mutagenesis following the QuikChange protocol (Stratagene). An N-ΔC86 expression plasmid truly lacking the last 86 codons of the N ORF was generated through PCR-based shortening of the plasmid and PstI-mediated religation of the amplicon. Both the tandem stop and true deletion N-ΔC86 constructs showed equal bioactivities in our assays. To randomize central N-tail sections, frameshifts were introduced through addition or deletion of one or two nucleotides by directed mutagenesis. Tetracysteine tags (SGGGFLNCCPGCCMEPGGGS) (30) were inserted into the N-tail through PCR amplification and religation of the amplicons using a SmaI restriction site engineered into the tag sequence. The (+) replicon was generated using a recombination PCR strategy based on the (−) construct that inverted the entire leader-luciferase-trailer cassette relative to the T7 promoter and the hepatitis delta virus ribozyme element. An expression plasmid encoding the amino-terminal section of P (PNT, amino acids 1–230) was generated through introduction of a FLAG epitope tag followed by a tandem stop codon at P residue 230. The corresponding carboxyl-terminal region of P (PCT) expression construct (amino acids 231–507) was generated through PCR amplification and religation of the shortened amplicon using an engineered EcoRI restriction site located upstream of the newly inserted start codon. The P-ΔXD expression construct was generated through insertion of tandem stop codons after P position 456 into the MeV Edm-P plasmid, abrogating expression of the X domain starting at P position 459.
Plasmids containing cDNA copies of the MeV Edm or MeV IC-B (31) genomes were further modified by replacing the N ORF with an Edm or IC-B-based N-ΔC86 ORF, respectively. RecMeV N-ΔC86-P-N and recMeV N-P-N were generated through doubling of the P-M intergenic junction using appropriate primers and transfer of an appropriately prepared cassette encoding standard N using the AatII restriction site in the P-M junction. All constructs generated in this study were sequence-confirmed. Sequences of oligonucleotide used in this study are available upon request.
Antibodies, SDS-PAGE, and Immunoblotting
BSR-T7/5 cells (4 × 105/well in a 12-well plate format) transfected with 2 μg of N-encoding plasmid DNA were washed in PBS and lysed in radioimmune precipitation assay buffer as described (32). Samples were fractionated on 8% SDS-PAGE gels, transferred to PVDF membranes (Millipore), and subjected to enhanced chemiluminescence detection (Pierce) using specific antibodies directed against MeV N (83KKKII; Millipore), MeV P (9H4; Abcam), GAPDH (6C5; Ambion), or FLAG (M2; Sigma) as specified. Immunoblots were developed using a ChemiDoc digital imaging system (Bio-Rad). The Image Lab package (Bio-Rad) was used for densitometry.
Minireplicon Luciferase Reporter Assays
BSR-T7/5 cells (4 × 105/well in a 12-well plate format) transfected with all MeV Edm or IC-B polymerase helper plasmids and the (+) or (−) reporter constructs, all under the control of the T7-polymerase promoter, were lysed after 40 h (unless otherwise specified) in Glo lysis buffer (Promega). Luciferase activities in cleared lysates (20,000 × g, 5 min, 4 °C) were determined using Bright-Glo firefly luciferase substrate (Promega) and an Envision Multilabel microplate reader (PerkinElmer Life Sciences) in top-count mode.
Co-immunoprecipitation
BSR-T7/5 cells (8 × 105/well in a 6-well plate format) transfected with 2 μg each of plasmid DNA encoding an MeV N construct and either MeV PNT or MeV PCT were harvested and subjected to co-immunoprecipitation as described (32). Following immunoprecipitation using α-FLAG or α-P antibodies, samples were fractionated on 10% SDS-PAGE gels, followed by immunoblotting and chemiluminescence detection as outlined.
Terminal RACE
Total RNA was isolated from infected Vero/hSLAM cells as outlined. For first strand synthesis, a positive polarity RNA-specific oligonucleotide primer (CAGTTATTGAGGAGAGTT) annealing in the F ORF was used to reduce contamination by polycistronic viral mRNA. The RACE System (Invitrogen) was used for cDNA end amplification with the following modifications: tailing reactions were performed in the presence of 10% Me2SO, 400 μm dCTP for 1-hour at 4 °C, followed by incubation at 37 °C for 30 min. PCR and nested PCR were performed with the gene specific primers (GATTCCTCTGATGGCTC; for standard N) or (GACGTAGCCTTCGGGCATGG; for N-ΔC86). PCR product was subcloned into the TOPO 2.1 vector (Invitrogen), and individual clones were amplified and sequence-analyzed.
Nucleocapsid Preparation
Vero/hSLAM cells (2.2 × 106 in 100-mm dishes) infected with recMeV N-P-N or recMeV N-ΔC86-P-N (multiplicity of infection of 0.005 TCID50/cell) were harvested in RNP lysis buffer (33) when maximal CPE was observed. Cleared lysates (5,000 × g, 5 min, 4 °C) were layered on CsCl density gradients (2.4 ml of 40% CsCl, 2.4 ml of 30% CsCl, 2.4 ml of 20% CsCl, and 0.8 ml of 30% glycerol) in TNE buffer (25 mm Tris/Cl, pH 7.4, 50 mm NaCl, 2 mm EDTA) and subjected to ultracentrifugation in an SW41 rotor (32,000 rpm, 16 h, 12 °C). Eight 1.5-ml gradient fractions were collected, and samples were concentrated by TCA precipitation and fractionated through SDS-PAGE as described.
Quantitative PCR
Total RNA was isolated from BSR-T7/5 cells (8 × 105/well in a 6-well format) transfected with plasmids encoding MeV RdRp components, standard N or N-ΔC86, and the (+) replicon or the (+) MeV-Luc N-ΔC86 genome as outlined. Contaminating DNA was removed through an on-column DNaseI digest and cDNA generated of 3 μg of total RNA using oligo(dT) primer. Quantitative PCR was performed in a 7500 fast real time PCR system (Applied Biosystems) using iTaq Fast SYBR Green Supermix with ROX (Bio-Rad) and specific primers annealing in the firefly luciferase (CGCCAAAAGCACTCTGATTGAC; CTCGGGTGTAATCAGAATAGCTG) or cellular GAPDH (CATGTTCCAGTATGACTCTACCC; GACCTTGCCCACAGCCTTGG) ORFs. ΔΔCt values were calculated using GAPDH as cellular standard, followed by calculating the ratios of relative mRNA levels obtained with each replicon construct in the presence of N-ΔC86 and standard N.
Statistical Analysis
To assess the statistical significance of differences between sample means, unpaired two-tailed t tests were applied using the Prism 5 (GraphPad) or Excel (Microsoft) software packages. When differences were compared relative to standard N in normalized assays, 95% confidence intervals were calculated based on sample numbers, data means, and standard deviations (34).
