Summary
Invadopodia-dependent degradation of the basement membrane plays a major role during metastasis of breast cancer cells. Basement membrane degradation is mediated by targeted secretion of various matrix metalloproteinases (MMPs). Specifically, MMP2 and MMP9 (MMP2/9) possess the ability to hydrolyze components of the basement membrane and regulate various aspects of tumor growth and metastasis. However, the membrane transport machinery that mediates targeting of MMP2/9 to the invadopodia during cancer cell invasion remains to be defined. Because Rab GTPases are key regulators of membrane transport, we screened a human Rab siRNA library and identified Rab40b GTPase as a protein required for secretion of MMP2/9. We also have shown that Rab40b functions during at least two distinct steps of MMP2/9 transport. Here, we demonstrate that Rab40b is required for MMP2/9 sorting into VAMP4-containing secretory vesicles. We also show that Rab40b regulates transport of MMP2/9 secretory vesicles during invadopodia formation and is required for invadopodia-dependent extracellular matrix degradation. Finally, we demonstrate that Rab40b is also required for breast cancer cell invasion in vitro. On the basis of these findings, we propose that Rab40b mediates trafficking of MMP2/9 during invadopodia formation and metastasis of breast cancer cells.
Key words: MMP2, MMP9, Rab40b, Invadopodia, Secretion, Invasion
Introduction
The basement membrane is made up of a network of extracellular matrix (ECM) proteins which serves as a barrier for cell motility and invasion. Loss of this barrier function is an important step in cancer cell invasion and metastasis (Roskelley and Bissell, 1995; Roskelley et al., 1995). Furthermore, ECM degradation can lead to increased adhesion of the migrating cells and also result in the release and/or activation of various growth factors required for angiogenesis, tumor growth and metastasis (Roskelley et al., 1995). Basement membrane disruption usually involves a localized degradation of the ECM through targeted secretion of matrix metalloproteinases (MMPs) (Polette et al., 2004; Visse and Nagase, 2003). MMPs are a large family of zinc-dependent endopeptidases that are capable of cleaving multiple ECM proteins. MMPs can be subdivided into multiple categories depending on their substrate specificity, such as collagenases, stromelysins and gelatinases (Visse and Nagase, 2003). MMP2 and MMP9, as well as membrane-type matrix metalloproteinase-1 (MT1-MMP, also known as MMP14), possess the ability to hydrolyze components of the basement membrane, and have recently emerged as key molecules involved in mediating various aspects of tumor growth and metastasis (Chambers and Matrisian, 1997; Egeblad and Werb, 2002; Polette et al., 2004; Shah et al., 2009). Furthermore, MMP2/9 and MT1-MMP are known to stimulate tumor angiogenesis as well as epithelial-to-mesenchymal transition (EMT), in large part through partial ECM proteolysis. Consistent with these observations, increased expression of MMP2/9 and MT1-MMP has been linked to increased invasiveness in tissue culture cells and increased metastasis in mice (Itoh et al., 1999; Itoh et al., 1998; Schmalfeldt et al., 2001).
Because increased levels of MMPs correlate with increased metastatic potential of tumors, the mechanisms regulating the production of MMPs have been an active area of research. MMP2/9 and MT1-MMP protein levels are transcriptionally tightly regulated. In addition to this, the activities of MMP2 and MMP9 are further controlled by extracellular activation of the pro-enzyme and by inhibition of the active MMPs by extracellular inhibitors, such as tissue inhibitors of MMPs (TIMPs) (Egeblad and Werb, 2002). Interestingly, MT1-MMP was shown to cleave and activate pro-MMP2 (Strongin et al., 1995). Finally, MMP2/9 and MT1-MMP undergo regulated targeting and secretion at the sites of forming invadopodia, the actin-rich finger-like cellular projections located at the ventral side of the cell (Murphy and Courtneidge, 2011; Poincloux et al., 2009; Stylli et al., 2008). Invadopodia are sites of localized ECM degradation and have been shown to be induced by Src kinase to mediate cancer cell invasion in vitro (Murphy and Courtneidge, 2011; Murphy and Gavrilovic, 1999). The role of invadopodia during cancer cell invasion in vivo is less well defined, but it has been shown that high expression levels of various invadopodia-forming proteins correlate with an increased metastatic potential (Blouw et al., 2008; Clark et al., 2009; Weaver, 2008). Furthermore, recent studies have demonstrated the formation of invadopodia-like structures in vivo using intravital imaging (Quintavalle et al., 2010).
Despite the importance of the targeting of MMPs to the invadopodia, the mechanisms regulating subcellular transport of MMPs are only beginning to emerge. MT1-MMP, MMP2 and MMP9 have been shown to be enriched at the invadopodia (Poincloux et al., 2009; Clark et al., 2008; Nakahara et al., 1997; Artym et al., 2006; Bourguignon et al., 1998; Monsky et al., 1993). It has been shown that endocytic recycling of MT1-MMP is important in targeting it to the plasma membrane and invadopodia (Bravo-Cordero et al., 2007; Remacle et al., 2003). Furthermore, selective endocytosis of MT1-MMP also plays a role in regulating its activity towards the ECM (Remacle et al., 2003). By contrast, almost nothing is known about the membrane transport machinery involved in targeted secretion of MMP2 and MMP9. Intracellular transport and targeting of membrane-bound organelles are regulated by multiple protein families. Rab GTPases have emerged as key regulators of membrane transport and were shown to be required for multiple membrane transport steps, such as cargo sorting, transport and fusion with the donor membranes. Thus, to start identifying the membrane transport and targeting machinery that regulates MMP2/9 secretion, we performed a Rab GTPase siRNA library screen. This screen identified Rab40b as a small monomeric GTPase required for the secretion of both MMP2 and MMP9. We have shown that, unlike MT1-MMP secretion, secretion of MMP2 and MMP9 is not dependent on endocytic transport, but instead relies on transport from the trans-Golgi Network (TGN) through VAMP4 and Rab40b-containing secretory vesicles. Rab40b knockdown results in mistargeting of MMP2 and MMP9 to lysosomes, where they are degraded. We also demonstrate that Rab40b regulates MMP2/9 trafficking during invadopodia formation and is required for invadopodia-dependent ECM degradation. Finally, we show that Rab40b knockdown inhibits in vitro invasion of MDA-MB-231 cells, while having no effect on cell motility. On the basis of these findings, we propose that Rab40b is the key GTPase required for MMP2/9 intracellular transport and targeting to the newly formed invadopodia, thus affecting the invasive capacity of breast cancer cells.
Results
Rab40b GTPase is required for MMP2 and MMP9 secretion
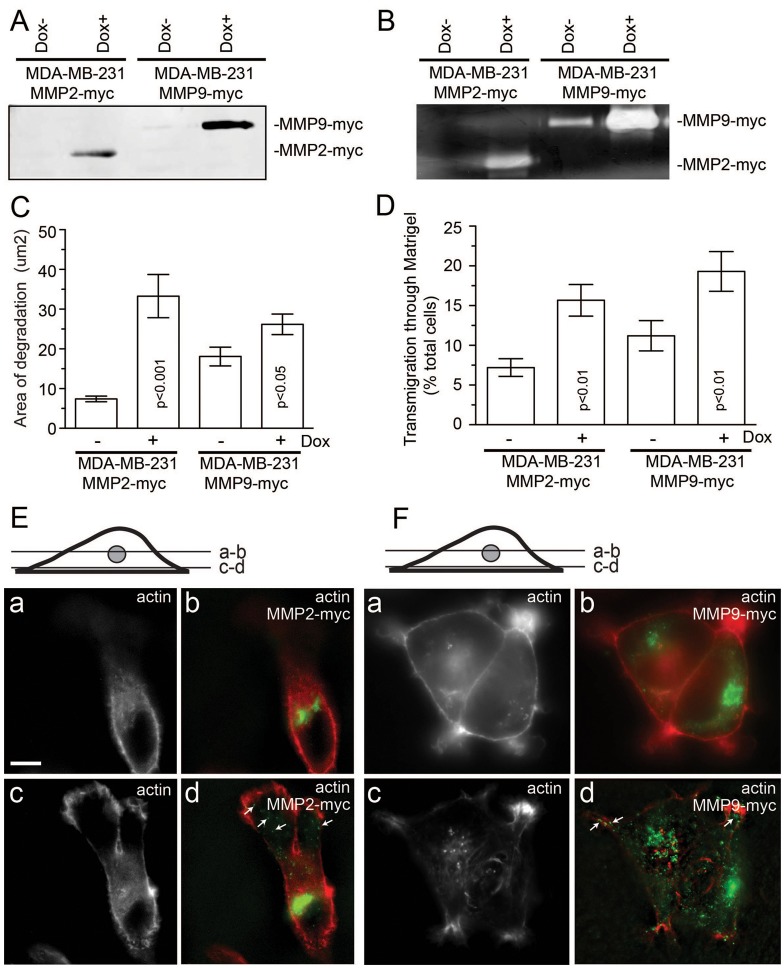
Given that little is known about the regulation of intracellular MMP2 and MMP9 transport, in this study we screened for Rab GTPases that regulate MMP2/9 transport and secretion. To that end, we created tet-inducible MDA-MB-231 cell lines expressing either MMP2–Myc (MDA-MMP2–Myc) or MMP9–Myc (MDA-MMP9–Myc). As shown in Fig. 1A,B, MDA-MMP2–Myc and MDA-MMP9–Myc cells express and secrete enzymatically active MMP2–Myc and MMP9–Myc in a doxycycline-dependent manner. Furthermore, doxycyline increased ECM degradation (Fig. 1C) and invasion (Fig. 1D) in these cells. We next analyzed the subcellular localization of MMP2–Myc and MMP9–Myc. As expected of secretory proteins, MMP2/9–Myc were enriched at the perinuclear region (Fig. 1E,F, a and b), where they colocalized with the trans-Golgi network (TGN) marker VAMP4 (supplementary material Fig. S1). Organelles containing MMP2/9–Myc were also found in the cytosol, especially in close proximity to the basal plasma membrane (Fig. 1E,F, c and d). Taken together, the above data suggest that these cells probably transport and secrete Myc-tagged MMP2/9 in a manner similar to endogenous MMP2/9.
