SUMMARY
The strength of synaptic communication at central synapses depends on the number of ionotropic glutamate receptors, particularly the class gated by the agonist AMPA (AMPARs). Cornichon proteins, evolutionarily conserved endoplasmic reticulum cargo adaptors, modify the properties of vertebrate AMPARs when coexpressed in heterologous cells. However, the contribution of cornichons to behavior and in vivo nervous system function has yet to be determined. Here, we take a genetic approach to these questions by studying CNI-1 – the sole cornichon homologue in C. elegans. cni-1 mutants hyper-reverse, a phenotype associated with increased glutamatergic synaptic transmission. Consistent with this behavior, we find larger glutamate-gated currents in cni-1 mutants with a corresponding increase in AMPAR number. Furthermore, we observe opposite phenotypes in transgenic worms that overexpress CNI-1 or vertebrate homologues. In reconstitution studies, we provide support for an evolutionarily conserved role for cornichons in regulating the export of vertebrate and invertebrate AMPARs.
INTRODUCTION
Rapid, excitatory synaptic communication that occurs in vertebrate brains, as well as in most other nervous systems, is primarily mediated by the neurotransmitter glutamate. The strength of this communication is dependent on the number of AMPA-class ionotropic glutamate receptors, as well as on their functional properties. Synaptic strength is plastic, and nervous system activity can change the number and properties of AMPARs (Anggono and Huganir, 2012; Kerchner and Nicoll, 2008; Kessels and Malinow, 2009).
While the properties of AMPARs can be modified by RNA editing (Wright and Vissel, 2012) and posttranslational modification of receptors (Lu and Roche, 2012), more recent genetic studies have demonstrated that AMPAR trafficking and function are critically dependent on auxiliary proteins, most prominently the TARP and SOL classes of transmembrane proteins (Jackson and Nicoll, 2011; Wang et al., 2012; Wang et al., 2008; Yan and Tomita, 2012; Zheng et al., 2004). Furthermore, proteomic studies have identified a host of new candidate auxiliary proteins (Schwenk et al., 2012; Schwenk et al., 2009; Shanks et al., 2012; von Engelhardt et al., 2010), including the vertebrate cornichon (CNIH) proteins CNIH-2 and CNIH-3 (Schwenk et al., 2009).
Cornichon was first identified in Drosophila, where the Cni protein binds to the EGF-like ligand Gurken, thus acting as a cargo receptor for recruitment of Gurken into COPII vesicles. In the absence of Cni, export from the endoplasmic reticulum (ER) and secretion of ligand are disrupted (Bokel et al., 2006; Roth et al., 1995). More recently, affinity purification of native AMPARs from rat brain followed by mass spectroscopy analysis identified CNIH-2 and CNIH-3 as AMPAR-interacting proteins. When coexpressed with AMPAR subunits in heterologous cells, these proteins slowed the kinetics of receptor desensitization and deactivation, and increased surface expression (Coombs et al., 2012; Kato et al., 2010; Schwenk et al., 2009; Shi et al., 2010).
Subsequent studies have revealed additional complexities. While CNIH-2 was found at the cell surface of hippocampal neurons, it did not reach the surface of cerebellar Purkinje neurons in stargazer mice, which lack the γ-2 TARP (Gill et al., 2011). Studies in HeLa cells found that overexpression of CNIH-2 altered the glycosylation pattern of the GluA2 AMPAR subunit, suggesting that CNIH-2 regulates AMPAR maturation in the ER, which may affect AMPAR function at synapses (Harmel et al., 2012). While studies in various heterologous cells have provided important insights into CNIH-2 function, these cells might express different proteins and use different trafficking pathways than those present in neurons. Consequently, studies to date have not fully resolved whether CNIHs primarily have a forward trafficking role in neurons, whether they are also recruited away from the ER-Golgi early secretory pathway to function at synapses, or whether they function as auxiliary proteins at synapses to modify the functional properties of AMPARs. Clearly, a deeper understanding of the contribution of CNIHs to nervous system function and behavior would benefit from in vivo studies. Genetic-based studies of cornichon function in neurons are a challenge in vertebrates, which express four cornichon homologues with possible redundant functions. In contrast, only one cornichon homologue is found in the C. elegans genome.
The study of synaptic signaling in the simple, well-defined nervous system of C. elegans has provided new insights into AMPAR function and behaviors regulated by glutamatergic signaling (de Bono and Maricq, 2005). In C. elegans, the GLR-1 AMPAR subunit is required for fast glutamate-gated current in specialized command interneurons that control the switch between forward and backward movement; thus, reversal frequency is disrupted in glr-1 loss-of-function or gain-of-function mutants (Hart et al., 1995; Maricq et al., 1995; Zheng et al., 1999). Genetic screens for modifiers of this behavior have led to the identification of three classes of auxiliary proteins that associate with GLR-1 and modify the kinetics of glutamate-gated current. These include the evolutionarily and functionally conserved TARP proteins, STG-1 and STG-2 (Walker et al., 2006a; Wang et al., 2008), and the two classes of CUB-domain transmembrane proteins, SOL-1 and SOL-2 (Walker et al., 2006a; Walker et al., 2006b; Wang et al., 2012; Zheng et al., 2006; Zheng et al., 2004).
Sequence analysis of the C. elegans genome revealed the presence of a single cornichon homologue (cni-1). We found that cni-1 mutants exhibited a hyper-reversal phenotype in contrast to the hypo-reversal phenotype observed in strains with loss-of-function mutations in either the GLR-1 subunit or in the auxiliary proteins (Brockie et al., 2001b; Mellem et al., 2002; Zheng et al., 2004). Cornichon’s well-characterized role in the export of specific proteins from the ER (Bokel et al., 2006; Castro et al., 2007; Herzig et al., 2012; Roth et al., 1995) prompted us to examine the in vivo trafficking of GLR-1. Compared to wild type, we found increased anterograde transport of GLR-1 in cni-1 mutants, with corresponding increases in synaptic GLR-1 expression and GLR-1-mediated currents. In contrast, trafficking and current were reduced with overexpression of CNI-1, the yeast homolog (Evr14p), or vertebrate CNIH-2. Reconstitution experiments revealed that cornichon proteins have evolutionarily conserved roles in limiting the export of AMPARs and modifying receptor function, thus contributing to the regulation of neuronal excitability.
RESULTS
CNI-1 is the sole cornichon homologue in C. elegans
Analysis of the C. elegans genome for genes that encode proteins with significant identity to members of the vertebrate cornichon family identified only one candidate homologue (T09E8.3; www.wormbase.org), which had significant identity to vertebrate and invertebrate corichon proteins as well as to the yeast protein Erv14p (Figures S1A and S1C). Analysis of the primary amino acid squence predicted three transmembrane domains (TMDs), as well as the topological arrangment of intra- and extracellular regions found in previously described cornichon proteins (Figure S1C). Invertebrate cornichon proteins as well as vertebrate CNIH-1 and CNIH-4 lack the signature 16 amino acid region found in the first extracellular domain of vertebrate CNIH-2 and CNIH-3. The T09E8.3 gene was originally named cnih-2 (Zhang et al., 2012); but because the protein lacks the signature region, a consensus decision was made to rename the gene cni-1 (Chris Rongo, personal communication). An available deletion mutation in cni-1 removes over half of the coding region and disrupts the 3’ UTR signal required for polyadenylation (Figure S1B).
Reversal frequency is increased in cni-1 mutants
C. elegans regulate the frequency of turning and reversals to mediate avoidance and attraction behaviors, and to aid in the exploration of their environs by restricting or expanding the search area (Hills et al., 2004). The neural circuitry that controls these reversals has been identified and includes the command interneurons that express the GLR-1 AMPAR (Brockie et al., 2001a; Chalfie et al., 1985; de Bono and Maricq, 2005; Piggott et al., 2011). Mutations in genes that disrupt glutamate release (eat-4) or the function of postsynaptic glutamate receptors (glr-1, sol-1 and stg-2) decrease reversal frequency (Figure 1A) (Brockie et al., 2001b; Mellem et al., 2002; Walker et al., 2006a; Wang et al., 2012; Wang et al., 2008; Zheng et al., 2004). In contrast, transgenic “lurcher” worms that express a gain-of-function mutation in GLR-1 have an increased reversal frequency (Zheng et al., 1999). Thus, reversal frequency provides a behavioral readout of the strength of AMPAR-mediated synaptic signaling.
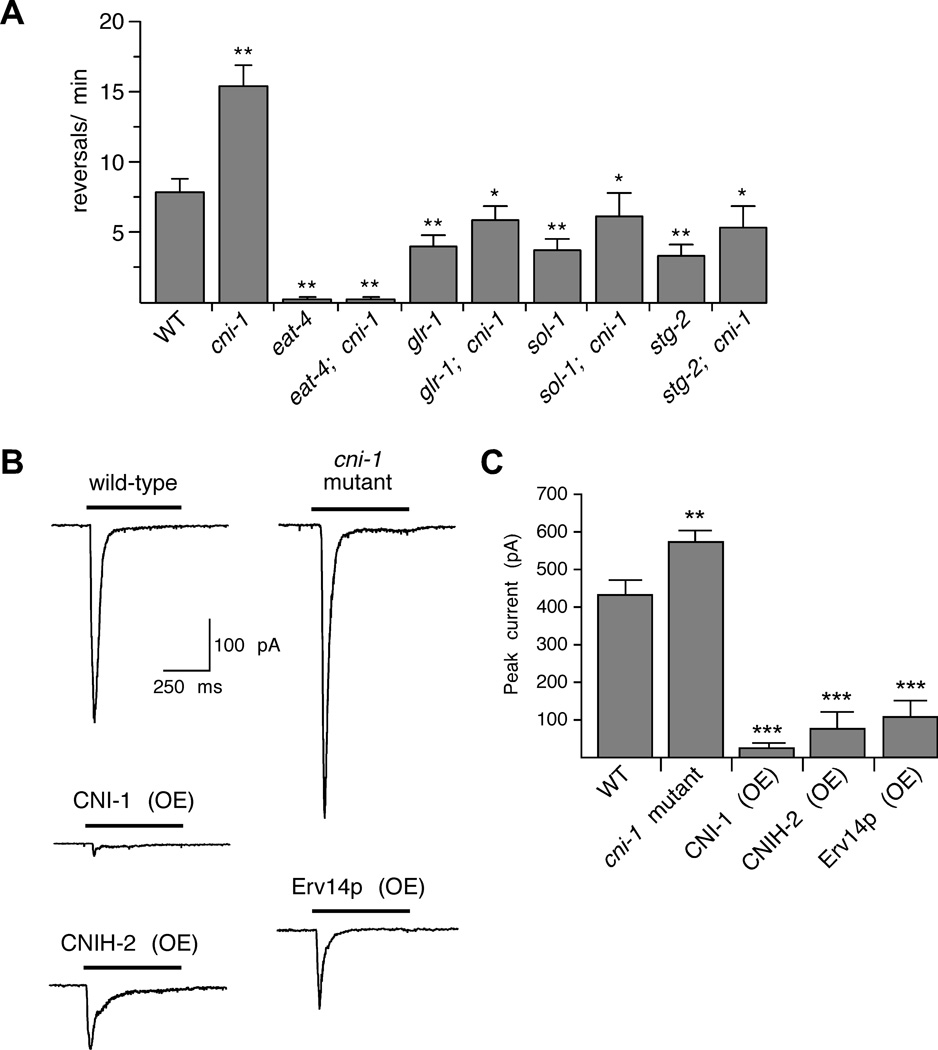
Figure 1.