RESULTS
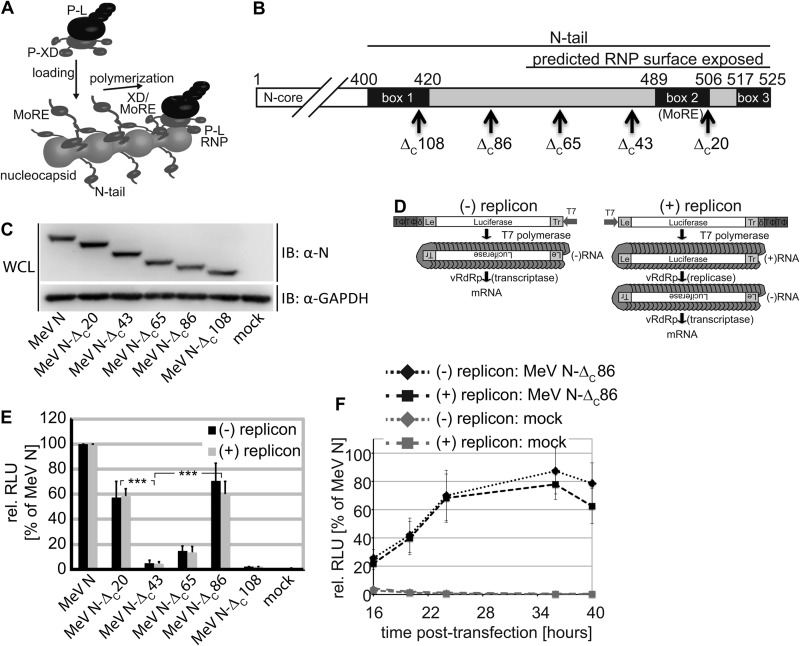
A current docking model of the MeV polymerase complex to the viral RNP template is shown in Fig. 1A. MeV RNP reconstructions posit N-tail residues 450–525 to be exposed and fully mobile on the nucleocapsid surface (8). If this is the case, we expect previously reported carboxyl-terminal tail truncations removing half of the MoRE domain (9) to generate highly flexible 40–50-residue tail stems that protrude from the RNPs. We hypothesized that a negative regulatory effect of these stems, rather than the absence of MoRE domain and carboxyl-terminal tail residues, may, in fact, account for the lack of template activity of RNPs with carboxyl-terminal P-binding domain truncations (9, 14). To test this idea experimentally, we designed a series of progressive N truncation constructs guided by the currently available MeV RNP reconstructions and bioactivity data (Fig. 1B). All N variants were expressed at levels comparable to unmodified MeV N and showed the anticipated mobility pattern in SDS-PAGE (Fig. 1C).
FIGURE 1.
Progressive MeV N-tail truncations significantly restore bioactivity. A, current model of MeV polymerase recruitment to the RNP template. The RdRp complex is thought to initially engage with RNP via the surface-exposed N-tail MoRE domain and the P-X domain and uses consecutive cycles of release and rebinding to cartwheel the polymerase along the template. B, schematic of the N-tail organization. Conserved microdomains (boxes 1–3) and the MoRE domain are highlighted. Arrows mark individual truncations generated, and numbers refer to amino acids. C, immunoblots (IB) of whole cell lysates (WCL) of cells transfected with N-encoding plasmids or vector DNA (mock). The blots were probed with specific antibodies for the MeV N protein and reprobed with antibodies directed against cellular GAPDH. D, schematic of the negative and positive polarity minireplicon reporter constructs used for RdRp activity assays. E, relative luciferase reporter activity (RLU) in cells expressing the (−) or (+) minireplicon construct, L, P, and the specified N variant. Otherwise identically transfected control cells received vector DNA in place of the N expression plasmid (mock). The values were normalized for those obtained in the presence of standard N and represent averages of at least four independent experiments ± S.D. ***, p < 0.001. F, kinetics of reporter expression in the presence of N-ΔC86. Cells were transfected as in D. For each time point, values were normalized for those obtained with standard N. Averages of at least five experiments ± S.D. are shown.
The N-tail MoRE Domain Is Dispensable for RdRp Activity
To assess bioactivity of these constructs, we first examined RdRp transcriptase activity in a conventional MeV minigenome reporter assay (27), which predominantly monitors primary RdRp-mediated mRNA synthesis after generation of a negative polarity RNA template by T7-polymerase (Fig. 1D, (−) replicon). RdRp activity was only partially affected by deletion of box 3 in the N-tail (MeV N-ΔC20), which was shown to be involved in N binding to viral matrix protein (35) and host factors such as hsp70 (36, 37). This observation was consistent with previous reports (9), which had even shown a slight enhancement of polymerase activity upon removal of the box 3 region; RNPs with N-tail truncations deleting the MoRE domain and carboxyl-terminal residues (MeV N-ΔC43) were biologically inactive (Fig. 1E).
Strikingly, however, even larger truncations removing part of the N-tail upstream of the MoRE domain (MeV N-ΔC86) substantially restored RdRp bioactivity. Equivalent results were obtained when we assessed our panel of N truncation constructs in a positive polarity minigenome system that requires both RdRp replicase and transcriptase activity for reporter expression (Fig. 1, D, (+) replicon, and E). Kinetic comparison with standard N revealed an initial delay in reporter expression in the presence of MeV N-ΔC86, followed by a plateau phase of nearly reference-like activity (Fig. 1F).
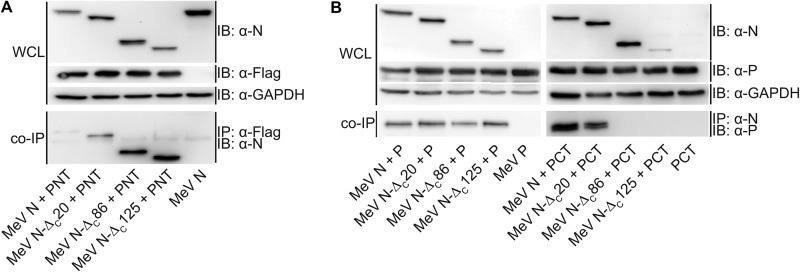
Protein interactions between the N protein core and a PNT, and the N protein tail and the PCT harboring the X domain were mapped biochemically in previous studies (3). However, none of the N protein tail modifications assessed supported RdRp activity. Because our bioactivity data for N-ΔC86 were rather unexpected, we generated MeV PNT and PCT expression plasmids and re-examined the interaction profile with bioactive MeV N-ΔC86 by co-immunoprecipitation. These experiments reiterated the efficient interaction of full-length P and PNT with N variants containing large tail deletions (MeV N-ΔC86 and N-ΔC125). In fact, the N tail truncations reproducibly enhanced co-immunoprecipitation efficiency compared with that observed with standard N. Our experiments furthermore confirmed that PCT interaction with the N protein requires the presence of the MoRE domain in the N-tail (Fig. 2, A and B).
FIGURE 2.
Bioactive MeV N-ΔC86 does not interact with a carboxyl-terminal PCT region harboring the P-X domain. Lysates of cells expressing the specified N construct and either the FLAG epitope-tagged amino-terminal PNT region (A) or full-length P or the carboxyl-terminal PCT region (B) (51) were subjected to direct immunoblotting (IB) or immunoprecipitation (IP) of PNT (α-Flag) or N (α-N). Co-precipitated (co-IP) N or P material, respectively, was detected in immunoblots using specific antibodies. Immunoblots shown are representative of two (PNT) or three (P, PCT) independent experiments. WCL, whole cell lysates.
