Background: Mycobacterium tuberculosis HbN detoxifies nitric oxide and protects its host under nitrosative stress.
Results: The HbN remains glycosylated and membrane-localized in M. tuberculosis and modulates host-pathogen interactions.
Conclusion: The HbN facilitates intracellular infection and cell survival by evading the immune system of the host.
Significance: This study unravels new knowledge about function(s) of HbN in biology and pathogenesis of M. tuberculosis.
Keywords: Hemoglobin, Hemoglobin Myoglobin, Host-Pathogen Interactions, Microbiology, Mycobacterium tuberculosis, Post-translational Modification
Abstract
Mycobacterium tuberculosis (Mtb) is a phenomenally successful human pathogen having evolved mechanisms that allow it to survive within the hazardous environment of macrophages and establish long term, persistent infection in the host against the control of cell-mediated immunity. One such mechanism is mediated by the truncated hemoglobin, HbN, of Mtb that displays a potent O2-dependent nitric oxide dioxygenase activity and protects its host from the toxicity of macrophage-generated nitric oxide (NO). Here we demonstrate for the first time that HbN is post-translationally modified by glycosylation in Mtb and remains localized on the cell membrane and the cell wall. The glycan linkage in the HbN was identified as mannose. The elevated expression of HbN in Mtb and M. smegmatis facilitated their entry within the macrophages as compared with isogenic control cells, and mutation in the glycan linkage of HbN disrupted this effect. Additionally, HbN-expressing cells exhibited higher survival within the THP-1 and mouse peritoneal macrophages, simultaneously increasing the intracellular level of proinflammatory cytokines IL-6 and TNF-α and suppressing the expression of co-stimulatory surface markers CD80 and CD86. These results, thus, suggest the involvement of HbN in modulating the host-pathogen interactions and immune system of the host apart from protecting the bacilli from nitrosative stress inside the activated macrophages, consequently driving cells toward increased infectivity and intracellular survival.
Introduction
Tuberculosis, the leading cause of nearly 2 million human deaths annually (1, 2), primarily develops after lung infection by Mycobacterium tuberculosis (Mtb),2 which has many manifestations affecting different organs, including the central nervous system. The progression of the disease is largely determined by the genetic background and physiological state of the pathogen along with suppression of the immune system of the host (3, 4). Many human epidemiological studies have demonstrated variations in the level of certain key components of virulence, such as stress survival, transmission, etc., among different strains of Mtb that may have dramatic consequences on the outcome of the infection (5, 6). It has been observed that some clinical isolates of Mtb display differences in the level of various cellular components and are more virulent than others, causing higher mortality in infected individuals (7). For example, a bioactive polyketide synthase-derived phenolic glycolipid (PKS) is produced by highly virulent strains of Mtb, HN878 and W4, but not by CDC1551 or H37Rv (8). This led to speculation that the differential level of some cellular components may play a crucial role in modulating the virulence and pathogenicity of Mtb.
The truncated hemoglobin, HbN, constitutes one of the vital components of the defense system in Mtb that may aid its persistence and intracellular survival due to its high oxygen affinity and efficient NO detoxification ability (9, 10, 11). The HbN-deficient strain of Mycobacterium bovis displays extremely low NO dioxygenase (NOD) activity and lacks respiratory protection from NO as compared with the native strain (12), substantiating the theory that the presence of HbN contributes to its survival ability in the NO-enriched environment of macrophages. This is also supported by the fact that the expression of Mtb HbN in the HMP mutant of Salmonella enterica Typhimurium enhances its growth and survival inside the THP-1 macrophages (11). Although HbN is produced at stationary phase of an aerobically growing culture of Mtb (9), transcriptional activities of the glbN gene increase nearly 2-fold within 48 h of macrophage infection by the tubercle bacillus (13), thereby indicating an increased requirement of HbN during its intracellular regime. Additionally, our primary studies indicated distinct variation in the expression level of the glbN gene in Mtb and M. smegmatis.3 Thus, it can be anticipated that changes in the cellular levels of HbN may have significant consequences on the pathogenic life cycle of Mtb.
Structural and biochemical studies done so far on the HbN of Mtb are mainly based on Escherichia coli-derived protein. The physiological state of the HbN is still not known in mycobacteria. Therefore, we attempted to investigate the subcellular localization and physiological state of the HbN in its native host, Mtb. We tested the assumption that the enhanced level of cellular HbN would influence the ability of Mtb to persist and survive better in its intracellular niche. Therefore, we cloned and overexpressed the glbN gene of Mtb in mycobacterial strains and studied its effect on infectivity, intracellular survival, and immune responses of the host. The present study demonstrates for the first time that HbN is post-translationally modified by glycosylation in its native host and remains localized on the cell membrane and the cell wall. The enhanced expression of HbN alters the membrane lipid profile of Mtb and changes the expression of co-stimulatory surface markers and the balance of pro- and anti-inflammatory cytokines during intracellular infection. These results, thus, unravel new functions of HbN that may be vital for the virulence and pathogenic life cycle of the tubercle bacillus.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Culture Conditions
E. coli strains JM109 and BL21DE3 were used routinely for the cloning and expression of recombinant genes. Cultures of E. coli strains were grown in Luria-Bertani (LB) or Terrific broth (containing 24 g of yeast extract, 12 g of Bacto-tryptone, 12.3 g of K2HPO4, 2.3 g of KH2PO4) at 37 °C and 180 rpm. Mycobacterial strains, M. tuberculosis H37Rv (MtbRv), M. tuberculosis H37 Ra (MtbRa), and M. smegmatis mc2 155, were used for the experimental studies and were grown in Middlebrook 7H10 agar (Difco) or 7H9 broth, supplemented with ADC (10% bovine serum albumin fraction V, dextrose, and sodium chloride), 0.2% glycerol, and 0.05% Tween 80. When required, ampicillin (Sigma) and hygromycin B (Sigma) were added at a concentration of 100 and 200 μg/ml, respectively, for E. coli. Hygromycin was added to 50 μg/ml for selective growth of mycobacteria. The plasmids p19Kpro (14) and pSC301 (15) were used for the cloning and the expression of recombinant genes in mycobacteria. Oligonucleotides were custom-synthesized by Essence Life Sciences (Chandigarh, India).
Mice C57BL/6 (female, 4–6 weeks) were procured from the National Institute of Pharmacological Education and Research (Mohali) and CSIR-Institute of Microbial Technology (Chandigarh, India). All experiments were approved by the Institutional Bio-safety and Animal Ethics Committees of the CSIR-Institute of Microbial Technology.
Expression of HbN in Mycobacteria
For the expression of HbN in mycobacteria, the glbN gene was amplified through PCR using gene-specific primers carrying a BamHI restriction site at the 5′-end and a PstI site at the 3′-end. The PCR product was cloned at the BamHI-PstI site of the mycobacterial expression vector, p19Kpro, under the promoter of the 19-kDa antigen of Mtb. The glbN gene with a His6 tag at the C terminus was also cloned in pSC301 under the superoxide dismutase promoter. The resulting expression plasmids were designated as p19kpro-HbN and pSC301-HbN. Expression of recombinant HbN protein was visualized by 15% SDS-PAGE after probing with HbN-specific polyclonal antibodies as described previously (10).
Microscopic Analysis of Cells
For scanning electron micrographs, mycobacterial cells were first suspended in buffer carrying 10 mm Tris·Cl (pH 7.5), 10 mm MgCl2, and 0.02% (v/v) Tween 80 and passed through the syringe to break clumps. After that, cells were first examined by conventional microscopy (Olympus BX51) and then visualized through scanning electron microscopy (Carl Zeiss).
Characterization of the HbN Expressed in Mtb
The recombinant HbN was expressed in E. coli and Mtb with a His6 tag at the C terminus and purified through metal affinity chromatography (nickel-nitrilotriacetic acid column; Invitrogen) following the manufacturer's instructions. Total protein concentration was determined using the bicinchoninic acid (BCA) kit (Pierce). Both species of HbN were analyzed by 15% SDS-PAGE after Western blotting using polyclonal anti-HbN and peroxidase-conjugated concanavalin A (ConA) antibodies (Sigma). Molecular mass of the intact protein was analyzed through matrix-assisted laser desorption/ionization (MALDI) following a standard procedure.
Analysis of ConA Binding
The whole cell lysate of wild type or HbN-expressing MtbRa or the purified/recombinant HbN from Mtb was resolved by 15% SDS-PAGE, transferred to a nitrocellulose membrane using standard procedures, and incubated with 5 μg/ml ConA-peroxidase (Sigma) in PBS (containing 1 mm CaCl2, 1 mm MnCl2, and 1 mm MgCl2) overnight at 20 °C. The unbound ConA was removed by washing the blot with PBS, and after that, it was probed with peroxidase-conjugated ConA antibodies using an ECL chemiluminescent kit (Pierce).
