Abstract
In this study, we conduct an in-depth analysis of annexin proteins from a diverse range of invertebrate taxa, including the major groups that contain the parasites and vector organisms that are harmful to humans and domestic animals. Using structure-based amino acid sequence alignments and phylogenetic analyses, we present a classification for this protein group and assign names to sequences with ambiguous annotations in public databases. Our analyses reveal six distinct annexin clades, and the mapping of genes encoding annexins to the genome of the human blood fluke Schistosoma mansoni supports the hypothesis of gene duplication as a major evolutionary event in annexin genesis. This study illuminates annexin diversity from a novel perspective using contemporary phylogenetic hypotheses of eukaryote evolution, and will aid the consolidation of annexin protein identities in public databases and provide a foundation for future functional analysis and characterisation of these proteins in parasites of socioeconomic importance.
Annexins are a large family of proteins which are widely expressed across all eukaryotes and play key roles in a range of fundamental biological activities, including calcium metabolism, cell adhesion, growth and differentiation and subcellular transport1, as well as membrane repair2. In parasites, annexins are considered to play critical roles in mechanisms linked to their survival, including the maintenance of cell structure integrity and modulation of the immune responses of the vertebrate hosts3. Due to their location at the host-parasite interface and their immunogenic properties, these parasite annexins have been proposed as potential targets for the development of novel drug and vaccine candidates3,4.
Structurally, annexins are characterised by a C-terminal domain comprised of four homologous repeats of ~70 amino acids in length. The homologous domains often contain the characteristic endonexin sequence (K-G-X-G-T), which structurally translates into a type II calcium binding site with a high affinity for calcium and phospholipids5. The variable N-terminal domain harbours sites for post-translational modifications and protein-protein interactions1. Previous studies6 have demonstrated that the evolution of annexins has been characterised by successive gene duplication events, which have led to the expansion and diversification of annexin-encoding genes in vertebrates, invertebrates, plants and protists. Despite the substantial amino acid sequence similarities, sequence variants in different groups of eukaryotes are associated with structural features and biochemical properties, resulting in functional differences that are specific to each eukaryote group3,7. Based on the classification proposed by Fernandez and Morgan8, which integrated the use of phylogenetic analyses of amino acid sequences with gene structural analyses and genetic linkage maps, annexins are grouped into distinct families that correspond to the evolutionary divisions of the eukaryotes. This classification system, endorsed by the 50th Harden Conference (First International Annexin Conference, Wye College, UK, Sept 1–5, 1999), led to the current annexin nomenclature, which includes ‘A’ (from vertebrates, including humans), ‘B’ (invertebrates, including parasitic helminths), ‘C’ (fungi), ‘D’ (plants), and ‘E’ (protists) annexins8. Within the A annexins, a total of 12 distinct sequences have been described and assigned the identifiers A1–A13 (annexin A12 being unassigned), while annexins in families B through E are numbered progressively based on their presumed evolutionary distance from the A annexins. However, newly identified annexin sequences are usually named based on identity with the first hit on a BLAST search and without consideration of the family as a whole, thus leading to ambiguity in identity and relationship to other annexins. The vast majority of these proteins described to date have been detected in animals, plants and fungi. Systematics in the last 15 years has shown these multicellular taxa to each have arisen independently from unicellular protists. As more genomic data for protists and multicellular eukaryotic groups become available, the annexin nomenclature will become increasingly complex.
There has been significant confusion and inconsistencies in classification and nomenclature for annexins within Group B, including those of parasitic helminths. Recent advances in high-throughput sequencing and bioinformatics have resulted in an explosion of large-scale genomic and transcriptomic studies of parasitic helminths9,10,11,12,13 and, in turn, of the sequence data deposited in public databases for a range of helminth species of medical and veterinary importance. These advances have resulted in the exacerbation of inconsistencies in classification of B annexins; for example, an annexin from the human blood fluke Schistosoma japonicum (gb:CAX82892) is currently designated as ‘annexin A13’ instead of carrying a ‘B’ identifier; and ‘annexin B2’ has been assigned to two distinct proteins, one from the human tapeworm Taenia solium (gb:AAY17503) and one from Schistosoma mansoni (up:G4VL6814). Given the biological significance of parasite annexins, implementing a rational and consistent nomenclature for these proteins will promote structural and functional investigations of individual members of this protein family, and thus assist future studies aimed at elucidating their role/s in host-parasite interactions and the modulation of the hosts' immune response.
In the present study, we (i) prepared a comprehensive, secondary structure-based sequence alignment of B annexins from a range of parasitic helminths of public health and veterinary importance (including the blood flukes Schistosoma spp., the carcinogenic liver flukes Clonorchis sinensis and Opisthorchis viverrini and the hookworm Necator americanus) and some parasite vectors available in public databases, (ii) inferred phylogenetic relationships and (iii) proposed a nomenclature of B annexins considering secondary structure, characteristic protein signature/motifs, taxonomic features and evolutionary distance from a corresponding vertebrate homolog (A annexin). We also mapped the various annexin groups to clades arising from the contemporary hypotheses of eukaryote evolution as presented by Walker and colleagues15.
Results
Identification of annexins
In order to construct a dataset of putative Group B annexin sequences, we searched genomic sequences of a total of 35 species from 12 invertebrate and protistan phyla. After identification and verification, amino acid sequences from 28 species were confirmed as putative functional annexins (see Table 1) with amino acid identities ranging from 0.17 to 0.84. The structure-based alignment of the amino acid sequences is provided in Supplementary Figure S2. The published sequence of annexin (Sm)5 (new name: annexin B7a) from S. mansoni (gi:256084742) lacked 75 amino acids corresponding to positions spanning D91-V165.