Taken together, these data demonstrate that an interaction between the N-tail MoRE domain and P-XD is not required for RdRp recruitment to, and/or movement along, the RNP template. In addition, they reveal a regulatory role in polymerase activity of the exposed central N-tail section upstream of the MoRE domain.
Regulatory Effect of the Unstructured, Central N-tail Section
To address the question of whether this effect depends on the primary sequence of the N-tail section present in MeV N-ΔC43 but absent in N-ΔC86 (N residues 440–482) or length of the exposed tail stem, we sequence randomized this region in the context of otherwise unchanged full-length N (MeV N-(rd440–481)) and the series of N-tail truncations (Fig. 3A). All constructs were efficiently expressed (Fig. 3B) and showed slightly increased bioactivity when compared with N-ΔC43, but none of the truncated N variants restored RdRp activity to the level observed for N-ΔC86 (Fig. 3C). In contrast, randomizing the central N-tail section between residues 439 and 482 in the background of full-length N or selectively deleting this N-tail region resulted in equivalent or slightly higher RdRp activity than that observed with standard N (Fig. 3, A–C).
FIGURE 3.
The central N-tail section down-regulates polymerase activity. A, sequences of full-length and truncated N constructs generated with randomized central tail sections. Areas shaded in gray denote scrambled sequence sections. B, immunoblots (IB) of whole lysates of cells expressing the N variants shown in A. C, minireplicon reporter assays to assess bioactivity of tail randomized N variants. D, immunoblot analysis of cells expressing full-length MeV N variants with tetracysteine (4xcys) epitope tags at the indicated positions. E, minireplicon reporter assays to determine bioactivity of tetracysteine-tagged N variants shown in D. In C and E, values and statistical analyses are relative to the minireplicon system containing standard N and represent averages of at least four experiments ± S.D. *, standard N (100%) outside the 95% confidence interval. RLU, relative luciferase reporter activity.
To test the effect of restricted tail flexibility on N bioactivity, we inserted structurally dominant tetracysteine tags (30, 38) at four different tail positions into otherwise unchanged full-length N. With the exception of an insertion at N position 446, tagged N proteins were efficiently expressed (Fig. 3D). However, bioactivity was reduced by ∼60–80% in each replicon system compared with that observed in the presence of standard N (Fig. 3E), demonstrating that structurally active elements are poorly tolerated in the central N-tail domain.
Taken together, these findings support that the central, unstructured N section does not engage in specific protein interactions with viral or host factors proteins. They reveal a regulatory effect of this N-tail domain on polymerase activity in the context of N truncations lacking the MoRE domain and box 3.
The Central N-tail Region and MoRE Domain Are Required for Virus Replication
Having demonstrated that removal of the central N-tail section efficiently restores bioactivity of an MeV N variant lacking the MoRE domain and box 3, we next asked whether MeV particles can be recovered that express N-ΔC86 in the place of full-length N. We exchanged the N open reading frame against that of the N-ΔC86 in a cDNA copy of the MeV-Edmonston (MeV-Edm) genome (31), but initial virus recovery attempts failed. This outcome could have been due to the ∼30% reduction in RdRp activity by N-ΔC86 that we observed in replicon assays or, alternatively, could indicate a fundamental role of the central N-tail region and intact MoRE domain and box 3 for virus replication. To distinguish between these possibilities, we rebuilt the N-ΔC86 construct in the background of the nonattenuated MeV IC-B isolate (31), which we had found to return ∼10-fold higher RdRp activities in transient replicon assays than the MeV Edm-based system (Fig. 4A). IC-B N-ΔC86 was generated by insertion of tandem stop codons at N positions 440 and 441. This construct was efficiently expressed and showed an electrophoretic mobility profile equivalent to that of Edm N-ΔC86 (Fig. 4B). Reproducing our experience with the Edm-based replicon system, RdRp activity of an IC-B replicon containing IC-B N-ΔC86 was reduced by ∼40% compared with standard IC-B N. Importantly, however, the IC-B replicon containing N-ΔC86 returned ∼6-fold higher reporter expression levels than the unchanged, standard Edm replicon (Fig. 4A).
FIGURE 4.
MeV N-ΔC86 does not sustain efficient virus replication. A, comparison of minireplicon reporter expression driven by wild type MeV-derived (P, N: MeV IC-B; L: MeV 9301B) versus MeV Edm-derived polymerase helper proteins. All results were normalized for those obtained with the wild type MeV system and in the presence of standard IC-B N. The values represent averages of three experiments ± S.D. *, standard Edm N outside the 95% confidence interval. B, immunoblot analysis of cells expressing MeV Edm- or IC-B-derived P and N variants. C, microphotographs of cell monolayers infected with newly recovered recMeV IC-B N-ΔC86. Representative fields of view are shown at a magnification of 200×. D, sequence analysis of recMeV IC-B N-ΔC86 at the time of recovery (day 0) and after 40 and 56 days of continued incubation. RLU, relative luciferase reporter activity; WCL, whole cell lysates.
We therefore transferred IC-B N-ΔC86 into the cloned IC-B genome (31). Individual infectious centers were identified after virus recovery transfection that could be passaged laterally (Fig. 4C), but virus spread was severely impaired for ∼40 days postrecovery. This period was followed by a phase of rapidly improving replication. Sequencing of the N protein tail after RT-PCR at different times postrecovery transfection revealed spontaneous viral adaptation, gradually replacing both stop codons in the IC-B N-tail with regular codons, as the molecular basis for regained viral growth (Fig. 4D).
These data indicate that the unsuccessful recovery attempts of recMeV-Edm N-ΔC86 did not reflect insufficient RdRp activity per se when N-ΔC86 is present. Rather, they highlight a fundamental mechanistic role of the central and carboxyl-terminal sections of the N protein tail in the context of virus replication that is distinct from basic RdRp recruitment to the nucleocapsid template.