Fig. 1.
Characterization of MDA-MB-231 cell lines expressing tet-inducible MMP2–Myc or MMP9–Myc. (A,B) MDA-MB-231 cells expressing dox-inducible MMP2–Myc or MMP9–Myc were incubated in Opti-MEM for 24 hours in the absence or presence of 1 µg/ml doxycycline. Opti-MEM medium was then collected and the amount of secreted MMP2–Myc or MMP9–Myc was analyzed by immunoblotting (A) or zymography (B). (C) MDA-MB-231 cells expressing dox-inducible either MMP2–Myc or MMP9–Myc were plated on gelatin and fibronectin-HiLyte Fluor488-coated coverslips and incubated in the presence or absence of 1 µg/ml doxycycline. After incubation for 20 hours, cells were fixed and invadopodia-associated ECM degradation was analyzed by in situ zymography (for more details see the Materials and Methods). Data shown are the means and s.e. of three independent experiments. (D) MDA-MB-231 cells expressing dox-inducible MMP2–Myc or MMP9–Myc were plated on matrigel-coated 8-µm-pore filters and incubated in the presence or absence of 1 µg/ml doxycycline. The ability of cells to invade through Matrigel matrix was analyzed using Crystal Violet staining. The data shown are the means and s.d. from three independent experiments. (E,F) MDA-MB-231 cells expressing MMP2–Myc (E) or MMP9–Myc (F) were fixed and stained with Rhodamine-phalloidin and mouse anti-Myc antibodies. Panels a and b show optical sections at the TGN level, whereas panels c and d show optical sections at the coverslip level. Arrows indicate cytosolic organelles containing MMP2–Myc or MMP9–Myc. Scale bar: 5 µm.
Next, we used an ELISA-based siRNA screen to identify Rab GTPases that regulate secretion of MMP2–Myc and MMP9–Myc (Fig. 2). In all cases, a pool of four different siRNAs was used (for more detailed description of screen see the Materials and Methods). Rab GTPases that either increased or decreased secretion of MMP2–Myc or MMP9–Myc were then re-screened using four individual siRNAs. Only candidates that affected MMP2/9 secretion after treatment with at least two out of four siRNAs, were considered for further characterization. As shown in Fig. 2, the screen identified several Rab GTPases that either increased or decreased secretion of MMP2–Myc or MMP9–Myc. Interestingly, most of the identified candidate proteins affected either MMP2–Myc or MMP9–Myc secretion, but knockdown of only Rab25 and Rab40b decreased secretion of both MMPs. Rab25 was previously identified as a GTPase that regulates cancer cell motility and invasion (Caswell et al., 2007; Dozynkiewicz et al., 2012). By contrast, little is known about the function of Rab40b. Therefore, we focused on understanding the cellular function of Rab40b GTPase for the remainder of this study.
Fig. 2.
MMP2 and MMP9 secretion-regulating proteins identified from siRNA screen. ELISA-based quantification of the secreted MMP2–Myc (A) and MMP9–Myc (B) in cells treated with various Rab siRNAs. For more details see the Materials and Methods. Data shown are the means and s.d. of three independent experiments. All the values above top grey line or below bottom grey line are significantly different to the control at P<0.01.
Rab40b is required for sorting and secretion of MMP2 and MMP9
All Rab GTPases function by binding and recruiting their respective effector proteins to the membranes. These effector proteins then regulate sorting, transport and targeting of specific membrane-bound organelles (Pfeffer, 2001). There is very limited information about the localization and function of Rab40b. Rab40b belongs to a sub-family of Rab40 GTPases that includes two other closely related members, Rab40a and Rab40c, and is characterized by the presence of a SOCS box at their C-terminal half (Fig. 3A) (Piessevaux et al., 2008; Stein et al., 2012). Interestingly, depletion of Rab40a did not have any effect on MMP2–Myc or MMP9–Myc secretion. By contrast, knockdown of Rab40c decreased secretion of MMP2–Myc, suggesting that there is some functional overlap between Rab40b and Rab40c GTPases. To validate the role of Rab40b in regulating MMP2/9 secretion, we tested the effects of individual siRNAs on secretion of MMP2–Myc and MMP9–Myc. As shown in Fig. 3B, all four Rab40b siRNAs resulted in a decrease in the levels of mRNA encoding Rab40b. Consistent with the involvement of Rab40b in regulating MMP2/9 intracellular transport, all four siRNAs also decreased the secretion of MMP2–Myc and MMP9–Myc (Fig. 3C,D). Similarly, Rab40b depletion also decreased secretion of endogenous MMP2 and MMP9 (Fig. 3E), but not MT1-MMP (Fig. 3F). Depletion of Rab40b also had little effect on LPS-induced release of IL6 (Fig. 3G) or GFP–hGH secretion (supplementary material Fig. S1H) suggesting that one of the functions of Rab40b is to regulate MMP2/9 intracellular transport.
Fig. 3.
Rab40b knockdown decreases MMP2 and MMP9 secretion in MDA-MB-231 cells. (A) Schematic representation of Rab40b domain structure. (B) The efficiency of Rab40b knockdown as determined by qPCR. (C,D) MDA-MB-231 cells expressing MMP2–Myc (C) or MMP9–Myc (D) were transfected with four different Rab40b siRNAs. Two days later, equal number of cells were plated in six-well dishes and incubated with 1 ml of medium for 24 hours. Medium was then collected and the effect of Rab40b knockdown on secretion of MMP2–Myc and MMP9–Myc analyzed by western blotting. Data shown are the means and s.d. of three independent experiments. (E) MDA-MB-231 cells were transfected with Rab40b siRNA#2. Two days later, equal number of cells were plated in six-well dishes and incubated with 1 ml of Opti-MEM for 24 hours. Opti-MEM was then collected and the effect of Rab40b knockdown on secretion of endogenous MMP2 and MMP9 was analyzed by zymography. Fetal bovine serum (rich in secreted MMP2/9) in the first lane was used as a positive control. Opti-MEM collected from a six-well dish without cells was used as negative control. (F) Mock-, Rab40b-siRNA- or VAMP4-siRNA-treated MDA-MB-231 cells were harvested and analyzed by flow cytometry to measure the levels of endogenous plasma membrane MT1-MMP. Data shown are the means and s.d. of three independent experiments. (G) Mock- or Rab40b-siRNA-treated MDA-MB-231 cells were plated in six-well dishes and stimulated with 1 mg/ml of LPS. After incubation for 16 hours, medium was collected and the levels of secreted IL-6 analyzed using ELISA. The data shown are the means and s.d. of three independent experiments.