Identification of Glycan Linkage with the HbN
The glycan linkage in the HbN was analyzed by releasing the carbohydrate after hydrolyzing the protein with trifluroacetic acid at 95 °C for 4 h, as described elsewhere (16). The hydrolysis reaction was dried under vacuum at 40 °C and then resuspended in 200 μl of water. The sample was then analyzed on an Aminex HPX-87H column on a high performance liquid chromatograph (Shimadzu CLASS-VP V6.14 SP1) equipped with a refractive index and UV detector. Sugars were eluted from the column with 5 mm sulfuric acid at a flow rate of 0.60 ml/min. Sugars were identified by comparing the column retention time with that of sugar standards (Sigma) prepared at known concentrations and run under identical conditions.
Extraction, Fractionation, and Lipid Analysis
Mycobacterial lipids and fatty acids were extracted and analyzed using the Microbial Identification System. Lipids were extracted by using a published procedure (17), and spots were detected by the following spray reagents: molybdatophosphoric acid (5% (w/v) in absolute ethanol), molybdenum blue spray reagent, ninhydrin (0.2% (w/v) in acetone), and anisaldehyde reagent for the detection of total lipids, phospholipids, aminolipids, and glycolipids, respectively. Further characterization of lipid components was done by fatty acid methyl ester analysis through GC/MS and fatty acid profiles were compared with the fragmentation profiles of the library of MIDI.
NOD Assay
NOD activities of cells were monitored polarographically by a Free Radica1 Analyzer using an NO microelectrode (WPI) as described previously (10). NO consumption buffer contained 60 mm K2HPO4, 33 mm KH2PO4, 7.5 mm (NH)2SO4, 1.5 mm sodium citrate, 10 mm glucose, and 200 μg/ml chloramphenicol. NO uptake rate of aerobically growing cells was checked for 1 μm NO and was corrected for the background NO decomposition. Total heme content of the cells was checked as mentioned previously (10).
Assays of Cytokine Levels
Supernatants from the infected THP-1 or mouse peritoneal cell macrophages were harvested at 48 h postinfection, and concentrations of tumor necrosis factor α (TNF-α), interleukins (IL-6 and IL-10), and various surface markers were measured by enzyme-linked immunosorbent assay (ELISA) using a commercially available kit (BD Pharmingen) following the manufacturer's instructions. Fluorochrome-labeled antibodies were used for flow cytometry, namely CD80-APC (16-10A1), CD86-PE (GL1), CD40-FITC (3/23), and IAb-biotin (KH74).
Macrophage Infection and Determination of Colony-forming Units (cfu)
Exponentially growing cultures of control and HbN-overexpressing cells of MtbRa were first washed three times with sterile phosphate buffer (pH 7.4). Prior to infection, MtbRa cells were dispersed by vortexing, sonicated in a sonicator, and then passed through a 30-gauge needle several times to obtain a single-cell suspension. Phorbol 12-myristate 13-acetate-differentiated THP-1 or mouse peritoneal macrophage cells, as required, were plated into 24-well flat bottom plates (Falcon) at 5 × 105 cells/well. The cells were infected with diluted and dispersed preparations of MtbRa to give an infection ratio of ∼20 bacilli/macrophage. After infection, extracellular bacteria were removed by washings with Hanks' balanced salt solution and treatment with gentamycin (200 μg/ml) for 1 h. The adequacy of dispersion and multiplicity of infection (MOI) were checked by performing acid-fast staining of washed infected cells. The cells were incubated at 37 °C in 95% air + 5% CO2 for 4 h. Infected macrophages were cultured in RPMI 1640 medium supplemented with 10% FBS (Invitrogen), l-glutamine (2 mm), HEPES (2.38 mg/ml), sodium bicarbonate (2.2 mg/ml), and 2-mercaptoethanol (0.05 mm). Bacteria were isolated at a specific time point, and viability of bacteria was determined after appropriate dilutions and plating on Middlebrook agar plates supplemented with ovalbumin, albumin, dextrose, and catalase and then counting the number of colonies. Data are expressed as mean cfu/ml of the lysate ± S.D.
Flow Cytometry for Monitoring the Expression of MHC-II and Co-stimulatory Molecules CD80, CD86, and CD40
Thioglycollate-elicited peritoneal exudate cells were infected with 15- and 30-day grown culture of wild type (H37Ra15 and H37Ra30) and HbN-expressing cells (HbN15 and HbN30) of MtbRa (1:20 MOI) for 48 h. Thereafter, cells were harvested and washed three times with 1% PBS-FCS (fetal calf serum in phosphate-buffered saline) and stained for surface expression of CD40, CD80, CD86, and MHC-II with their respective fluorochrome-tagged antibodies by incubating at 4 °C for 30 min. The usual steps of washing with 1% PBS-FCS were followed after each incubation. The cells were fixed with 1% paraformaldehyde and acquired on a FACSCalibur flow cytometer and analyzed by FACSDiva software.
RESULTS
Implications of Enhanced Expression of HbN on Physiology of Mtb
Because the expression levels of several genes differ in clinical and laboratory strains of MtbRv (5) and transcriptional activity of the glbN gene increases significantly under nitrosative stress and hypoxia and during macrophage infection (13), we checked the effects of an increased level of HbN for the physiological properties of its native host. The glbN gene of MtbRv has been cloned individually under the constitutive promoter of 19-kDa antigen as a native gene, and the superoxide dismutase promoter of Mtb has been cloned as a His-tagged protein (see “Experimental Procedures”) and transformed into mycobacterial strains M. smegmatis and MtbRa (having the glbN gene and its upstream regulatory region identical to MtbRv). Western blot analysis, using anti-HbN polyclonal antibodies, confirmed constitutive expression of HbN at all stages of growth in transformed mycobacterial cells as opposed to control cells, where HbN was detected only at the late stationary phase of aerobically grown cultures of MtbRa.
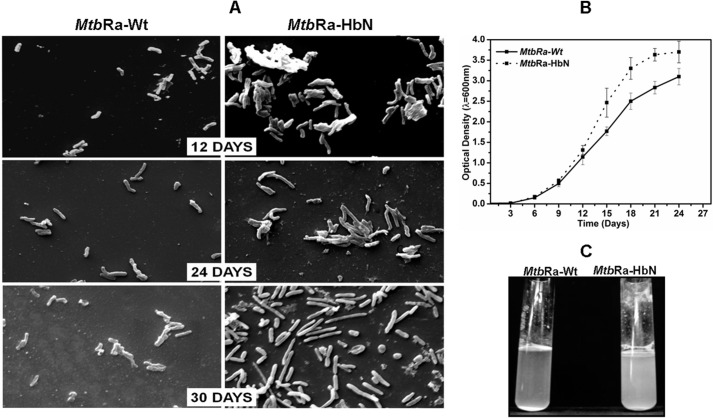
MtbRa transformed with HbN expression plasmid formed colonies with smooth texture as compared with rough colonies of the control cells. Scanning electron microscopy of these cells indicated that HbN-overexpressing cells of MtbRa are highly aggregated and relatively elongated compared with the control cells (Fig. 1A). Maximum aggregation and elongation were observed in 25–30-day cultures. The growth profile of the control and the HbN-overexpressing cells of MtbRa followed a similar pattern except that the HbN-expressing cells grew slightly better and attained higher cell mass at stationary phase (25–30 days), showing increased clumping and adherence at the side of the flasks or tubes (Fig. 1, B and C).
FIGURE 1.
Overexpression of glbN gene in MtbRa. A, scanning electron micrograph of wild type (MtbRa-Wt) and HbN-overexpressing (MtbRa-HbN) cells of MtbRa at different phases of growth. B, growth profile of wild type (continuous line) and HbN-overexpressing (dotted line) cells of MtbRa. C, growth properties of 25-day grown culture of HbN-overexpressing (MtbRa-HbN) and wild type (MtbRa-Wt) cells of MtbRa. Error bars, S.D.