Generation of a Replication Competent recMeV N-ΔC86 Variant
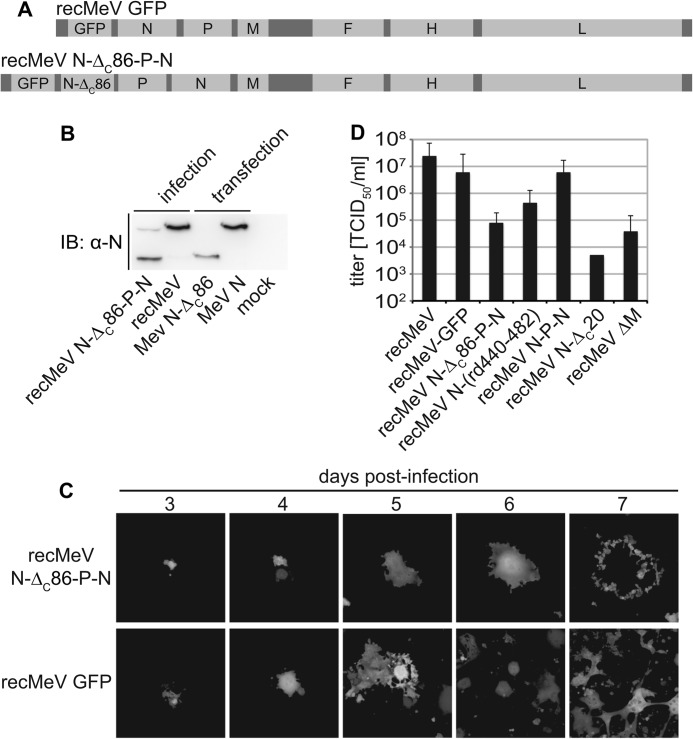
To characterize the mechanistic role of N-tail in the context of virus replication, we generated a recMeV variant encoding the N-ΔC86 protein in addition to standard N in a recMeV-GFP background, also harboring an eGFP open reading frame as an additional transcription unit (39). Taking advantage of the transcription gradient of paramyxovirus gene expression (40), we moved the standard N reading frame into a post-P position (Fig. 5A), resulting in substantially higher N-ΔC86 than full-length N protein levels in infected cells after virus recovery (Fig. 5B). Despite productive passaging of recMeV N-ΔC86-P-N virions, virus growth was severely impaired as evidenced by limited lateral virus spread through cell monolayers (Fig. 5C) and reduced viral titers compared with standard recMeV and a recMeV N-P-N variant harboring two copies of the unchanged N reading frame (Fig. 5D). In contrast to recMeV N-ΔC86, recombinant viruses lacking only the box 3 region of the N-tail (recMeV N-ΔC20) or harboring the randomized N-(rd440–481) protein could be recovered without additional full-length N complementation. Although recMeV N-ΔC20 reached substantially lower titers than standard recMeV, yields of recMeV N-(rd440–481) showed an intermediate reduction (Fig. 5D). These phenotypes confirm a negative regulatory effect of the N-ΔC86 construct in the context of virus replication.
FIGURE 5.
recMeV encoding both N-ΔC86 and standard N are replication competent. A, schematic of the genomes of standard recMeV GFP and newly generated recMeV N-ΔC86-P-N. B, immunoblot (IB) analysis of N protein expression in cells infected with recMeV N-ΔC86-P-N or standard recMeV. For comparison, total lysates of cells transfected with N-ΔC86 or standard N expression plasmids were analyzed in parallel. Control cells received infection media only (mock). C, cytopathic effect and lateral spread of cells infected with recMeV N-ΔC86-P-N or recMeV GFP, monitored by following eGFP expression in infected cells. Representative fields of view are shown at a magnification of 200×. D, stock titers of cell-associated particles of a panel of four recombinants generated for this study in comparison with standard recMeV or recMeV lacking the M protein (recMeV ΔM). Error bars represent the titer range observed.
Binding of P-XD to the N-tail Is Required to Stabilize RdRp-RNP Interaction during Virus Replication
In comparison to RdRp activity in minireplicon assays, self-sustained viral genome replication adds at least two additional functional requirements to helper plasmid-driven (+) replicon reporter expression: the correct inaugural positioning of the RdRp replicase complex on the template for generation of complete antigenomic and genomic RNA copies, and sufficient stability of the RdRp-RNP complex during polymerization to ensure faithful replication of RNA sequences substantially longer than the minireplicon reporter constructs.
To test whether these RdRp functionalities are affected by the N-ΔC86 truncation, we first assessed the molecular nature of the terminal leader sequence of viral genomes in cells infected with standard MeV (recMeV-Edm or recMeV-IC-B) or recMeV N-ΔC86-P-N, using rapid amplification of cDNA ends (RACE) and sequencing of 29–40 independent subclones/virus examined. Of a combined 53 genomic sequences obtained for the standard MeV strains, 51 represented complete genome copies, whereas 2 showed partial terminal truncations (Table 1). These data closely matched those obtained for recMeV N-ΔC86-P-N: 34 genomic sequences were obtained; of these, 33 represented complete genome copies, and 1 a truncated sequence. These results indicate successful replication of the genome termini in the presence of the N-ΔC86 variant.
TABLE 1.
RACE analysis of viral genome termini present in cells infected with the specified MeV recombinants
The values represent the total number of independent genomes subcloned and analyzed and the subset of these that featured complete leader sequences, termini-truncated sequences, and first open reading frame viral mRNA.
| Virus | Clones analyzed | Complete leader sequences | Termini-truncated sequences | Viral mRNA |
|---|---|---|---|---|
| recMeV-Edm | 36 | 27 | 0 | 9 |
| recMeV-IC-B | 29 | 24 | 2 | 3 |
| recMeV N-ΔC86-P-N | 40 | 33 | 1 | 6 |
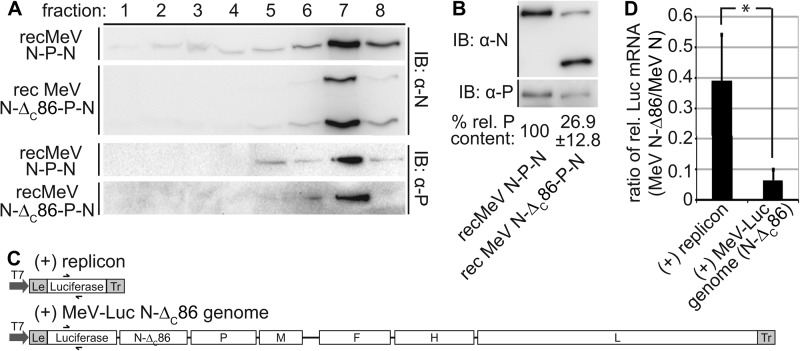
Second, we purified intact RNPs from cells infected with recMeV N-P-N or recMeV N-ΔC86-P-N through flotation in cesium chloride gradients. Consistent with previous studies (3, 5), RNPs containing full-length or truncated N proteins accumulated predominantly in higher gradient fractions (Fig. 6A). P protein antigenic material was likewise concentrated in these fractions. When we adjusted the nucleocapsid-containing fraction seven for equal N signal intensity and thus overall nucleocapsid content for subsequent immunoblots, quantification of the relative P protein content revealed an ∼4-fold lower amount of P in RNP samples derived from recMeV N-ΔC86-P-N compared with recMeV N-P-N-infected cells (Fig. 6B).
FIGURE 6.
Presence of N-ΔC86 in nucleocapsids affects the stability of P binding to RNPs. A, purification of viral nucleocapsids through cesium chloride gradient fractionation. Gradient fractions were loaded from top (1) to bottom (8), and immunoblots (IB) were decorated with specific antibodies directed against the MeV N or P protein, respectively. B, fraction 7 material from A was densitometrically adjusted for ± 10% equal amounts of nucleocapsid material by N signal intensity and subjected to SDS-PAGE, and the relative amounts of co-floating P material were determined. Numbers represent averages of densitometric quantifications of three independent experiments ± S.D. C, schematic of the (+) replicon and the (+) MeV-Luc N-ΔC86 genome construct, drawn to scale. D, quantitative PCR analysis of relative luciferase mRNA levels obtained after co-transfection of cells with MeV P and L expression plasmids, the replicon constructs shown in C, and either standard N or N-ΔC86-encoding plasmids. The columns show ratios of relative mRNA levels obtained with each replicon construct in the presence of N-ΔC86 and standard N. The values represent averages of at least three independent experiments, each quantified in duplicate ± S.D. *, p < 0.05.
