Abstract
Aggregation of monoclonal antibodies is often a multi-step process involving structural alterations in monomeric proteins and subsequent formation of soluble or insoluble oligomers. The role of local conformational stability and dynamics of native and/or partially altered structures in determining the aggregation propensity of monoclonal antibodies, however, is not well understood. Here, we investigate the role of conformational stability and dynamics of regions with distinct solvent exposure in determining the aggregation propensity of an IgG1 and IgG2 monoclonal antibody. The temperatures employed span the pre-unfolding range (10–40°C) and the onset temperatures (Tonset) for exposure of apolar residues (∼50°C), alterations in secondary structures (∼60°C) and initiation of visible aggregate formation (∼60°C). Solvent-exposed regions were found to precede solvent-shielded regions in an initiation of aggregation for both proteins. Such a process was observed upon alterations in overall tertiary structure while retaining the secondary structures in both the proteins. In addition, a greater dynamic nature of solvent-shielded regions in potential intermediates of IgG1 and the improved conformational stability increased its resistance to aggregation when compared to IgG2. These results suggest that local conformational stability and fluctuations of partially altered structures can influence the aggregation propensity of immunoglobulins.
Keywords: monoclonal antibody, conformational stability, dynamics, aggregation, fluorescence spectroscopy, circular dichroism, differential scanning calorimetry, light scattering
Introduction
The native state conformational stability of proteins is governed by a combination of intra- and inter-molecular covalent and non-covalent interactions.1,2 The interdependent nature of these interactions often results in highly cooperative unfolding of proteins. The unfolding of larger multi-domain proteins, such as natural immunoglobulins or recombinant monoclonal antibodies (mAbs), however, is accompanied by non-cooperative processes resulting from formation of intermediates.3,4 The population, relative stability, dynamics, and native/non-native interactions of such partly structured intermediates not only reflect the stability of subdomains,5–7 but may also be related to exposure of aggregation prone regions.8,9
Native proteins are commonly kinetically and thermodynamically resistant to local unfolding of aggregation-prone regions10,11, which maintains their chemical, biochemical, and conformational stability. The aggregation-prone region(s) in a three-dimensional native structure of immunoglobulins can exist in regions with different degrees of solvent exposure. Such regions in immunoglobulins can manifest differences in their conformational stability12–14 and dynamic15–19 properties in response to changes in temperature and the presence of stabilizing or destabilizing cosolutes.19 These variables have the potential to induce partially folded intermediate structures, which are less stable than the native state and more prone to aggregation. For many proteins, such partially folded intermediates are known to proceed via competing pathways to retain elements of the native state and can lead to the formation of native/non-native aggregates.20–29 Higher resolution analytical and computational methods such as phi-analysis,30 small-angle X-ray scattering (SAXS)31 and nuclear magnetic resonance (NMR)32 coupled with molecular dynamics (MD) simulations33 have been used to provide detailed structural information concerning native states as well as partially altered intermediate conformations of many proteins. Experimental limitations such as the size of proteins, high material requirements, and low throughput, however, limit their application to routine analysis of such structures in large proteins such as mAbs. Limited studies characterizing such aggregation prone intermediates of immunoglobulins and/or their fragments have been reported.34–36
Recently, the role of intramolecular dynamics of regions with distinct solvent exposures in modulating global conformational stability was investigated using an IgG1 monoclonal antibody.19 A fluorescence spectroscopy based approach employing red-edge excitation (REE) and fluorescence quenching was employed in these studies. REE, an approach in which the excitation wavelength was increased towards the red edge of the absorption band, was used to sample and monitor a fraction of tryptophan (Trp) residues in environments that were increasingly exposed to solvent. A wide range of temperatures at relatively short intervals (2.5°C) were studied to evaluate changes in local conformational stability with increasing temperature. Fluorescence quenching studies, used to probe protein matrix dynamics, were performed at selected temperatures including a pre-unfolding temperature range, an onset temperature (Tonset), and at the apparent thermal melting temperature (TMA) to obtain a snapshot of local dynamics of the antibody as a function of temperature.19 The effect of sucrose or arginine as representative stabilizing and destabilizing excipients,18,37 respectively, were also evaluated under these conditions.19
The current study is aimed at (1) characterizing and comparing the conformational stability and dynamic properties of regions with distinct solvent exposure at various temperatures using two additional monoclonal antibodies (an IgG1 and IgG2) and (2) understanding the relevance of local conformational stability and dynamics to the aggregation propensity of representative IgG1 and IgG2 monoclonal antibodies.
Experimental
Materials
The purified IgG1 and IgG2 monoclonal antibody (mAb) samples, both containing variable domains that bind to Streptavidin, were supplied by Amgen Inc. (Seattle, WA) and were stored in the formulation buffer at 4°C. The mAb solutions used for characterization were prepared by dialyzing the protein stock into 20 mM citrate-phosphate buffer at pH 5 (I = 0.1, adjusted using NaCl). All buffer components and chemicals were purchased from Sigma (St. Louis, MI).