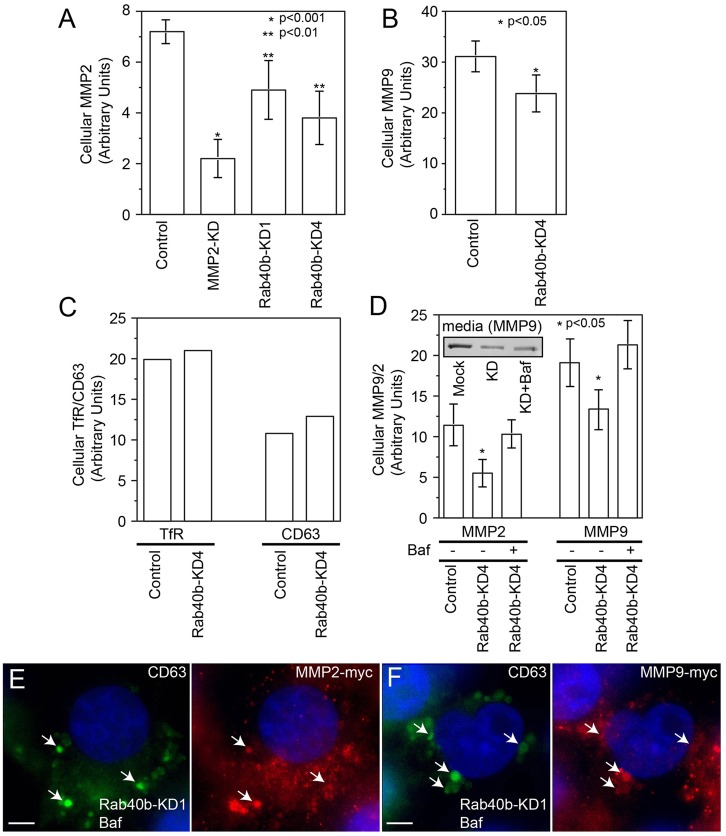
Because Rab40b depletion leads to a decrease in MMP2 and MMP9 secretion, one would predict that this would result in the accumulation of MMP2 and MMP9 within the cell. To determine this, we measured the intracellular levels of MMP2–Myc and MMP9–Myc using FACS analysis. Surprisingly, knockdown of Rab40b decreased intracellular MMP2/9 levels (Fig. 4A,B), while having no effect on levels of other post-TGN proteins, such as transferrin receptor, CD63 or IL6 (Fig. 4C and data not shown). Given that Rab40b depletion resulted in simultaneous decrease in the secretion and intracellular levels of MMP2/9, it raises the possibility that MMP2/9 are mis-sorted to lysosomes and degraded in Rab40b-depleted cells. To test this hypothesis, we incubated Rab40b-siRNA-treated cells in the presence or absence of the lysosomal inhibitor bafilomycin. As shown in Fig. 4D, bafilomycin treatment reversed the effects of Rab40b siRNA on intracellular levels of MMP2 and MMP9. Interestingly, bafilomycin did not rescue the secretory block caused by depletion of Rab40b, suggesting that in addition to MMP2/9 sorting, Rab40b also directly affects the targeting and fusion of MMP2/9 secretory vesicles with the cellular plasma membrane. To further test this possibility, we stained Rab40b-knockdown and bafilomycin-treated cells with anti-Myc and anti-CD63 (lysosomal marker) antibodies. As shown in Fig. 4E,F, MMP2–Myc and MMP9–Myc were present in the small organelles scattered through the cytosol as well as in bafilomycin-induced enlarged lytic organelles (see arrows).
Fig. 4.
Rab40b increases lysosomal degradation of MMP2 and MMP9. (A–C) Mock- or Rab40b-siRNA-treated MDA-MB-231 cells were harvested and analyzed by flow cytometry to measure the levels of intracellular MMP2–Myc (A), MMP9–Myc (B), transferrin receptor (C) and CD63 (C). Data shown in A and B are the means and s.d. of three independent experiments. Data shown in C are the means of two independent experiments. (D) MDA-MB-231 cells stably expressing MMP2–Myc or MMP9–Myc were transfected with Rab40b siRNA. Two days later, cells were incubated in the presence of absence of bafilomycin for 12 hours and medium was collected to measure the amounts of secreted MMP2–Myc and MMP9–Myc (inset). Cells were then analyzed by flow cytometry to measure the levels of intracellular MMP2–Myc and MMP9–Myc. Data shown are the means and s.d. of three independent experiments. (E,F) MDA-MB-231 cells stably expressing MMP2–Myc (E) or MMP9–Myc (F) were transfected with Rab40b siRNA. Two days later, cells were treated with bafilomycin for 12 hours, then fixed and co-stained with anti-Myc (red) or anti-CD63 (green) antibodies. Nuclei are stained blue with DAPI. Scale bar: 5 µm.
Rab40b is localized to VAMP4-containing secretory vesicles
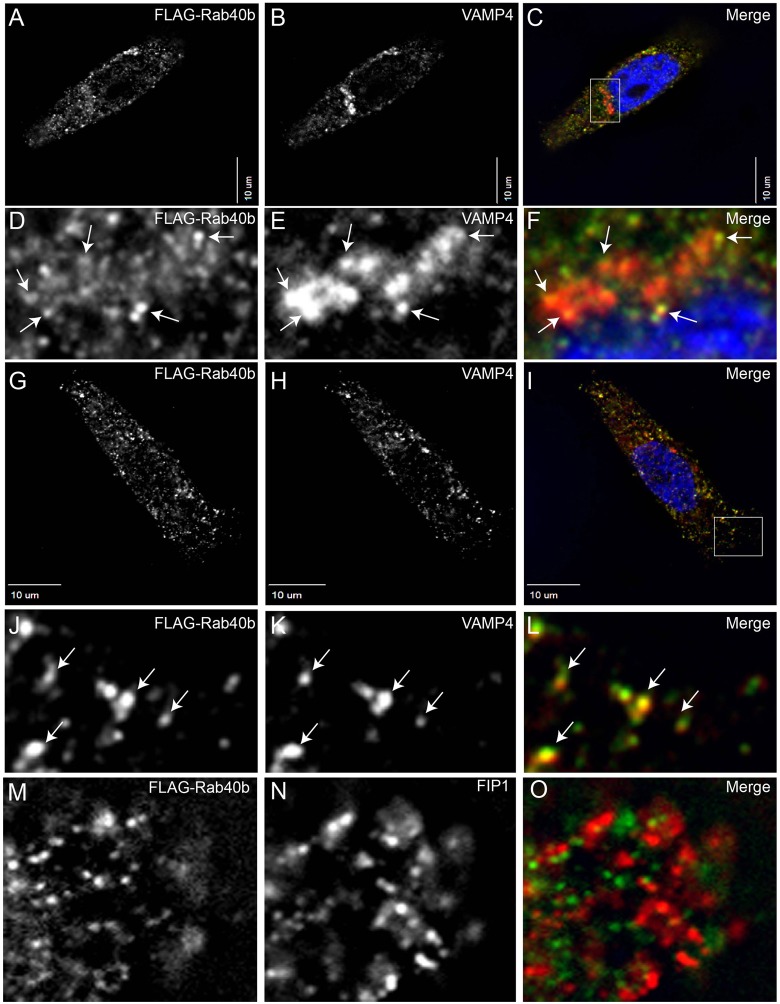
Subcellular localization of a protein can often provide clues about its cellular function. To this end, we compared the localization of FLAG–tagged Rab40b with the localization of VAMP4, a well-established marker for secretory vesicles and TGN (Steegmaier et al., 1999). As shown in Fig. 5A–F, FLAG–Rab40b was present at VAMP4-containing vesicles located at the edges of the TGN (marked by arrows). Although, the resolution of the images does not allow us to unequivocally determine the identity of these organelles, it is possible that these are VAMP4-containing secretory vesicles budding from the TGN. Consistently, FLAG–Rab40b colocalizes with VAMP4-secretory vesicles at the periphery of the cell (Fig. 5G–L, arrows), suggesting that Rab40b organelles contain VAMP4 (Kakhlon et al., 2006; Krzewski et al., 2011; Steegmaier et al., 1999). Similarly, VAMP4 also colocalized with MMP2–Myc and MMP9–Myc (supplementary material Fig. S1A–F). Because MT1-MMP is present in the recycling endosomes (Bravo-Cordero et al., 2007), we next compared the localization of FLAG–Rab40b and Rab11-FIP1/RCP, a known recycling endosome marker (Peden et al., 2004). As shown in Fig. 5M–O, FLAG–Rab40b was not present in recycling endosomes, suggesting that MMP2/9 and MT1-MMP are probably targeted to plasma membrane by distinct membrane transport pathways. Consistent with this, depletion of various Rab11 effector proteins (known to regulate recycling endosomes) or the Exocyst complex (known to mediate MT1-MMP targeting) did not have any effect on MMP2/9 secretion (data not shown).
Fig. 5.
FLAG–Rab40b colocalizes with VAMP4-containing secretory vesicles. MDA-MB-231 cells were transfected with FLAG–Rab40b and plated on collagen-coated glass coverlips. Cells were then fixed and stained with anti-FLAG (A,C,D,F,G,I,J,L,M,O), anti-VAMP4 (B,C,E,F,H,I,K,L) and anti-FIP1 (N,O) antibodies. In D–F, arrows indicate VAMP4 and Rab40b-containing vesicles at the edges of the TGN. In J–L, arrows indicate peripheral organelles containing VAMP4 and Rab40b. In C and I, boxed region marks area shown as higher magnification images in D–F and J–L. Nuclei in C,F,I are stained blue with DAPI.
Our data suggest that VAMP4 is the R-SNARE responsible for fusion of MMP2/9 secretory vesicles with the plasma membrane. To examine the role of VAMP4, we analyzed the effect of VAMP4 knockdown (supplementary material Fig. S2A) on secretion of endogenous MMP2 and MMP9. As shown in supplementary material Fig. S2B–D, VAMP4 depletion reduced the amounts of MMP2 and MMP9 in the medium. By contrast, the depletion of other vSNAREs, namely VAMP3 and VAMP7, did not have any effect on MMP2/9 secretion (supplementary material Fig. S2A–D).