Reversal frequency and glutamate-gated currents are increased in cni-1 mutants. (A) Reversal frequency in wild-type worms and various single and double mutants. For wild-type and cni-1 mutants, n=8; for glr-1,sol-1 and stg-2 single and double mutants, n=7; and for eat-4 single and double mutants, n=5. ** Significantly different from both wild type and cni-1 mutants (p<0.01). * Significantly different from cni-1 mutants (p<0.05). (B) Currents measured in AVA neurons in response to pressure application of 3 mM glutamate. Cells were voltage-clamped at −60 mV. (C) Average peak glutamate-gated current in wild-type worms (n=11), cni-1 mutants (n=11) and transgenic mutants that overexpressed (OE) either CNI-1 (n=6), CNIH-2 (n=8) or Erv14p (n=7). Significantly different from wild type (** p<0.01 and *** p<0.001). Error bars indicate SEM.
See also Figures S1, S2 and S3.
We found that the average forward time in cni-1 mutants was considerably shorter than in wild type, with a corresponding increase in the frequency of reversals (Figure 1A). This increase suggested that the strength of AMPAR-mediated signaling was increased in cni-1 mutants. In contrast, we did not observe phenotypes associated with loss of GLR-1-mediated signaling, such as altered avoidance responses to tactile or osmotic stimuli (Figure S2), nor did the cni-1 mutation suppress the behavioral defects of glr-1 mutants (Figure 1A and Figure S2A).
To further address whether the increased reversal frequency observed in cni-1 mutants was dependent on glutamatergic signaling, we examined relevant single and double mutants (Figure 1A). Double mutants with cni-1 and eat-4, which encodes the vesicular glutamate transporter, were indistinguishable from eat-4 mutants alone, indicating that the hyper-reversal behavior in cni-1 mutants was dependent on glutamatergic neurotransmission. We next asked whether the hyper-reversal behavior was dependent on the GLR-1 AMPAR. Compared to wild type, reversal frequency was decreased in glr-1 mutants (Brockie et al., 2001b), and the glr-1 mutation suppressed the hyper-reversal phenotype of cni-1 mutants (Figure 1A). These results indicate that the effects of cni-1 on reversal behavior primarily depend on AMPAR-mediated synaptic signaling. In support of this hypothesis, we found that the cni-1 reversal phenotype was also dependent on AMPAR auxiliary proteins. Thus, reversals were suppressed in the double mutants sol-1; cni-1 and stg-2; cni-1 (Figure 1A).
The amplitude of glutamate-gated currents is increased in cni-1 mutants
To evaluate the contributions of CNI-1 to AMPAR-mediated currents, we turned to in vivo patch-clamp electrophysiological analysis. Compared to wild type, the amplitude of the glutamate-gated and GLR-1-dependent current in the AVA command interneurons was significantly increased in cni-1 mutants (Figures 1B and 1C). We found the same relative increase when we used the partial agonist kainate and there was no difference in the efficacy of the two agonists (Figure S3A). In contrast, overexpression of CNI-1 in AVA dramatically reduced the amplitude of the glutamate-gated current. In these transgenic worms, the amount of CNI-1 expressed from the multi-copy transgene is predicted to be much greater than that from the endogenous cni-1 gene. We next asked whether currents were also modified in transgenic cni-1 mutants that overexpressed the related vertebrate CNIH-2 or yeast Erv14p cornichon proteins. We found dramatically reduced currents in these transgenic worms, consistent with an evolutionarily conserved role for cornichon proteins in the regulation of AMPAR-mediated synaptic signaling (Figures 1B and 1C).
We also evaluated whether NMDA receptors (NMDARs) were dependent on CNI-1. Peak NMDA-gated currents were increased in cni-1 mutants and reduced with overexpression of either CNI-1 or CNIH-2 (Figure S3B and S3C). In contrast to what we observed for AMPARs, overexpression of CNI-1 or CNIH-2 caused much smaller changes in NMDA-gated current. These effects are not likely secondary to changes in GLR-1 given that NMDAR-mediated currents in glr-1 mutants are indistinguishable from those in wild-type worms (Brockie et al., 2001b; Mellem et al., 2002; Zheng et al., 2004). These results, together with our behavioral data, demonstrate that CNI-1’s most prominent role in AVA appears to be the regulation of AMPARs. Our findings are consistent with an earlier study in yeast demonstrating that Erv14p regulates multiple classes of proteins (Herzig et al., 2012).
The number of synaptic GLR-1 AMPARs is increased in cni-1 mutants
The increased current that we observed in cni-1 mutants might be secondary to an increase in the number of synaptic AMPARs. To evaluate whether the number of receptors at synapses is modified in cni-1 mutants, we used confocal microscopy to image transgenic strains that expressed a functional GLR-1::GFP fusion protein. Consistent with our earlier behavioral and electrophysiological data, we found that the fluorescence intensity of GLR-1::GFP was greater in transgenic cni-1 mutants and reduced with overexpression of CNI-1, CNIH-2 or Erv14p (Figures 2A and 2B). We obtained similar results with overexpression of CNI-1 in transgenic wild-type worms (Figure S4A). To address whether the surface expression of GLR-1 was modified by the mutation in cni-1, we tagged GLR-1 with the pH-sensitive reporter superecliptic phluorin (SEP). The fluorescence of SEP is quenched by the relatively acidic pH of intracellular vesicles, but exhibits greatly enhanced fluorescence at extracellular pH, thus distinguishing intracellular proteins from those at the surface (Miesenbock et al., 1998; Wang et al., 2012). We also found that the surface expression of GLR-1 was increased in cni-1 mutants (Figure S4B), which is again consistent with the observed increase in GLR-1-mediated current (Figures 1B and 1C).
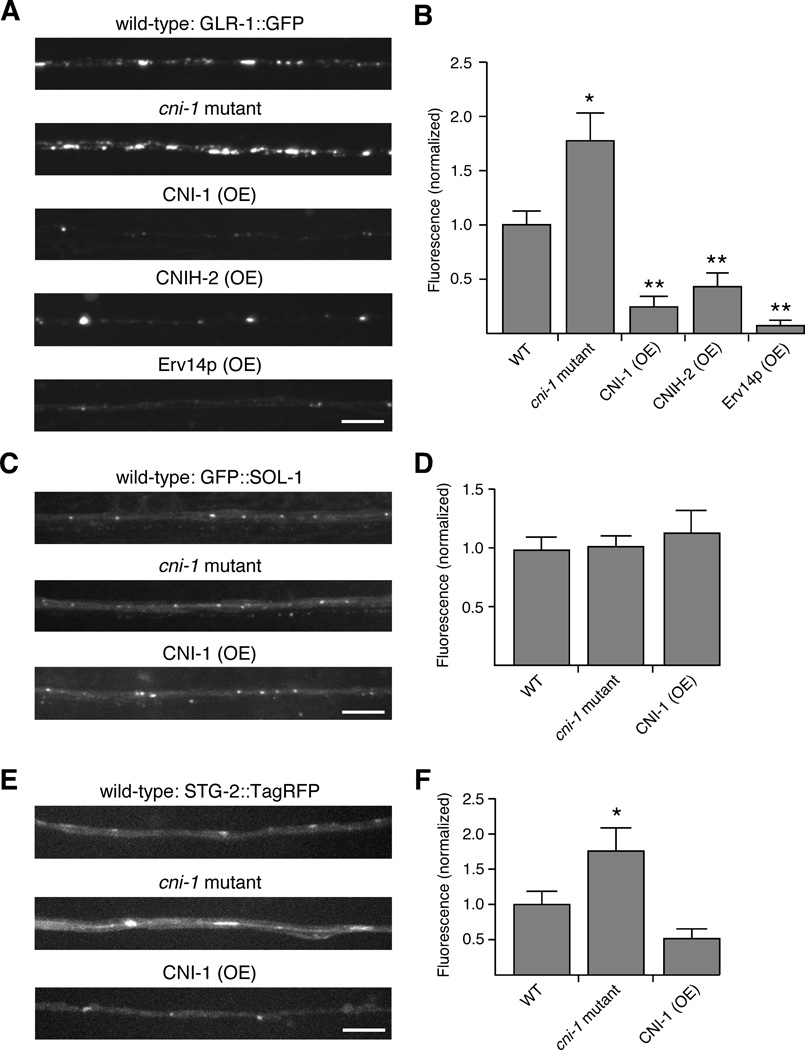
Figure 2.
Synaptic levels of GLR-1::GFP are increased in cni-1 mutants and decreased by overexpression of cornichon proteins. (A and B) Confocal images of GLR-1::GFP (A) and the total GFP fluorescence (B) in the AVA processes of wild-type worms (n=11), cni-1 mutants (n=11) and transgenic mutants that overexpressed CNI-1 (n=10), CNIH-2 (n=9) or Erv14p (n=7). (C and E) Confocal images of either GFP::SOL-1 (C) or STG-2::TagRFP (E) in the AVA neurons of transgenic worms. (D and F) GFP (D) or TagRFP (F) fluorescence in transgenic wild-type worms (GFP, n=12; TagRFP, n=15), cni-1 mutants (GFP, n=11; TagRFP, n=10) and transgenic mutants that overexpressed CNI-1 (GFP, n=5; TagRFP, n=8).
Significantly different from wild type (* p<0.05 and ** p<0.01). Scale bars represent 5 µm. Error bars represent SEM.
See also Figure S4.