Table 1. Organisms searched for annexin proteins.
| Phylum | Class | Family | Organism | Full genome sequenced | No of full-length annexin sequences | No of partial annexin sequences | Full-length annexin proteins (no of putative isoforms) | URL |
|---|---|---|---|---|---|---|---|---|
| Apicomplexa | Aconoidasida | Plasmodiidae | Plasmodium falciparum* | yes | 0 | 0 | 1 | |
| Arthropoda | Arachnida | Ixodidae | Ixodes scapularis | yes | 1 | 0 | B28 | 1 |
| Insecta | Bombycidae | Bombyx mori | yes | 4 | 0 | B11 (2), B17 (2) | 1 | |
| Insecta | Culicidae | Aedes aegypti | yes | 6 | 0 | B9 (2), B17 (4) | 1 | |
| Insecta | Culicidae | Anopheles gambiae | yes | 4 | 1 | B9, B17 (3) | 1 | |
| Insecta | Culicidae | Culex quinquefasciatus | yes | 5 | 0 | B9 (2), B17 (3) | 1 | |
| Insecta | Drosophilidae | Drosophila melanogaster | yes | 5 | 0 | B9 (3), B11 (2) | 1 | |
| Insecta | Pediculidae | Pediculus humanus | yes | 5 | 0 | B9 (2), B11, B17 (2) | 1 | |
| Cnidaria | Hydrozoa | Hydridae | Hydra vulgaris | yes | 1 | 0 | B12 | 1 |
| Hydrozoa | Hydridae | Hydra magnipapillata | yes | 3 | 0 | B4, B12 (2) | 1 | |
| Mollusca | Gastropoda | Pomatiopsidae | Oncomelania hupensis | no | 0 | 0 | 1 | |
| Nematoda | Adenophorea | Trichinellidae | Trichinella spiralis | yes | 0 | 0 | 1 | |
| Chromadorea | Ascarididae | Ascaris suum* | yes | 7 | 0 | B8, B19 (3), B21 (3) | 1 | |
| Chromadorea | Onchocercidae | Brugia malayi* | yes | 2 | 1 | B37, B40 | 1 | |
| Secernentea | Heteroderidae | Heterodera glycines | no | 1 | 0 | B19 | 1 | |
| Secernentea | Rhabditidae | Caenorhabditis elegans | yes | 5 | 0 | B8 (2), B19, B21, B36 | 1 | |
| Secernentea | Strongyloididae | Strongyloides ratti* | yes | 0 | 1 | 4 | ||
| Secernentea | Trichostrongylidae | Haemonchus contortus | yes | 0 | 1 | 2 | ||
| Secernentea | Uncinariidae | Necator americanus | yes | 0 | 5 | 3 | ||
| Placozoa | Tricoplacia** | Trichoplacidae** | Trichoplax adhaerens | yes | 3 | 0 | B4, B6, B26 | 1 |
| Platyhelminthes | Cestoda | Taeniidae | Echinococcus granulosus* | yes | 12 | 0 | B1, B2, B3, B5, B15, B18, B20, B23, B24, B25, B33, B38 | 6 |
| Cestoda | Taeniidae | Taenia solium* | no | 3 | 0 | B1, B2, B3 | 1 | |
| Monogenea | Microcotylidae | Microcotyle sebastis | no | 1 | 0 | B34 | 1 | |
| Rhabditophora | Dugesiidae | Schmidtea mediterranea | yes | 3 | 0 | B5, B16, B27 | 8 | |
| Trematoda | Fasciolidae | Fasciola gigantica | yes | 3 | 4 | B22, B30, B39 | 5 | |
| Trematoda | Fasciolidae | Fasciola hepatica | yes | 3 | 6 | B5 (2), B7 | 5 | |
| Trematoda | Opisthorchiidae | Clonorchis sinensis | yes | 7 | 0 | B5 (2), B7, B14, B22, B30, B35 | 5 | |
| Trematoda | Opisthorchiidae | Opisthorchis viverrini | yes | 5 | 2 | B5 (3), B22, B30 | 5 | |
| Trematoda | Schistosomatidae | Schistosoma haematobium* | yes | 7 | 0 | B5 (2), B7, B13, B22, B30, B32 | 7 | |
| Trematoda | Schistosomatidae | Schistosoma japonicum* | yes | 6 | 0 | B5, B7, B22, B30, B32, B39 | 1 | |
| Trematoda | Schistosomatidae | Schistosoma mansoni | yes | 13 | 0 | B5 (2), B7 (2), B10, B13, B22, B29, B30, B31, B32, B39 (2) | 1 | |
| Porifera | Demospongiae | Spongillidae | Ephydatia fluviatilis | no | 1 | 0 | B4 | 1 |
| Sarcomastigophora | Kinetoplastea | Trypanosomatidae | Leishmania braziliensis* | yes | 0 | 0 | 1 | |
| Kinetoplastea | Trypanosomatidae | Trypanosoma brucei* | yes | 0 | 0 | 1 | ||
| Lobosea | Entamoebidae | Entamoeba histolytica** | yes | 0 | 0 | 1 |
Sequences accessed at:
All taxonomy checked in Catalogue of Life at http://www.catalogueoflife.org/col, 11th March 2013 edition.
*not in Catalogue of Life; taxonomy thus taken from NCBI Taxonomy Browser.
**listed as not assigned.
Annexin sequences were detected in all species of invertebrates surveyed except the gastropod mollusc Oncomelania hupensis, and the nematode Trichinella spiralis. No molluscan annexin sequences have yet been identified, although large-scale genomic datasets are available for two gastropods, Oncomelania hupensis, the pulmonate intermediate host (vector) of the trematode Opisthorchis viverrini, and the marine gastropod, the sea hare Aplysia californicus16.
Many invertebrates have multiple annexins. The human blood fluke S. mansoni (Platyhelminthes: Digenea) has 13 and for the liver fluke Fasciola hepatica, there are currently 10 annexin sequences known, although some of them are only partial.