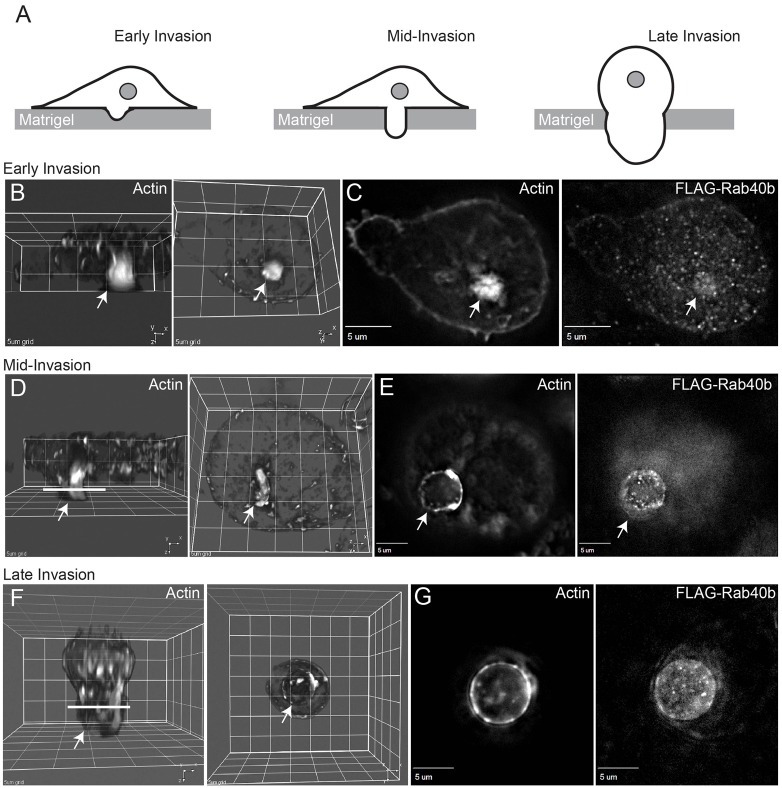
Previous findings from several laboratories have demonstrated that MMP2 and MMP9 are transported to the forming invadopodia during cancer cell invasion (Murphy and Courtneidge, 2011; Poincloux et al., 2009). Thus, we examined the localization of Rab40b-containing vesicles during cell invasion in vitro. To that end, FLAG–Rab40b-expressing MDA-MB-231 cells were seeded on Matrigel-coated Transwell filters containing 8 µm pores (for more details see the Materials and Methods). After 24 or 36 hours cells were fixed and stained with Rhodamine-phalloidin to visualize cells at different stages of invasion through Matrigel and filter pores (Fig. 6A). During early stages of invasion, cells formed an actin-rich invadopodia at the ventral side of the cell (Fig. 6A–C). FLAG–Rab40b-containing organelles were observed to be enriched at the site of forming invadopodia (Fig. 6C, arrow). At the mid-stage of the invasion, MDA-MB-231 extended pseudopodia (probably derived from invadopodia) into the filter pore (Fig. 6A,D), which also contained Rab40b-organelles clustered close to the pseudopodia plasma membrane (Fig. 6E). Finally, these pseudopodia thickened and filled the entire filter pore as cells started migrating through the filter to the bottom chamber (Fig. 6A,F,G). Similarly, organelles containing MMP2–Myc and MMP9–Myc also accumulated within these pseudopodia during cell invasion through the 8 µm pore (supplementary material Fig. S3). Taken together, all these data suggest that Rab40b-containing vesicles mediate the delivery of the MMP2/9 to the invadopodia/pseudopodia during cancer cell invasion.
Fig. 6.
Localization of Rab40b-containing organelles during cell invasion in vitro. MDA-MB-231 cells stably expressing FLAG–Rab40b were seeded on Matrigel-coated filters containing 8 µm pores. Cells were incubated for either 24 hours (B,D) or 36 hours (F). Cells were then fixed and stained with Rhodamine-phalloidin or anti-FLAG antibodies. Drawings in A depict the invasion stages imaged in panels B,D,F. Arrows in all images indicate invadopodia or pseudopodia (probably derived from invadopodia). B,D,F are 3D rendering of images shown in C,E,G and show cells from Z-Y (left panels) and X-Y (right panels) planes. Lines in D,F indicate the level of the optical sections depicted in E,G.
Rab40b is required for breast cancer cell invasion and invadopodia-dependent ECM degradation in vitro
Previous research has suggested that MMP2 and MMP9 play key roles in degradation of the ECM during breast cancer cell metastasis. Thus, we tested the effect of Rab40b knockdown on the ability of MDA-MB-231 cells to invade in vitro using Transwell filters (with 8 µm pores) coated with Matrigel. Consistent with the involvement of Rab40b in MMP2/9 secretion, depletion of Rab40b, but not Rab40a, resulted in a significant decrease in cell invasion, while having no effect on cell motility (Fig. 7A,B). Because VAMP4 is present on Rab40b-containing organelles, we also tested the effect of VAMP4 depletion on cell invasion. As shown in supplementary material Fig. S2E, depletion of VAMP4 inhibited the invasion ability of MDA-MB-231 cells.
Fig. 7.
Rab40b is required for MDA-MB-231 cell invasion in vitro. (A,B) Mock- or siRNA-treated MDA-MB-231 cells were plated on matrigel-coated (A) or uncoated (B) 8-µm-pore filters. Cells were then incubated for 8 hours (B) or 16 hours (A) and the extent of cell migration to the bottom side of the filter was analyzed by Crystal Violet staining (see the Materials and Methods). Data shown are the means and s.d. of three independent experiments; *P>0.05. (C–H) MDA-MB-231 cells were plated on gelatin and fibronectin-HiLyte Fluor488-coated coverslips (D,E,G and H). After incubation for 20 hours, cells were fixed and stained with Rhodamine-phalloidin (C,E,F,H). Arrows indicate invadopodia. Scale bars: 5 µm (C–E), 1 µm (F–H). (I–K) MDA-MB-231 cells were plated on gelatin-coated coverslips. After incubation for 20 hours, cells were fixed and stained with Rhodamine-phalloidin (I,K) and rabbit anti-Tks5 antibodies (J,K). Scale bar: 1 µm.
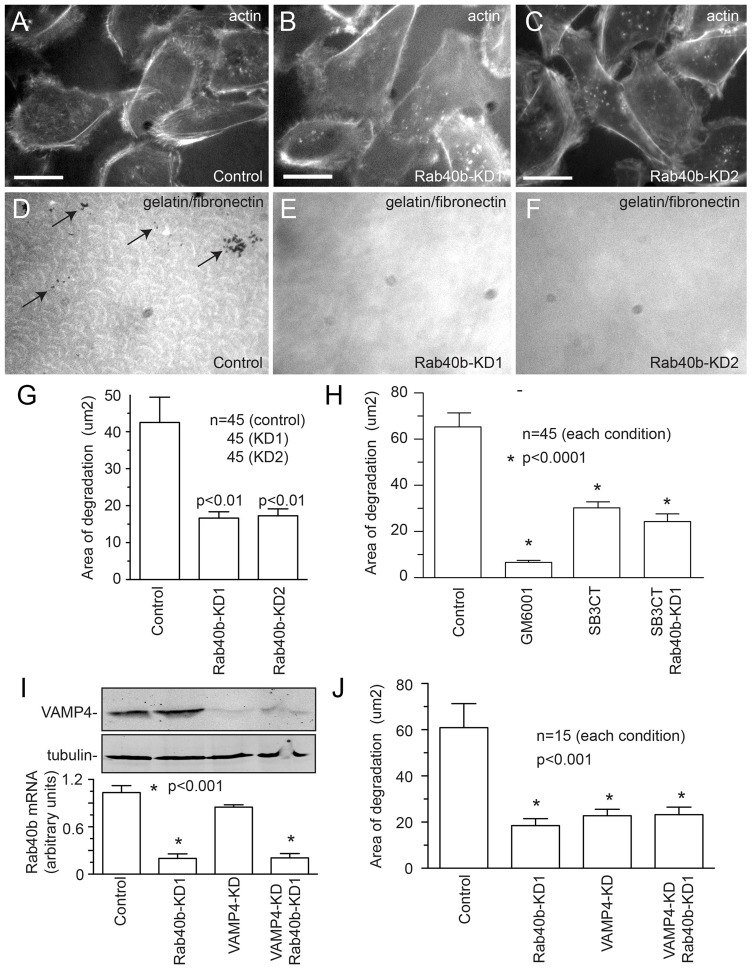
Our cellular localization data suggested that Rab40b is involved in targeting vesicles containing MMP2/9 to the invadopodia. To test this hypothesis, we investigated whether Rab40b is required for invadopodia-associated MMP secretion and ECM degradation. MDA-MB-231 cells were plated on gelatin-coated glass coverslips overlaid with fibronectin conjugated to HiLyte Fluor488. As shown in Fig. 7C–H, MDA-MB-231 cells formed actin-rich punctate structures, which associated with localized degradation of the fibronectin and gelatin matrix. These actin puncta can be identified as invadopodia, because they contained the invadopodia marker Tks5 (Fig. 7I–K) (Courtneidge et al., 2005; Murphy et al., 2011). Next, we tested whether Rab40b is required for invadopodia formation and invadopodia-associated ECM degradation. Rab40b depletion resulted in a decrease in invadopodia-associated ECM degradation (Fig. 8A–G), but had no effect on invadopodia formation (supplementary material Fig. S4).
Fig. 8.