Methods
Steady-state intrinsic (Trp) fluorescence and acrylamide quenching upon red-edge excitation
The detailed methodology for these measurements is described elsewhere.19 Briefly, intrinsic Trp fluorescence spectra were acquired using a peltier controlled PTI Quanta Master Spectrophotometer (Lawrenceville, NJ) using 0.1 mg/mL (∼6.7 × 10−4 mM) of mAb (IgG1 and IgG2) over a temperature range of 10–40°C (10°C intervals) and 40–90°C (5°C intervals). An equilibration time of 3 min was used at each temperature. Samples were excited from 292 to 304 nm at 3 nm intervals (to sample Trp residues from environments with different degrees of solvent exposure) and the emission spectra were collected from 310 to 380 nm. The peak position and peak intensity were determined by fitting the emission spectra using a mean spectral center of mass (MSM) method. The apparent thermal melting temperatures (TMA) were determined using a Sigmoidal-Boltzmann fit of Trp peak position versus temperature plots at each excitation wavelength. The mid-point of such fits was considered to define the TMA value. The Trp peak intensity in the presence of varying concentrations of acrylamide (0–0.5 M) was used to construct Stern-Volmer plots at all of the excitation wavelengths and temperatures employed in these studies. Any inner filter effects due to the presence of acrylamide were corrected as described earlier.19,38
Static light scattering, extrinsic (1-anilino naphthalene-8-sulphonate) fluorescence, far-UV circular dichroism and differential scanning calorimetry
Static light scattering intensity (SLS) and extrinsic (1-anilino naphthalene-8-sulphonate; ANS) fluorescence were both performed as a function of temperature (10–90°C at 2.5°C intervals) using 0.1 mg/mL of mAb using a PTI Quanta Master Spectrophotometer. The excitation and emission slit width were set at 3 nm. Furthermore, the excitation wavelength, 295 nm for SLS and 375 for ANS, and emission wavelengths of 280–380 for SLS and 425–575 nm for ANS experiments were used. The molar ratio of ANS:protein was maintained at 20:1. An equilibration time of 3 min was used at each temperature level prior to each measurement. Far-UV circular dichroism (CD) signals were acquired on a peltier equipped Jasco J-815 spectrometer (Tokyo, Japan) using 0.2 mg/mL protein. The CD signal was monitored at a constant wavelength of 217 nm over a temperature range of 10–90°C at 2.5°C intervals. The temperature ramp rate for CD measurements were 15°C/h. The differential scanning calorimetric (DSC) experiments were performed using a VP-capillary DSC equipped with an autosampler (MicroCal, Northampton, MA) at 1 mg/mL mAb concentration using a scan rate of 60°C/h. The thermograms were processed by MicroCal routines to determine the baseline, normalize the trace to the mAb concentration and fit the processed trace using a non-two-state unfolding model. Detailed SLS, ANS fluorescence, CD, and DSC characterization protocols are described earlier.16,18
Results
Intrinsic Trp fluorescence (peak position and fluorescence intensity) as a function of temperature and excitation wavelength
The tertiary structure stability of IgG1 and IgG2 mAbs as a function of temperature at pH 5.0 was evaluated by monitoring Trp peak position (emission maxima) shifts at various excitation wavelengths (292–304 nm at 3 nm intervals) [Fig. 1(A-D)]. Within the pre-unfolding temperature range (10 to ∼40°C), the Trp emission peak position was shifted to a higher wavelength upon red-edge excitation for both mAbs [Fig. 1(A,B)]. In addition, the fluorescence intensity was progressively lowered as the excitation wavelength was increased from 292 to 304 nm [Fig. 1(C,D)]. The higher peak position and lower fluorescence intensity at 304 versus 292 nm suggests that red-edge excitation can probe Trp present in environments with different extents of solvent exposure (i.e., 304 nm probes more solvent-exposed Trp residues while 292 nm would detect Trp residues relatively more shielded). Furthermore, the apparent thermal melting temperatures (TMA) obtained from sigmoidal fitting of Trp peak position versus temperature plots were lower for 304 nm than 292 nm excitation for both mAbs [Fig. 1(E)]. The TMA values for the IgG1, however, were higher than the IgG2 for both solvent-exposed and solvent-shielded regions, suggesting higher overall stability of the IgG1. Such unfolding behavior for regions with distinct solvent exposures as a function of temperature was reported previously for a different IgG1 monoclonal antibody.19 In addition, within the pre-unfolding temperature range, the Trp peak position shift as a function of temperature manifested a red-shifted, curvilinear profile upon excitation at 292 nm (solvent-shielded region) for both IgG1 and IgG2 [Fig. 1(A,B)]. No such curvilinear plots in the pre-unfolding region were observed for 304 nm excitation (solvent-exposed region) [Fig. 1(A,B)].