The searchable database of annexin proteins on the existing Annexin Website (http://www.annexins.org/) has been updated to include the sequences surveyed in the present study.
Phylogenetic analyses of group B annexins
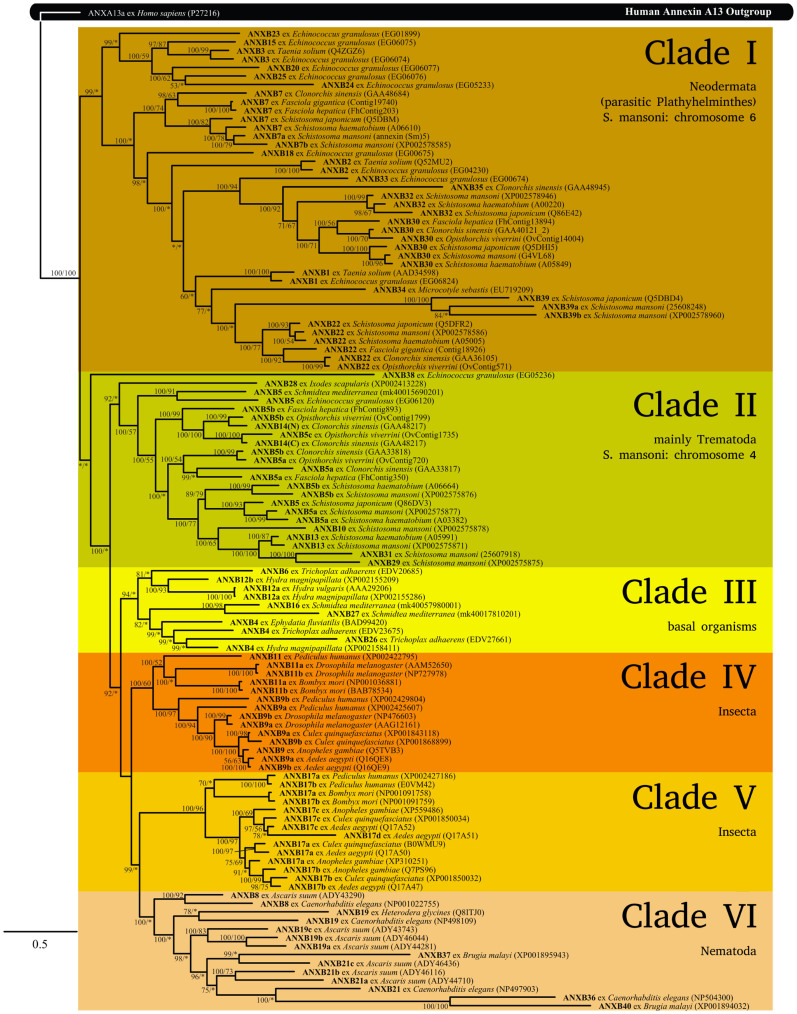
Bayesian inference analysis of the structure-based amino acid sequence alignment (Supplementary Figure S2) resulted in a consensus tree with most of the putative B annexins forming clades with relatively high posterior probability (Figure 1). The maximum likelihood tree had similar topology, however, confidence based on ML bootstrapping for many of the basal nodes was low.
Figure 1. Consensus Bayesian phylogram of B annexins based on Bayesian inference analysis of the structure-based amino acid sequence alignment.
Posterior probabilities and maximum likelihood bootstrap values, respectively, are shown at the nodes. ‘*’ indicates values <50%.
The Bayesian inference and maximum likelihood analyses differed in two instances. Maximum likelihood analysis placed the annexin from Microcotyle sebastis (gb:EU719209) with low bootstrap support in a clade together with annexins from T. solium (up:Q52MU2; B2) and Echinococcus granulosus (EG_04230; B2). In the Bayesian analysis, the M. sebastis sequence was not within the annexin B2 group, but was placed external to a clade containing B22 and B39 annexins with moderate support from posterior probability (Figure 1). A similar situation was encountered with an annexin from E. granulosus (EG_00675). Maximum likelihood analysis placed this annexin in the clade formed by annexin B7 sequences, but again with low bootstrap support (Figure 1). Bayesian inference, in contrast, placed the E. granulosus sequence distant from annexin B7, with strong posterior probability, and separate to other annexin clades. Since the Bayesian inference analysis offered stronger support for these groupings, we assigned these two sequences to their own clades, i.e. annexin B18 (E. granulosus, EG_00675) and annexin B34 (M. sebastis, gb:EU719209). Our present results indicate that two annexins, namely “AnxB13” from B. mori and “B2” from S. mansoni (up:C3VEV017), should indeed be renamed as B11 and B30, respectively.
The phylogenetic clades grouped strongly according to phyla (e.g., Arthropoda, Nematoda, Platyhelminthes) and in most cases according to class (e.g., Insecta, Cestoda, Trematoda), both at individual B-number groupings and in more basal major clades (see Figure 1). There are two basal clades of annexins (I and II) restricted to the phylum Platyhelminthes. In the phylum Arthropoda, two major clades (IV and V) are restricted to class Insecta and these clades grouped relatively close in both the Bayesian inference and maximum likelihood analyses. The only other arthropod annexin included in these analyses, from the tick Ixodes scapularis (annexin B28, class Arachnida), grouped well outside of these two clades, more closely related to the platyhelminth clade II (Figure 1).
A couple of notable exceptions included annexins B4 and B5. The annexin B4 group contained sequences derived from members of the Cnidaria (Hydra magnipapillata), Porifera (Ephydatia fluviatilis), and the basal metazoan phylum Placozoa (Trichoplax adhaerens) (see Figure 1). However, visual examination of the structure-based amino acid sequence alignment and the close relationship inferred between these annexin sequences in the phylogenetic analyses supported the grouping despite the relatively distant taxonomic relationships of these organism over three phyla. The annexin sequences from these three phyla were grouped together in clade III comprising of basal organisms with two annexins from S. mediterranea (see Figure 1), which may indicate an older origin for these latter sequences. The annexin B5 group contained sequences obtained from platyhelminths of the classes Cestoda, Trematoda and Turbellaria, strongly supporting the orthology of these sequences.