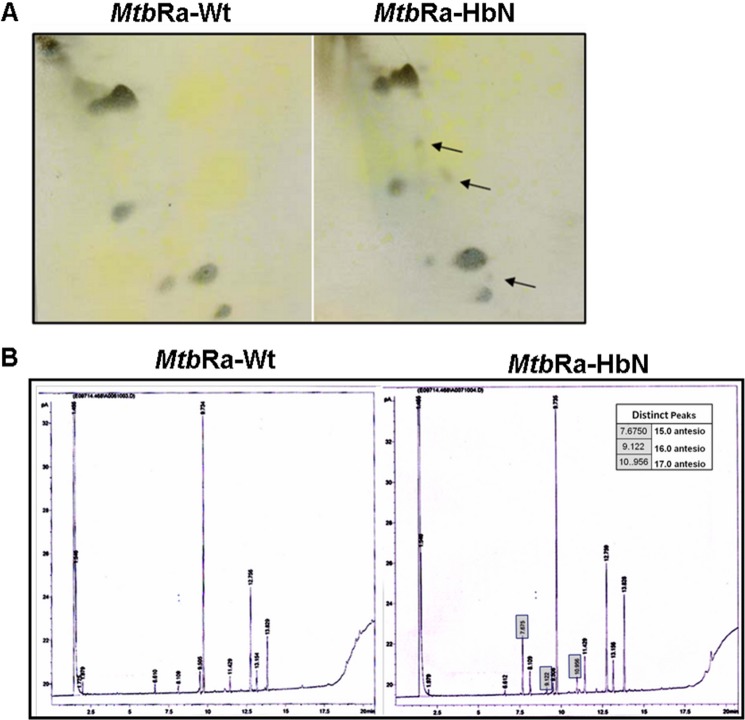
To further investigate whether the changes in the cell aggregation in the HbN-overexpressing cells of MtbRa are in any way correlated with any biochemical changes in the cell wall or the plasma membrane, we checked and compared the lipid content of the HbN-expressing cells with that of isogenic wild type Mtb. Total lipid content of the HbN-expressing cells appeared relatively higher than that of the control cells, specifically at stationary phase (25–30 days), when cells displayed maximum cell aggregation and accumulation of HbN. TLC analysis of mycobacterial cell wall lipids substantiated distinct changes in the membrane polar lipids and revealed the appearance of a few new species of lipids in the HbN-overexpressing Mtb (Fig. 2A). Fatty acid methyl ester analysis of membrane lipids subsequently confirmed the changes in the fatty acids of membrane lipids in the HbN-expressing cells, showing the appearance of new species of lipids and distinct changes in the level of some fatty acids (Fig. 2B). Some of the prominently up-regulated fatty acids in the HbN-overexpressing cells were 15.0 anteiso (pentadecanoic acid), 16.0 anteiso (palmitic acid), and 17.0 anteiso (methyl hexadecanoic acid). These results suggested that increased expression of HbN in Mtb may have some impact on fatty acid biosynthesis in the cell, specifically the anteiso-fatty acids that have been found up-regulated.
FIGURE 2.
Lipid profile of native and HbN-overexpressing cells of MtbRa. A, two-dimensional TLC analysis of polar lipids of cell membrane in wild type and HbN-expressing MtbRa. The arrow indicates the up-regulated/newly appeared lipids in the cell wall. B, fatty acid methyl ester analysis of wild type (MtbRa-Wt) (left) and HbN-overexpressing (MtbRa-HbN) (right) cells of MtbRa. Some of the prominently up-regulated new fatty acids in the membrane lipids of HbN-overexpressing cells of MtbRa are shown in boxes.
Subcellular Localization of HbN in Mtb
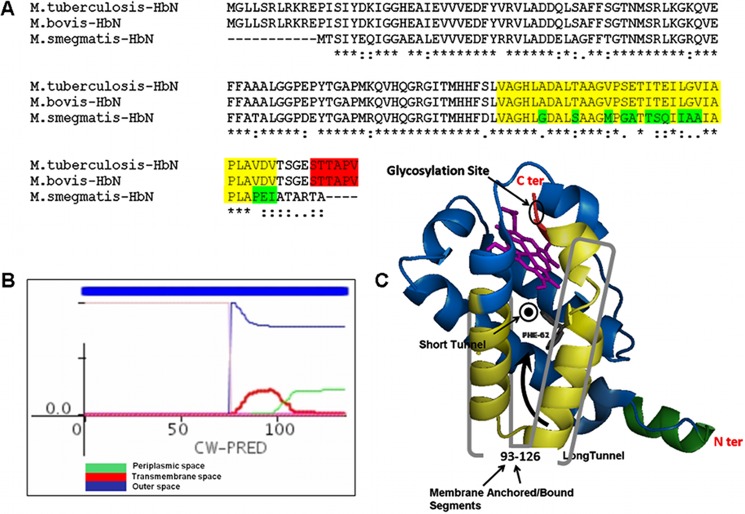
To gain insight into the implications of increased HbN expression on the physiology of Mtb, we checked cellular localization of the HbN in the wild type and the HbN-expressing cells of MtbRa after cell fractionation and immunoblotting. The HbN appeared localized mainly on the membrane and the cell wall in the wild type and the HbN-overexpressing cells of Mtb (Fig. 3A) as opposed to E. coli, where it is expressed as a cytoplasmic protein (9, 10). A small amount of HbN was also detected occasionally in the cytoplasmic fraction of the HbN-overexpressing cells but not in the case of wild type cells. To further check whether the cell wall-associated HbN is exposed on the mycobacterial cell surface, we performed a trypsin sensitivity test using HbN-expressing cells of Mtb and comparing them with isogenic GFP-expressing cells as a control for the cytoplasmic protein. When whole cells of HbN-expressing Mtb were subjected to trypsin treatment, a distinct decrease in the level of HbN was observed within 10 min as compared with the untreated control cells (Fig. 3B). Conversely, no change in the level of cytoplasmic GFP was observed when exposed to similar trypsin treatment. These results suggest that a significant amount of HbN remains exposed on the cell surface in Mtb. Because HbN lacks a conventional signal peptide but carries a positively charged Pre-A region at the N terminus, we tested if it is responsible for its membrane targeting. When the Pre-A-deleted mutant of HbN was checked, it still exhibited membrane localization in Mtb similar to the wild type HbN (results not shown), thus ruling out its role in membrane localization of HbN and suggesting the involvement of other factors in this process. Bioinformatic analysis of the HbN primary sequences, using the CW-Pred (18) and DAS (19) programs, suggested that the C-terminal region of HbN, spanning amino acid residues 93–123, has a high probability of being a transmembrane region (Fig. 5, A and B) and might be contributing in cellular localization of HbN in mycobacteria. Interestingly, the corresponding region of M. smegmatis HbN displayed several changes and lower probability for the extracellular region when compared with the HbN of Mtb.
FIGURE 3.
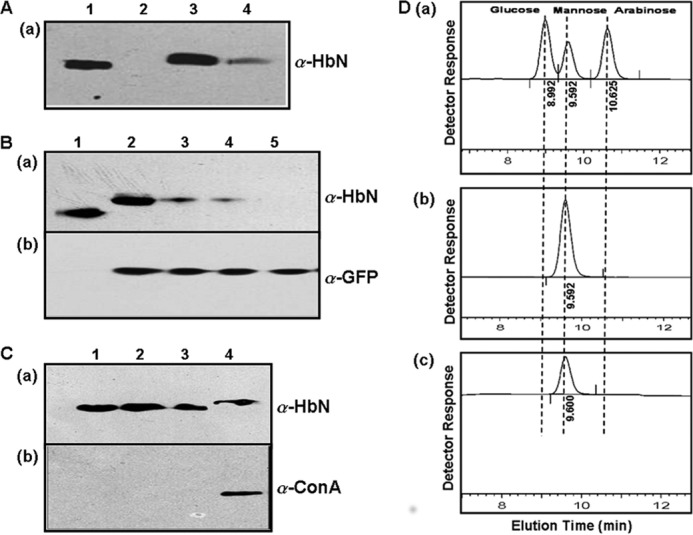
Cellular localization and post-translational modification of HbN in MtbRa. A, Western blot analysis of cellular fractions of MtbRa probed with HbN-specific polyclonal antibodies (α-HbN). Lane 1, whole cell lysate; lane 2, cytoplasmic fraction; lane 3, membrane fraction; lane 4, cell wall fraction. B, trypsin sensitivity test for the extracellular localization of HbN in MtbRa. Whole cells of HbN of MtbRa-HbN were exposed at 4 °C with 100 μg/ml trypsin for 0 (lane 2), 5 (lane 3), 10 (lane 4), and 15 (lane 5) min and probed with HbN-specific antibodies (a) and GFP-specific antibodies (α-GFP) (b). GFP was taken as control for cytoplasmic protein. The treatment was given for different time points as in A. C, Western blot analysis of HbN mutants altered at the glycosylation site and probed with HbN-specific polyclonal antibodies (a) and binding to ConA antibody (α-ConA) (b). Lane 1, wild type HbN expressed in E. coli; lane 2, HbN mutant carrying a replacement of Thr-132 and Thr-133 with alanine and expressed in MtbRa; lane 3, HbN mutants carrying a 4-residue (TAPV) deletion at the C terminus and expressed in MtbRa; lane 4, wild type HbN expressed in MtbRa. D, analysis of the sugar linked with HbN overexpressed in MtbRa. Sugar attached with the HbN was released after acid hydrolysis and identified by Dionex ion chromatography after comparing with standard sugars. Shown are elution profiles of standard sugars having a mixture of glucose, mannose, and arabinose (a); mannose (b); and sugar extracted from the HbN (c).
FIGURE 5.