We did not find any apparent changes in AVA morphology in cni-1 mutants, or in transgenic worms that overexpressed CNI-1. In addition, changing the expression of CNI-1 did not affect all components of the GLR-1 signaling complex. Thus, in cni-1 mutants or transgenic mutants that overexpressed CNI-1, the intensity of GFP::SOL-1 was relatively unaffected compared to the pronounced changes in GLR-1 (Figures 2C and 2D). We also generated transgenic worms that coexpressed GLR-1::GFP and STG-2::TagRFP. Interestingly, we observed a significant increase in STG-2::TagRFP in cni-1 mutants and a decrease with CNI-1 overexpression (Figures 2E and 2F). However, these changes might be secondary to changes in GLR-1 given that CNI-1 overexpression dramatically reduced GLR-1::GFP fluorescence in the ventral cord (Figure S4C). In contrast, the major effect of CNI-1 overexpression on STG-2::TagRFP was a shift from punctate to more diffuse expression.
Transport of GLR-1 AMPARs is increased in cni-1 mutants
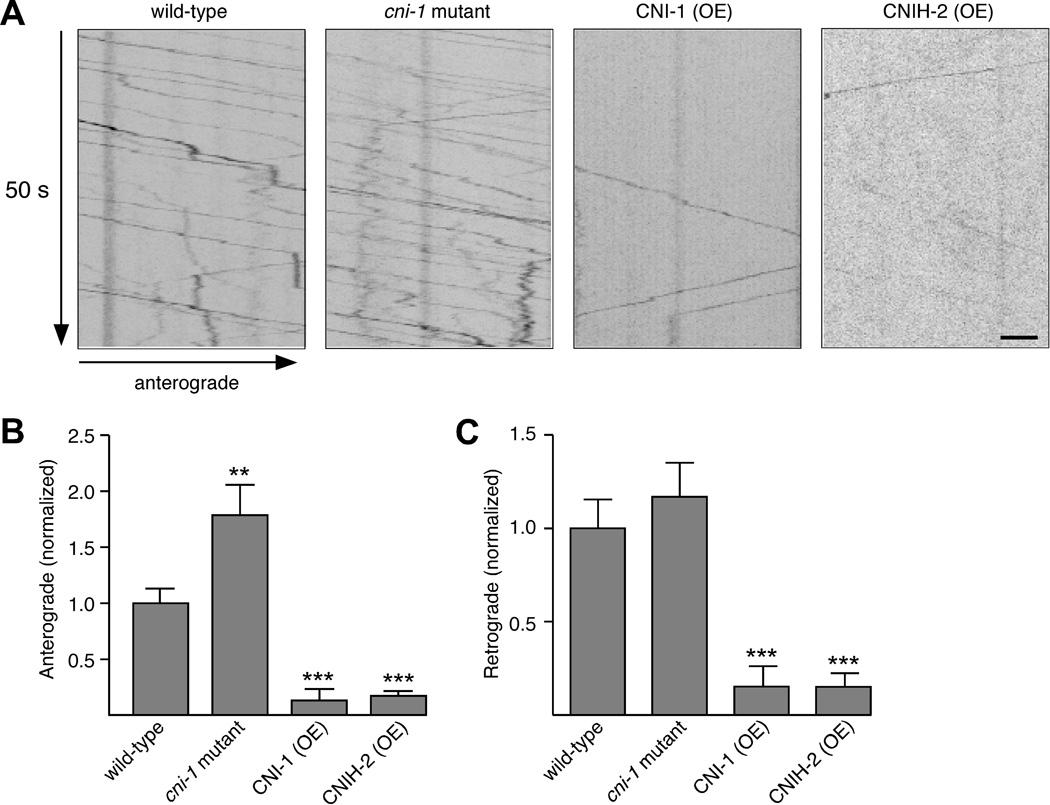
The increase in synaptic GLR-1 in cni-1 mutants might be caused by changes in either the anterograde or retrograde transport of GLR-1. To address these possibilities, we obtained streaming confocal images of GLR-1::GFP (Figure 3A). In cni-1 mutants, the frequency of anterograde transport events in the AVA process was significantly increased compared to wild type (Figure 3B), whereas these events were dramatically decreased in transgenic worms that overexpressed CNI-1 or CNIH-2. Interestingly, the cni-1 mutation had less effect on retrograde transport (Figure 3C). However, retrograde events were significantly reduced by overexpression of either CNI-1 or CNIH-2, which we suggest was most likely consequent to the dramatic reduction in anterograde transport of AMPARs. Thus, the behavioral and electrophysiological changes in cni-1 mutants appear secondary to the increased export of AMPARs from the cell body.
Figure 3.
The frequency of GLR-1 anterograde transport is increased in cni-1 mutants. (A) Kymographs showing the movement of GLR-1::GFP in the AVA interneurons. Scale bar represents 2 µm. (B and C) Quantification of the number of anterograde (B) and retrograde (C) transport events in wild-type worms (n=13), cni-1 mutants (n=9), and cni-1 transgenic mutants that overexpressed either CNI-1 (n=10) or CNIH-2 (n=10). Significantly different from wild type (** p<0.01 and *** p<0.001). Error bars represent SEM.
CNI-1 is expressed in AMPAR-expressing neurons and localizes to the ER
To determine the cellular distribution of cni-1, we used confocal microscopy to examine transgenic strains in which the cni-1 promoter drove expression of GFP (Figure S5A). We found expression in many tissues, including widespread distribution in the nervous system (Figure 4A and Figure S5B). To address whether CNI-1 is expressed in the same neurons as GLR-1, we coexpressed CNI-1::GFP and mCherry driven by the glr-1 promoter and found that CNI-1 was expressed in all GLR-1-expressing neurons (Figure S5B). In independent experiments, we confirmed that CNI-1 is expressed in the AVA interneurons using the flp-18 promoter to identify AVA (Feinberg et al., 2008) (Figure S5C). In neuronal cell bodies, CNI-1::GFP appeared punctate and distinctively clustered at perinuclear sites, suggestive of localization to the ER (Figure 4A).
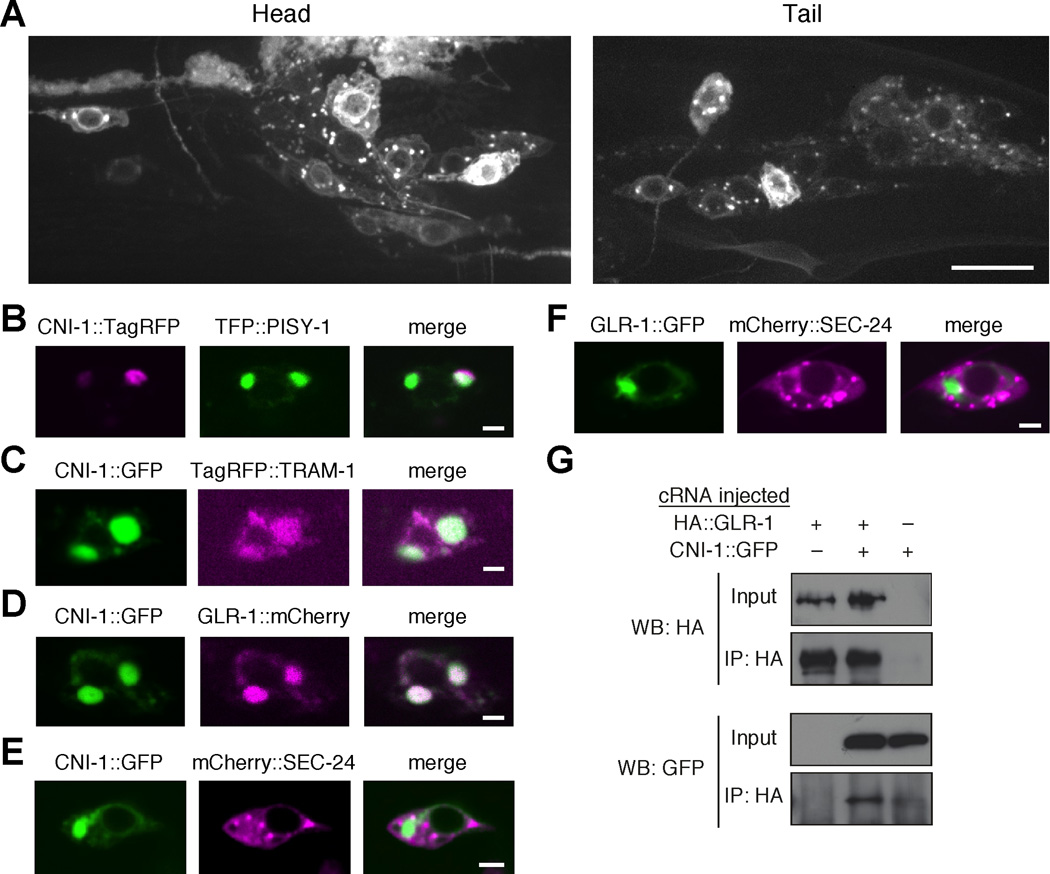
Figure 4.
CNI-1 is widely expressed in the nervous system where it colocalizes with GLR-1 in the ER. (A) Confocal images of the head and tail region of transgenic worms that expressed the CNI-1 ::GFP reporter shown in Figure S5A . Scale bar represents 10 µm. (B–F) Single plane, confocal images of the AVA cell bodies in transgenic worms that expressed various combinations of fluorescently labeled proteins. Scale bars represent 2 µm. (G) Immunoprecipitation of HA::GLR-1 and CNI-1::GFP coexpressed in Xenopus oocytes.
See also Figure S5.
To better examine the subcellular localization of CNI-1, we coexpressed CNI-1 with either TRAM-1, a marker of rough ER, or PISY-1, a general ER marker that also labels Golgi structures (Leber et al., 1995; Lofke et al., 2008; Rolls et al., 2002). While the distribution of PISY-1 (Figure 4B) was more localized compared to the diffuse reticular distribution of TRAM-1 (Figure 4C), we found that CNI-1 colocalized with both markers, and that CNI-1 and GLR-1 colocalized (Figure 4D). To address whether accumulations of CNI-1 and GLR-1 were near ER exit sites (ERES), we coexpressed either CNI-1::GFP or GLR-1::GFP with the COPII protein, SEC-24 (F12F6.6), tagged with mCherry. In the cell body, both CNI-1::GFP and GLR-1::GFP were found localized adjacent to mCherry::SEC-24 puncta (Figures 4E and 4F) in a pattern similar to that described previously for localization of nicotinic acetylcholine receptors to ERES (Srinivasan et al., 2011). These data show that CNI-1 is expressed in discrete regions of the ER/Golgi where it colocalized with GLR-1.