To test the implication of this phenotype on polymerase function, we generated an MeV-luciferase genomic plasmid ((+) MeV-Luc N-ΔC86 genome). This construct is equivalent in organization to the (+) replicon reporter plasmid but is ∼10-times larger in overall size. In addition, it contains an N to N-ΔC86 exchange, because we consider it imperative that polymerase activities measured in the presence of standard or truncated N are based on the same RNA template and that transfected cells are void of any full-length N protein material at all times (Fig. 6C). Upon co-transfection with the MeV RdRp helper plasmids, we noted through quantitative real time PCR analysis that in the case of the (+) replicon, the presence of N-ΔC86 reduced relative luciferase mRNA levels to ∼40% of that observed for full-length N. When the larger (+) MeV-Luc N-ΔC86 genome plasmid was examined under otherwise identical experimental conditions, however, we found relative luciferase mRNA levels of only 5% that generated in the presence of full-length N (Fig. 6D). Taken together, these data indicate that P-XD binding to the N-tail is not required for initial RdRp positioning on the RNP template but critically stabilizes the RNP-RdRp complex as RNA polymerization proceeds.
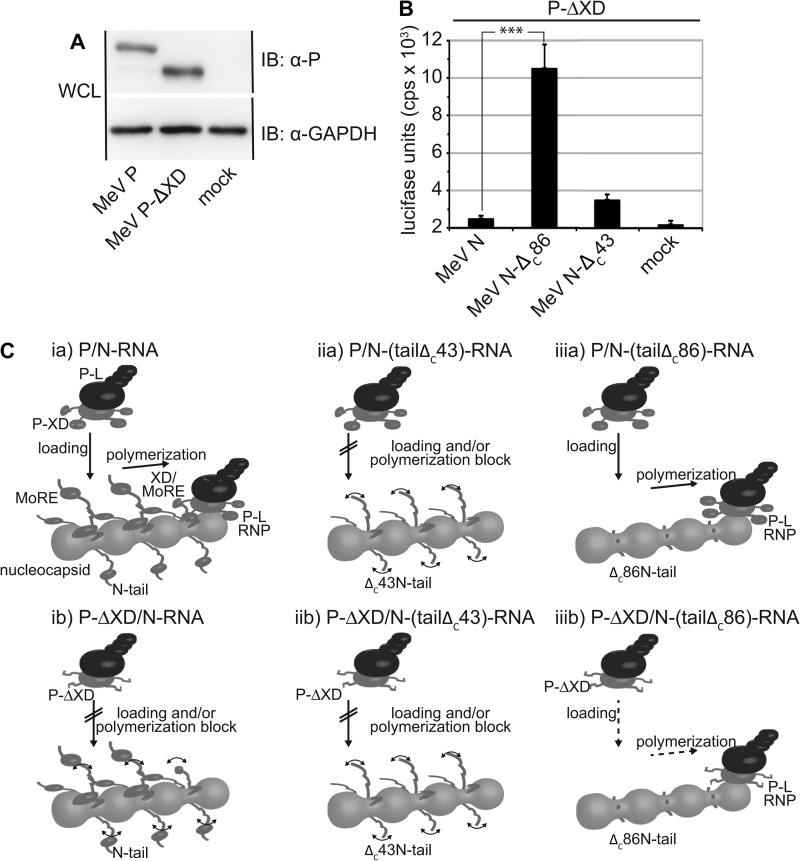
Truncated P-ΔXD Partially Regains Bioactivity When Combined with an N-ΔC86-RNP Template
The P-XD assembles into an antiparallel three-helix bundle that provides the binding site for the N-MoRE domain (17). For counteranalysis of the N-ΔC86 bioactivity data, we generated a carboxyl-terminally truncated MeV P variant lacking the entire X domain. Immunodetection confirmed efficient expression of the resulting P-ΔXD construct (Fig. 7A). When subjected to replicon assays, P-ΔXD lacked all bioactivity when combined with standard nucleocapsid templates composed of full-length N subunits (Fig. 7B). In the presence of N-ΔC86-RNP nucleocapsids, however, we observed a significant increase in reporter expression, indicating that polymerase activity of P-ΔXD-L was partially restored in the absence of the freely flexible N-tail domains. This finding reveals that the presence of either the N-MoRE or the P-X domain alone blocks polymerase activity in the absence of the binding partner. However, simultaneous deletion of both domains restores access of the polymerase complex to the template and bioactivity.
FIGURE 7.
Polymerase activity of P-ΔXD-L is partially restored when combined with an N-ΔC86-RNP template. A, immunoblots (IB) of whole lysates of cells transfected with P-encoding plasmids or vector DNA (mock). The blots were probed with specific antibodies for the MeV P protein and reprobed with antibodies directed against cellular GAPDH. B, luciferase reporter activity in cells expressing the (−) MeV-Edm replicon, L, truncated P-ΔXD, and the specified N variant. Otherwise identically transfected control cells received vector DNA in place of the N expression plasmid (mock). The values represent averages of 13 independent experiments ± S.E. ***, p < 0.001. C, revised model of MeV polymerase recruitment. The P-L polymerase complex directly engages the nucleocapsid for N-tail-independent loading and progress. If nucleocapsids are composed of full-length N (panel ia), the P-XD interaction with N-MoRE rearranges the central N-tail sections to facilitate polymerase binding to the core and stabilizes binding of the advancing complex to the template. In the absence of the P-X domain (panel ib) or in the case of a MoRE-deleted N-ΔC43 and P or P-ΔXD (panels iia and iib), the exposed, freely mobile central tail sections prevent polymerase binding and/or progress. The absence of these mobile tail sections in N-ΔC86 nucleocapsids (panels iiia and iiib) allows productive interaction of the polymerase complex with, and advancement along, the template. WCL, whole cell lysates.
DISCUSSION
The current model of MeV RdRp recruitment to the RNP template for genome replication and mRNA synthesis assumes an inaugural interaction between a high affinity MoRE domain in the N-tail and the X domain in the viral P protein (8, 17). After polymerization is initiated, recurring release and rebinding of X domains in tetrameric P to MoRE elements in nucleocapsid is thought to allow progress of the P-L polymerase along the RNP template.
Based on the insight gained in our study, we propose that central elements of this model need to be redefined (Fig. 7C). Specifically, we found that the recruitment of the MeV polymerase complex to the RNP template occurs independently of any interaction between the N-tail MoRE/box 3 region and P-XD. This initiation mechanism stands in stark contrast to the current paradigm that paramyxovirus P proteins must interact with the carboxyl-terminal tail region of N for RNP binding (10, 14, 41). The previous polymerase binding models were heavily influenced by experiments showing that progressive carboxyl-terminal truncations of the MeV N-tail, which partially or completely remove the MoRE domain and downstream residues, largely eliminate polymerase function in replicon assays (9).