In silico analysis of the primary sequence of HbN of Mtb. A, multiple-sequence alignment of the amino acid sequence of HbN from M. tuberculosis, M. bovis, and M. smegmatis. The segment shown in fluorescent yellow depicts extracellular and membrane-anchored/bound regions predicted in the HbN of M. tuberculosis and M. bovis. The glycosylation site in HbN is highlighted in red. The differences within the membrane-anchored/bound region in the HbN of M. smegmatis are highlighted in green. B, prediction of membrane-bound/anchored region in Mtb HbN using CW-PRED that assigns a score of 1.0 to such proteins. The output here depicts three different colors to define the triplet state in which a residue can be present, namely periplasmic, extracellular, or transmembrane. Predictions for all of the states are combined in the output, and the analysis indicated that the region spanning amino acid residues 93–126 at the C terminus may constitute a potential extracellular association of MtbRa-HbN. C, a hypothetical schematic model of HbN depicting the site of glycosylation and the potential membrane-anchored segment shown in yellowish green.
Post-translational Modification of HbN in Mtb
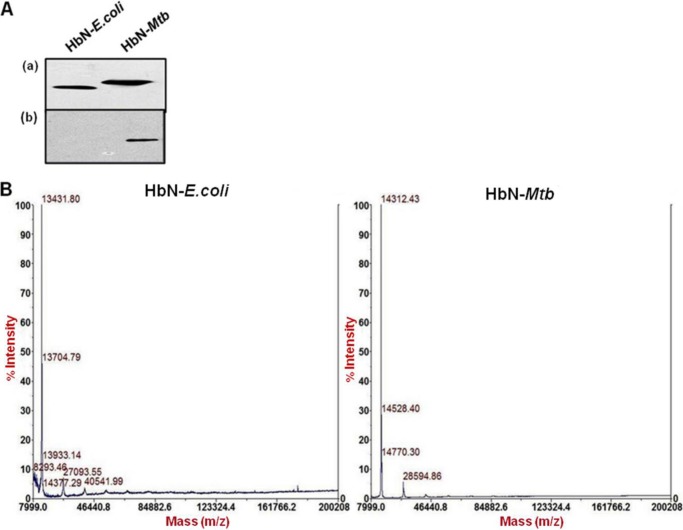
While analyzing the HbN expressed in E. coli and Mtb, we observed a distinct difference in the protein size on the Western blot that cross-reacted with HbN-specific antibodies (Figs. 3 and 4A). Slower migration of HbN expressed in Mtb suggested that it may be post-translationally modified. Because several cell wall- and membrane-associated proteins of Mtb have been found glycosylated (20), we first checked the possibility of a glycan linkage with the HbN. When two species of HbN, obtained from the E. coli and Mtb, were probed with ConA and HbN antibodies, both species of HbN cross-reacted with anti-HbN antibodies, but ConA reactivity was observed only with the HbN isolated from the Mtb (Fig. 4A). MALDI analysis (Fig. 4B) further substantiated the difference in the size of the HbN produced in E. coli (13.4 kDa) and Mtb (14.3 kDa). Furthermore, a putative O-mannosylation site (STTAP) was identified at the C-terminal region of HbN (Fig. 5A). The nature of the sugar residue linked with the HbN of Mtb was identified on HPLC after extracting the carbohydrate from the protein and comparing its elution profile with the standard having a mixture of sugars (Fig. 3D). The elution pattern of carbohydrate released from the HbN confirmed the linkage of mannose with the protein (Fig. 3D). It is noteworthy that the O-mannosylation site, present in the HbN, is restricted to the members of only the Mtb complex (supplemental Fig. S1) and is not present in the HbN of non-pathogenic strains, such as M. smegmatis (Fig. 5A).
FIGURE 4.
Characteristics of the HbN expressed in MtbRa. A, Western blot analysis of HbN expressed in E. coli and MtbRa and probed with HbN-specific antibodies (a) and binding to peroxidase-conjugated ConA (b). B, MALDI analysis of the molecular mass of HbN expressed in E. coli and MtbRa.
To validate the site of glycosylation in the HbN, two site-directed mutants were generated. In the first case, two threonine residues of the glycosylation motif in HbN were substituted with an alanine (HbNT132A,T133A), and in the second case, the glycosylation motif was disrupted by deleting last 4 amino acid residues from the C terminus of the HbN (HbN C4del). Both of these HbN mutants displayed faster migration than the wild type HbN on SDS-PAGE and lacked ConA reactivity when expressed in Mtb (Fig. 3C). These results confirmed the site of glycan linkage with the HbN. Unglycosylated mutants of the HbN were localized mainly on the cell membrane but not on the cell wall and displayed lower trypsin sensitivity than the wild type HbN when overexpressed in Mtb (data not shown). These results suggest the requirement of glycosylated HbN for the cell surface exposure in Mtb.
NO Scavenging by M. smegmatis Expressing Wild Type and Glycosylation-deficient Mutant of HbN
Because a lack of glycosylation in HbN reduced its cell surface exposure and the HbN homolog in M. smegmatis is attenuated in NOD function (21) and lacks a glycosylation motif (Fig. 5A), we checked the NOD activity of M. smegmatis expressing wild type and the glycosylation mutant of the HbN. Aerobically growing cells of wild type M. smegmatis displayed extremely slow NO scavenging. Expression of Mtb HbN in M. smegmatis elevated the NOD activity of these cells more than 100-fold, although the cellular level of plasmid-encoded (Table 1) HbN constituted nearly 2–3% of total protein, indicating that glycosylated HbN is functioning as an efficient NO scavenger. When the glycosylation-deficient mutant of HbN was similarly expressed in M. smegmatis, NOD activity of these cells appeared to be nearly half of that expressing wild type HbN, suggesting that post-translational modification of HbN may be important for the optimal NO scavenging and detoxification by HbN-expressing cells of the tubercle bacillus. In contrast, no distinct difference in the NOD activity of recombinant E. coli expressing wild type and the glycosylation-lacking mutant of HbN was observed (Table 1).
TABLE 1.
NO uptake by recombinant E. coli and M. smegmatis expressing glycosylation-deficient mutant of Mtb HbN
Cultures of E. coli and M. smegmatis mc2 155 were grown in vigorously aerated LB and Middlebrook 7H9 (Difco) medium, respectively, and harvested at the late exponential phase of growth when the culture A600 reached 0.8. Cells were washed with NO consumption buffer as described previously (10) and resuspended in the same buffer at a density of 5 × 107 cells/ml. NO consumption activity of cells was checked by NO microelectrode (WPI) following the procedure described previously (10). Data represent the mean of three independent results.
| Strains | Heme content | NO consumption activity |
|---|---|---|
| pmol/1010 cells | nmol−1 1010 cells | |
| E. coli BL21DE3 | 3.4 ± 0.9 | 0.05 ± 0.0 |
| E. coli BL21DE3 (HbN Wt) | 34.7 ± 4.8 | 17.84 ± 4.1 |
| E. coli BL21DE3 (HbNT132A,T133A) | 39.1 ± 5.6 | 19.4 ± 3.5 |
| M. smegmatis | 2.6 ± 1.1 | 0.11 ± 0.06 |
| M. smegmatis (HbN WT) | 24.6 ± 3.8 | 36.8 ± 4.1 |
| M. smegmatis (HbNT132A,T133A) | 29.4 ± 3.3 | 20.9 ± 2.9 |
Macrophage Infectivity and Intracellular Survival of HbN-overexpressing Mtb
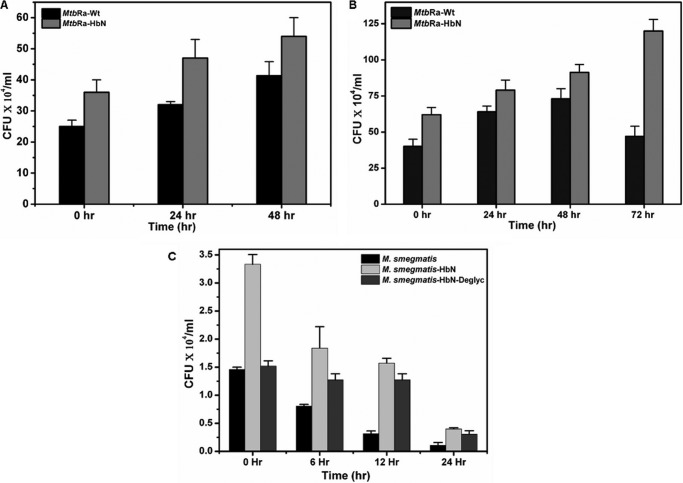
Because HbN remained localized on the cell membrane and the cell wall in a glycosylated state and caused significant changes in the membrane lipids during overexpression in MtbRa, we tested whether enhanced level of HbN in the cell provides any benefit during intracellular infection and survival as compared with isogenic control cells. The mouse peritoneal macrophages were infected with well dispersed cells of wild type and HbN-overexpressing MtbRa in a ratio of 1:20 (20 bacterial cells/macrophage), and the intracellular survival of tubercle bacilli was checked at different time intervals. As compared with control cells, a distinct increase in cfu of the HbN-overexpressing cells was observed (Fig. 6A) in mouse peritoneal cell lines at 0 h postinfection (cfu taken immediately after infection and washing), suggesting that the HbN-overexpressing cells are able to infect macrophages better than the wild type cells. Additionally, numbers of viable cells (cfu) were significantly higher in the case of HbN-overexpressing cells than the control when checked at 24, 48, and 72 h postinfection. Similar results were obtained in THP-1 macrophages (Fig. 6B).