The colocalization of CNI-1 and GLR-1 also suggests that the two proteins might interact with each other. To test this possibility, we coexpressed HA::GLR-1 and CNI-1::GFP in Xenopus oocytes and found an association between the proteins using an immunoprecipitation strategy (Figure 4G). Together, the biochemical and cell biological data indicate a close association between CNI-1 and GLR-1, a finding that is also consistent with studies that demonstrate an association between CNIH-2 and the vertebrate GluA1 AMPAR subunit (Kato et al., 2010; Schwenk et al., 2009; Shi et al., 2010).
CNI-1 colocalizes with surface GLR-1 in the processes of AVA
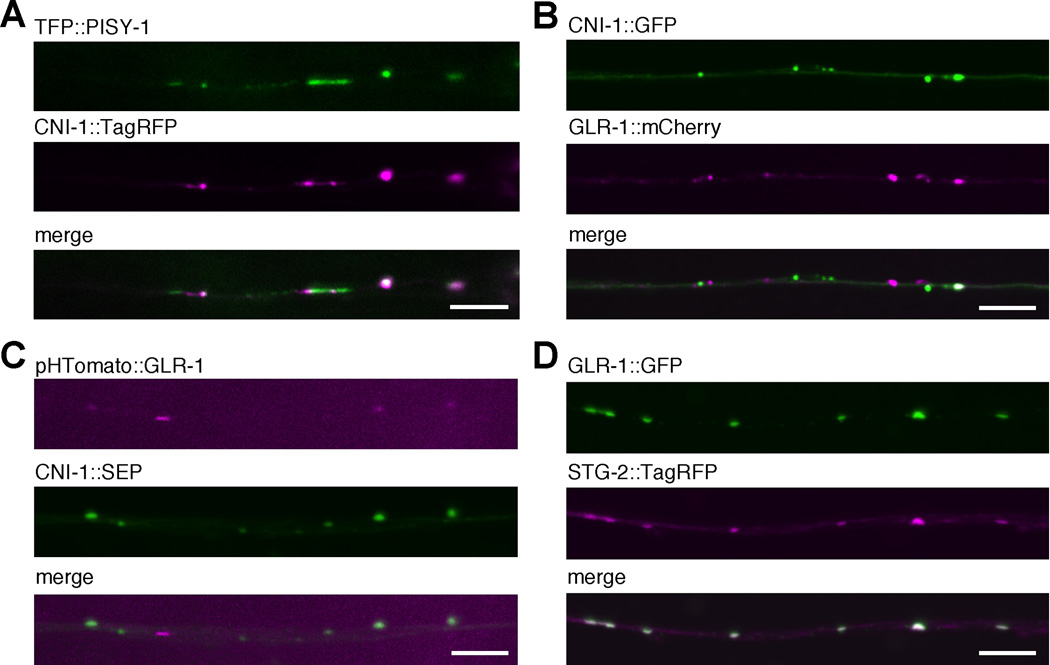
In C. elegans, ER extends into neural processes (Rolls et al., 2002). We found that CNI-1 was also expressed in the processes of AVA, where it was distributed in a punctate pattern that colocalized with the ER/Golgi marker, PISY-1 (Figure 5A). When we examined the localization of CNI-1::GFP and GLR-1::mCherry in the AVA neural processes, we noted a prominent punctate distribution of both proteins where a subset of GLR-1 puncta colocalized with CNI-1 puncta (Figure 5B). While these experiments suggested that the majority of CNI-1 was intracellular and associated with organelles, they did not distinguish between intracellular and cell-surface localization of CNI-1. Therefore, we also tagged CNI-1 with SEP and GLR-1 with pHTomato, a red-shifted pH-sensitive reporter (Li and Tsien, 2012) and coexpressed these tagged proteins in AVA (Figure 5C). Because of reduced intracellular fluorescence using these pH-sensitive fluorophores, we were able to observe extensive colocalization of CNI-1 and GLR-1 with approximately 40% – 80% of the GLR-1 puncta colocalizing with CNI-1 puncta. Although less dramatic than the colocalization of GLR-1 with STG-2 (Figure 5D) (Wang et al., 2008), the punctate colocalization of GLR-1 with CNI-1 suggests that CNI-1 might also function at synapses. Surface expression of CNI-1 was also supported by in vivo antibody labeling experiments (Figure S6). These results indicate that CNI-1 is not restricted to the cell body or intracellular organelles and that CNI-1 is present at synapses.
Figure 5.
Surface expressed CNI-1 colocalizes with synaptic GLR-1. (A–D) Confocal images of the AVA processes in transgenic worms that expressed various combinations of fluorescently labeled proteins. Scale bars represent 5 µm.
See also Figure S6.
CNI-1 limits ER export of GLR-1
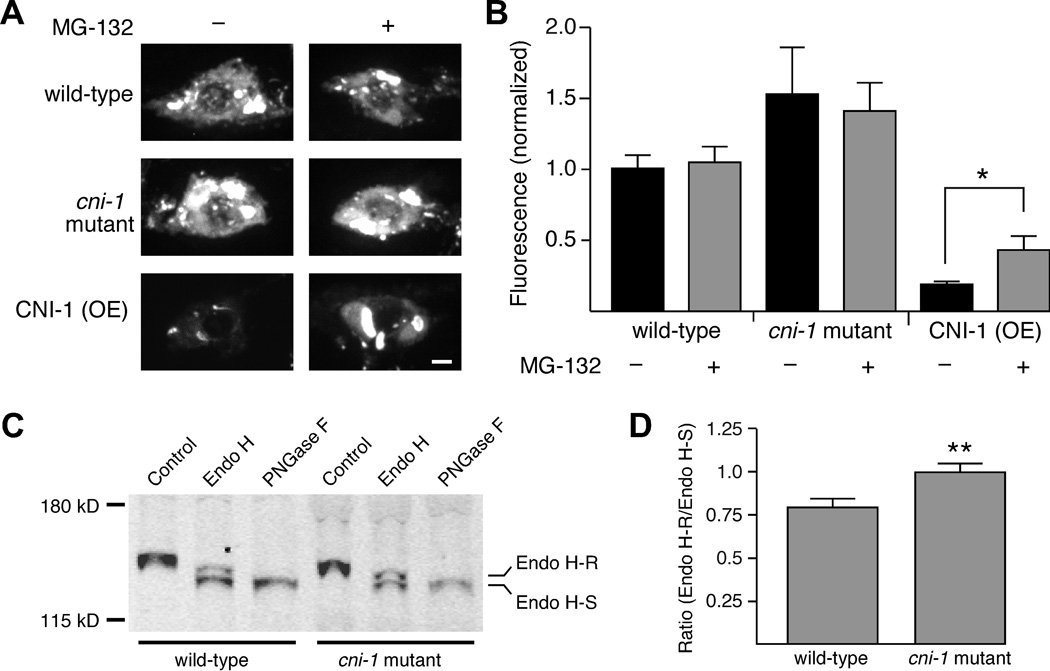
Our data are consistent with a model in which CNI-1 functions to limit export of GLR-1 to synapses. However, in transgenic worms that overexpressed CNI-1 we also found that the fluorescence intensity of GLR-1::GFP was considerably reduced in cell bodies (Figures 6A and 6B). We reasoned that overexpression of CNI-1 might lead to the shunting of retained GLR-1 to the endoplasmic-reticulum-associated protein degradation pathway (ERAD) and ultimately the proteasome. To test this hypothesis, we treated worms with the proteasome inhibitor MG-132, a drug that is commonly used to evaluate ERAD-mediated degradation of transmembrane proteins (Altier et al., 2011). In transgenic worms that overexpressed CNI-1, incubation with MG-132 markedly increased GLR-1 fluorescence. In contrast, we did not observe an increase in fluorescence in wild-type controls or cni-1 mutants (Figures 6A and 6B). These results are consistent with the hypothesis that CNI-1 overexpression blocks export of GLR-1, leading to subsequent degradation by the proteasome.
Figure 6.
Overexpression of CNI-1 results in GLR-1::GFP accumulation in neuronal cell bodies and its subsequent degradation. (A) Confocal images of GLR-1::GFP in AVA cell bodies in worms either with or without MG-132 treatment. Scale bar represents 2 µm. (B) Average GFP fluorescence intensity in worms either with (wild type, n=20; cni-1 mutant, n=12; CNI-1 (OE), n=15) or without (wild type, n=22; cni-1 mutant, n=13; CNI-1 (OE), n=16) MG-132 treatment. * p<0.05. (C) Western blot showing the relative amounts of Endo H sensitive (Endo H-S) and resistant (Endo H-R) GLR-1::GFP isolated from transgenic wild-type worms and cni-1 mutants. (D) The ratio of Endo H-R to Endo H-S GLR-1::GFP in wild type (n=11) and cni-1 mutants (n=10). ** p<0.01. Error bars represent SEM.
If CNI-1 has a role in ER export then the glycoslylation of GLR-1 might be altered in cni-1 mutants. The glycosylation state of proteins in the ER is distinct from those in the Golgi, and can be distinguished by differential sensitivity to the enzyme Endoglycosidase H (Endo H) (Chun et al., 2008). We found that the relative amount of GLR-1 that was resistant to Endo H was increased in cni-1 mutants compared to wild-type worms (Figures 6C and 6D). This result is consistent with the increased GLR-1 anterograde trafficking observed in cni-1 mutants (Figure 3).