Mapping of genes coding annexins in the genome of Schistosoma mansoni - implications for annexin evolution
In most clades (Figure 1) there was a substantial number of putative paralogs or isoforms, indicated by letters appended to the annexin group number. However, the Platyhelminthes clade I is comprised mostly of annexin groups containing only or mainly orthologs. For example, there were large numbers of sequences retrieved for E. granulosus and S. mansoni. In clade I, all nine E. granulosus sequences were considered orthologs and for S. mansoni, three sequences were inferred as orthologs and four as paralogs. In contrast, there were seven paralogs but only one ortholog for Ascaris suum in clade VI. This pattern may indicate that there was significant gene duplication prior to the divergence of the Platyhelminthes, with each duplicated gene lineage passed into a range of species. However, not all lineages are represented in all species, arguing for a birth-and-death model of gene evolution. Alternatively, gene duplications or isoform development within species may be older in the Platyhelminthes than the other phyla studied here. Older duplications will accumulate mutations with time, giving greater p-distances and hence be assigned different annexin numbers using the present approach.
By 2011, 81% of the S. mansoni genome could be mapped to the seven autosomal and one Z/W sex-determining chromosomes18. The majority of annexins are located on autosomal chromosomes 4 and 6 (Table 2). From our present data (Figure 1), it is clearly visible that sequences from the upper Platyhelminthes clade (I) are associated with S. mansoni chromosome 6, and sequences from the lower clade (II) are associated with chromosome 4. One could thus infer that multiple copies within each of these clades have arisen by successive duplication events.
Table 2. Genomic mapping of Schistosoma mansoni annexins.
| Annexin | GeneDB | Location | Chromosome | Clade |
|---|---|---|---|---|
| B5a | Smp_045560 | 4519381–4536000 | 4 | II |
| B5b | Smp_045550 | 4548453–4568563 | 4 | II |
| B10 | Smp_146690 | 4476102–4503979 | 4 | II |
| B13 | Smp_045500 | 4628571–4637840 | 4 | II |
| B29 | Smp_207040 | 4583330–4595243 | 4 | II |
| B31 | Smp_045490 | 4611818–4625468 | 4 | II |
| B7a | Smp_074140 | 286104–308545 | 6 | I |
| B7b partial | Smp_162160 | 258618–271457 | 6 | I |
| B22 | Smp_074150 | 330560–360327 | 6 | I |
| B30 | Smp_077720 | 19430888–19460055 | 6 | I |
| B32 | Smp_164100 | 19468674–19494199 | 6 | I |
| B39b | Smp_077880 | 19921307–19935211 | 6 | I |
| B39b partial | Smp_201250 | 48936–58086 | SC_0076 (chromosome 1) | I |
| B39b partial | Smp_201340 | 249520–254223 | SC_0153 (chromosome 1) | I |
| B39a | Smp_155580 | 115733–134818 | SC_0154 | I |
| B39a partial | Smp_194120 | 504–4350 | SC_0542 | I |
| B39a partial | Smp_178820 | 1962573–1962761 | SC_0041 | I |
| B39b partial | Smp_205300 | 20951–26123 | SC_0276 | I |
| B39b partial | Smp_173300 | 79778–80578 | SC_0154 | I |
| B39b partial | Smp_173290 | 81573–82755 | SC_0154 | I |
Sequences accessed at: http://www.genedb.org/Homepage/Smansoni.
The patterns in different species may provide clues as to when in evolution the duplication events occurred, although this is somewhat confounded by differing amounts of data in each species. The present tree (Figure 1) suggests that, for example, annexins B30 and B32 represent a duplication event only in trematodes. From the species involved, we hypothesise that clade II was the original platyhelminth annexin clade, given that Ixodes and Schmidtea are at its base, and that clade I arose from a duplication early in platyhelminth evolution.
Discussion
The annexin nomenclature and diversity largely reflects the early investigations of these molecules in “advanced” multicellular organisms, and the focus on the roles of these molecules in humans and mammalian models. As a result, the current annexin nomenclature scheme has an implicit understanding that the substantial diversity of annexin structure and function occurs within animals, fungi and plants, as four of five annexin groups are seen in these multicellular taxa. The ‘protistan’ annexins are grouped together as Group E annexins. Such categorisation, while convenient, cannot possibly be supported by modern concepts of eukaryotic phylogeny. Phylogenetic systematic studies15,19 have broken down the traditional concepts of relationships of single-celled eukaryotes, resulting in a new system of highly divergent clades, thereby changing concepts from primitive stem-group protozoa and algae as precursors of ‘crown eukaryotes’, to diverse ‘supergroups’, two of which contain multicellular animals and fungi, and plants.