FIGURE 6.
Intracellular infection and survival of wild type and HbN-overexpressing mycobacteria. Shown are cfu counts at different time intervals after infection with MtbRa-Wt and MtbRa-HbN. Shown are wild type (black bar) and HbN-overexpressing (gray bar) MtbRa in mouse peritoneal (A) and THP-1 cell lines (B). C, cfu counts at different time intervals after infection with M. smegmatis wild type (black bar) and M. smegmatis overexpressing HbN (light gray) and its unglycosylated mutant (gray bar) in RAW cell line. The data shown are representative of three independent experiments. Error bars, S.D.
Glycosylation-deficient Mutant of HbN Displays Lower Infectivity during Macrophage Infection
Our extensive attempt to generate a HbN knockout strain of MtbRa did not succeed; therefore, we selected M. smegmatis as an alternate system due to the fact that the HbN homolog in M. smegmatis is not expressed in aerobically growing cells3 and lacks a glycosylation site. To check whether glycosylation of HbN has any effect on the efficiency of intracellular infection, wild type and the glycosylation mutant of HbN (HbNT132A,T133A) was expressed in M. smegmatis. Wild type M. smegmatis infected macrophages with lower efficiency and was unable to survive for a longer duration within the macrophages (Fig. 6C). Interestingly, M. smegmatis expressing Mtb HbN exhibited a nearly 2.5–3-fold increase in cfu counts (0 h postinfection) and displayed increased intracellular survival as compared with the control cells, indicating that these cells are able to infect macrophages more efficiently than the wild type cells (Fig. 6C). In contrast, macrophage infection with M. smegmatis expressing the glycosylation deficient mutant of HbN did not show any significant change at 0 h postinfection. An increase in the cfu of HbN-expressing cells at 0 h postinfection in different cell lines suggests that these cells are able to adhere to and/or invade macrophages better than the control cells, and this effect may not be cell line-specific. Thus, glycosylation of Mtb HbN may be vital for establishing efficient contact with the macrophage during intracellular infection.
HbN-overexpressing Mtb Alters Secretion of Pro- and Anti-inflammatory Cytokines during Macrophage Infection
Because cell wall lipids of Mtb are known to modulate host-immune responses (22, 23) and HbN overexpression brings about distinct changes in the cell wall lipids of Mtb, we attempted to check the levels of pro- and anti-inflammatory cytokines after infecting mouse peritoneal cells with the wild type and the HbN-overexpressing cells of Mtb. We used both 15- and 30-day grown cells of wild type and HbN-expressing cells to check the levels of cytokines after infecting mouse peritoneal macrophages. The levels of IL-6, IL-10, and TNF-α were measured in the culture supernatant of macrophages. The HbN-expressing cells of Mtb elicited a 2–3-fold increase in the levels of inflammatory cytokine, IL-6 and TNF-α, in comparison with the control cells (Fig. 7, A and B). The level of IL-6 increased maximally in the case of 30-day HbN-expressing cells of Mtb. In comparison, the change in the level of anti-inflammatory cytokine, Il-10, was not very significant (data not shown). These results suggested the involvement of HbN in modulating the immune responses by altering the balance of pro- and anti-inflammatory cytokines of the host. It is likely that these effects are an outcome of HbN-mediated changes in the membrane lipids and/or some other unknown factors of Mtb.
FIGURE 7.
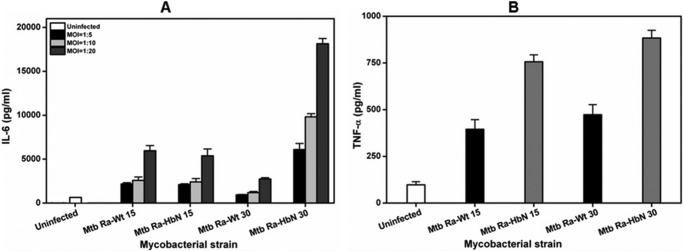
Levels of pro- and anti-inflammatory cytokines in macrophages infected with HbN-expressing MtbRa. A, levels of IL-6 secretion in Mtb-infected mouse peritoneal macrophages. 15- and 30-day grown cells of wild type and HbN-overexpressing cells of MtbRa were infected in mouse peritoneal macrophages at MOIs (shown in the inset) of 1:5, 1:10, and 1:20. Levels of IL-6 were measured in culture supernatant after 48 h. The data are represented as bars in the following order: uninfected, wild type MtbRa-Wt (15 days), MtbRa-HbN (15 days), MtbRa-Wt (30 days), and MtbRa-HbN (30 days). B, levels of TNF-α in Mtb-infected mouse peritoneal macrophages. 15- and 30-day grown cells of wild type and HbN-expressing cells of MtbRa were infected into mouse peritoneal macrophages at an MOI of 1:20, and the level of TNF-α in Mtb-infected macrophages was measured in culture supernatant. The data are represented as bars in the following order: uninfected, wild type MtbRa-Wt (15 days), MtbRa-HbN (15 days), MtbRa-Wt (30 days), and MtbRa-HbN (30 days). Results shown as mean ± S.D. (error bars) are representative of three independent experiments and are expressed as pg/ml.
Changes in the Expression of Co-stimulatory Surface Molecules of Macrophages upon Infection with HbN-overexpressing Mtb
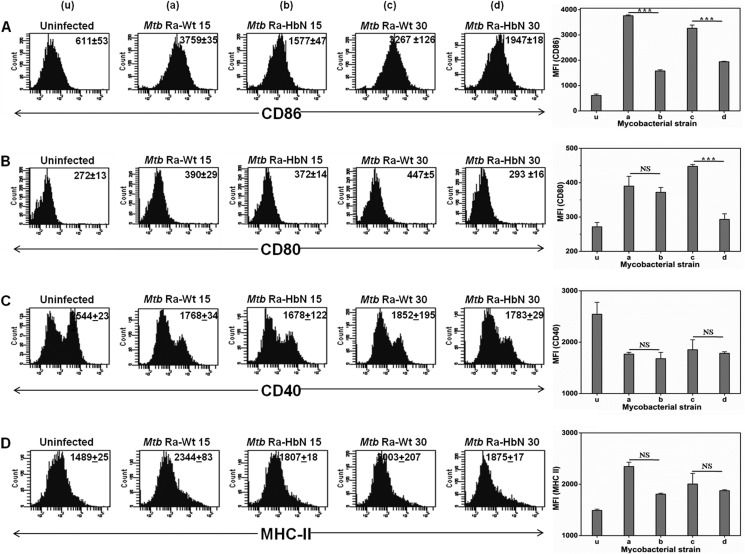
Macrophages respond to bacterial infection by expressing co-stimulatory surface markers that regulate pathogen recognition and ingestion, cell adhesion, cell-cell interactions, antigen presentation, and co-stimulation of T cells (23, 24). Therefore, the changes in the T cell-stimulating surface expression markers, such as CD40, CD80, and CD86, were estimated by FACS analysis after infection with the wild type and the HbN-overexpressing cells of MtbRa. We used both exponentially growing (15 days) and stationary phase (30 days) cultures of Mtb for the infection into mouse peritoneal macrophages. Interestingly, HbN-overexpressing cells (both 15 and 30 days old) significantly down-regulated the expression of CD80 (p < 0.001) and CD86 (p < 0.001) when compared with their respective wild type strains (Fig. 8, A and B). These changes were observed in a dose-dependent manner with respect to different MOIs (i.e. 1:5, 1:10, and 1:20) (data not shown). No significant changes in the exhibition of CD40 and MHC II were detected (Fig. 8, C and D).
FIGURE 8.
Macrophage infection with MtbRa overexpressing HbN suppresses the exhibition of co-stimulatory surface markers. The mouse peritoneal macrophages were infected with 15- and 30-day-old cells of wild type and HbN-overexpressing cells of MtbRa at an MOI of 1:20 and incubated for 48 h. Bar diagrams and flow cytometry histograms indicate the expression of CD86 (A), CD80 (B), CD40 (C), and MHC-II (D). The x axis in the bar diagram represents the strains used for the infection, and the y axis indicates mean fluorescent intensity (MFI). Data represent mean ± S.E. (error bars) of two independent experiments. Numbers in the inset of flow cytometry histograms correspond to mean fluorescent intensity ± S.E. Statistical analysis was done by a one-way analysis of variance, Tukey-Kramer multiple comparison test. u, a, b, c, and d represent uninfected, Mtb Ra-Wt 15, Mtb Ra-HbN 15, Mtb Ra-Wt 30, and Mtb Ra-HbN 30, respectively.