Of note, because minireplicon activity depends on initial, random RNA encapsidation by N proteins (42), an inability of these MoRE domain truncated N variants to encapsidate RNA could be responsible for this phenotype. However, previous studies have confirmed that carboxyl-terminal truncations in paramyxoviurs N do not interfere with RNP formation (5, 33).
Our study reproduced earlier activity data for MoRE domain-deficient MeV N (9) but revealed that a substantially larger truncation, removing the unstructured central N-tail section in addition to the MoRE domain and box 3, significantly restores polymerase activity. This finding illuminates a novel regulatory function of the central N-tail region and demonstrates that P-XD interaction with the carboxyl-terminal N-tail section is dispensable for productive template binding and RNA synthesis. Supported by essentially equivalent RdRp activity results obtained with (−) and (+)-type replicon constructs, N-tail-independent polymerase loading appears not to be limited to RdRp in transcriptase configuration but also applies to the replicase complex.
We note that even larger tail truncations (i.e. N-ΔC108) again abolished nucleocapsid bioactivity. Because these deletions remove N residues posited in the interstitial space between RNP turns (8), loss of bioactivity may likely reflect transition from native, loosely coiled MeV nucleocapsids to condensed, rigid structures (7). Sequence-randomizing the central N-tail section did not substantially alter RdRp bioactivity in the context of full-length (remained active) or MoRE/box 3-truncated (remained largely inactive) N. However, introduction of structurally dominant tetracysteine tags into the central tail region significantly reduced polymerase activity. We conclude that the regulatory effect of the central N-tail section is unlikely based on direct binding of host co-factors to this region, which would be expected to be sensitive to sequence randomization. Indeed, sequence comparison between N proteins of different members of the morbillivirus genus to which MeV belongs revealed a particularly low degree of sequence conservation of this area (43). Rather, P-XD docking to the N-tail may rearrange and/or organize the tails, giving the polymerase complex access to productive interaction with the RNP template through a direct, previously unappreciated interaction of MeV P-L polymerase complexes with the nucleocapsid core, which may be mediated by residues located in the PNT half of the P protein (44).
Strong support for this model arises from the bioactivity profile of truncated P protein lacking the X domain (Fig. 7C). The inability of P-ΔXD-L complexes to organize the N-tails through XD-MoRE domain interactions may account for the loss of bioactivity when combined with standard nucleocapsids. In contrast, ΔC86N-tail truncated nucleocapsids are directly accessible by the polymerase without a need for tail reorganization, resulting in partially restored bioactivity of P-ΔXD-L polymerase complexes. Tetracysteine tags in the N-tail may likewise impede this P-XD binding-induced preparation of the RNP core for polymerase docking.
Direct binding of polymerase complexes to the ∼40-residue tail stems present in N-ΔC86 appears unlikely, because of the proposed position of these tail residues in the interstitial space between nucleocapsid turns (8) and our co-immunoprecipitation results. Also, a conserved box 1 located at the beginning of the tail (amino acids 401–420) was implicated in binding to a cellular N receptor of unknown molecular nature (45, 46). Earlier truncation studies of SeV and MeV N reached the conclusions that (i) N proteins with carboxyl-terminal deletions are capable of encapsidating RNA (3, 5, 14), but (ii) the resulting RNPs do not support P binding (5, 47), and (iii) the resulting RNPs cannot serve as template for the polymerase (9, 14, 47). In light of our present findings, we interpret these experiments for MeV, and possibly also SeV, to likely reflect obscured polymerase complex access to the RNP core by the remaining central tail sections in partially truncated N variants. Interestingly, a recent characterization of the related mumps virus P protein revealed that its interaction with nucleocapsid does not fully require the N-tail but can be mediated by other contact domains in P and N (15, 44).
Our molecular analysis of the genome termini of over 30 independent clones each of recombinant MeVs harboring truncated or standard N demonstrated accurate replication of RNPs in both cases. Simultaneous analysis of gradient-purified nucleocapsid confirmed that RNPs of recMeV N-ΔC86-P-N were predominantly composed of truncated N variants, indicating that the full-length N is not preferentially incorporated into nascent RNPs during replication of this virus. These results underscore that correct, initial positioning of the polymerase on the nucleocapsid template is independent of the P-XD and N-MoRE interaction. Although we cannot entirely rule out that, alternatively, two distinct nucleocapsid populations form in cells infected with this recombinant, several considerations render this hypothesis highly unlikely: the abundance of truncated versus standard N in infected cells; the efficient co-immunoprecipitation of truncated N-ΔC86 with PNT; and the reportedly efficient RNA encapsidation by truncated N protein variants (5, 33).
The severely restricted growth phenotype of the recMeV N-ΔC86-P-N dual-N recombinant virus, the unsuccessful rescue attempts of recMeV-Edm N-ΔC86, and the spontaneous adaptation of recMeV-IC-B N-ΔC86 prove, however, that all tail-truncated N variants are incapable of supporting virus replication. Through regulatory functions and/or interaction with the viral matrix protein (35–37), the box 3 section of the MeV N-tail facilitates efficient genome replication and incorporation into nascent particles. Conceivably, the elimination of this region 3, rather than the deletion of the N-MoRE domain, may prevent the recovery of recombinant MeV N-ΔC86 virions. However, M-deleted MeV recombinants replicate in cell culture (48), and we recovered an MeV N-ΔC20 recombinant that lacks box 3 but leaves the MoRE domain intact.
Considering ∼70% bioactivity of N-ΔC86-containing RNPs in replicon assays, we are confident that the inability of N-ΔC86 to support virus replication is based on a fundamental difference between the replicon system and self-sustained replication rather than an overall reduction in RdRp activity. The successful recovery of the MeV recombinant harboring the N-ΔC20 construct, which showed a similar reduction in bioactivity in replicon assays, corroborates this interpretation. Fulfilling an auxiliary role, dynamic cycles of N-tail to P-XD docking and release may be required to stabilize the RdRp-RNP complex as the polymerase moves along the template (20, 49, 50). This view is supported by our observation that increasing RNP template length in transient replicon assays results in an overproportional decline in efficiency of successful replication in the presence of N-ΔC86. Purified nucleocapsids of recMeV N-ΔC86-P-N show a reduced, but appreciable, level of co-floating P compared with standard MeV RNPs. Under these conditions, the full-length N subunits present in the mixed RNPs may contribute to the biochemically detectable association.
In conclusion, we propose that the tightly orchestrated N-MoRE interaction with P-XD is not required to recruit or position the polymerase complex on the RNP template but may rather arrange the central N-tail sections to allow access for direct polymerase docking to the nucleocapsid core. As such, the interaction of N-MoRE with P-XD may help to overcome a negative regulatory effect of central N-tail domains that limit polymerase loading on, or advancing along, the template. After successful initiation of polymerization, iterative cycles of release and rebinding between N-MoRE and P-XD are not essential for polymerase activity per se but reduce the risk of premature chain termination by dynamically stabilizing the interaction between P and the template.