Figure 1.
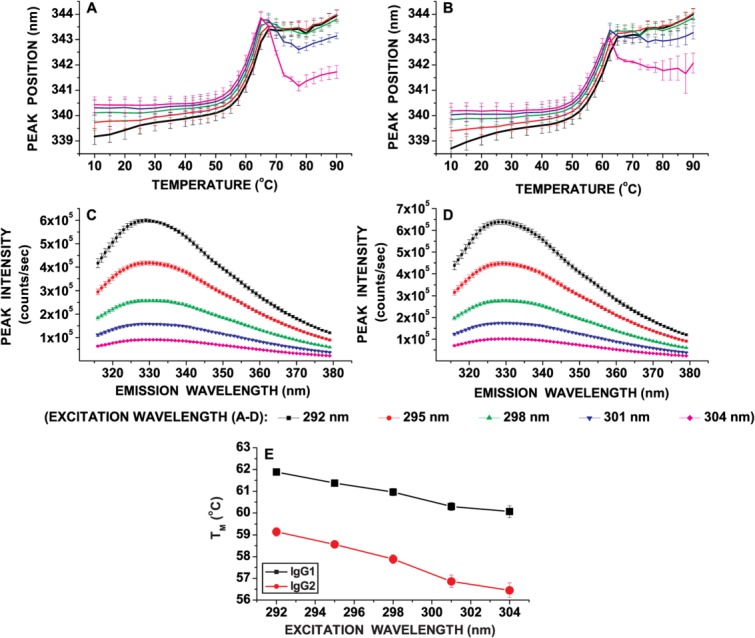
Local conformational stability of an IgG1 and IgG2 mAb as a function of temperature as measured by REE fluorescence spectroscopy. Intrinsic Trp fluorescence peak position shift as a function of temperature and excitation wavelength for (A) the IgG1 and, (B) the IgG2. Intrinsic Trp fluorescence intensity at 20°C and pH 5.0 as a function of excitation wavelength for (C) IgG1 and (D) IgG2. Sub-figure (E) shows the TMA obtained by a sigmoidal fit of peak position versus temperature plots for both the IgG1 and IgG2 mAbs as a function of excitation wavelength. Each data point represents mean and standard deviation of three (n = 3) replicates. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Static light scattering, ANS fluorescence, circular dichroism and differential scanning calorimetry as a function of temperature
The exposure of apolar patches/regions, changes in secondary structures and overall conformational stability of the IgG1 and IgG2 mAbs were determined as a function of temperature by ANS fluorescence, circular dichroism and DSC, respectively [Fig. 2(A-C)]. The aggregation propensity of the IgG1 and IgG2 as a function of temperature were then compared using static light scattering [Fig. 2(D)]. A well-defined structural transition was detected for both the IgG1 and IgG2 molecules between 50°C and 75°C with an onset temperature of ∼50°C by monitoring changes in ANS fluorescence as a function of temperature. The initial ANS fluorescence intensity in the pre-unfolding temperature range and extent of ANS fluorescence after the major transition were both greater for the IgG1 than the IgG2 [Fig. 2(A)]. Furthermore, an increase (more negative) in CD ellipticity above 60°C for both IgG1 and IgG2 indicates secondary structure transitions, which may further result in the formation of intermolecular structures [Fig. 2(B)]. No major structural changes in overall secondary structure were detected for both proteins in response to increases in temperature below 60°C using CD. A decrease in CD signal (more positive) was observed at ≥65°C for IgG2 and ≥70°C for the IgG1, suggesting differences in dissociation of the thermally induced intermolecular β-structure. The SLS data [Fig. 2(D)] does not suggest loss of aggregates from solution in this temperature range. The formation of intermolecular β-structure-rich species spanned a wider range of temperature for the IgG1 (57.5–70°C) than the IgG2 (60–65°C). The temperatures at which initiation of exposure (accessibility) of apolar regions (ANS data; ∼50°C) and alterations in secondary structure (CD data; ∼60°C) occur, however, is comparable between the two mAbs. Despite these similarities, the aggregation propensities upon thermal unfolding of the two proteins are different when monitored by static light scattering measurements [Fig. 2(D)]. The Tonset and Taggregation (from the midpoints of static light scattering intensity versus temperature plots) are greater for the IgG1 than the IgG2 [Fig. 2(C)]. In comparison, three distinct transitions were observed for the IgG1 while the IgG2 thermal unfolding proceeds via two apparent transitions as seen in the DSC thermograms [Fig. 2(C)].
Figure 2.