DISCUSSION
The outcome of infection by Mtb is primarily determined by the complex and dynamic interactions between the physiological state of the pathogen and immune responses of the host. The level of intracellular expression of mycobacterial stress response genes, upon infection, has been found to play a vital role in modulating the immune system of the host (26, 27). The hemoglobin, HbN, of Mtb has been proposed to play a protective role under nitrosative stress and hypoxia by virtue of its ability to sustain bacterial respiration under low oxygen and to protect it from macrophage-generated NO (9, 10). The present study unravels some important features of Mtb HbN that are classically considered virulence factors of pathogenic organisms. In this work, we have demonstrated for the first time that HbN is post-translationally modified via glycosylation and remains cell surface-localized in Mtb, which may be important for optimal NO-scavenging by Mtb. An increase in the cellular level of HbN enhances the ability of Mtb to establish effective infection and modulate host-immune responses, thereby suggesting its pivotal role in the host-pathogen interactions. Results presented herein, thus, provide evidence for new functions of HbN in Mtb apart from conferring protection to its host from macrophage-generated NO and nitrosative stress.
The HbN of Mtb has been expressed as a cytoplasmic protein in E. coli, and its NO-scavenging activity is well demonstrated in various heterologous hosts (9–11). Unlike E. coli, HbN remains localized on the cell membrane and the cell surface of its native host, Mtb. Extracytoplasmic localization of HbN in Mtb may be functionally relevant for protecting its cellular respiration under the NO-enriched environment of macrophages. However, it is intriguing how HbN is targeted on the membrane and the cell surface in the absence of conventional N-terminal signal sequences. Involvement of the 12-residue-long positively charged N-terminal pre-A region (28) of HbN in membrane targeting and anchorage was also ruled out. This pattern is very similar to the subcellular distribution of l-alanine dehydrogenase and Eis proteins of Mtb that lack a conventional transport signal but associate with the cell envelope (29, 30). The C-terminal region, spanning residues 93–123, constitutes a transmembrane region in HbN and may possibly be involved in membrane targeting, very similar to membrane-localized SecE of Mtb that lacks an N-terminal transport signal and has a single C-terminal transmembrane region (31). Many tail-anchored membrane proteins with a single C-terminal transmembrane region, facing the cytosol or projecting out on the cell surface, have been recognized in bacteria (32). A trypsin sensitivity assay suggested that a significant part of HbN may be located on the cell surface. The molecular mechanism by which HbN is targeted to the extracellular locations in Mtb is currently unknown. Nevertheless, the membrane and surface localization of HbN in Mtb suggest that it may interact with the host and play a role in bacterial pathogenesis.
Surface-localized and secreted proteins of Mtb are crucial for establishing a parasitic relationship with eukaryotic host (33). The majority of exported proteins of Mtb are glycosylated and play an important role in host-pathogen interactions (33, 34). The HbN exists as a glycosylated and cell membrane/surface-localized protein in Mtb. The presence of an O-mannosylation site at the C-terminal region of the HbN was validated by site-directed mutagenesis, and the carbohydrate linked with the HbN was identified as mannose. The structure of the 7–8 amino acid residues of the C-terminal region of HbN, where the glycan linkage occurs, could not be resolved in the x-ray structure (28). The glycosylation-deficient mutant of HbN, although it appeared to be associated with the cell membrane, could not be detected on the cell wall/surface. Interestingly, NOD activity of M. smegmatis expressing the glycosylation-lacking mutant of HbN was relatively slower than that for M. smegmatis expressing wild type HbN, whereas no distinct difference in NO dioxygenation was observed for E. coli expressing wild type or the mutant HbN. These results indicate that glycan linkage of HbN at the C terminus is crucial for modulating its topology for the cell surface localization and optimal NO scavenging in mycobacteria.
A substantial alteration in lipid content and its metabolic pathway has been observed in Mtb in response to oxygen availability (35). The fatty acid and lipid profile of Mtb changes remarkably after HbN overexpression. The increased concentration of HbN on the cell membrane of Mtb is likely to change the local oxygen concentration and the redox environment of the cell, which may affect the lipid repertoire of the cell membrane. This is also supported by studies showing that changes in the redox environment of the cell due to expression of the whiB3 gene in Mtb affect its lipid anabolism, resulting cell elongation in Mtb (36). Thus, phenotypic changes in Mtb might be the result of altered membrane lipids due to changes in the redox environment of the cell after HbN overexpression. Being positioned at the bacterial surface, lipids are able to interact with cells and can contribute to the interplay between host and pathogen (37). Therefore, extracellular localization of glycan-linked HbN and changes in the lipid components during HbN overexpression in Mtb may have significant implications for the host-pathogen interactions. The HbN-overexpressing cells of MtbRa displayed increased infectivity and intracellular survival over wild type cells. Notably, cfu counts of HbN-expressing Mtb and M. smegmatis were significantly higher at 0 h postinfection, irrespective of the cell lines used, suggesting increased entry of bacilli within the macrophages. M. smegmatis expressing glycosylation-deficient HbN of Mtb was unable to show any significant change in cfu counts over wild type, indicating that glycan-linked HbN may be important for the infectivity of Mtb. This is substantiated by the fact that many glycosylated proteins are vital and contribute significantly to the infectiousness of Mtb by binding with the lectin receptors of the host macrophages that are preferentially used by the bacilli to enter and evade the host defense mechanism (37–39). Additionally, mycobacterial cell envelope lipids are also known to modulate the initial step of cell entry and control the outcome of bacterial infection (37). Therefore, it is likely that alterations in the membrane lipids together with increased level of glycan-linked extracellular HbN in Mtb are contributing to the enhanced adhesion and macrophage infection.
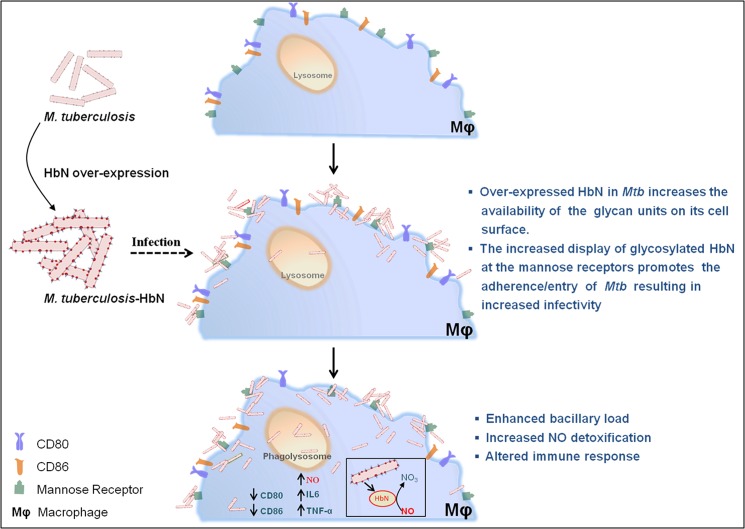
The cell wall lipids and glycosylated proteins of Mtb are known to activate macrophages and confer strong immune responses by generating several protective mechanisms, leading to generation of NO and a wide variety of cytokines and inflammatory mediators (40–44). Thus, increased accumulation of lipids and glycosylated HbN on the cell membrane and the cell surface might elicit increased immune responses during intracellular infection. This is evident from the elevated level of the inflammatory cytokines IL-6 and TNF-α, which are known to regulate expression of inducible nitric-oxide synthase and NO production (42). Additionally, HbN-overexpressing cells of Mtb appeared to paralyze the function of T cells by down-regulating the co-stimulatory surface markers CD80 and CD86. Macrophages are known to respond against bacterial infection by modulating the expression of the co-stimulatory molecule and major histocompatibility complex (MHC) on the surface of antigen-presenting cells, which play an important role in providing optimum activation of T cells. Down-regulation in the display of some of these molecules may induce anergy, a state of unresponsiveness in the T cells (45, 46). Among these, CD80 and CD86 expressed on the surface of antigen-presenting cells play a decisive role in the activation of T cells (25, 46, 47). Our results signify that HbN-overexpressing cells of Mtb can suppress the expression of CD80 and CD86 co-stimulatory molecules and thereby may impair the function of macrophages to optimally activate T cells. It is anticipated that due to effective modulation of host-immune responses and efficient NO scavenging ability, HbN may allow the tubercle bacillus to survive better inside the host macrophages. A schematic diagram displaying the interactions between Mtb, HbN, and macrophage is shown in Fig. 9.
FIGURE 9.