Acknowledgments
We thank A. L. Hammond and B. Bankamp for discussion and comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants AI071002, AI085328, and AI083402 (to R. K. P.).
- MeV
- measles virus
- RNP
- ribonucleoprotein
- N
- nucleoprotein
- MoRE
- molecular recognition element
- P
- phosphoprotein
- RdRp
- RNA-dependent RNA polymerase
- L
- polymerase
- SeV
- Sendai virus
- X domain
- MoRE-binding domain in P protein
- MeV-Edm
- measles virus Edmonston strain
- PNT
- amino-terminal N protein region
- PCT
- carboxyl-terminal N protein region
- RACE
- rapid amplification of cDNA ends.
REFERENCES
- 1. Ruigrok R. W., Crépin T., Kolakofsky D. (2011) Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr. Opin. Microbiol. 14, 504–510 [DOI] [PubMed] [Google Scholar]
- 2. Longhi S. (2009) Nucleocapsid structure and function. Curr. Top. Microbiol. Immunol. 329, 103–128 [DOI] [PubMed] [Google Scholar]
- 3. Bankamp B., Horikami S. M., Thompson P. D., Huber M., Billeter M., Moyer S. A. (1996) Domains of the measles virus N protein required for binding to P protein and self-assembly. Virology 216, 272–277 [DOI] [PubMed] [Google Scholar]
- 4. Curran J., Pelet T., Kolakofsky D. (1994) An acidic activation-like domain of the Sendai virus P protein is required for RNA synthesis and encapsidation. Virology 202, 875–884 [DOI] [PubMed] [Google Scholar]
- 5. Buchholz C. J., Retzler C., Homann H. E., Neubert W. J. (1994) The carboxy-terminal domain of Sendai virus nucleocapsid protein is involved in complex formation between phosphoprotein and nucleocapsid-like particles. Virology 204, 770–776 [DOI] [PubMed] [Google Scholar]
- 6. Kingston R. L., Hamel D. J., Gay L. S., Dahlquist F. W., Matthews B. W. (2004) Structural basis for the attachment of a paramyxoviral polymerase to its template. Proc. Natl. Acad. Sci. U.S.A. 101, 8301–8306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Desfosses A., Goret G., Farias Estrozi L., Ruigrok R. W., Gutsche I. (2011) Nucleoprotein-RNA orientation in the measles virus nucleocapsid by three-dimensional electron microscopy. J. Virol. 85, 1391–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jensen M. R., Communie G., Ribeiro E. A., Jr., Martinez N., Desfosses A., Salmon L., Mollica L., Gabel F., Jamin M., Longhi S., Ruigrok R. W., Blackledge M. (2011) Intrinsic disorder in measles virus nucleocapsids. Proc. Natl. Acad. Sci. U.S.A. 108, 9839–9844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang X., Glendening C., Linke H., Parks C. L., Brooks C., Udem S. A., Oglesbee M. (2002) Identification and characterization of a regulatory domain on the carboxyl terminus of the measles virus nucleocapsid protein. J. Virol. 76, 8737–8746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Longhi S., Receveur-Bréchot V., Karlin D., Johansson K., Darbon H., Bhella D., Yeo R., Finet S., Canard B. (2003) The C-terminal domain of the measles virus nucleoprotein is intrinsically disordered and folds upon binding to the C-terminal moiety of the phosphoprotein. J. Biol. Chem. 278, 18638–18648 [DOI] [PubMed] [Google Scholar]
- 11. Schoehn G., Mavrakis M., Albertini A., Wade R., Hoenger A., Ruigrok R. W. (2004) The 12 A structure of trypsin-treated measles virus N-RNA. J. Mol. Biol. 339, 301–312 [DOI] [PubMed] [Google Scholar]
- 12. Bhella D., Ralph A., Yeo R. P. (2004) Conformational flexibility in recombinant measles virus nucleocapsids visualised by cryo-negative stain electron microscopy and real-space helical reconstruction. J. Mol. Biol. 340, 319–331 [DOI] [PubMed] [Google Scholar]
- 13. Tawar R. G., Duquerroy S., Vonrhein C., Varela P. F., Damier-Piolle L., Castagné N., MacLellan K., Bedouelle H., Bricogne G., Bhella D., Eléouët J. F., Rey F. A. (2009) Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science 326, 1279–1283 [DOI] [PubMed] [Google Scholar]
- 14. Curran J., Homann H., Buchholz C., Rochat S., Neubert W., Kolakofsky D. (1993) The hypervariable C-terminal tail of the Sendai paramyxovirus nucleocapsid protein is required for template function but not for RNA encapsidation. J. Virol. 67, 4358–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kingston R. L., Baase W. A., Gay L. S. (2004) Characterization of nucleocapsid binding by the measles virus and mumps virus phosphoproteins. J. Virol. 78, 8630–8640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bourhis J. M., Canard B., Longhi S. (2006) Structural disorder within the replicative complex of measles virus. Functional implications. Virology 344, 94–110 [DOI] [PubMed] [Google Scholar]
- 17. Longhi S. (2012) The measles virus N(TAIL)-XD complex. An illustrative example of fuzziness. Adv. Exp. Med. Biol. 725, 126–141 [DOI] [PubMed] [Google Scholar]
- 18. Communie G., Crépin T., Maurin D., Jensen M. R., Blackledge M., Ruigrok R. W. (2013) Structure of the tetramerization domain of measles virus phosphoprotein. J. Virol. 87, 7166–7169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Houben K., Blanchard L., Blackledge M., Marion D. (2007) Intrinsic dynamics of the partly unstructured PX domain from the Sendai virus RNA polymerase cofactor P. Biophys. J. 93, 2830–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Curran J. (1998) A role for the Sendai virus P protein trimer in RNA synthesis. J. Virol. 72, 4274–4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Longhi S. (2011) Structural Disorder within the Measles Virus Nucleoprotein and Phosphoprotein: Functional Implications for Transcription and Replication, in Negative Strand RNA Virus (Luo M., ed.) pp. 95–125, World Scientific Publishing, Singapore [Google Scholar]
- 22. Shu Y., Habchi J., Costanzo S., Padilla A., Brunel J., Gerlier D., Oglesbee M., Longhi S. (2012) Plasticity in structural and functional interactions between the phosphoprotein and nucleoprotein of measles virus. J. Biol. Chem. 287, 11951–11967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Buchholz U. J., Finke S., Conzelmann K. K. (1999) Generation of bovine respiratory syncytial virus (BRSV) from cDNA. BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73, 251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ono N., Tatsuo H., Hidaka Y., Aoki T., Minagawa H., Yanagi Y. (2001) Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J. Virol. 75, 4399–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Doyle J., Prussia A., White L. K., Sun A., Liotta D. C., Snyder J. P., Compans R. W., Plemper R. K. (2006) Two domains that control prefusion stability and transport competence of the measles virus fusion protein. J. Virol. 80, 1524–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paal T., Brindley M. A., St Clair C., Prussia A., Gaus D., Krumm S. A., Snyder J. P., Plemper R. K. (2009) Probing the spatial organization of measles virus fusion complexes. J. Virol. 