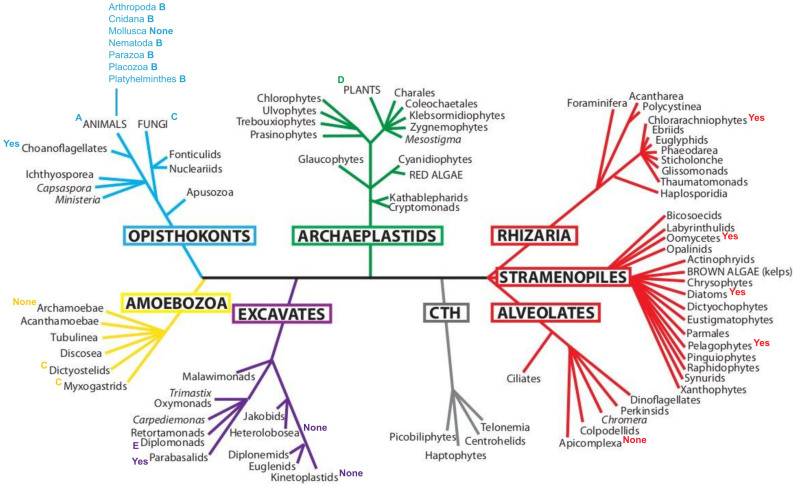
The current view of eukaryotic systematics recognises six distinct lineages; in five of these, annexin sequences have been identified (Figure 2). The lineages are comprised of the opisthokonts (in which one finds Group A, B and C annexins), the archaeplastids (Group D annexins), the SAR (Stramenopile, Alveolate, Rhizarian) clade (annexins not yet categorised), the centrohelid-telonemid-haptophyte (CTH) clade (no annexins described), the excavates (Group E annexins) and the amoebozoa (Group C annexins). The CTH supergroup constitutes the only major clade in which annexin molecules are apparently absent. This observation is gaining support by the recent draft genomes for two of these groups, the haptophytes (Emiliania huxleyi) and the picobilophytes20, which have not yielded any annexin sequences.
Figure 2. Eukaryote scheme and occurrence of annexins as known to date.
Denotation of ‘A’-‘E’ indicates presence of annexins in the groups listed. ‘Yes’ indicates presence of (full-length or partial) uncategorised annexin sequences. ‘None’ indicates absence of annexins to current knowledge.
Records of annexins in other supergroup members (see Figure 2) are fragmentary and reflect research attention paid to particular species of major significance to humans. Some interesting patterns emerge that raise questions about annexin and protist phylogeny.
Firstly, it is obviously well supported that the metazoans are monophyletic. The Group B annexins separate strongly into clades reflective of the phylogenetic relationships of the organisms in which they are found. Thus, clades I and II are found largely in platyhelminths, while other clades are dominant in ecdysozoan lineages. Therefore, there is strong support for the three major animal lineages, lophotrochozoan, deuterostome and ecdysozoan, in annexin phylogeny. Annexins are also present in closest relatives of the animals, the choanoflagellates.
Secondly, in the current nomenclature, the fungal annexins are classed along with those of dictyostelid and myxogastrid amoebozoans, as Group C annexins. Phylogenetic inferences based on numerous genes and cellular ultrastructure place the dictyostelids and myxogastrids firmly within the Amoebozoa and not within the opisthokonts with the fungi15. The prima facie case for including annexins from Dictyostelium discoideum or Physarum polycephalum with fungi in the Group C annexins is thus not supported by phylogeny. Interestingly, although the genome of the archamoeban Entamoeba histolytica has been described21, no annexin sequences have been located in that species.
Group D annexins are those of plants and their phylogeny has recently been investigated in more detail22,23. Annexins are present in green algae including both the chlorophytes and streptophytes, but apparently are not in the red algae, a major clade of the supergroup Archaeplastida. The genomic sequences available for one cryptomonad, Guillardia theta, indicate the presence of annexins in this photosynthetic protist. G. theta is the product of secondary endosymbiosis, a process whereby the original non-photosynthetic cell incorporated a red algal cell24. Detailed phylogenetic analyses of group D annexins in relation to other holders of secondary red algal endosymbioses should unravel whether the annexins in that cell arose with the host cell or the endosymbiont.
The stramenopiles, alveolates and rhizarians (SAR) supergroup is a monophyletic but structurally and ecologically diverse clade. As the alveolates contain some very important human pathogens, genomic data on this clade is abundantly available. The human cerebral malaria parasite, Plasmodium falciparum, lacks annexin sequences and such sequences have not yet been detected among other alveolates, indicating that this clade may have lost these proteins. Other lineages in the SAR group do contain annexins. A single annexin sequence known for the Rhizaria occurs in the enigmatic Bigelowiella natans, a chlorarachniophyte that has also undergone secondary endosymbiosis. The hypothesis to test here is whether annexins have transferred between supergroups through secondary endosymbiosis.
Finally, the Group E annexins are found in two disparate groups of excavates, the diplomonads and parabasalids. Phylogeny of the Excavata indicates an early bifurcation into two lineages, the metamonads (including Giardia) and the Discoba (including the kinetoplastids and the amoeboflagellate Naegleria gruberi). Interestingly, although genomes for a number of parasitic species (e.g. trypanosomes) in the Discoba have been well described, annexins have only been detected in the metamonads, suggesting loss of these proteins from the discoban lineage.
The Bayesian inference and maximum likelihood analyses agree with respect to topology and nodal support for the majority of the clades containing the assigned Group B numbers (Figure 1). Within the framework of the current annexin nomenclature, we have assigned novel annexins from parasitic organisms and parasite vectors to remedy ambiguous annexin names found in databases. The phylogenetic analyses conducted in this context shows clearly that annexin diversity follows the phyla, and that within groups, there have been successive gene duplication events, as previously proposed8,23.
Individual effects of diversification of the annexin and species phylogenies is difficult to determine. Clearly, the demarcation cannot be attributed to variable regions of these proteins, such as the N-terminal domain, which is divergent both within and between clades. In contrast, variations in canonical features are more suitable to study effects of diversification, and one such feature, that is accessible at the level of primary structure, is the presence or absence of the endonexin sequence. This motif, at the level of three-dimensional structure, is responsible for the canonical type II calcium binding in annexins. Indeed, common, but inconsistent patterns of presence or absence of these motifs emerge when examining Group B annexins (see Supplementary Figure S4).