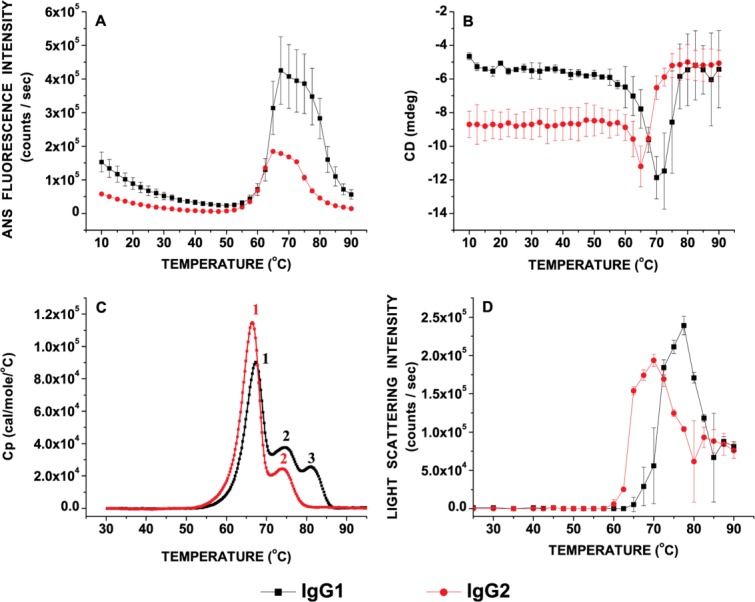
Characterization of the IgG1 and IgG2 conformational stability at pH 5.0 as a function of temperature as measured by (A) Extrinsic (ANS) fluorescence intensity, (B) Circular dichroism signal at 217 nm, (C) DSC thermograms, and (D) static light scattering intensity monitored at 295 nm. Each data point represents mean and standard deviation of three (n = 3) replicates. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Acrylamide quenching of tryptophan fluorescence as a function of excitation wavelength and temperature
Fluorescence quenching experiments are commonly used to study alterations in local environments in proteins surrounding a fluorophore of interest (Trp in this case). Stern-Volmer plots are constructed by plotting the extent of quenching of Trp fluorescence versus increasing concentrations of the quencher (here acrylamide). A standard Stern-Volmer plot usually results in a positive slope and such positive slopes were observed for all of the Stern-Volmer plots generated for the IgG1 and IgG2 (Fig. 3). Stern-Volmer plots can be employed qualitatively and quantitatively to monitor dynamics in Trp environments of proteins.19,39–41 Figure 3 presents Stern-Volmer plots for the IgG1 [Fig. 3(A-G)] and the IgG2 [Fig. 3(H–N)] at various excitation wavelengths (292–304 nm at 3 nm intervals) and temperatures (10°C, 20°C, 40°C, 50°C, 60°C, 70°C, 80°C). These span a wide range of temperatures and structural alterations in the IgG1 and IgG2 over the pre-unfolding temperature range (for instance, 10°C and 20°C), Tonset (40°C, 50°C, and 60°C) and TMA (60°C, 70°C, and 80°C). The Tonset and TMA values are temperature points at which tertiary structural changes (intrinsic Trp fluorescence and ANS data), alterations in secondary structure (circular dichroism) and formation of aggregates (static light scattering) were detected for both the IgG1 and IgG2 as described in the preceding section. The wide range of temperatures employed should aid in evaluating the characteristics of fluctuations in structure that are formed and stable at individual temperatures. The exact nature of these structures, however, cannot be defined using the lower resolution techniques employed here.
Figure 3.
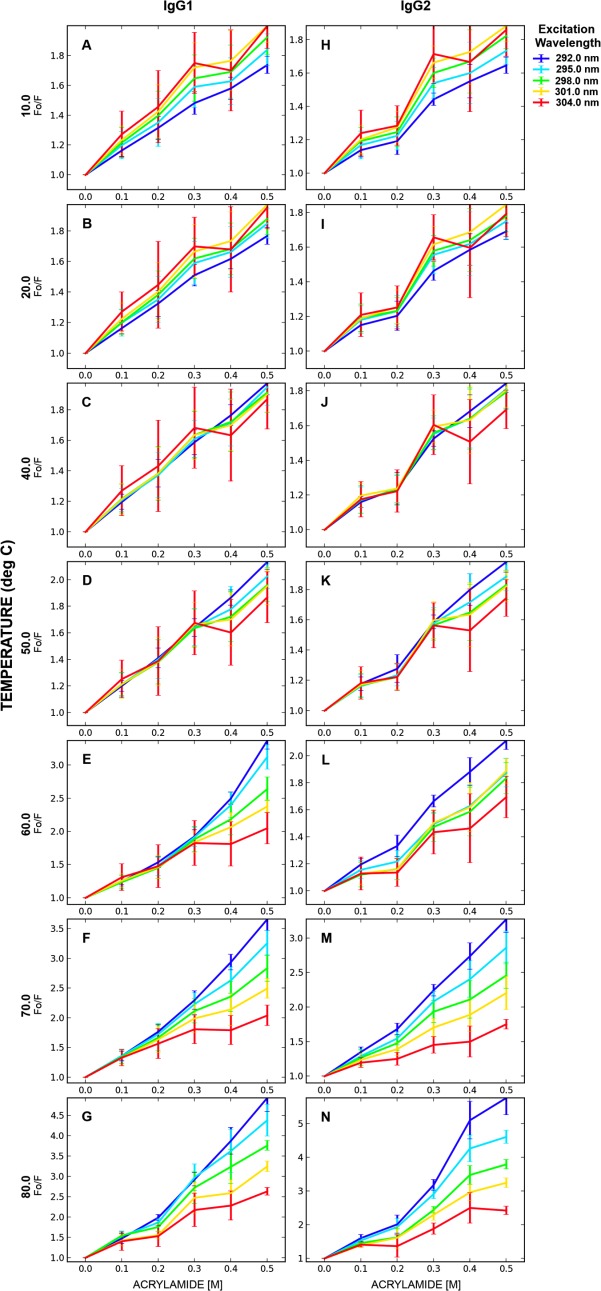
Stern-Volmer plots for the IgG1 and IgG2 mAbs as a function of excitation wavelengths at different temperatures. The line connecting the data points are not mathematical fits but simply connect the mean of the experimental data points and are used for visual aid purposes only. Samples were at 0.1 mg/mL in 20 mM citrate-phosphate buffer, pH 5.0. Each data point represents mean and standard deviation of three (n = 3) replicates. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