Rab40b is required for invadopodia-associated ECM degradation. (A–F) Untreated or Rab40b-siRNA-treated MDA-MB-231 cells were plated on gelatin and fibronectin HiLyte Fluor488-coated coverslips (D–F). After 20 hours of incubation, cells were fixed and stained with Rhodamine-phalloidin (A–C). Scale bars: 25 µm. (G) Quantification of ECM degradation in untreated and Rab40b-siRNA-treated MDA-MB-231 cells. Data shown are the means and s.e. of three independent experiments. n is the total number of cells analyzed. (H) Quantification of ECM degradation in MDA-MB-231 cells treated with either GM6001 (broad-spectrum MMP inhibitor) or SB3CT (MMP2/MMP9 inhibitor). Where indicated, cells were also treated with Rab40b siRNA. Data shown are the means and s.e. of three independent experiments. n is the total number of cells analyzed. (I) Mock-, Rab40b-siRNA- or VAMP4 siRNA-treated cells were harvested and the levels of mRNA encoding Rab40b analyzed by qPCR (graph). The data shown are the means and s.d. The levels of VAMP4 were analyzed by western blotting (top panels) with tubulin used as a loading control. (J) Quantification of ECM degradation in mock-, Rab40b-siRNA- or VAMP4-siRNA-treated MDA-MB-231 cells. Data shown are the means and s.e. of three independent experiments. n is the total number of cells analyzed.
Although Rab40b knockdown decreased invadopodia-associated ECM degradation, it remains unclear whether this effect is due to a decrease in MMP2/9 targeting. Some reports have questioned whether MMP2/9 are required for invadopodia-associated ECM degradation, or whether invadopodia are dependent only on MT1-MMP activity (Hotary et al., 2006; Poincloux et al., 2009). To validate the role of MMP2/9 in invadopodia-associated ECM degradation, cells were treated with a broad-spectrum MMP inhibitor GM6001 or with the MMP2/9-specific inhibitor SB3CT (supplementary material Fig. S1G). As previously reported (Clark et al., 2007), GM6001 almost completely blocked invadopodia-associated ECM degradation, while having no effect on invadopodia number (Fig. 8H; supplementary material Fig. S4B,F). By contrast, treatment with SB3CT (Ikejiri et al., 2005; Overall and Kleifeld, 2006) only partially inhibited invadopodia-induced ECM degradation (Fig. 8H; supplementary material Fig. S4C,F). Taken together, these data indicate that in MDA-MB-231 cells, MMP2/9 secretion is at least partially responsible for invadopodia-dependent ECM degradation. To further test whether Rab40b is required for MMP2/9 targeting to the invadopodia, we co-treated MDA-MB-231 cells with Rab40b siRNA and SB3CT. As shown in Fig. 8H, simultaneous inhibition of MMP2/9 and depletion of Rab40b did not further inhibit ECM degradation as compared with knockdown of Rab40b or inhibition of MMP2/9 alone (Fig. 8H; supplementary material Fig. S4D,F), demonstrating that Rab40b and MMP2/9 probably act in the same pathway. Similarly, dual knockdown of Rab40b and VAMP4 also did not further inhibit ECM degradation compared with depletion of Rab40b or VAMP4 alone (Fig. 8I,J). These data combined with our imaging experiments suggest that Rab40b and VAMP4 are responsible for targeting of MMP2/9-containing transport vesicles to the invadopodia during invasion of cancer cells.
To test whether Rab40b plays a similar role in other cancers, we next tested the effect of Rab40b knockdown on invadopodia-associated ECM degradation in human melanoma HMCB cells. HMCB cells also formed invadopodia reminiscent to MDA-MB-231 breast cancer cells (supplementary material Fig. S5A,B). Importantly, Rab40b knockdown also led to a decrease in the area of invadopodia-associated ECM degradation (supplementary material Fig. S5C–F), suggesting that Rab40b also regulates targeted MMP2/9 secretion in non-breast cancers.
Discussion
Multiple studies have demonstrated that MMP2/9 play an important role during invasion and metastasis of various cancers (Chambers and Matrisian, 1997; Egeblad and Werb, 2002; Polette et al., 2004; Shah et al., 2009), yet the machinery that regulates MMP2/9 transport and fusion with the plasma membrane remains elusive. In this study, we used an siRNA screen to identify Rab40b as a small monomeric GTPase that is required for secretion of both MMP2 and MMP9. All Rab GTPases act as master regulators of various membrane transport steps, by regulating cargo sorting, vesicle budding, vesicle transport and vesicle targeting to the appropriate acceptor compartment (Pfeffer, 2001). Before this study, little was known about the cellular function of Rab40b GTPase, or the identity of Rab40b-interacting proteins. Thus, to further understand the mechanisms of MMP2/9 transport, we investigated the localization and function of Rab40b during MDA-MB-231 breast cancer cell invasion in vitro. Interestingly, we have shown that Rab40b-containing vesicles colocalize with R-SNARE, VAMP4, which has been shown to mediate protein transport from the TGN to the plasma membrane (Steegmaier et al., 1999). Furthermore, we demonstrated that Rab40b and VAMP4 depletion by siRNA results in a decrease in MMP2/9 secretion and MDA-MB-231 cell invasion. The data described above indicate that MMP2/9 are transported to the plasma membrane by secretory vesicles containing Rab40b and VAMP4. Interestingly, MT1-MMP, the other MMP that mediates ECM degradation during cell invasion, was shown to be transported to the plasma membrane by recycling endosomes (Hotary et al., 2006; Poincloux et al., 2009). Furthermore, recent work demonstrated that in addition to Rab8, targeting of MT1-MMP to the plasma membrane requires the Exocyst complex, a protein complex known to function as a transport-vesicle tethering factor (Sakurai-Yageta et al., 2008). We demonstrate that siRNA-dependent knockdown of Rab8 or of components of the Exocyst complex has no effect on MMP2/9 transport and secretion (Fig. 2 and data not shown). Thus, MMP2/9 and MT1-MMP appear to be transported and secreted by distinct membrane transport pathways.
One of the effects of Rab40b knockdown was a decrease in MMP2/9 secretion. This decrease was, at least in part, caused by increased lysosomal degradation of MMP2 and MMP9. Because Rab40b localizes to the VAMP4-containing budding sites at the TGN, it is likely that Rab40b is required for appropriate sorting of MMP2 and MMP9 to the vesicles destined to be transported to the plasma membrane. Interestingly, inhibition of lysosomal degradation did not rescue the MMP2/9 secretion defect, suggesting that Rab40b is also required for MMP2/9 targeting and fusion with the plasma membrane. Consistent with this observation, Rab40b knockdown inhibited invadopodia-associated ECM degradation and in vitro invasion of MDA-MB-231 cells, suggesting that Rab40b regulates MMP2/9 sorting at the TGN as well as MMP2/9 vesicle targeting to the invadopodia.
Invadopodia are actin-rich, finger-like cellular projections that have been implicated in mediating ECM degradation and cell invasion (Murphy and Courtneidge, 2011). Invadopodia have been primarily studied in tissue culture cells, and are sometimes referred to as podosomes. The functional differences between podosomes and invadopodia remain unclear, but generally actin structures in cancer cells are called invadopodia, whereas in non-cancerous cells they are called podosomes. It has been demonstrated that MMP2/9 and MT1-MMP are all enriched at the invadopodia where they mediate ECM degradation in vitro and in vivo (Murphy and Courtneidge, 2011). Interestingly, although Rab40b knockdown had only a moderate effect on MMP2/9 secretion into the medium, it significantly decreased MMP2/9-dependent invadopodia-associated ECM degradation. Taken together, our data suggest that Rab40b is required for MMP2/9 secretion at the newly formed invadopodia. However, knockdown of Rab40b decreased ECM degradation at invadopodia by only 50%. The rest of the degradation is presumably mediated by MT1-MMP. This is consistent with previous observations that MT1-MMP is transported to invadopodia by Exocyst-dependent recycling endosomes (Sakurai-Yageta et al., 2008). The question that remains is why do cancer cells need two distinct pathways of transporting MMPs to invadopodia? Because VAMP4 and the Exocyst complex are also present at the plasma membrane outside invadopodia, it is possible that overlapping targeting mechanisms by two different transport pathways are required to ensure the fidelity of ECM degradation at the invadopodia. In addition to degrading collagen, MT1-MMP has been shown to cleave and activate MMP2 (Strongin et al., 1995). Thus co-targeting of MMP2/9 and MT1-MMP to the same location would result in an amplification of ECM degradation associated with invadopodia.
Although this study demonstrates that Rab40b is required for MMP2/9 sorting and targeting to invadopodia, it is still unknown what effector proteins bind to Rab40b. All Rab GTPases function by recruiting multiple effector proteins, which then mediate transport vesicle formation, transport and targeting (Pfeffer, 2001). Furthermore, Rab40b has a SOCS box domain, a feature unique to the Rab40 sub-family of proteins (Stein et al., 2012). Interestingly, the SOCS box motif within other proteins is implicated in regulating cytokine secretion (Piessevaux et al., 2008). With the establishment of Rab40b as an important regulator of MMP2/9 targeting to invadopodia, the identification of Rab40b canonical effector proteins, as well as Rab40b-SOCS-interacting proteins will be future steps to identify the mechanisms that regulate MMP2/9 targeting during cancer cell invasion.