The most frequent lack of the endonexin sequence appears in repeat III (30 times), as compared to repeats I, II and IV (19, 13 and 10 times, respectively). Interestingly, the calcium-dependent membrane binding mechanisms of some invertebrate annexins may engage exclusively the canonical membrane binding site of the I/IV module (Leow et al., submitted). There is a trend towards loss of endonexin motifs in one or more annexin repeats in clades I (trematode) and VI (nematode), whereas the clades of insect annexins (clades IV and V) retain endonexin domains in all annexin repeats. Endonexin sequences are generally present in all four repeats in the basal invertebrates, although individual repeats of some basal annexins may have lost the motifs. The trend towards partial or complete loss of the endonexin motif may reflect the early changes in cellular structure that led to evolution of the unique cellular architecture of helminths, notably those of the parasite groups.
Current genome data is biased towards species with direct implications for humans, but future studies dissecting uncategorised annexins in supergroups such as the Rhizaria and Excavates (see Figure 2) will advance our understanding of molecular evolution. Intriguingly, instances of secondary endosymbiosis may be potentially complicating, but highly informative.
Contemporary phylogenies in the past decade have postulated highly divergent eukaryotic clades, different from the traditional top-down concept with a “ladder” from amitochondriate parasites “up” to multicellular organisms15. This has led to parts of the traditional annexin nomenclature being unnecessarily confusing. A prominent example of the current annexin nomenclature resulting in complicated relationships is the case of Group C annexins, which appear in both the Amoebozoa (Dictyostelids, Myxogastrids) and the Opisthokonts (Fungi). With increasing amounts of genomic data becoming available, the nomenclature of annexins might benefit from some modifications, particularly considering changing inferences of eukaryotic evolution.
Methods
Sequence identification and secondary structure-based alignment
Putative annexin amino acid sequences (available in public databases) from 34 organisms representing 13 phyla (Table 1) were identified using the BLASTp and tBLASTn algorithms25, and the corresponding nucleotide sequences were subsequently retrieved. Two search patterns were used (see Supplementary Table S1), namely the C-terminal domain of Anx(Sm)1 (gb:XP_002578586; 330 residues), as well as its first repeat only (71 residues). The selection focused on parasitic organisms and their vectors, but also included non-parasitic organisms representing annexins that have already been established in the literature. Secondary structure elements for each amino acid sequence were predicted using the software PSIPRED26. A secondary structure-based sequence alignment was generated automatically using the software SBAL27, visually inspected and manually adjusted (see Supplementary Figure S2). For each annexin protein sequence, the corresponding cDNA was retrieved from public databases (see Table 1); subsequently, all cDNA sequences were aligned using ClustalW28 with default parameters.
The phyla from which representative annexins were obtained included a range of protistan (Amoeboza: Archamoebae, Alveolata: Apicomplexa, Excavata: Euglenozoa) and animal (Placozoa, Radiata: Cnidaria, Lophotrochozoa: Mollusca and Platyhelminthes, Ecdysozoa: Nematoda and Arthropoda, Deuterostomia: Chordata) groups.
The annexin (Sm)5 from S. mansoni (new name: annexin B7a) gene was amplified from a cDNA library obtained from seven different life cycle stages (eggs through to adult worms) by polymerase chain reaction (PCR) using Pfu polymerase (Stratagene), buffers and nucleotides as recommended by the manufacturer, and 0.25 μM of each forward (5′-CATGCCATGGGCATGGGAAGAGATAAATCACAAATAA-3′) and reverse primer (5′-CCGCTCGAGTTGCCATTCAGCACCAATTA-3′), and a cycling protocol of 1 min at 95°C followed by 30 cycles of denaturation at 95°C for 10 sec, annealing at 53°C for 30 sec and extension at 68°C for 3 min. The final extension step was 68°C for 7 min. DNA sequencing was performed with BigDye (Applied Biosystems) terminator chemistry as per the manufacturer's instructions.
Phylogenetic analyses and prediction of orthologous/paralogous relationships
A non-redundant data set, including full-length annexin sequences, was extracted from the structure-based sequence alignment. Best-fit evolutionary models for maximum-likelihood (ML) phylogenetic analyses of both annexin amino acid and nucleotide sequences were predicted using ProtTest29 and jModelTest30, respectively. The best-fit model inferred from the Akaike Information Criteria (AIC) was used in the amino acid dataset analyses and the Bayesian Information Criteria (BIC) for the nucleotide dataset. For each amino acid and nucleotide sequence alignment, ML and Bayesian Inference (BI) trees were derived using MEGA v.531 and MrBayes 3.1.232, respectively. All trees were rooted using the human annexin A13 (GenBank accession numbers NP_004297 and NM_004306, for the amino acid and nucleotide sequence, respectively) as the outgroup. The ML phylogenetic trees of amino acid and nucleotide sequences were constructed using the Jones-Taylor-Thornton (JTT) model assuming uniform rates among sites (+G + I; i.e. including gamma, proportion of invariant sites) and the General Time Reversible model (GTR), respectively. For each ML analysis, the bootstrapped confidence interval was based on 100 replicates. BI analyses for both nucleotide and amino acid sequence alignments were run over 1,000,000 generations (‘ngen = 1,000,000’) with two runs each containing four simultaneous Markov Chain Monte Carlo (MCMC) chains (‘nchains = 4’) and every 100th tree being saved (‘samplefreq = 100’). The parameters used were as follows: ‘nst = 6’, ‘rates = invgamma’, with MCMC left at default settings, ‘ratepr = variable’ and ‘burnin = 100’. Consensus trees were constructed, with ‘contype = allcompat’ nodal support being determined using consensus posterior probabilities. An initial Bayesian inference analysis of the amino acid dataset which excluded sequences of Schmidtea mediterranea was performed at 10,000,000 generations. The overall topology and posterior probability values did not vary significantly from the final analysis conducted at 1,000,000 generations as the likelihood probabilities stabilised well before 1,000,000 generations when examined in Tracer v1.5 software (Tracer v.1.5; http://beast.bio.ed.ac.uk/Tracer). All trees were displayed using FigTree v1.4 (http://tree.bio.ed.ac.uk/software/figtree/).