83, 10480–10493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krumm S. A., Ndungu J. M., Yoon J. J., Dochow M., Sun A., Natchus M., Snyder J. P., Plemper R. K. (2011) Potent host-directed small-molecule inhibitors of myxovirus RNA-dependent RNA-polymerases. PLoS One 6, e20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Radecke F., Spielhofer P., Schneider H., Kaelin K., Huber M., Dötsch C., Christiansen G., Billeter M. A. (1995) Rescue of measles viruses from cloned DNA. EMBO J. 14, 5773–5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Seki F., Yamada K., Nakatsu Y., Okamura K., Yanagi Y., Nakayama T., Komase K., Takeda M. (2011) The SI strain of measles virus derived from a patient with subacute sclerosing panencephalitis possesses typical genome alterations and unique amino acid changes that modulate receptor specificity and reduce membrane fusion activity. J. Virol. 85, 11871–11882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Madani F., Lind J., Damberg P., Adams S. R., Tsien R. Y., Gräslund A. O. (2009) Hairpin structure of a biarsenical-tetracysteine motif determined by NMR spectroscopy. J. Am. Chem. Soc. 131, 4613–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Takeda M., Takeuchi K., Miyajima N., Kobune F., Ami Y., Nagata N., Suzaki Y., Nagai Y., Tashiro M. (2000) Recovery of pathogenic measles virus from cloned cDNA. J. Virol. 74, 6643–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dochow M., Krumm S. A., Crowe J. E., Jr., Moore M. L., Plemper R. K. (2012) Independent structural domains in the paramyxovirus polymerase protein. J. Biol. Chem. 287, 6878–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Buchholz C. J., Spehner D., Drillien R., Neubert W. J., Homann H. E. (1993) The conserved N-terminal region of Sendai virus nucleocapsid protein NP is required for nucleocapsid assembly. J. Virol. 67, 5803–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gardner M. J., Altman D. G. (1986) Confidence intervals rather than P values. Estimation rather than hypothesis testing. Br. Med. J. (Clin. Res. Ed.) 292, 746–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Iwasaki M., Takeda M., Shirogane Y., Nakatsu Y., Nakamura T., Yanagi Y. (2009) The matrix protein of measles virus regulates viral RNA synthesis and assembly by interacting with the nucleocapsid protein. J. Virol. 83, 10374–10383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Couturier M., Buccellato M., Costanzo S., Bourhis J. M., Shu Y., Nicaise M., Desmadril M., Flaudrops C., Longhi S., Oglesbee M. (2010) High affinity binding between Hsp70 and the C-terminal domain of the measles virus nucleoprotein requires an Hsp40 co-chaperone. J. Mol. Recognit. 23, 301–315 [DOI] [PubMed] [Google Scholar]
- 37. Zhang X., Bourhis J. M., Longhi S., Carsillo T., Buccellato M., Morin B., Canard B., Oglesbee M. (2005) Hsp72 recognizes a P binding motif in the measles virus N protein C-terminus. Virology 337, 162–174 [DOI] [PubMed] [Google Scholar]
- 38. Griffin B. A., Adams S. R., Tsien R. Y. (1998) Specific covalent labeling of recombinant protein molecules inside live cells. Science 281, 269–272 [DOI] [PubMed] [Google Scholar]
- 39. Duprex W. P., McQuaid S., Hangartner L., Billeter M. A., Rima B. K. (1999) Observation of measles virus cell-to-cell spread in astrocytoma cells by using a green fluorescent protein-expressing recombinant virus. J. Virol. 73, 9568–9575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Collins P. L., Wertz G. W. (1983) cDNA cloning and transcriptional mapping of nine polyadenylylated RNAs encoded by the genome of human respiratory syncytial virus. Proc. Natl. Acad. Sci. U.S.A. 80, 3208–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Houben K., Marion D., Tarbouriech N., Ruigrok R. W., Blanchard L. (2007) Interaction of the C-terminal domains of sendai virus N and P proteins. Comparison of polymerase-nucleocapsid interactions within the paramyxovirus family. J. Virol. 81, 6807–6816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rennick L. J., Duprex W. P., Rima B. K. (2007) Measles virus minigenomes encoding two autofluorescent proteins reveal cell-to-cell variation in reporter expression dependent on viral sequences between the transcription units. J. Gen. Virol. 88, 2710–2718 [DOI] [PubMed] [Google Scholar]
- 43. Diallo A., Barrett T., Barbron M., Meyer G., Lefèvre P. C. (1994) Cloning of the nucleocapsid protein gene of peste-des-petits-ruminants virus. Relationship to other morbilliviruses. J. Gen. Virol. 75, 233–237 [DOI] [PubMed] [Google Scholar]
- 44. Cox R., Green T. J., Purushotham S., Deivanayagam C., Bedwell G. J., Prevelige P. E., Luo M. (2013) Structural and functional characterization of the mumps virus phosphoprotein. J. Virol. 87, 7558–7568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Laine D., Trescol-Biémont M. C., Longhi S., Libeau G., Marie J. C., Vidalain P. O., Azocar O., Diallo A., Canard B., Rabourdin-Combe C., Valentin H. (2003) Measles virus (MV) nucleoprotein binds to a novel cell surface receptor distinct from FcγRII via its C-terminal domain. Role in MV-induced immunosuppression. J. Virol. 77, 11332–11346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Laine D., Bourhis J. M., Longhi S., Flacher M., Cassard L., Canard B., Sautès-Fridman C., Rabourdin-Combe C., Valentin H. (2005) Measles virus nucleoprotein induces cell-proliferation arrest and apoptosis through NTAIL-NR and NCORE-FcγRIIB1 interactions, respectively. J. Gen. Virol. 86, 1771–1784 [DOI] [PubMed] [Google Scholar]
- 47. Cevik B., Kaesberg J., Smallwood S., Feller J. A., Moyer S. A. (2004) Mapping the phosphoprotein binding site on Sendai virus NP protein assembled into nucleocapsids. Virology 325, 216–224 [DOI] [PubMed] [Google Scholar]
- 48. Cathomen T., Mrkic B., Spehner D., Drillien R., Naef R., Pavlovic J., Aguzzi A., Billeter M. A., Cattaneo R. (1998) A matrix-less measles virus is infectious and elicits extensive cell fusion. Consequences for propagation in the brain. EMBO J. 17, 3899–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Curran J., Kolakofsky D. (1999) Replication of paramyxoviruses. Adv. Virus Res. 54, 403–4222 [DOI] [PubMed] [Google Scholar]
- 50. Kolakofsky D., Le Mercier P., Iseni F., Garcin D. (2004) Viral DNA polymerase scanning and the gymnastics of Sendai virus RNA synthesis. Virology 318, 463–473 [DOI] [PubMed] [Google Scholar]
- 51. Johansson K., Bourhis J. M., Campanacci V., Cambillau C., Canard B., Longhi S. (2003) Crystal structure of the measles virus phosphoprotein domain responsible for the induced folding of the C-terminal domain of the nucleoprotein. J. Biol. Chem. 278, 44567–44573 [DOI] [PubMed] [Google Scholar]