Materials and Methods
Antibodies and constructs
Mouse monoclonal anti-Myc (9E10) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse anti-MMP2 and mouse anti-MMP9 were purchased from Millipore (Temecula, CA). Anti-Myc antibody conjugated with HRP (GTX21261) was obtained from Gene Tex. Anti-FLAG antibody was purchased from Sigma (St Louis, MO). Rabbit polyclonal anti-Tks5 antibody was generated using recombinant human Tks5-SH3-1/Tks5-SH3-4 domains as previously described (Blouw et al., 2008; Seals et al., 2005). Anti-VAMP4 antibodies were previously described (Gordon et al., 2010). Alexa-Fluor-594- and Alexa-Fluor-488-conjugated anti-rabbit and anti-mouse secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA).
Cell culture and generation of tet-inducible MDA-MB-231 cell lines
MDA-MB-231 cells were cultured in 50% RPMI-1640 and 50% DMEM with 4.5 g/l glucose, 5.84 g/l L-glutamine and 10% heat-inactivated fetal bovine serum (FBS), and supplemented with 100 IU/ml penicillin and 100 µg streptomycin. To create tet-inducible MDA-MB-231 MMP2–Myc and MMP9–Myc stable cell lines, MMP2–Myc or MMP9–Myc were cloned into the pHUSH retroviral expression vector (Genentech, South San Francisco, CA). Stable, clonal cell lines were then selected using 1 µg/ml of puromycin and grown using the above described medium [MDA-MB-231 medium supplemented with tet-free FBS (Clontech Laboratories, Mountain View, CA)]. To induce either MMP2–Myc or MMP9–Myc expression, stable cells lines were incubated in the presence of 1 µg/ml of doxycycline for 24 hours.
siRNA library screen
The siRNA screen was performed with a pool of four different siRNAs against each protein of interest. MDA-MB-231 stable cell lines expressing either MMP9–Myc or MMP2–Myc were plated on 96-well plates at 1.2×104 density and co-transfected with siRNA oligonucleotides using RNAi Max (Invitrogen, Carlsbad, CA), as described in the manufacturer's protocol. Transfected cells were incubated for 36 hours. Complete growth medium was then replaced with serum-free medium and cells incubated for another 32 hours. The cell viability was monitored by Perkin Elmer Envision reader using Cell Titer Glow reagent (Promega, Madison, WI). Medium was then collected and levels of secreted MMP2–Myc or MMP9–Myc analyzed by ELISA.
ELISA assays
For the siRNA screen, 96-well microplates were coated with 100 µl of diluted anti-MMP2 (2 µg/ml in PBS) or diluted anti-MMP9 (0.5 µg/ml in PBS) antibody solution. This was followed by a wash, after which plates were blocked with 200 µl of blocking buffer. Plates were then washed and incubated with 100 µl of medium collected from siRNA-treated cells. After 2 hours of incubation, plates were washed and incubated with 90 µl of mouse anti-Myc antibody conjugated to HRP. Plates were again incubated for 1 hour at room temperature (22°C), followed by a wash. The amount of MMP2–Myc or MMP9–Myc was measured using HRP substrate solution and optical density at 450 nm using a NanoDrop ND-1000 Spectrophotometer.
To measure levels of secreted IL-6, mock- or Rab40b-siRNA-treated cells were seeded into six-well dishes at a density of 8×105 cells. IL-6 secretion was induced with 1 mg/ml of LPS. After incubation for 16 hours, medium was collected for ELISA analysis and cells were harvested for Bradford assay to normalize samples. IL-6 ELISA analysis was performed using IL-6 DuoSet ELISA Development system as described in the manufacturer's protocol. The optical density at 450 nm was determined using a NanoDrop ND-1000 Spectrophotometer.
In vitro invasion and motility assays
MDA-MB-231 cell motility or invasion was measured using Transwell filter assays. To measure motility, mock- or siRNA-treated cells were resuspended in fresh serum-containing medium and added to the top chamber of six Transwell filter insets (6.5 mm filters with 8 µm pores, Corning, NY) at a density of 100,000 cells per filter. MDA-MB-231 conditioned medium (the medium after 48 hours of incubation with cells) was added to the bottom chamber. In invasion assays, filter was coated with Matrigel matrix. After a 16 hours (invasion assays) or 8 hours (motility assays) cells were stained with 0.1% Crystal Violet. After three washes, cells that remained on the upper surface of the filter were removed with a cotton swab. The dye from cells remaining on the bottom side of the filter was extracted with 2.5% acetic acid and quantified by measuring optical density at 570 nm.
Fluorescent microscopy
MDA-MB-231 cells were plated on collagen-coated glass coverslips, grown overnight and fixed with 4% paraformaldehyde for 15 minutes, permeabilized for 10 minutes in phosphate-buffered saline (PBS) containing 0.4% saponin, and non-specific sites were blocked with PBS containing 0.2% BSA and 1% fetal bovine serum. Cells were then incubated with specific antibodies, washed in PBS, and mounted in VectaShield (Vector Laboratories, Burlingame, CA). Cells were imaged with an inverted Zeiss Axiovert 200M deconvolution microscope with a 63× oil-immersion lens and Sensicam QE CCD camera. Image processing was performed using Intelligent Imaging Innovations (Denver, CO) three-dimensional rendering and exploration software.
To visualize invadopodia, Transwell filter insets (6.5 mm filters with 8 µm pores, Corning) were coated with Matrigel. Briefly, filters were coated with 60 µl of Matrigel for 1 hour at 4°C, after which 55 µl of Matrigel was removed and filters were incubated at 37°C for 30 minutes to allow Matrigel to polymerize. Medium was then added to both sides of the filter for 15 minutes to rehydrate the Matrigel. MDA-MB-231 cells stably expressing FLAG–Rab40b were then seeded at a density of 50,000 cells per filter. MDA-MB-231 conditioned medium (the medium after 48 hours of incubation with cells) was added to the bottom chamber. Cells were incubated for either 24 or 36 hours, fixed and stained with Rhodamine-phalloidin, anti-Myc or anti-FLAG antibodies. Z-stacks of invading cells were imaged using 100 nm z-step. The 3D rendering was performed using Intelligent Imaging Innovations (Denver, CO) three-dimensional rendering and exploration software.
FACS analysis
Mock or Rab40b-siRNA-treated MDA-MB-231 cells were incubated at 37°C for 64 hours. Cells were then trypsinized and fixed with 4% paraformaldehyde, followed by quenching with 0.1 M glycine. Cells were then permeabilized with FACS buffer (PBS containing 0.4% saponin, 1% BSA and 2% FBS) for 30 minutes, followed by incubation with primary antibodies for 1 hour. Cells were washed three times with FACS buffer and incubated for 30 minutes with FITC-conjugated secondary antibodies. After another set of three washes, cells were resuspended in 500 ml of PBS and analyzed by flow cytometry using Cytomics FC500 flow cytometer (Beckman Coulter, Brea, CA).
To analyze plasma membrane MMP14, mock-, Rab40b-siRNA- or VAMP4-siRNA-treated MDA-MB-231 cells were incubated at 37°C for 64 hours. Cells were then trypsinized and resuspended in HEPES-buffered and serum-supplemented media. Cells were then incubated with anti-MMP14-APC antibodies at 37°C for 1 hour. Cells were washed three times with medium, resuspended in 500 ml of PBS and analyzed by flow cytometry using Cytomics FC500 flow cytometer (Beckman Coulter, Brea, CA).
Zymography assays
Mock- or Rab40b-siRNA-treated MDA-MB-231 cells were incubated in the complete medium at 37°C. After 48 hours of incubation, medium was replaced with Opti-MEM (Invitrogen, Carlsbad, CA) and cells were incubated at 37°C for another 24 hours. Opti-MEM medium was collected and cell lysates were harvested using PBS containing 1% Triton X-100. The levels of secreted MMP2 and MMP9 in Opti-MEM were then analyzed by zymography. Fetal bovine serum (rich in MMP2 and MMP9) was used as positive control. Opti-MEM collected from cell-free wells was used as a negative control. Briefly, Opti-MEM samples were diluted in the standard SDS-PAGE sample buffer without a reducing agent. Samples were then separated using 7.5% polyacrylamide gels containing 3 µg/ml of gelatin. Sample loading was normalized based on cell lysate protein concentrations. Following electrophoresis, gels were washed twice with 2.5% Triton X-100 to remove SDS. Gels were then incubated in digestion buffer (50 mM Tris-HCl, pH 8.0 containing 5 mM CaCl2) at 37°C for 24–48 hours. Gels were subsequently stained with Coomassie Brilliant Blue dye. The extent of gelatin proteolysis by endogenous MMP2 (bottom band) and MMP9 (top band) was detected as white bands on a dark background.