The backtrans feature of TreeBeST (http://treesoft.sourceforge.net/treebest.shtml) was used to create a protein-guided codon alignment of the nucleotide sequences using the present protein sequence alignment. A species-guided ML tree using the Hasegawa-Kishino-Yano (HKY) model was constructed in TreeBeST and viewed in FigTree v1.4. The species tree was constructed with reference to relevant molecular phylogenies33,34,35,36,37,38,39 and the Tree of Life web project (http://tolweb.org/tree/phylogeny.html and references therein). For these analyses only, the human annexin A13 was removed as the outgroup as it was not consistent with the species tree constraint and instead the annexin B sequence from the freshwater sponge Ephydatia fluviatilis was used as the outgroup.
Nomenclature strategy
The B Group naming convention implemented here for the new sequence data sought to conform to the framework of nomenclature proposed by Fernandez and Morgan8, who suggested that new names should be assigned based on their level of amino acid sequence identity (‘closeness’) to the authoritative human annexins. Initial alignment and phylogenetic analyses of the amino acid sequence data reported here with that of the human annexins ANXA1−ANXA13 resulted in phylograms that were markedly polyphyletic, with the human annexins interspersed within various clades of B annexin sequences (data not shown). This observed polyphyly made determining ‘closeness’ of the new B annexin amino acid sequence data to the human annexins as a whole for naming purposes ambiguous. Therefore, we chose to exclude the human annexins ANXA1−ANXA11 from these analyses and use the human annexin ANXA13 as the functional outgroup in all subsequent analyses and for naming purposes. Since the annexin A13 gene is the probable common ancestor of all vertebrate annexins40, it is the appropriate outgroup sequence and root for the non-vertebrate phyla presented here.
The determination of ‘closeness’ of the B annexins to the A13 sequence for naming purposes was initially undertaken using the maximum likelihood and Bayesian inference phylogenies. However, due to the large number of new sequences included in the present study (n = 115), we selected p-distances of B amino acid sequences relative to the human annexin A13 sequence as a more robust and objective method for assigning names because this yields the actual proportion of amino acid sites which differ between two sequences rather than inferring genetic distance based on a model of evolution41. The p-distances were calculated in MEGA v.531, with the setting ‘pairwise deletion of gaps/missing data’ selected. The data were then exported into a spreadsheet and sorted (see Supplementary Table S3). B annexin amino acid identifiers were assigned respecting those that have already been established in the literature (i.e. B1, B2, B3 from Taenia solium; B9 and B11 from Drosophila melanogaster; B12 from Hydra vulgaris) beginning with B4.
The same B annexin number identifier was assigned to sequences proposed to be orthologs (shared ancestry through speciation) or paralogs (shared ancestry through duplication) based on clades with shared similarity (i.e. putative isoforms) as assessed by a combination of inferred relationships from the phylogenetic analyses, orthology/paralogy and secondary structure. Clades containing sequences from different species were considered putative orthologs and were assigned the same identifier. Where sequences from the same species were present in a clade, letter designations (‘a’, ‘b’, ‘c’, etc) were appended after the B numbers to indicate either putative isoforms or putative paralogs. The number assigned to a group of putative isoforms or paralogs was determined based on the sequence with the shortest p-distance to annexin A13. The subsequent letter designations for the putative isoforms or paralogs within the B annexins were then assigned in descending order based on p-distance from annexin A13.
Author Contributions
C.C., J.M.S., T.L.M., G.W., M.K.J. and A.H. designed, performed, and analysed computational work. C.Y.L., L.T., L.M., C.W., M.K.J. and A.H. designed, performed, and analysed experimental work. A.L., R.B.G. and M.K.J. provided essential datasets. All authors wrote and reviewed the manuscript.
Supplementary Material
Supplementary Material
Acknowledgments
We gratefully acknowledge funding of our laboratories by the National Health and Medical Research Council (A.H., M.K.J., R.B.G.) and the Australian Research Council (A.H., R.B.G.). C.C. and A.L. are supported by an NHMRC early career and principal research fellowship, respectively. T.L.M. is supported by a Queensland Government Smart Futures Fellowship. C.Y.L. is supported by a Malaysian Government and Universiti Sains Malaysia ASTS scholarship.
References
- Gerke V. & Moss S. E. Annexins: from structure to function. Physiol. Rev. 82, 331–371 (2002). [DOI] [PubMed] [Google Scholar]
- Draeger A., Monastyrskaya K. & Babiychuk E. B. Plasma membrane repair and cellular damage control: the annexin survival kit. Biochem Pharmacol 81, 703–12 (2011). [DOI] [PubMed] [Google Scholar]
- Hofmann A. et al. Parasite annexins - new molecules with potential for drug and vaccine development. BioEssays 32, 967–976 (2010). [DOI] [PubMed] [Google Scholar]
- Jenikova G. et al. A1-giardin based live heterologous vaccine protects against Giardia lamblia infection in a murine model. Vaccine 29, 9529–9537 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisow M., Fritsche U., Hexham J., Dash B. & Johnson T. A consensus amino acid sequence repeat in Torpedo and mammalian calcium-dependent membrane binding proteins. Nature 320, 636–638 (1986). [DOI] [PubMed] [Google Scholar]
- Moss S. E. & Morgan R. O. The annexins. Genome Biol 5, 219 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka-Postupolska D., Clark G. & Hofmann A. Structure, function and membrane interactions of plant annexins: an update. Plant Sci. 181, 230–241 (2011). [DOI] [PubMed] [Google Scholar]
- Fernandez M. P. & Morgan R. O. in Annexins: biological importance and annexin-related pathologies. (ed. Bandorowicz-Pikula J.) pp. 21–37 (Landes Bioscience, Georgetown, TX, 2003).