In the pre-unfolding temperature range (for instance, 10°C and 20°C), it was found that the slope of Stern-Volmer plots were greater as the excitation wavelength was increased from 292 to 304 nm [Fig. 3(A,B,H, and I)] for both proteins. No major differences were observed between the two mAb's quenching patterns as a function of excitation wavelength in the pre-unfolding temperature range (10°C and 20°C). At 40°C [representing the Tonset using Trp peak position versus temperature data, Fig. 1(A,B)] and 50°C [representing Tonset from ANS fluorescence vs. temperature data, Fig. 2(A)], tertiary structure perturbations were detected using static (time-averaged) techniques as discussed earlier. At these temperatures, the magnitude of acrylamide quenching for the solvent-exposed regions (304 nm excitation) is reduced relative to the solvent-shielded regions (292 nm excitation) for both the IgG1 [Fig. 3(C,D)] and IgG2 [Fig. 3(J,K)]. These effects can be detected when higher acrylamide concentrations are used in the experiments, the potential reason for which is discussed later. The magnitude of quenching, and hence the dynamic characteristics, of solvent-shielded regions (292 nm excitation) are subtly increased at the onset temperature of tertiary structure alterations (40°C or 50°C). Above 50°C, notably at 60°C (Tonset from CD data), however, the magnitude of quenching for solvent-shielded regions (292 nm excitation) was found to increase considerably compared to 40°C or 50°C [Fig. 3(E,L)] for both mAbs, albeit to different extents. This temperature (60°C) represents the initiation of secondary structure perturbations. No major change was observed in the dynamics of solvent-exposed (304 nm excitation) regions between 40°C and 60°C. The dynamics/accessibility of solvent-shielded regions (292 nm excitation) continues to increase as the temperature is raised above 60°C [Fig. 3(F,G,M,N)].
The effect of temperature on the dynamics/accessibility of solvent-exposed and solvent-shielded regions for the IgG1 and IgG2 are plotted separately in Figure 4 for better representation of the data. It was observed that as the temperature increases from 10°C to 55°C, the local dynamics of solvent-shielded indole sidechains (probed by 292 nm excitation) were progressively increased for both mAbs [Fig. 4(A,C)], as represented by increases in magnitude of quenching with increases in temperature. In contrast, the solvent-exposed regions (304 nm excitation) were found to decrease in their accessibility as indicated by a progressively lower magnitude of acrylamide quenching with increases in temperature from 10°C to 45°C [Fig. 4(B,D); compare Supporting Information Fig. S1 with S6]. Supporting Information Figures S1–S6 can aid in trend analysis of each of the triplicate individual runs in Figure 4 separately as a function of excitation wavelengths. These results suggests that the solvent-exposed and solvent-shielded regions in both the IgG1 and IgG2 respond differently to changes in temperature in the range where major aggregation was not detected by light scattering (10–55°C).
Figure 4.