Reconstitution experiments support a conserved role for cornichons in limiting AMPAR export from the ER
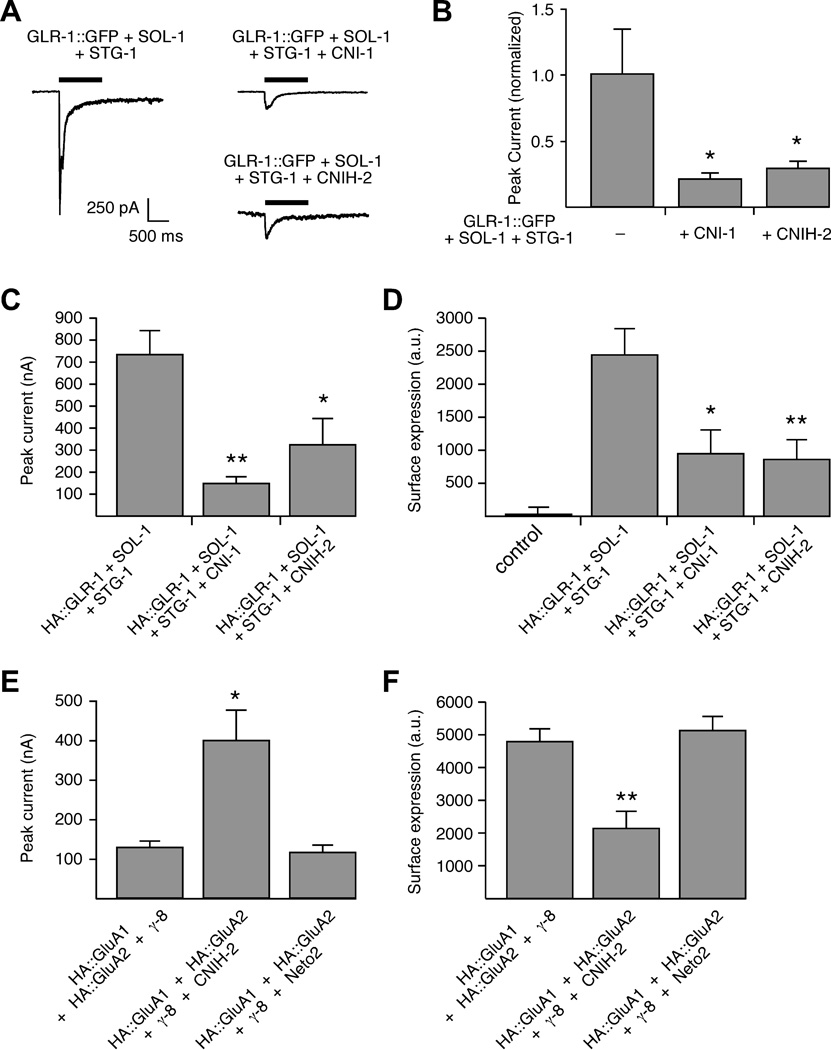
In C. elegans, muscle cells do not express glutamate receptor subunits or known auxiliary proteins for iGluRs, thus providing an ideal system for genetic-based reconstitution experiments. We recorded glutamate-gated currents from muscle cells in transgenic worms that ectopically expressed GLR-1, SOL-1 and STG-1 and compared them to currents from muscle cells that expressed these three proteins along with either CNI-1 or CNIH-2. Similar to what we observed with overexpression studies in the AVA neurons, the amplitude of glutamate-gated current in muscle was significantly decreased with overexpression of either CNI-1 or CNIH-2 (Figures 7A and 7B). We next asked whether CNI-1 modified glutamate-gated currents mediated by vertebrate AMPARs expressed in C. elegans muscle, and found that CNI-1 also reduced the amplitude of currents mediated by GluA1 (Figure S7A), and reduced SEP::GluA1 fluorescence (Figure S7B).
Figure 7.
Overexpressing CNI-1 or CNIH-2 modifies glutamate-gated current and AMPAR surface expression. (A) Glutamate-gated current in transgenic muscle cells in response to pressure application of 3 mM glutamate. Cells were voltage clamped at −60 mV. (B) Average peak glutamate-gated current in muscle cells that expressed GLR-1::GFP, SOL-1 and STG-1 (n=6); GLR-1::GFP, SOL-1, STG-1 and CNI-1 (n=5); or GLR-1::GFP, SOL-1, STG-1 and CNIH-2 (n=11). * Significantly different from GLR-1::GFP + SOL-1 + STG-1 (p<0.05). (C and D) Glutamate-gated current in Xenopus oocytes (C) and GLR-1 surface expression (D) in non-injected control oocytes or in oocytes that expressed HA::GLR-1, SOL-1 and STG-1 either with or without coexpression of CNI-1 or CNIH-2 (n=6 for all conditions). Significantly different from HA::GLR-1 + SOL-1 + STG-1 (* p<0.05 and ** p<0.01). (E and F) Glutamate-gated current (E) and AMPAR surface expression (F) in Xenopus oocytes (n=6 for all conditions). Significantly different from HA::GluA1 + HA::GluA2 + γ-8 (* p<0.05 and ** p<0.01).
Error bars represent SEM.
See also Figure S7.
Our evaluation of surface AMPARs in transgenic worms relied on measurements of fluorescence intensity. To more directly compare surface expression and receptor mediated currents, we turned to reconstitution studies in Xenopus oocytes. Overexpression of CNI-1 or CNIH-2 reduced GLR-1-mediated currents as well as surface expression of GLR-1 (Figures 7C and 7D). Similarly, overexpression of CNI-1 reduced GluA1-mediated currents and surface expression of GluA1 (Figures S7C and S7D). While coexpression of CNIH-2 with GluA1, GluA2 and the γ-8 TARP auxiliary subunit reduced AMPAR surface expression, it increased the peak glutamate-gated current, suggesting an additional effect of cornichon proteins on AMPAR function (Figures 7E and 7F). In contrast, coexpression of the Neto2 CUB-domain protein had no effect on either AMPAR-mediated current or surface expression (Figures 7E and 7F).
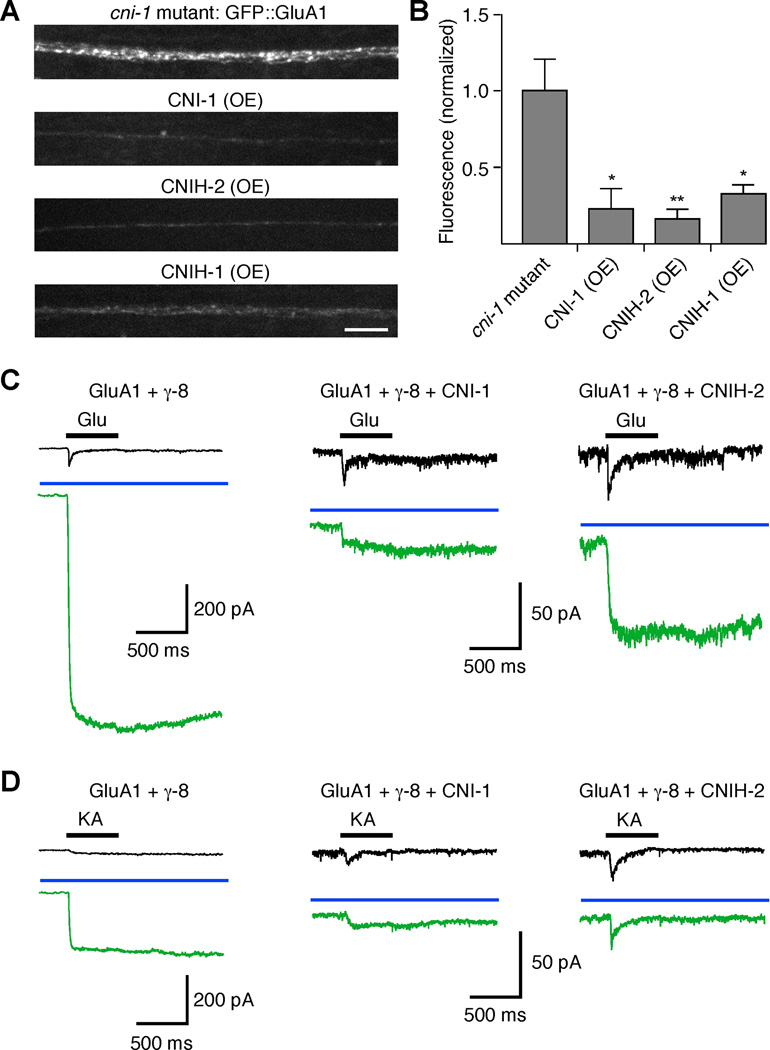
We next extended our reconstitution studies to address whether invertebrate and vertebrate cornichons have conserved roles in limiting the export of vertebrate AMPARs in neurons. Therefore, we coexpressed GFP-tagged GluA1 and γ-8 either with or without a cornichon protein in the AVA neurons. We found that overexpression of CNI-1, or vertebrate CNIH-2 or CNIH-1, dramatically reduced the fluorescence intensity of GFP::GluA1 in transgenic worms (Figures 8A and 8B). Electrophysiological analysis showed a reduction in peak glutamate- or kainate-gated current with overexpression of cornichon proteins (Figures 8C and 8D, black traces).
Figure 8.
Reconstitution of vertebrate GluA1 receptor function in transgenic C. elegans. Cornichon proteins decrease GluA1-mediated current and synaptic GluA1 levels when coexpressed in C. elegans AVA neurons. (A and B) Confocal images (A) and quantification (B) of GFP::GluA1 fluorescence in the AVA neurons of transgenic cni-1 mutants (n=9), or transgenic mutants that also overexpressed CNI-1 (n=4), CNIH-2 (n=5) or CNIH-1 (n=5). Scale bar represents 5 µm; error bars represent SEM. Significantly different from cni-1 mutants (* p<0.05, ** p<0.01). (C and D) GluA1-mediated glutamate- (C) and kainate- (D) gated current in the AVA neurons of various transgenic worms both before (black) and after (green) treatment with 100 µM cyclothiazide. The blue bar indicates the presence of cyclothiazide.
The reduced currents that we observed in transgenic worms that overexpressed either CNI-1 or CNIH-2 are consistent with cornichon’s putative role in the export of R-1 from the ER. However, these reconstitution experiments did not address whether cornichon proteins might have additional effects on receptor function. To address this question, we determined the relative efficacy of cyclothiazide (CTZ), a drug that blocks the desensitization of vertebrate AMPARs, thereby causing potentiation of the peak current in response to the relatively slow speed of pressure application of agonist (Partin et al., 1993). In preliminary studies, we found CTZ strongly potentiated glutamate- and kainate-gated currents in transgenic worms that overexpressed vertebrate GluA1 and γ-8 in the AVA neurons. However, we observed far less potentiation in strains that coexpressed either worm CNI-1 or vertebrate CNIH-2 (Figures 8C and 8D). One interpretation of these data is that coexpression of cornichons slowed AMPAR desensitization or otherwise modified AMPAR properties(Gill et al., 2011; Gill et al., 2012; Schwenk et al., 2009), thus reducing the potentiation by CTZ.
The results from our genetic studies together with our reconstitution experiments in C. elegans neurons and muscle cells, and Xenopus oocytes demonstrate an evolutionarily conserved role for cornichon proteins in regulating the ER export of AMPARs. Furthermore, cornichon proteins colocalized with AMPARs at synapses and, when overexpressed, modified receptor function either directly of indirectly.
DISCUSSION
CNI-1 regulates ER export of AMPARs in C. elegans
The number of functional AMPARs at central excitatory synapses is a critical determinant of synaptic strength, and the strength of synaptic transmission is determined in part by the balance between delivery and removal of synaptic AMPARs. Our study has demonstrated that invertebrate and vertebrate cornichon proteins have conserved roles in the control of AMPAR export from the ER. In cni-1 mutants, the export of AMPARs is unregulated causing increased transport of receptors, larger synaptic currents, neuronal hyper-excitability and disrupted foraging behavior secondary to an increased reversal frequency. While CNI-1 might affect many proteins (Bokel et al., 2006; Castro et al., 2007; Herzig et al., 2012; Roth et al., 1995), the hyper-reversal phenotype observed in cni-1 mutants is primarily dependent on synaptic AMPARs, as demonstrated by the strong suppression of the cni-1 mutant phenotype by mutations in either glr-1, sol-1 or stg-2.