In situ zymography assays
The in situ zymography/matrix degradation assay was done by coating 18 mm round coverslips in 12-well plates with 2.5% gelatin, 2.5% sucrose in PBS at 37°C. The gelatin was allowed to set at 4°C before crosslinking with 0.5% glutaraldehyde by incubation at 4°C for 15 minutes. A 50 µg/ml solution of FITC-fibronectin (Cytoskeleton, Denver, CO) was then overlaid on top of crosslinked gelatin and incubated in the dark for 1 hour at 4°C. The dish was sterilized with 70% ethanol, washed with DMEM, and equilibrated with invadopodia medium [DMEM supplemented with 20% FBS (Atlanta Biologicals) and 10% Nu-Serum] for 30 minutes. MDA-MB-231 cells (4×104 cells in 2 ml of invadopodia medium) were then added to each well and incubated for 20 hours. The cells were then fixed in 3% paraformaldehyde, permeabilized with 0.4% Triton X-100 in PBS and stained with Rhodamine-phalloidin (Invitrogen, Carlsbad, CA). To quantify invadopodia formation and localized matrix degradation, ten randomly chosen fields were imaged (using 63× objective) per treatment for each experiment. A total of 260–330 cells were counted in at least three independent experiments. To measure the number of cells with invadopodia, cells were counted based on the presence of actin puncta and degradation spots seen underneath the cells within the cell boundaries. To measure invadopodia-associated area of degradation, the areas lacking FITC-fibronectin fluorescence were measured using Intelligent Imaging Innovations (Denver, CO) three-dimensional rendering and exploration software. Only degradation areas associated within cell boundaries were analyzed.
Reverse transcriptase polymerase chain reaction (RT-PCR) and quantitative PCR (qPCR)
Total RNA was extracted from 2×107 MDA-MB-231 cells using TRIzol (Invitrogen) according to the manufacturer's protocol. Reverse transcription to cDNA was performed with SuperScript III (Invitrogen) using random hexamer primers. PCR was performed using Taq polymerase (Invitrogen). To quantify the percent of knockdown, cDNA from mock- or siRNA-treated cells was analyzed in triplicate by qPCR amplification using Sybr Green qPCR Master Mix using Applied Biosystems ViiA7 Real-Time PCR System. The qPCR amplification conditions were: 50°C (2 minutes), 95°C (10 minutes), 40 cycles at 95°C (15 seconds), 60°C (1 minute). Primer pairs were designed to amplify mRNA-specific fragments and unique products were tested by melt-curve analysis. Relative quantification was calculated by the ΔΔCT method. Data shown as the fold change (averaged from two independent experiments) in Rab40b-knockdown cells compared with mock-treated cells. β-actin served as normalizing control (sense, 5′-AAAGACCTGTACGCCAACAC-3′; anti-sense, 5′-GTCATACTCCTGCTTGCTGA-3′). MMP2 was amplified using following primers: sense, 5′-CCTGATGGCACCCATTTACACC-3′; anti-sense, 5′-CGACGGCATCCAGGTTATCG-3′. MMP9 was amplified using following primers: sense, 5′-CCCTTCTACGGCCACTACTGTG-3′; anti-sense, 5′-GCACTGCAGGATGTCATAG-3′.
Flow cytometry-based GFP–hGH secretion assay
To measure hGH secretion we used HeLa cells stably expressing GFP–FKBP-hGH as previously described (Gordon et al., 2010) (supplementary material Fig. S1H). This assay is based on the property of FKBP to form ligand-reversible aggregates, thus trapping GFP–FKBP-hGH in the ER. Addition of FKBP ligand AP21998 results in solubilization of these aggregates and synchronous secretion of GFP–hGH from the cells. Because hGH is tagged with GFP (Gordon et al., 2010), secretion can be quantified in live cells by measuring the cell-associated GFP fluorescence (not secreted) by flow cytometry.
To measure the effect of Rab40b knockdown on hGH secretion, HeLa cells stably expressing GFP–FKBP-hGH were plated at 25% confluence on six-well plates and transfected with siRNA targeted to either Rab40b or Syntaxin 5. Syntaxin 5 siRNA was used as a positive control, because it was previously reported that Synatxin 5 depletion inhibits hGH secretion (Gordon et al., 2010). 72 hours after transfection, cells were trypsinised and counted before plating at 50,000 cells per well on a 24-well plate. Cells were incubated overnight before use in the secretion assay. Secretion of the GFP–FKBP-hGH initiated using AP21998 at a concentration of 1 µM in prewarmed culture medium. To halt secretion, samples were placed on ice, washed with cold PBS and trypsinized on ice for 2 hours. To measure the amount of cargo remaining in cells after secretion, samples were analysed using a BD Fortessa flow cytometer with live cells gated by forward and side scatter. Ten thousand live cells were analyzed for each sample. The mean GFP fluorescence was calculated using the software FLOWJO (TreeStar). The values of intracellular GFP remaining after secretion were normalized using GFP fluorescence before addition of AP21998.
Statistical analyses
Two-tailed independent Student's t-tests were used to analyze the results of MMP2/9 and IL6 secretion, FACS analysis, invasion and motility assays. All statistical calculations were done using GraphPad Prism 5.0d software (La Jolla, CA). Unless otherwise noted, all data shown are the means and s.d. of three independent experiments.
Supplementary Material
Acknowledgments
We are grateful to Dr John Tentler (UC Denver, AMC) for the HMCB melanoma cell line.
Footnotes
Author contributions
A.J. was responsible for most of the experiments as well as participated in writing the manuscript. J.Jing generated tet-inducible MMP2–Myc and MMP9–Myc cell lines and performed the siRNA screen. J.L. and J.Junutula helped with the siRNA screen. P.S. provided us with cell lines and helped with MMP secretion assays. S.G. and A.P. completed HGF secretion assays and provided Vamp4 reagents. R.P. oversaw and directed the entire project and participated in writing the manuscript.
Funding
This work was supported by grants from the Susan G. Komen foundation [grant number BCTR0706749 to R.P.]; and the Cancer League of Colorado to R.P. This project was also partially funded by the National Institutes of Health [grant number DK064380 to R.P.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.126573/-/DC1
References
- Artym V. V., Zhang Y., Seillier-Moiseiwitsch F., Yamada K. M., Mueller S. C. (2006). Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 66, 3034–3043 10.1158/0008-5472.CAN-05-2177 [DOI] [PubMed] [Google Scholar]
- Blouw B., Seals D. F., Pass I., Diaz B., Courtneidge S. A. (2008). A role for the podosome/invadopodia scaffold protein Tks5 in tumor growth in vivo. Eur. J. Cell Biol. 87, 555–567 10.1016/j.ejcb.2008.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguignon L. Y., Gunja-Smith Z., Iida N. (1998). CD44v3,8�–10 is involved in cytoskeleton-mediated tumor cell migration and matrix metalloproteinase (MMP-9) association in metastatic breast cancer cells. J. Cell Physiol. 176, 206–215 [DOI] [PubMed] [Google Scholar]
- Bravo-Cordero J. J., Marrero-Diaz R., Megías D., Genís L., García-Grande A., García M. A., Arroyo A. G., Montoya M. C. (2007). MT1-MMP proinvasive activity is regulated by a novel Rab8-dependent exocytic pathway. EMBO J. 26, 1499–1510 10.1038/sj.emboj.7601606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell P. T., Spence H. J., Parsons M., White D. P., Clark K., Cheng K. W., Mills G. B., Humphries M. J., Messent A. J., Anderson K. I. et al. (2007). Rab25 associates with alpha5beta1 integrin to promote invasive migration in 3D microenvironments. Dev. Cell 13, 496–510 10.1016/j.devcel.2007.08.012 [DOI] [PubMed] [Google Scholar]
- Chambers A. F., Matrisian L. M. (1997). Changing views of the role of matrix metalloproteinases in metastasis. J. Natl. Cancer Inst. 89, 1260–1270 10.1093/jnci/89.17.1260 [DOI] [PubMed] [Google Scholar]
- Clark E. S., Whigham A. S., Yarbrough W. G., Weaver A. M. (2007). Cortactin is an essential regulator of matrix metalloproteinase secretion and extracellular matrix degradation in invadopodia. Cancer Res. 67, 4227–4235 10.1158/0008-5472.CAN-06-3928 [DOI] [PubMed] [Google Scholar]
- Clark E. S., Weaver A. M. (2008). A new role for cortactin in invadopodia:regulation of protease secretion. Eur. J. Cell Biol. 87, 581–590 10.1016/j.ejcb.2008.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark E. S., Brown B., Whigham A. S., Kochaishvili A., Yarbrough W. G., Weaver A. M. (2009). Aggressiveness of HNSCC tumors depends on expression levels of cortactin, a gene in the 11q13 amplicon. Oncogene 28, 431–444 10.1038/onc.2008.389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtneidge S. A., Azucena E. F., Pass I., Seals D. F., Tesfay L. (2005). The SRC substrate Tks5, podosomes (invadopodia), and cancer cell invasion. Cold Spring Harb. Symp. Quant. Biol. 70, 167–171 10.1101/sqb.2005.70.014 [DOI] [PubMed] [Google Scholar]