A schematic diagram displaying interactions of Mtb, HbN, and macrophages during intracellular infection. HbN overexpression in Mtb changes the membrane lipid profile and increases the level of mannose-linked HbN at the cell wall/surface, allowing increased adhesion/recognition of Mtb via macrophage receptors (lectin/mannose binding), thus increasing the entry/infectivity of Mtb. During macrophage infection, Mtb is exposed to combined stresses of hypoxia, nitric oxide, and reactive toxic species along with host immune responses that may inhibit its intracellular survival. Surface exposure of the HbN may allow it to interact directly with the macrophage-generated NO and detoxify it via an NO dioxygenase reaction and protect the cellular respiration of Mtb, allowing it to grow better. Additionally, an increase in surface-exposed glycan-linked HbN and changes in membrane-associated lipids in Mtb may facilitate its interactions with macrophages and alter host-immune responses. Thus, due to its efficient NO-scavenging activity and the ability to evade the immune system of the host during intracellular infection, HbN may assist Mtb in establishing the infection effectively and survive better inside the macrophages.
It is interesting to note that an increase in the cellular level of HbN confers distinct changes in the ability of MtbRa to alter host immune responses and establish effective intracellular infection and survival that may be vital for the pathogenicity of Mtb. Intraspecies genetic diversity and heterogeneity in the level of gene expression have been demonstrated to be important in the pathogenesis and epidemiology of several pathogens (5, 48). A significant change in the expression level of some genes and lipid contents has been observed among clinical isolates of Mtb as well as laboratory strains of Mtb37Rv and Mtb37Ra (48). Although variation in the cellular level of HbN has not been explored among various clinical and laboratory strains of mycobacteria, our study suggests that an increase in the level of HbN may contribute significantly to the pathogenicity of Mtb. Our extensive efforts for generating an HbN knock-out strain of MtbRa were unsuccessful to further explore this point, although an HbN knockout has been reported in an animal pathogen, M. bovis (12). It is likely that these two species of mycobacteria, having distinct species specificity, respond differently with respect to genetic recombination. To overcome this problem, we used M. smegmatis as a model to substantiate the function of Mtb HbN because it carries an HbN homolog that is attenuated in NO detoxification (49) and lacks a glycosylation site. Moreover, its expression was not detected on Western blot in aerobically growing cells of M. smegmatis.3 Overexpression of Mtb HbN in M. smegmatis led it to localize on the cell wall/surface and allowed it to infect macrophages more efficiently than the isogenic strain expressing an unglycosylated mutant of HbN. These results further supported the involvement of glycan-linked HbN in modulating its cellular localization and interactions of Mtb with the host during intracellular infection.
Taken together, the present study reveals the following major findings. (i) HbN carries an O-mannosylation site at the C-terminal region and is post-translationally modified by glycan linkage in Mtb. (ii) HbN remains localized on the cell membrane and the cell surface of Mtb, and a lack of glycosylation interrupts its extracytoplasmic localization in M. smegmatis and reduces its NO uptake ability compared with M. smegmatis expressing wild type HbN. (iii) Expression of HbN facilitates intracellular infection and survival of Mtb and effectively changes the immune responses of the host by altering the balance of pro- and anti-inflammatory cytokines and co-stimulatory surface markers. It is therefore likely that, due to its efficient NO scavenging activity and its ability to evade the immune system of the host during intracellular infection, HbN may assist Mtb in establishing the infection effectively and drives cells toward better survival inside the host macrophages. The elucidation of post-translational modification and immunomodulation of the host by HbN-expressing Mtb during intracellular infection, thus, unravels new aspects of HbN functionality and points toward its multifaceted role in the biology and pathogenesis of Mtb.
This work was supported by the European Commission under the FP7 program on health and CSIR Supra-institutional Project Grants SIP10 and BSC0210. This work was also supported by CSIR and the Department of Biotechnology (to S. A., D. S., and S. S.).

This article contains supplemental Fig. S1.
S. Arya, D. Sethi, S. Singh, M. D. Hade, and K. L. Dikshit, unpublished observations.
- Mtb
- M. tuberculosis
- MtbRv
- M. tuberculosis H37Rv
- MtbRa
- M. tuberculosis H37Ra
- NOD
- nitric-oxide dioxygenase
- MOI
- multiplicity of infection
- MFI
- mean fluorescence intensity
- ConA
- concanavalin A.
REFERENCES
- 1. World Health Organization (2008) Tuberculosis Facts. World Health Organization, Geneva, Switzerland [Google Scholar]
- 2. Dye C. (2006) Global epidemiology of tuberculosis. Lancet 367, 938–940 [DOI] [PubMed] [Google Scholar]
- 3. Manca C., Tsenova L., Barry C. E., 3rd, Bergtold A., Freeman S., Haslett P. A., Musser J. M., Freedman V. H., Kaplan G. (1999) Mycobacterium tuberculosis CDC1551 induces a more vigorous host response in vivo and in vitro, but is not more virulent than other clinical isolates. J. Immunol. 162, 6740–6746 [PubMed] [Google Scholar]
- 4. Smith I. (2003) Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 16, 463–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Homolka S., Niemann S., Russell D. G., Rohde K. H. (2010) Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates. Delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS Pathog. 6, e1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hershberg R., Lipatov M., Small P. M., Sheffer H., Niemann S., Homolka S., Roach J. C., Kremer K., Petrov D. A., Feldman M. W., Gagneux S. (2008) High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 6, e311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koo M. S., Subbian S., Kaplan G. (2012) Strain specific transcriptional response in Mycobacterium tuberculosis infected macrophages. Cell Commun. Signal. 10, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ordway D., Henao-Tamayo M., Harton M., Palanisamy G., Troudt J., Shanley C., Basaraba R. J., Orme I. M. (2007) The Hypervirulent Mycobacterium tuberculosis strain HN878 induces a potent TH1 response followed by rapid down-regulation. J. Immunol. 179, 522–531 [DOI] [PubMed] [Google Scholar]
- 9. Couture M., Yeh S. R., Wittenberg B. A., Wittenberg J. B., Ouellet Y., Rousseau D. L., Guertin M. (1999) A cooperative oxygen-binding hemoglobin from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 96, 11223–11228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pathania R., Navani N. K., Gardner A. M., Gardner P. R., Dikshit K. L. (2002) Nitric oxide scavenging and detoxification by the Mycobacterium tuberculosis haemoglobin, HbN in Escherichia coli. Mol. Microbiol. 45, 1303–1314 [DOI] [PubMed] [Google Scholar]
- 11. Pawaria S., Rajamohan G., Gambhir V., Lama A., Varshney G. C., Dikshit K. L. (2007) Intracellular growth and survival of Salmonella enterica serovar Typhimurium carrying truncated hemoglobins of Mycobacterium tuberculosis. Microb. Pathog. 42, 119–128 [DOI] [PubMed] [Google Scholar]
- 12. Ouellet H., Ouellet Y., Richard C., Labarre M., Wittenberg B., Wittenberg J., Guertin M. (2002) Truncated hemoglobin HbN protects Mycobacterium bovis from nitric oxide. Proc. Natl. Acad. Sci. U.S.A. 99, 5902–5907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pawaria S., Lama A., Raje M., Dikshit K. L. (2008) Responses of Mycobacterium tuberculosis hemoglobin promoters to in vitro and in vivo growth conditions. Appl. Environ. Microbiol. 74, 3512–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garbe T. R., Barathi J., Barnini S., Zhang Y., Abou-Zeid C., Tang D., Mukherjee R., Young D. B. (1994) Transformation of mycobacterial species using hygromycin resistance as selectable marker. Microbiology 140, 133–138 [DOI] [PubMed] [Google Scholar]
- 15. Cowley S. C., Av-Gay Y. (2001) Monitoring promoter activity and protein localization in Mycobacterium spp. using green fluorescent protein. Gene 264, 225–231 [DOI] [PubMed] [Google Scholar]
- 16. Michell S. L., Whelan A. O., Wheeler P. R., Panico M., Easton R. L., Etienne A. T., Haslam S. M., Dell A., Morris H. R., Reason A. J. (2003) The MPB83 antigen from Mycobacterium bovis contains O-linked mannose and (1→3)-mannobiose moieties. J. Biol. Chem. 278, 16423–16432 [DOI] [PubMed] [Google Scholar]