Effect of temperature on the acrylamide quenching profiles for the IgG1 and IgG2 plotted at 292 nm (A and C) and 304 nm (B and D) excitation wavelengths. The lines are drawn for visual aid purposes only. Samples were at 0.1 mg/mL in 20 mM citrate-phosphate buffer, pH 5.0. Each data point represents mean and standard deviation of three (n = 3) replicates. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Since the IgG1 and IgG2 molecules were determined to have different heat-induced aggregation propensities, the dynamic nature and accessibility of different solvent-exposed structures formed in the temperature range between 55°C and 75°C (at 5°C intervals) were also studied using the acrylamide quenching approach. The corresponding Stern-Volmer plots are compared in Figure 5. This temperature range represents different stages of unfolding and/or initiation of aggregation processes for both proteins. At 55°C [before the Tonset of aggregation as determined by static light scattering, Fig. 2(D)], the acrylamide quenching patterns were comparable for both mAbs at all excitation wavelengths employed [Fig. 5(A–E)]. At 60°C, however, the magnitude of the positive slope in the SV plot was greater for IgG1 than IgG2 [Fig. 5(F–J)], especially for the solvent-shielded regions (292 nm excitation) [Fig. 5(F)]. This suggests a greater dynamic nature of such regions in IgG1 than in IgG2 at this temperature. This is also the temperature at which light scattering data indicates initiation of significant aggregation for IgG2 but not IgG1 [Fig. 2(D)]. As the temperature was raised from 60°C to 65°C (the Tonset for IgG1 aggregation), the dynamics of more solvent-shielded regions (292, 295, 298 nm excitation) in the IgG1 was dampened relative to IgG2 as indicated by the lower magnitude of the acrylamide quenching profile [Fig. 5(F–H) compared to Fig. 5(K–M)]. These effects are less pronounced for solvent-exposed regions [Fig. 5(I-J, N-O)]. At 75°C, the magnitude of quenching is significantly greater than that at 65°C for both the IgG1 and IgG2 in all of the regions monitored by the varying excitation wavelengths [Fig. 5(U–Y)]. This may be due to a greater number of exposed indole sidechains upon extensive aggregation, as suggested by the light scattering intensity at 75°C [Fig. 2(D)].
Figure 5.
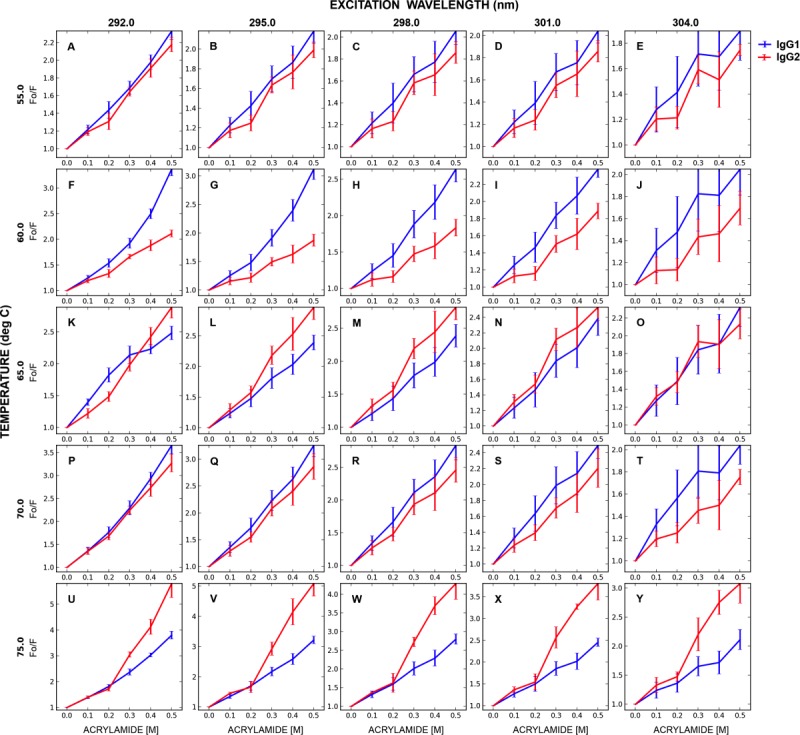
Stern-Volmer plots comparing the IgG1 versus the IgG2 at various excitation wavelength and temperatures (°C). The lines are for visual aid only. Samples were at 0.1 mg/mL in 20 mM citrate-phosphate buffer, pH 5.0. Each data point represents mean and standard deviation of three (n = 3) replicates. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Discussion
In complex, multi-domain proteins such as immunoglobulins, many different intermediate conformations can be populated, which possess different kinetic and thermodynamic stabilities. In addition to known experimental limitations of higher resolution techniques such as NMR and SAXS, the short lifetime and the dynamic nature of energetically similar conformational microstates within a native state ensemble (and/or partially altered structures) further limit their comprehensive characterization over a variety of solution conditions. Therefore, a complementary approach using lower resolution techniques (Table I) was employed to better understand the contributions of regions with distinct solvent exposure to the overall stability of natively folded and/or partially altered structures of an IgG1 and IgG2 monoclonal antibody. Temperature was used to accelerate the generation of detectable intermediate species.
Table I.