- Jex A. R. et al. Ascaris suum draft genome. Nature 479, 529–33 (2011). [DOI] [PubMed] [Google Scholar]
- Cantacessi C., Campbell B. E. & Gasser R. B. Key strongylid nematodes of animals - Impact of next-generation transcriptomics on systems biology and biotechnology. Biotechnol Adv 30, 469–488 (2012). [DOI] [PubMed] [Google Scholar]
- Young N. D. et al. Whole-genome sequence of Schistosoma haematobium. Nat Genet 44, 221–5 (2012). [DOI] [PubMed] [Google Scholar]
- Mitreva M. The genome of a blood fluke associated with human cancer. Nat Genet 44, 116–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva L. L. et al. The Schistosoma mansoni phylome: using evolutionary genomics to gain insight into a parasite's biology. BMC Genomics 13, 617 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protasio A. V. et al. A systematically improved high quality genome and transcriptome of the human blood fluke Schistosoma mansoni. PLoS Negl Trop Dis 6, e1455 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G., Dorrell R. G., Schlacht A. & Dacks J. B. Eukaryotic systematics: a user's guide for cell biologists and parasitologists. Parasitology 138, 1638–1663 (2011). [DOI] [PubMed] [Google Scholar]
- Heyland A., Vue Z., Voolstra C. R., Medina M. & Moroz L. L. Developmental transcriptome of Aplysia californica. J Exp Zool B Mol Dev Evol 316B, 113–34 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tararam C. A., Farias L. P., Wilson R. A. & Leite L. C. Schistosoma mansoni Annexin 2: molecular characterization and immunolocalization. Exp Parasitol 126, 146–155 (2010). [DOI] [PubMed] [Google Scholar]
- Swain M. T. et al. Schistosoma comparative genomics: integrating genome structure, parasite biology and anthelmintic discovery. Trends Parasitol 27, 555–564 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adl S. M. et al. The revised classification of eukaryotes. J Eukaryot Microbiol 59, 429–493 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Yoon H. et al. Single-Cell Genomics Reveals Organismal Interactions in Uncultivated Marine Protists. Science 332, 714–717 (2011). [DOI] [PubMed] [Google Scholar]
- Loftus B. et al. The genome of the protist parasite Entamoeba histolytica. Nature 433, 865–8 (2005). [DOI] [PubMed] [Google Scholar]
- Jami S. K., Clark G. B., Ayele B. T., Ashe P. & Kirti P. B. Genome-wide comparative analysis of annexin superfamily in plants. PLoS One 7, e47801 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark G. B., Morgan R. O., Fernandez M. & Roux S. J. Evolutionary adaptation of plant annexins has diversified their molecular structures, interactions and functional roles. New Phytol 196, 695–712 (2012). [DOI] [PubMed] [Google Scholar]
- McFadden G. I. Primary and secondary endosymbiosis and the origin of plastids. J Phycol 37, 951–959 (2001). [Google Scholar]
- Altschul S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson K. et al. Protein structure prediction servers at University College London. Nucl Acids Res 33, W36–W38 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. K. et al. SBAL: a practical tool to generate and edit structure-based amino acid sequence alignments. Bioinformatics 28, 1026–1027 (2012). [DOI] [PubMed] [Google Scholar]
- Larkin M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–8 (2007). [DOI] [PubMed] [Google Scholar]
- Abascal F., Zardoya R. & Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105 (2005). [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol 25, 1253–1256 (2008). [DOI] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F. & Huelsenbeck J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003). [DOI] [PubMed] [Google Scholar]
- Lockyer A. E. et al. The phylogeny of the Schistosomatidae based on three genes with emphasis on the interrelationships of Schistosoma Weinland, 1858. Parasitology 126, 203–224 (2003). [DOI] [PubMed] [Google Scholar]
- Olson P. D., Littlewood D. T., Bray R. A. & Mariaux J. Interrelationships and evolution of the tapeworms (Platyhelminthes: Cestoda). Mol Phylogenet Evol 19, 443–467 (2001). [DOI] [PubMed] [Google Scholar]
- Olson P. D., Cribb T. H., Tkach V. V., Bray R. A. & Littlewood D. T. Phylogeny and classification of the Digenea (Platyhelminthes: Trematoda). Int J Parasitol 33, 733–755 (2003). [DOI] [PubMed] [Google Scholar]
- Reidenbach K. R. et al. Phylogenetic analysis and temporal diversification of mosquitoes (Diptera: Culicidae) based on nuclear genes and morphology. BMC Evol Biol 9, 298 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riutort M., Álvarez-Presas M., Lázaro E., Solà E. & Paps J. Evolutionary history of the Tricladida and the Platyhelminthes: an up-to-date phylogenetic and systematic account. Int J Dev Biol 56, 5–17 (2012). [DOI] [PubMed] [Google Scholar]
- Rohde K. et al. Contributions to the phylogeny of Platyhelminthes based on partial sequencing of 18S ribosomal DNA. Int J Parasitol 23, 705–724 (1993). [DOI] [PubMed] [Google Scholar]
- Zarowiecki M. Z., Huyse T. & Littlewood D. T. Making the most of mitochondrial genomes--markers for phylogeny, molecular ecology and barcodes in Schistosoma (Platyhelminthes: Digenea). Int J Parasitol 37, 1401–1418 (2007). [DOI] [PubMed] [Google Scholar]
- Iglesias J. M. et al. Comparative genetics and evolution of annexin a13 as the founder gene of vertebrate annexins. Mol Biol Evol 19, 608–18 (2002). [DOI] [PubMed] [Google Scholar]
- Nei M. & Kumar S. Molecular Evolution and Phylogenetics. (Oxford University Press, 2000).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material