- 17. Bligh E. G., Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 18. Fimereli D., Tsirigos K., Litou Z., Liakopoulos T., Bagos P., Hamodrakas S. (2012) CW-PRED. A HMM-based method for the classification of cell wall-anchored proteins of Gram-positive bacteria. in Artificial Intelligence: Theories and Applications (Maglogiannis I., Plagianakos V., Vlahavas I., eds) pp. 285–290, Springer, Berlin [Google Scholar]
- 19. Cserzö M., Wallin E., Simon I., von Heijne G., Elofsson A. (1997) Prediction of transmembrane α-helices in prokaryotic membrane proteins. The dense alignment surface method. Protein. Eng. 10, 673–676 [DOI] [PubMed] [Google Scholar]
- 20. Herrmann J. L., Delahay R., Gallagher A., Robertson B., Young D. (2000) Analysis of post-translational modification of mycobacterial proteins using a cassette expression system. FEBS Lett. 473, 358–362 [DOI] [PubMed] [Google Scholar]
- 21. Lama A., Pawaria S., Dikshit K. L. (2006) Oxygen binding and NO scavenging properties of truncated hemoglobin, HbN, of Mycobacterium smegmatis. FEBS Lett. 580, 4031–4041 [DOI] [PubMed] [Google Scholar]
- 22. Karakousis P. C., Bishai W. R., Dorman S. E. (2004) Mycobacterium tuberculosis cell envelope lipids and the host immune response. Cell Microbiol. 6, 105–116 [DOI] [PubMed] [Google Scholar]
- 23. Mendelson M., Walters S., Smith I., Kaplan G. (2005) Strain-specific mycobacterial lipids and the stimulation of protective immunity to tuberculosis. Tuberculosis 85, 407–413 [DOI] [PubMed] [Google Scholar]
- 24. Khan N., Gowthaman U., Pahari S., Agrewala J. N. (2012) Manipulation of costimulatory molecules by intracellular pathogens. Veni, Vidi, Vici!! PLoS Pathog. 8, e1002676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Suvas S., Singh V., Sahdev S., Vohra H., Agrewala J. N. (2002) Distinct role of CD80 and CD86 in the regulation of the activation of B cell and B cell lymphoma. J. Biol. Chem. 277, 7766–7775 [DOI] [PubMed] [Google Scholar]
- 26. Schnappinger D., Ehrt S., Voskuil M. I., Liu Y., Mangan J. A., Monahan I. M., Dolganov G., Efron B., Butcher P. D., Nathan C., Schoolnik G. K. (2003) transcriptional adaptation of Mycobacterium tuberculosis within macrophages. Insights into the phagosomal environment. J. Exp. Med. 198, 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dutta N. K., Mehra S., Martinez A. N., Alvarez X., Renner N. A., Morici L. A., Pahar B., Maclean A. G., Lackner A. A., Kaushal D. (2012) The stress-response factor SigH modulates the interaction between Mycobacterium tuberculosis and host phagocytes. PLoS One 7, e28958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Milani M., Pesce A., Ouellet Y., Ascenzi P., Guertin M., Bolognesi M. (2001) Mycobacterium tuberculosis hemoglobin N displays a protein tunnel suited for O2 diffusion to the heme. EMBO J. 20, 3902–3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Giffin M. M., Modesti L., Raab R. W., Wayne L. G., Sohaskey C. D. (2012) ald of Mycobacterium tuberculosis encodes both the alanine dehydrogenase and the putative glycine dehydrogenase. J. Bacteriol. 194, 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dahl J. L., Wei J., Moulder J. W., Laal S., Friedman R. L. (2001) Subcellular localization of the intracellular survival-enhancing Eis protein of Mycobacterium tuberculosis. Infect. Immun. 69, 4295–4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Craney A., Tahlan K., Andrews D., Nodwell J. (2011) Bacterial transmembrane proteins that lack N-terminal signal sequences. PLoS One 6, e19421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Borgese N., Brambillasca S., Colombo S. (2007) How tails guide tail-anchored proteins to their destinations. Curr. Opin. Cell Biol. 19, 368–375 [DOI] [PubMed] [Google Scholar]
- 33. Ragas A., Roussel L., Puzo G., Rivière M. (2007) The Mycobacterium tuberculosis cell-surface glycoprotein apa as a potential adhesin to colonize target cells via the innate immune system pulmonary C-type lectin surfactant protein A. J. Biol. Chem. 282, 5133–5142 [DOI] [PubMed] [Google Scholar]
- 34. González-Zamorano M., Mendoza-Hernández G., Xolalpa W., Parada C., Vallecillo A. J., Bigi F., Espitia C. (2009) Mycobacterium tuberculosis glycoproteomics based on ConA-lectin affinity capture of mannosylated proteins. J. Proteome Res. 8, 721–733 [DOI] [PubMed] [Google Scholar]
- 35. Galagan J. E., Minch K., Peterson M., Lyubetskaya A., Azizi E., Sweet L., Gomes A., Rustad T., Dolganov G., Glotova I., Abeel T., Mahwinney C., Kennedy A. D., Allard R., Brabant W., Krueger A., Jaini S., Honda B., Yu W. H., Hickey M. J., Zucker J., Garay C., Weiner B., Sisk P., Stolte C., Winkler J. K., Van de Peer Y., Iazzetti P., Camacho D., Dreyfuss J., Liu Y., Dorhoi A., Mollenkopf H. J., Drogaris P., Lamontagne J., Zhou Y., Piquenot J., Park S. T., Raman S., Kaufmann S. H., Mohney R. P., Chelsky D., Moody D. B., Sherman D. R., Schoolnik G. K. (2013) The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 499, 178–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Singh A., Crossman D. K., Mai D., Guidry L., Voskuil M. I., Renfrow M. B., Steyn A. J. (2009) Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 5, e1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Neyrolles O., Guilhot C. (2011) Recent advances in deciphering the contribution of Mycobacterium tuberculosis lipids to pathogenesis. Tuberculosis 91, 187–195 [DOI] [PubMed] [Google Scholar]
- 38. Astarie-Dequeker C., Le Guyader L., Malaga W., Seaphanh F.-K., Chalut C., Lopez A., Guilhot C. (2009) Phthiocerol dimycocerosates of Mycobacterium tuberculosis participate in macrophage invasion by inducing changes in the organization of plasma membrane lipids. PLoS Pathog. 5, e1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Astarie-Dequeker C., N'Diaye E.-N., Le Cabec V., Rittig M. G., Prandi J., Maridonneau-Parini I. (1999) The mannose receptor mediates uptake of pathogenic and nonpathogenic mycobacteria and bypasses bactericidal responses in human macrophages. Infect. Immun. 67, 469–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schlesinger L. (1993) Macrophage phagocytosis of virulent but not attenuated strains of Mycobacterium tuberculosis is mediated by mannose receptors in addition to complement receptors. J. Immunol. 150, 2920–2930 [PubMed] [Google Scholar]
- 41. Brightbill H. D., Libraty D. H., Krutzik S. R., Yang R. B., Belisle J. T., Bleharski J. R., Maitland M., Norgard M. V., Plevy S. E., Smale S. T., Brennan P. J., Bloom B. R., Godowski P. J., Modlin R. L. (1999) Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science 285, 732–736 [DOI] [PubMed] [Google Scholar]
- 42. Brennan P. J. (2003) Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis 83, 91–97 [DOI] [PubMed] [Google Scholar]
- 43. Herbst S., Schaible U. E., Schneider B. E. (2011) Interferon γ activated macrophages kill mycobacteria by nitric oxide induced apoptosis. PLoS One 6, e19105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ragno S., Romano M., Howell S., Pappin D. J., Jenner P. J., Colston M. J. (2001) Changes in gene expression in macrophages infected with Mycobacterium tuberculosis. A combined transcriptomic and proteomic approach. Immunology 104, 99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wei J., Dahl J. L., Moulder J. W., Roberts E. A., O'Gaora P., Young D. B., Friedman R. L. (2000) Identification of a Mycobacterium tuberculosis gene that enhances mycobacterial survival in macrophages. J. Bacteriol. 182, 377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Agrewala J. N., Suvas S., Verma R. K., Mishra G. C. (1998) Differential effect of anti-B7–1 and anti-M150 antibodies in restricting the delivery of costimulatory signals from B cells and macrophages. J. Immunol. 160, 1067–1077 [PubMed] [Google Scholar]
- 47. Balkhi M. Y., Latchumanan V. K., Singh B., Sharma P., Natarajan K. (2004) Cross-regulation of CD86 by CD80 differentially regulates T helper responses from Mycobacterium tuberculosis secretory antigen-activated dendritic cell subsets. J. Leukoc. Biol. 75, 874–883 [DOI] [PubMed] [Google Scholar]
- 48. Gao Q., Kripke K. E., Saldanha A. J., Yan W., Holmes S., Small P. M. (2005) Gene expression diversity among Mycobacterium tuberculosis clinical isolates. Microbiology 151, 5–14 [DOI] [PubMed] [Google Scholar]
- 49. Lama A., Pawaria S., Bidon-Chanal A., Anand A., Gelpí J. L., Arya S., Martí M., Estrin D. A., Luque F. J., Dikshit K. L. (2009) Role of Pre-A motif in nitric oxide scavenging by truncated hemoglobin, HbN, of Mycobacterium tuberculosis. J. Biol. Chem. 284, 14457–14468 [DOI] [PMC free article] [PubMed] [Google Scholar]