In support of our hypothesis that CNI-1 regulates GLR-1 export from the ER, we found that CNI-1 colocalized with ER markers in the AVA neuronal cell bodies. Of particular interest was the colocalization of CNI-1 with GLR-1, and with the ER/Golgi marker PISY-1. We also found that CNI-1 and GLR-1 were in close apposition to SEC-24/COPII puncta, which mark putative ER export sites (Srinivasan et al., 2011). These data suggest that CNI-1 might have a spatially restricted role and act to control the export of GLR-1 at the ER-Golgi interface. In support of this hypothesis, we found a larger percentage of Endo H resistant GLR-1 in cni-1 mutants suggesting ER export was increased in the mutants compared to wild type. These data are consistent with reconstitution experiments in HeLa cells that demonstrated a CNIH-2-dependent increase in immature-glycosylated GluA2 receptors (Harmel et al., 2012).
Cornichon proteins have a conserved role in limiting the ER export of AMPARs
To address whether vertebrate cornichons had conserved roles in limiting the export of vertebrate AMPARs, we reconstituted GluA1 function in C. elegans neurons. We generated transgenic glr-1; cni-1; stg-2 triple mutants that coexpressed the vertebrate GluA1 AMPAR and the vertebrate γ-8 auxiliary protein. glr-1 and stg-2 mutants lack fast glutamate-gated currents in AVA (Brockie and Maricq, 2006; Zheng et al., 2004), thereby facilitating the interpretation of our reconstitution studies. Interestingly, we found punctate expression of GluA1 in neuronal processes and we could record glutamate-gated current, indicating that the vertebrate receptors were transported to the surface and were functional. In these transgenic worms, coexpression of vertebrate CNIH-2 or CNIH-1, or worm CNI-1 dramatically reduced GFP::GluA1 fluorescence in neuronal processes. Furthermore, CNIH-2 and CNI-1 reduced glutamate- and kainate-gated currents, indicating that an evolutionarily conserved role of cornichon proteins is to limit the export of AMPARs.
While our manuscript was in revision, a study was published that evaluated the contribution of CNIH-2 and CNIH-3 to AMPAR-mediated synaptic transmission in CA1 neurons of the vertebrate hippocampus (Herring et al., 2013). In this study, the authors found that genetic perturbation of CNIH-2 and CNIH-3, a subset of the four cornichon proteins expressed in the brain that appear predominant in the hippocampus, was associated with reduced numbers of GluA1 containing AMPARs along with a corresponding decrease in peak glutamate-gated currents. Given the results from our reconstitution studies of vertebrate GluA1 in C. elegans neurons, the finding of reduced current in the conditional knockout mice suggests that vertebrate CA1 neurons might express additional quality control machinery. For example, GluA1 receptors not associated with a cornichon protein might be more susceptible to degradation in vertebrate neurons.
CNI-1 differentially regulates the components of the GLR-1 signaling complex
Auxiliary proteins are known to contribute to AMPAR function (Jackson and Nicoll, 2011; Yan and Tomita, 2012). In C. elegans, the auxiliary proteins SOL-1, SOL-2 and STG-2 colocalize with GLR-1 at synapses (Wang et al., 2012; Wang et al., 2008; Zheng et al., 2004). These transmembrane proteins did not show the same dependence on cornichon as that observed for GLR-1. Thus, the levels of SOL-1 were not appreciably altered in the processes of AVA in cni-1 mutants or with overexpression of CNI-1. Although we did find an increase in STG-2 in cni-1 mutants and a minor decrease when CNI-1 was overexpressed, we suspect that these effects were secondary to changes in GLR-1.
Recent analysis of the yeast cornichon homologue, Erv14p, provides a possible mechanism for cornichon’s cargo specificity (Herzig et al., 2012). Erv14p associates with many transmembrane proteins and target specificity appears to be associated with the length of the transmembrane domain(s). However, other mechanisms also contribute to ER export and cargo can be made independent of Erv14p by the addition of different trafficking domains (Herzig et al., 2012).
CNI-1 modifies neuronal excitability by regulating the number of synaptic AMPARs
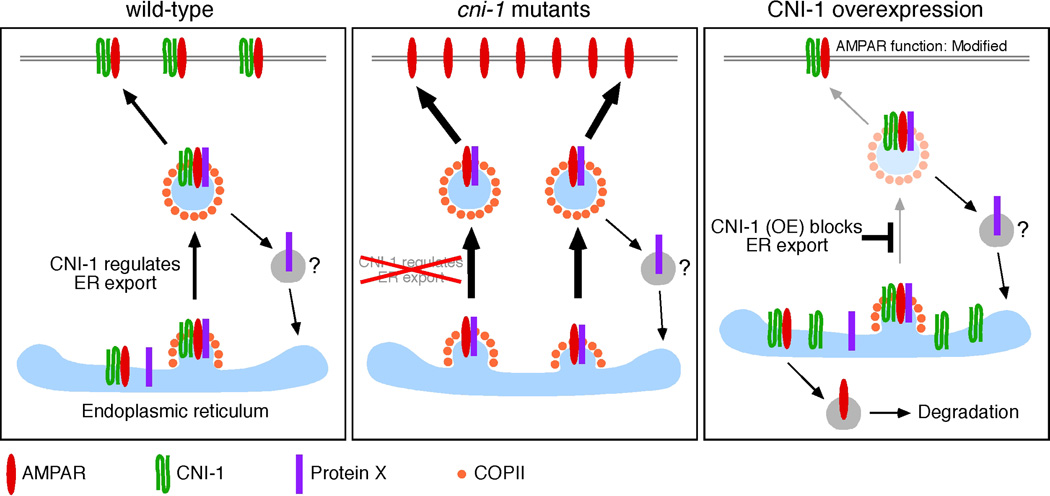
The study of the cni-1 mutant along with transgenic rescue and AMPAR reconstitution experiments demonstrate that an ancient evolutionarily conserved role for cornichon proteins is to regulate the ER export of AMPARs. We envision two possible mechanisms. In the first model, CNI-1 binds to AMPAR subunits and blocks export from the ER –perhaps by masking an ER export signal. Binding of a putative Protein X exposes the export signal thereby allowing the complex to exit the ER (Figure 9). In cni-1 mutants, AMPARs more readily exit the ER, resulting in greater delivery of receptors to the cell surface and synapses. This model is similar to one postulated for the regulation of TGF-α from the ER (Castro et al., 2007). Here, cornichon binds to TGF-α, thus limiting ER export until it associates with the transmembrane protein Star.
Figure 9.
CNI-1 modifies neuron excitability by regulating the export of AMPARs from the ER. In this model, GLR-1 binds to CNI-1, which masks an ER export signal and prevents export. Association of a putative Protein X modifies the complex such that the ER export signal is exposed and the AMPAR can exit the ER. Thus, in cni-1 mutants AMPARs more readily exit the ER. In contrast, AMPAR exit from the ER is slowed by overexpression of CNI-1. Furthermore, the function of AMPARs that do reach synapses is modified.
In the second model, AMPARs directly bind to COPII proteins for export. Overexpression of CNI-1 limits export of AMPARs by competing for COPII binding sites. Conversely, in the absence of cni-1 more COPII sites are available for GLR-1 binding resulting in increased ER export. In both models, ER-retained receptors are susceptible to subsequent degradation. Although we favor the first model because we found that AMPARs and their auxiliary proteins do not have the same dependence on CNI-1, discrimination of the two models will require a systematic study of the molecular requirements for ER export of AMPARs.
CNI-1 and CNIH-2 overexpression modifies export and function of vertebrate AMPARs
Importantly, we showed in reconstitution studies in transgenic worms that vertebrate and C. elegans cornichon proteins similarly decreased the ER export of vertebrate AMPARs, and decreased glutamate-gated currents. These data provide further evidence for the conservation of cornichon function. Our results also highlight the utility of reconstituting vertebrate receptors in C. elegans. This strategy not only reveals conserved functions, but also points out significant differences. For example, the interesting difference between our results and those of Herring et al., suggests that additional machinery contributes to the regulation of AMPAR signaling complexes in vertebrates.
Our data showing that CTZ efficacy was decreased by overexpression of cornichon proteins suggests that cornichons might also function as classical auxiliary proteins to modify either the kinetics of AMPAR desensitization or receptor sensitivity to CTZ. Alternatively, cornichon might modify AMPAR function by interacting with associated auxiliary proteins, or exert indirect effects by modifying the maturation of synaptic receptors, e.g., the glycosylation state or stoichiometry of the receptor complex. In preliminary studies, we did not find an obvious difference in GLR-1 kinetics in wild type and cni-1 mutants (data not shown), suggesting that the effects of wild-type CNI-1 on AMPAR function might be subtle compared to those observed with overexpression of cornichon proteins. Unfortunately, we were not able to determine the relative effects of CTZ on GLR-1-mediated currents given that C. elegans AMPARs are insensitive to CTZ treatment (data not shown). A third possibility is raised by a recent proteomic study that identified many potential auxiliary proteins (Schwenk et al., 2012). Perhaps cornichon proteins help recruit or assemble these or yet to be identified proteins to the receptor complex.
CNI-1 colocalizes with GLR-1 in neural processes
We also found substantial CNI-1 in neuronal processes. Much of CNI-1 appeared to be intracellular and associated with organelles marked by PISY-1. However, more refined analysis revealed that CNI-1 was also at the surface of neuronal processes and colocalized with surface GLR-1, similar to what we have observed for STG-2, SOL-1 and SOL-2 auxiliary subunits (Wang et al., 2012; Wang et al., 2008; Zheng et al., 2004). Although the changes in peak glutamate-gated current in cni-1 mutants, and with overexpression of cornichon proteins, can be explained by changes in surface expression, the presence of CNI-1 at synapses is intriguing. Our reconstitution studies demonstrating changes in CTZ sensitivity are consistent with a possible auxiliary role for CNI-1 at synapses. Alternatively, CNI-1 might regulate local trafficking of AMPARs between endosomal compartments and synapses – a possible role that might be analogous to its function as a regulator of GLR-1 export from the ER.