Structural Transitions Observed by Various Biophysical Techniques Employed in This Study
| Sr. no. | Technique | Type(s) of structural transitions monitored |
|---|---|---|
| 1 | Fluorescence spectroscopy | Tertiary structure stability by monitoring Trp peak position shifts as a function of an experimental variable(s) |
| 2 | Red-edge excitation (REE) | Distinct solvent-exposed regions by monitoring Trp peak position shifts as a function of excitation wavelength and an experimental variable(s) |
| 3 | ANS binding | Exposure of apolar regions/patches in proteins by measuring alterations in ANS fluorescence |
| 4 | Circular dichroism spectroscopy | Alterations in secondary structures by monitoring molar ellipticity |
| 5 | Differential scanning calorimetry | Overall conformational (thermal) stability by monitoring heat absorbed or released during transitions/structural alterations |
| 6 | Static light scattering | Aggregation propensity (time-averaged measurement) by monitoring changes light scattering intensity |
| 7 | Stern-Volmer analysis | Structural alterations in local environments of fluorophore; qualitative and quantitative analysis of slopes of plots constructed by plotting extent of quenching versus quencher concentration |
Local conformational stability of native and partially altered structures
Higher peak positions and lower fluorescence quantum yields upon red-edge excitation [Fig. 1(A-D)] suggest that Trp containing regions with different degrees of solvent-exposure can be probed for the IgG1 and IgG2 used in this study. The stability of the tertiary structure within different regions with distinct indole solvent exposure in both mAbs can therefore be probed by monitoring changes in Trp peak position with changes in temperature. In the pre-unfolding temperature range from 10°C to ∼40°C (before any detectable unfolding), no major differences were detected in the unfolding pattern based on Trp peak position shifts. For both mAbs, however, the solvent-shielded regions (292 nm excitation data) were found to have a subtle red-shifted curvilinear shape in peak position shift versus temperature plots in the pre-unfolding temperature range [Fig. 1(A,B)]. Such a red-shift was not observed in solvent-exposed regions (304 nm excitation) with increases in temperature. This response (red-shift in Trp peak position) to temperature in the solvent-shielded regions of the IgG1 and IgG2 is in contrast (blue-shifted Trp peak position) to that reported for a different IgG1 studied earlier.19 Increases in apolar interactions with temperature in the low temperature regime were thought to be responsible for the blue-shifted peak position reported previously.19 The small red-shifts observed here suggest that even in the pre-unfolding temperature range, the local disorder of solvent-shielded regions may increase. Such an effect can give rise to an ensemble of temperature induced alterations in the native structure. It is possible that these structures are different in their local conformational stability and/or dynamic behavior especially in their solvent-shielded regions. The overall secondary structure of the IgG1 and IgG2, however, was not perturbed in this temperature range as suggested by circular dichroism [Fig. 2(B)]. Overall, these results suggest that in the pre-unfolding temperature range, the tertiary structure stability of solvent-shielded regions in these antibodies is more sensitive to changes in temperature compared to solvent-exposed residues.
Upon increases in temperature from 40°C to 55°C (and above), an initiation in the overall exposure of apolar regions in both mAbs was observed as suggested by intrinsic Trp fluorescence [Fig. 1(A,B)] and ANS studies [Fig. 2(A)]. This suggests major perturbations in tertiary structure stability in all regions (solvent-exposed and solvent-shielded). No change in the secondary structure, however, was observed for either mAb. In contrast, upon major conformational alterations, the solvent-exposed regions (excitation at 304 nm) in both mAbs were found to be less stable compared to solvent-shielded regions (292 nm excitation) as shown in Figure 1(E). Such changes in tertiary structure stability accompanied by an overall increase in the exposure of apolar regions with no significant change in secondary structure is indicative of formation of molten globule-like intermediates. Upon further increases in temperature to 60°C, alterations in secondary structure were observed for both the IgG1 and IgG2 [Fig. 2(B)]. Initiation of aggregation, however, was only observed for the IgG2 [Fig. 2(D)] at this temperature. In addition, formation of intermolecular β-structure-rich species was observed for both mAbs by CD analysis and the IgG1 transition spanned a wider range of temperatures compared to the IgG2. In addition, DSC studies also indicate the presence of an additional transition for the IgG1 in this temperature range compared to the IgG2 [Fig. 2(C)]. It is therefore possible that the conformational stability and/or dynamics (and hence the accessibility) of different regions in structures formed between 55°C and 65°C may determine the distinct aggregation propensity of the IgG1. To test this hypothesis, acrylamide quenching studies accompanied by red-edge excitation measurements were performed over the temperature range of 55–75°C. The goal here is to better understand the relative contributions of dynamics and accessibility of regions with distinct solvent exposures in the IgG1 and IgG2 and their relevance to differences in their aggregation behavior.
Effect of temperature on the local dynamic properties of regions with distinct solvent-exposures
The local dynamic properties of regions with distinct solvent-exposures were probed using acrylamide quenching of Trp fluorescence. Acrylamide is a neutral quencher that can diffuse into and through a protein complex and quench indole fluorescence. This approach can probe the relative accessibility of Trp residues, which in turn depends upon the dynamic nature of the protein's matrix and local fluorophore environment. The magnitude of quenching of Trp fluorescence can therefore be related to the dynamics of the protein, and when coupled with red-edge excitation, should enable the monitoring of different solvent-exposed regions. The deviation from linearity of Stern-Volmer plots towards the x-axis (Fig. 3) suggests the expected heterogeneous distribution of Trp residues in immunoglobulins. Quantitative parameters such as Stern-Volmer constants and/or bimolecular quenching constants cannot be reliably determined from such non-linear plots. Positive slopes in Stern-Volmer plots, however, can still be used as a qualitative indicator of relative dynamic behavior.