- Dozynkiewicz M. A., Jamieson N. B., Macpherson I., Grindlay J., van den Berghe P. V., von Thun A., Morton J. P., Gourley C., Timpson P., Nixon C. et al. (2012). Rab25 and CLIC3 collaborate to promote integrin recycling from late endosomes/lysosomes and drive cancer progression. Dev. Cell 22, 131–145 10.1016/j.devcel.2011.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egeblad M., Werb Z. (2002). New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2, 161–174 10.1038/nrc745 [DOI] [PubMed] [Google Scholar]
- Gordon D. E., Bond L. M., Sahlender D. A., Peden A. A. (2010). A targeted siRNA screen to identify SNAREs required for constitutive secretion in mammalian cells. Traffic 11, 1191–1204 10.1111/j.1600-0854.2010.01087.x [DOI] [PubMed] [Google Scholar]
- Hotary K., Li X. Y., Allen E., Stevens S. L., Weiss S. J. (2006). A cancer cell metalloprotease triad regulates the basement membrane transmigration program. Genes Dev. 20, 2673–2686 10.1101/gad.1451806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikejiri M., Bernardo M. M., Bonfil R. D., Toth M., Chang M., Fridman R., Mobashery S. (2005). Potent mechanism-based inhibitors for matrix metalloproteinases. J. Biol. Chem. 280, 33992–34002 10.1074/jbc.M504303200 [DOI] [PubMed] [Google Scholar]
- Itoh T., Tanioka M., Yoshida H., Yoshioka T., Nishimoto H., Itohara S. (1998). Reduced angiogenesis and tumor progression in gelatinase A-deficient mice. Cancer Res. 58, 1048–1051 [PubMed] [Google Scholar]
- Itoh T., Tanioka M., Matsuda H., Nishimoto H., Yoshioka T., Suzuki R., Uehira M. (1999). Experimental metastasis is suppressed in MMP-9-deficient mice. Clin. Exp. Metastasis 17, 177–181 10.1023/A:1006603723759 [DOI] [PubMed] [Google Scholar]
- Kakhlon O., Sakya P., Larijani B., Watson R., Tooze S. A. (2006). GGA function is required for maturation of neuroendocrine secretory granules. EMBO J. 25, 1590–1602 10.1038/sj.emboj.7601067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzewski K., Gil-Krzewska A., Watts J., Stern J. N., Strominger J. L. (2011). VAMP4- and VAMP7-expressing vesicles are both required for cytotoxic granule exocytosis in NK cells. Eur. J. Immunol. 41, 3323–3329 10.1002/eji.201141582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsky W. L., Kelly T., Lin C. Y. (1993). Binding and localization of M(r) 72,000 matrix metalloproteinase at cell surface invadopodia. Cancer Res. 53, 3159–3164 [PubMed] [Google Scholar]
- Murphy D. A., Courtneidge S. A. (2011). The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12, 413–426 10.1038/nrm3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy G., Gavrilovic J. (1999). Proteolysis and cell migration: creating a path? Curr. Opin. Cell Biol. 11, 614–621 10.1016/S0955-0674(99)00022-8 [DOI] [PubMed] [Google Scholar]
- Murphy D. A., Diaz B., Bromann P. A., Tsai J. H., Kawakami Y., Maurer J., Stewart R. A., Izpisúa-Belmonte J. C., Courtneidge S. A. (2011). A Src-Tks5 pathway is required for neural crest cell migration during embryonic development. PLoS ONE 6, e22499 10.1371/journal.pone.0022499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara H., Howard L., Thompson E. W., Sato H., Seiki M., Yeh Y., Chen W. T. (1997). Transmembrane/cytoplasmic domain-mediated membrane type1-matrix metalloprotease docking to invadopodiais required for cell invasion. Proc. Natl. Acad. Sci. USA 94, 7959–7964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overall C. M., Kleifeld O. (2006). Towards third generation matrix metalloproteinase inhibitors for cancer therapy. Br. J. Cancer 94, 941–946 10.1038/sj.bjc.6603043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden A. A., Schonteich E., Chun J., Junutula J. R., Scheller R. H., Prekeris R. (2004). The RCP-Rab11 complex regulates endocytic protein sorting. Mol. Biol. Cell 15, 3530–3541 10.1091/mbc.E03-12-0918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S. R. (2001). Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol. 11, 487–491 10.1016/S0962-8924(01)02147-X [DOI] [PubMed] [Google Scholar]
- Piessevaux J., Lavens D., Peelman F., Tavernier J. (2008). The many faces of the SOCS box. Cytokine Growth Factor Rev. 19, 371–381 10.1016/j.cytogfr.2008.08.006 [DOI] [PubMed] [Google Scholar]
- Poincloux R., Lizárraga F., Chavrier P. (2009). Matrix invasion by tumour cells: a focus on MT1-MMP trafficking to invadopodia. J. Cell Sci. 122, 3015–3024 10.1242/jcs.034561 [DOI] [PubMed] [Google Scholar]
- Polette M., Nawrocki-Raby B., Gilles C., Clavel C., Birembaut P. (2004). Tumour invasion and matrix metalloproteinases. Crit. Rev. Oncol. Hematol. 49, 179–186 10.1016/j.critrevonc.2003.10.008 [DOI] [PubMed] [Google Scholar]
- Quintavalle M., Elia L., Condorelli G., Courtneidge S. A. (2010). MicroRNA control of podosome formation in vascular smooth muscle cells in vivo and in vitro. J. Cell Biol. 189, 13–22 10.1083/jcb.200912096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacle A., Murphy G., Roghi C. (2003). Membrane type I-matrix metalloproteinase (MT1-MMP) is internalised by two different pathways and is recycled to the cell surface. J. Cell Sci. 116, 3905–3916 10.1242/jcs.00710 [DOI] [PubMed] [Google Scholar]
- Roskelley C. D., Bissell M. J. (1995). Dynamic reciprocity revisited: a continuous, bidirectional flow of information between cells and the extracellular matrix regulates mammary epithelial cell function. Biochem. Cell Biol. 73, 391–397 10.1139/o95-046 [DOI] [PubMed] [Google Scholar]
- Roskelley C. D., Srebrow A., Bissell M. J. (1995). A hierarchy of ECM-mediated signalling regulates tissue-specific gene expression. Curr. Opin. Cell Biol. 7, 736–747 10.1016/0955-0674(95)80117-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai-Yageta M., Recchi C., Le Dez G., Sibarita J. B., Daviet L., Camonis J., D'Souza-Schorey C., Chavrier P. (2008). The interaction of IQGAP1 with the exocyst complex is required for tumor cell invasion downstream of Cdc42 and RhoA. J. Cell Biol. 181, 985–998 10.1083/jcb.200709076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalfeldt B., Prechtel D., Härting K., Späthe K., Rutke S., Konik E., Fridman R., Berger U., Schmitt M., Kuhn W. et al. (2001). Increased expression of matrix metalloproteinases (MMP)-2, MMP-9, and the urokinase-type plasminogen activator is associated with progression from benign to advanced ovarian cancer. Clin. Cancer Res. 7, 2396–2404 [PubMed] [Google Scholar]
- Seals D. F., Azucena E. F., Jr., Pass I., Tesfay L., Gordon R., Woodrow M., Resau J. H., Courtneidge S. A. (2005). The adaptor protein Tks5/Fish is required for podosome formation and function, and for the protease-driven invasion of cancer cells. Cancer Cell 7, 155–165 10.1016/j.ccr.2005.01.006 [DOI] [PubMed] [Google Scholar]
- Shah F. D., Shukla S. N., Shah P. M., Shukla H. K., Patel P. S. (2009). Clinical significance of matrix metalloproteinase 2 and 9 in breast cancer. Indian J. Cancer 46, 194–202 10.4103/0019-509X.52953 [DOI] [PubMed] [Google Scholar]
- Steegmaier M., Klumperman J., Foletti D. L., Yoo J. S., Scheller R. H. (1999). Vesicle-associated membrane protein 4 is implicated in trans-Golgi network vesicle trafficking. Mol. Biol. Cell 10, 1957–1972 10.1091/mbc.10.6.1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M., Pilli M., Bernauer S., Habermann B. H., Zerial M., Wade R. C. (2012). The interaction properties of the human Rab GTPase family—comparative analysis reveals determinants of molecular binding selectivity. PLoS ONE 7, e34870 10.1371/journal.pone.0034870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strongin A. Y., Collier I., Bannikov G., Marmer B. L., Grant G. A., Goldberg G. I. (1995). Mechanism of cell surface activation of 72-kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J. Biol. Chem. 270, 5331–5338 10.1074/jbc.270.10.5331 [DOI] [PubMed] [Google Scholar]
- Stylli S. S., Kaye A. H., Lock P. (2008). Invadopodia: at the cutting edge of tumour invasion. J. Clin. Neurosci. 15, 725–737 10.1016/j.jocn.2008.03.003 [DOI] [PubMed] [Google Scholar]
- Visse R., Nagase H. (2003). Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ. Res. 92, 827–839 10.1161/01.RES.0000070112.80711.3D [DOI] [PubMed] [Google Scholar]
- Weaver A. M. (2008). Cortactin in tumor invasiveness. Cancer Lett. 265, 157–166 10.1016/j.canlet.2008.02.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.