Our results help provide a new mechanistic view of the global regulation of receptor numbers at the postsynaptic membrane. Neurons rely on compensatory homeostatic mechanisms that regulate synaptic strength and optimize neuronal excitability (Davis, 2006; Goold and Nicoll, 2010; Turrigiano, 2008). In cni-1 mutants, the neural circuit that regulates reversals used for avoidance and foraging behaviors is hyper-excitable secondary to an increase in the number of synaptic AMPARs. We propose that CNI-1 regulates the export of AMPARs in response to external or internal cues, thus contributing to homeostatic processes such as global regulation of neuronal excitability.
Experimental procedures
General methods, genetics and plasmids
All C. elegans strains were raised at 20 °C under standard laboratory conditions. Transgenic strains carrying multi-copy transgene arrays were generated using microinjection into the gonad of adult hermaphrodite lin-15(n765ts) mutants, wild-type worms, or relevant mutant worms. Transgenic worms were selected by rescue of the lin-15(n765ts) mutant phenotype, or by expression of a co-injected fluorescent marker. Single copy transgenic strains were generated following an established protocol (Frokjaer-Jensen et al., 2008). Fluorescently labeled CNI-1 was found to be functional in transgenic experiments that showed the fusion protein had a similar effect on the localization of fluorescently tagged GLR-1 as that of untagged CNI-1. Function of fluorescently labeled STG-2 was confirmed by rescue of stg-2(ak134) suppression of the lurcher worm hyper-reversal phenotype. Plasmids, transgenes and mutant strains are described in Supplemental experimental procedures.
Confocal microscopy
Confocal images were acquired using a Nikon Ti-eclipse equipped with a WaveFX-X1 spinning disc confocal system (Quorum Technologies), and captured by a Cascade 1024B EMCCD camera (Photometrics). Streaming movies (100 ms exposure) of GLR-1::GFP transport were acquired in a single focal plane in the AVA processes. Image acquisition and kymographs were generated using MetaMorph 7.7.10 (Molecular Devices). Anterograde and retrograde events were quantified by counting the number of trajectories in each direction with the experimenter blind to the genotype.
Electrophysiological studies
Electrophysiological recordings from AVA interneurons and muscle cells from dissected transgenic worms were performed as described (Jensen et al., 2012; Mellem et al., 2002).
Behavioral analysis
Reversal frequency, nose touch response and osmotic avoidance assays were performed as previous described (Brockie et al., 2001b; Mellem et al., 2002). A reversal was defined as a switch from forward to backward or from backward to forward movement. All behavioral assays were performed blind. Statistical significance was determined by using the standard Student’s t test.
MG-132 treatment
Proteasome inhibition assays were performed as described (Orsborn et al., 2007). Briefly, adult worms were placed in liquid medium containing 1 μM M −132 for 3 hr. Controls were incubated in liquid medium without MG-132. Immediately following incubation, GLR-1::GFP in the AVA cell bodies was imaged by confocal microscopy.
Quantification of fluorescence
Total fluorescence in neuronal processes was measured using a linescan measurement in MetaMorph 7.7.10 (Molecular Devices) and analyzed with a custom written MATLAB script (based on http://terpconnect.umd.edu/~toh/spectrum/PeakFindingandMeasurement.htm). A detailed description of the quantification of fluorescence signals can be found in Supplemental experimental procedures
Electrophysiology, surface labeling and coimmunoprecipitation studies in Xenopus laevis oocytes
Two-electrode voltage-clamp recordings, and surface labeling using HA::GLR-1, HA::GluA1 and anti-HA antibodies were performed as described (Morimoto-Tomita et al., 2009). For coimmunoprecipitations, oocyte membranes were suspended in lysis buffer containing TED (25 mM Tris-Cl pH 7.4, 2 mM EDTA, 1 mM DTT), 1% Triton X-100, Halt protease inhibitors (Pierce) and centrifuged at 100,000 x g for 30 min (Morimoto-Tomita et al., 2009). The supernatants were then incubated with 5 µg of anti HA antibody and 30 µl of protein G sepharose beads. The beads were then washed five times with 1% Triton in TEEN (25 mM Tris-Cl pH 7.4, 1 mM EDTA, 1 mM EGTA, 150 mM NaCl). Bound proteins were eluted by heating the resin in 40 µl of 1X SDS-PAGE sample buffer and analyzed by SDS-PAGE.
Antibody staining in transgenic worms
Transgenic worms that expressed CNI-1::GFP in the AVA neurons were immunolabeled as previously described (Gottschalk and Schafer, 2006; Zheng et al., 2004). Briefly, anti-GFP polyclonal sera (Molecular Probes) was diluted (1:1000) in injection buffer and injected into the pseudocoelome of transgenic worms. Worms were allowed to recover for two hours before imaging.
Endo H digestion and western blots
Mixed-stage worms were washed off plates with M9 buffer. After an additional wash in M9, excess buffer was removed and worms were resuspended in 2 volumes of protein sample buffer and frozen in liquid nitrogen. Samples were heated at 95 °C for 10 min and the resulting lysates were diluted in reaction buffer for either Endo H or PNGase F (as per protocol, New England BioLabs) and digested with 250 units for 30 min at 37 °C. Products were run on 7% TGX precast gels (Bio-Rad) and transferred to nitrocellulose membranes. Blots were incubated with monoclonal anti-GFP antibodies (Santa Cruz Biotechnology) in Odyssey Blocking Buffer (LI-COR). Bands were detected with IRDye 800CW on the Odyssey Imager (LI-COR). Endo H sensitivity was quantified in ImageJ.
Statistical analysis
The results were analyzed using an unpaired Student’s T-test.
Supplementary Material
Acknowledgements
We thank members of the Maricq laboratory and Marcus Babst for comments on the manuscript, Craig Walker for preliminary analysis of the cni-1 mutant, Aleksander Maricq for assistance writing the MATLAB script, Linda Hauth for generating transgenic strains, and the Caenorhabditis Genetics Center (funded by the National Institutes of Health [NIH]) for providing worm strains. This research was made possible by support from NIH Grant NS35812 (A.V.M.) and MH077939 (S.T.). T.Y. is supported by the Program for Young Researcher Overseas Visits, Graduate School of Pharmaceutical Science, The University of Tokyo.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altier C, Garcia-Caballero A, Simms B, You H, Chen L, Walcher J, Tedford HW, Hermosilla T, Zamponi GW. The Cavbeta subunit prevents RFP2-mediated ubiquitination and proteasomal degradation of L-type channels. Nat Neurosci. 2011;14:173–180. doi: 10.1038/nn.2712. [DOI] [PubMed] [Google Scholar]
- Anggono V, Huganir RL. Regulation of AMPA receptor trafficking and synaptic plasticity. Curr Opin Neurobiol. 2012;22:461–469. doi: 10.1016/j.conb.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokel C, Dass S, Wilsch-Brauninger M, Roth S. Drosophila Cornichon acts as cargo receptor for ER export of the TGFalpha-like growth factor Gurken. Development. 2006;133:459–470. doi: 10.1242/dev.02219. [DOI] [PubMed] [Google Scholar]
- Brockie PJ, Madsen DM, Zheng Y, Mellem J, Maricq AV. Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC-42. J Neurosci. 2001a;21:1510–1522. doi: 10.1523/JNEUROSCI.21-05-01510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockie PJ, Maricq AV. Building a synapse: genetic analysis of glutamatergic neurotransmission. Biochem Soc Trans. 2006;34:64–67. doi: 10.1042/BST0340064. [DOI] [PubMed] [Google Scholar]
- Brockie PJ, Mellem JE, Hills T, Madsen DM, Maricq AV. The C. elegans glutamate receptor subunit NMR-1 is required for slow NMDA-activated currents that regulate reversal frequency during locomotion. Neuron. 2001b;31:617–630. doi: 10.1016/s0896-6273(01)00394-4. [DOI] [PubMed] [Google Scholar]
- Castro CP, Piscopo D, Nakagawa T, Derynck R. Cornichon regulates transport and secretion of TGFalpha-related proteins in metazoan cells. Journal of cell science. 2007;120:2454–2466. doi: 10.1242/jcs.004200. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Sulston JE, White JG, Southgate E, Thomson JN, Brenner S. The neural circuit for touch sensitivity in Caenorhabditis elegans. J Neurosci. 1985;5:956–964. doi: 10.1523/JNEUROSCI.05-04-00956.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun DK, McEwen JM, Burbea M, Kaplan JM. UNC-108/Rab2 regulates postendocytic trafficking in Caenorhabditis elegans. Mol Biol Cell. 2008;19:2682–2695. doi: 10.1091/mbc.E07-11-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs ID, Soto D, Zonouzi M, Renzi M, Shelley C, Farrant M, Cull-Candy SG. Cornichons modify channel properties of recombinant and glial AMPA receptors. J Neurosci. 2012;32:9796–9804. doi: 10.1523/JNEUROSCI.0345-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis GW. Homeostatic control of neural activity: from phenomenology to molecular design. Annu Rev Neurosci. 2006;29:307–323. doi: 10.1146/annurev.neuro.28.061604.135751. [DOI] [PubMed] [Google Scholar]
- de Bono M, Maricq AV. Neuronal substrates of complex behaviors in C. elegans. Annu Rev Neurosci. 2005;28:451–501. doi: 10.1146/annurev.neuro.27.070203.144259. [DOI] [PubMed] [Google Scholar]
- Feinberg EH, Vanhoven MK, Bendesky A, Wang G, Fetter RD, Shen K, Bargmann CI. GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57:353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nature genetics. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill MB, Kato AS, Roberts MF, Yu H, Wang H, Tomita S, Bredt DS. Cornichon-2 modulates AMPA receptor-transmembrane AMPA receptor regulatory protein assembly to dictate gating and pharmacology. J Neurosci. 2011;31:6928–6938. doi: 10.1523/JNEUROSCI.6271-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill MB, Kato AS, Wang H, Bredt DS. AMPA receptor modulation by cornichon-2 dictated by transmembrane AMPA receptor regulatory protein isoform. Eur J Neurosci. 2012;35:182–194. doi: 10.1111/j.1460-9568.2011.07948.x. [DOI] [PubMed] [Google Scholar]
- Goold CP, Nicoll RA. Single-cell optogenetic excitation drives homeostatic synaptic depression. Neuron. 2010;68:512–528. doi: 10.1016/j.neuron.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A, Schafer WR. Visualization of integral and peripheral cell surface proteins in live Caenorhabditis elegans. J Neurosci Methods. 2006;154:68–79. doi: 10.1016/j.jneumeth.2005.11.016. [DOI] [PubMed] [Google Scholar]
- Harmel N, Cokic B, Zolles G, Berkefeld H, Mauric V, Fakler B, Stein V, Klocker N. AMPA receptors commandeer an ancient cargo exporter for use as an auxiliary subunit for signaling. PLoS One. 2012;7:e30681. doi: 10.1371/journal.pone.0030681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AC, Sims S, Kaplan JM. Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature. 1995;378:82–85. doi: 10.1038/378082a0. [DOI] [PubMed] [Google Scholar]
- Herring BE, Shi Y, Suh YH, Zheng CY, Blankenship SM, Roche KW, Nicoll RA. Cornichon proteins determine the subunit composition of synaptic AMPA receptors. Neuron. 2013;77:1083–1096. doi: 10.1016/j.neuron.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig Y, Sharpe HJ, Elbaz Y, Munro S, Schuldiner M. A systematic approach to pair secretory cargo receptors with their cargo suggests a mechanism for cargo selection by Erv14. PLoS Biol. 2012;10:e1001329. doi: 10.1371/journal.pbio.1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hills T, Brockie PJ, Maricq AV. Dopamine and glutamate control area-restricted search behavior in Caenorhabditis elegans. J Neurosci. 2004;24:1217–1225. doi: 10.1523/JNEUROSCI.1569-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AC, Nicoll RA. The expanding social network of ionotropic glutamate receptors: TARPs and other transmembrane auxiliary subunits. Neuron. 2011;70:178–199. doi: 10.1016/j.neuron.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M, Hoerndli FJ, Brockie PJ, Wang R, Johnson E, Maxfield D, Francis MM, Madsen DM, Maricq AV. Wnt signaling regulates acetylcholine receptor translocation and synaptic plasticity in the adult nervous system. Cell. 2012;149:173–187. doi: 10.1016/j.cell.2011.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato AS, Gill MB, Ho MT, Yu H, Tu Y, Siuda ER, Wang H, Qian YW, Nisenbaum ES, Tomita S, et al. Hippocampal AMPA receptor gating controlled by both TARP and cornichon proteins. Neuron. 2010;68:1082–1096. doi: 10.1016/j.neuron.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerchner GA, Nicoll RA. Silent synapses and the emergence of a postsynaptic mechanism for LTP. Nat Rev Neurosci. 2008;9:813–825. doi: 10.1038/nrn2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessels HW, Malinow R. Synaptic AMPA receptor plasticity and behavior. Neuron. 2009;61:340–350. doi: 10.1016/j.neuron.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber A, Hrastnik C, Daum G. Phospholipid-synthesizing enzymes in Golgi membranes of the yeast, Saccharomyces cerevisiae. FEBS letters. 1995;377:271–274. doi: 10.1016/0014-5793(95)01361-x. [DOI] [PubMed] [Google Scholar]
- Li Y, Tsien RW. pHTomato, a red, genetically encoded indicator that enables multiplex interrogation of synaptic activity. Nat Neurosci. 2012;15:1047–1053. doi: 10.1038/nn.3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofke C, Ischebeck T, Konig S, Freitag S, Heilmann I. Alternative metabolic fates of phosphatidylinositol produced by phosphatidylinositol synthase isoforms in Arabidopsis thaliana. The Biochemical journal. 2008;413:115–124. doi: 10.1042/BJ20071371. [DOI] [PubMed] [Google Scholar]
- Lu W, Roche KW. Posttranslational regulation of AMPA receptor trafficking and function. Curr Opin Neurobiol. 2012;22:470–479. doi: 10.1016/j.conb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricq AV, Peckol E, Driscoll M, Bargmann CI. Mechanosensory signalling in C. elegans mediated by the GLR-1 glutamate receptor. Nature. 1995;378:78–81. doi: 10.1038/378078a0. [DOI] [PubMed] [Google Scholar]
- Mellem JE, Brockie PJ, Zheng Y, Madsen DM, Maricq AV. Decoding of Polymodal Sensory Stimuli by Postsynaptic Glutamate Receptors in C. elegans. Neuron. 2002;36:933–944. doi: 10.1016/s0896-6273(02)01088-7. [DOI] [PubMed] [Google Scholar]
- Miesenbock G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- Morimoto-Tomita M, Zhang W, Straub C, Cho CH, Kim KS, Howe JR, Tomita S. Autoinactivation of neuronal AMPA receptors via glutamate-regulated TARP interaction. Neuron. 2009;61:101–112. doi: 10.1016/j.neuron.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsborn AM, Li W, McEwen TJ, Mizuno T, Kuzmin E, Matsumoto K, Bennett KL. GLH-1, the C. elegans P granule protein, is controlled by the JNK KGB-1 and by the COP9 subunit CSN-5. Development. 2007;134:3383–3392. doi: 10.1242/dev.005181. [DOI] [PubMed] [Google Scholar]
- Partin KM, Patneau DK, Winters CA, Mayer ML, Buonanno A. Selective modulation of desensitization at AMPA versus kainate receptors by cyclothiazide and concanavalin A. Neuron. 1993;11:1069–1082. doi: 10.1016/0896-6273(93)90220-l. [DOI] [PubMed] [Google Scholar]
- Piggott BJ, Liu J, Feng Z, Wescott SA, Xu XZ. The neural circuits and synaptic mechanisms underlying motor initiation in C. elegans. Cell. 2011;147:922–933. doi: 10.1016/j.cell.2011.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls MM, Hall DH, Victor M, Stelzer EH, Rapoport TA. Targeting of rough endoplasmic reticulum membrane proteins and ribosomes in invertebrate neurons. Mol Biol Cell. 2002;13:1778–1791. doi: 10.1091/mbc.01-10-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S, Neuman-Silberberg FS, Barcelo G, Schupbach T. cornichon and the EGF receptor signaling process are necessary for both anterior-posterior and dorsal-ventral pattern formation in Drosophila. Cell. 1995;81:967–978. doi: 10.1016/0092-8674(95)90016-0. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Harmel N, Brechet A, Zolles G, Berkefeld H, Muller CS, Bildl W, Baehrens D, Huber B, Kulik A, et al. High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron. 2012;74:621–633. doi: 10.1016/j.neuron.2012.03.034. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Harmel N, Zolles G, Bildl W, Kulik A, Heimrich B, Chisaka O, Jonas P, Schulte U, Fakler B, et al. Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science. 2009;323:1313–1319. doi: 10.1126/science.1167852. [DOI] [PubMed] [Google Scholar]
- Shanks NF, Savas JN, Maruo T, Cais O, Hirao A, Oe S, Ghosh A, Noda Y, Greger IH, Yates JR, 3rd, et al. Differences in AMPA and Kainate Receptor Interactomes Facilitate Identification of AMPA Receptor Auxiliary Subunit GSG1L. Cell reports. 2012;1:590–598. doi: 10.1016/j.celrep.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Suh YH, Milstein AD, Isozaki K, Schmid SM, Roche KW, Nicoll RA. Functional comparison of the effects of TARPs and cornichons on AMPA receptor trafficking and gating. Proc Natl Acad Sci U S A. 2010;107:16315–16319. doi: 10.1073/pnas.1011706107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R, Pantoja R, Moss FJ, Mackey ED, Son CD, Miwa J, Lester HA. Nicotine up-regulates alpha4beta2 nicotinic receptors and ER exit sites via stoichiometry-dependent chaperoning. The Journal of general physiology. 2011;137:59–79. doi: 10.1085/jgp.201010532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano GG. The self-tuning neuron: synaptic scaling of excitatory synapses. Cell. 2008;135:422–435. doi: 10.1016/j.cell.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Engelhardt J, Mack V, Sprengel R, Kavenstock N, Li KW, Stern-Bach Y, Smit AB, Seeburg PH, Monyer H. CKAMP44: a brain-specific protein attenuating short-term synaptic plasticity in the dentate gyrus. Science. 2010;327:1518–1522. doi: 10.1126/science.1184178. [DOI] [PubMed] [Google Scholar]
- Walker CS, Brockie PJ, Madsen DM, Francis MM, Zheng Y, Koduri S, Mellem JE, Strutz-Seebohm N, Maricq AV. Reconstitution of invertebrate glutamate receptor function depends on stargazin-like proteins. Proc Natl Acad Sci U S A. 2006a;103:10781–10786. doi: 10.1073/pnas.0604482103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CS, Francis MM, Brockie PJ, Madsen DM, Zheng Y, Maricq AV. Conserved SOL-1 proteins regulate ionotropic glutamate receptor desensitization. Proc Natl Acad Sci U S A. 2006b;103:10787–10792. doi: 10.1073/pnas.0604520103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Mellem JE, Jensen M, Brockie PJ, Walker CS, Hoerndli FJ, Hauth L, Madsen DM, Maricq AV. The SOL-2/Neto Auxiliary Protein Modulates the Function of AMPA-Subtype Ionotropic Glutamate Receptors. Neuron. 2012;75:838–850. doi: 10.1016/j.neuron.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Walker CS, Brockie PJ, Francis MM, Mellem JE, Madsen DM, Maricq AV. Evolutionary conserved role for TARPs in the gating of glutamate receptors and tuning of synaptic function. Neuron. 2008;59:997–1008. doi: 10.1016/j.neuron.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A, Vissel B. The essential role of AMPA receptor GluR2 subunit RNA editing in the normal and diseased brain. Front Mol Neurosci. 2012;5:34. doi: 10.3389/fnmol.2012.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Tomita S. Defined criteria for auxiliary subunits of glutamate receptors. J Physiol. 2012;590:21–31. doi: 10.1113/jphysiol.2011.213868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Isack NR, Glodowski DR, Liu J, Chen CC, Xu XZ, Grant BD, Rongo C. RAB-6.2 and the retromer regulate glutamate receptor recycling through a retrograde pathway. J Cell Biol. 2012;196:85–101. doi: 10.1083/jcb.201104141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Brockie PJ, Mellem JE, Madsen DM, Maricq AV. Neuronal Control of Locomotion in C. elegans is Modified by a Dominant Mutation in the GLR-1 Ionotropic Glutamate Receptor. Neuron. 1999;24:347–361. doi: 10.1016/s0896-6273(00)80849-1. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Brockie PJ, Mellem JE, Madsen DM, Walker CS, Francis MM, Maricq AV. SOL-1 is an auxiliary subunit that modulates the gating of GLR-1 glutamate receptors in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2006;103:1100–1105. doi: 10.1073/pnas.0504612103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Mellem JE, Brockie PJ, Madsen DM, Maricq AV. SOL-1 is a CUB-domain protein required for GLR-1 glutamate receptor function in C. elegans. Nature. 2004;427:451–457. doi: 10.1038/nature02244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.