In the pre-unfolding temperature range, the solvent-exposed regions (monitored by excitation at 304 nm) in both mAbs were found to be more dynamic relative to solvent-shielded (probed by 292 nm excitation) regions. This is indicated by trends in Stern-Volmer plots as a function of excitation wavelength, in which a greater (more positive) slope was observed for solvent-exposed regions (304 nm data) when compared to solvent-shielded regions at 10°C and 20°C [Fig. 3(A,B,H,I)]. The Trp peak position data at 10°C and 20°C [Fig. 1(A,B)] also suggests that Trp residues in solvent-shielded regions are relatively more buried compared to higher temperatures within the pre-unfolding temperature range. Above 40°C (or 50°C), the progressive decrease in the magnitude of quenching observed primarily for surface-exposed regions [304 nm data, Fig. 3(C,D,J,K)] suggests that the surface dynamics is dampened for both mAbs with increases in temperature above 40°C. The decrease in magnitude of quenching for surface-exposed regions suggests that Trp residues (and apolar residues in this region) are progressively buried (shielded from solvent) in structures that are formed at temperatures >40°C. Further changes in the magnitude of quenching (and hence dynamics) was not observed for solvent-exposed regions, until the mAbs form aggregates that can be detected by static light scattering [Fig. 4(B,D)]. In contrast, a subtle increase in the magnitude of quenching was observed for solvent-shielded regions [Fig. 4(A,C)] in both the IgG1 and IgG2 molecules as the solution temperature was raised to 50°C. The need for higher acrylamide concentrations to observe these effects is presumably due to the complex (opposite) dynamic behavior of solvent-exposed versus solvent-shielded regions. It does not appear to be due to any of the known unfolding properties of acrylamide itself. The increased dynamics of solvent-shielded regions in conjunction with red-shifted curvilinear plots of Trp peak position versus temperature [Fig. 1(A,B)] support the idea that even in the pre-unfolding temperature range minor alterations in the structure of solvent-shielded regions may give rise to an ensemble of structures that may be subtly different from the native structural ensemble of the protein. Comparison of the dynamic properties of solvent-exposed and solvent-shielded regions in the temperature range up to 50°C suggests that the solvent-exposed regions may precede the solvent-shielded regions in initiation and subsequent formation of structures that are potential precursors for aggregation. For the IgG1 and IgG2 of this study, such a process is initiated in the temperature range where changes in tertiary structure were observed whereas no secondary structural alterations were detected, at least using the techniques employed in this study [Figs. 1(A,B) and 2(A,B)].
At temperature ≥60°C, the dynamics of solvent-shielded regions were considerably increased for both mAbs, as indicated by a progressively increased magnitude of fluorescence quenching upon increases in temperature [Fig. 3(E–G,L–N)]. This temperature (∼60°C) also represents the Tonset for secondary structure alterations [Fig. 2(B)]. It is possible that increases in the dynamics of solvent-shielded regions may predispose the antibody to alterations in secondary structure or vice-versa. It is interesting to note that at 60°C [Tonset of IgG2 aggregation, Fig. 2(D)], the solvent-shielded regions in the IgG1 structures thus formed were relatively more dynamic than the corresponding IgG2 structures [see, e.g., Fig. 5(F)]. Such an increased dynamic nature of IgG1 intermediates may prevent it from nucleating and subsequently aggregating when compared to its IgG2 counterpart [Fig. 5(F–H)]. At 65°C [Tonset of IgG1 aggregation, Fig. 2(C)], the dynamics of IgG1 intermediates, however, were relatively dampened compared to that at 60°C. Such a dampening effect may reflect the behavior of more solvent-shielded regions in the IgG1 involved in aggregation formation. These results suggest that the greater dynamic nature of solvent-shielded regions in aggregation-prone intermediates of IgG1 cause it to be more resistant to initiation and formation of visible aggregates when compared to that of the IgG2. At temperature >65°C, formation of higher order aggregates, as suggested by increased light scattering intensity [Fig. 2(D)], may expose Trp residues to the solvent. This can result in a greater magnitude of fluorescence quenching as seen at 70°C and 75°C in Figure 5 [Fig. 5(U–Y) compared to corresponding sub-figures at lower temperatures in Fig. 5(A–O)].
Relevance of local conformational stability and dynamics to the aggregation of the IgG1 and IgG2
The characterization of local conformation stability and dynamics of the mAbs using a wide range of temperatures highlight a few important findings with regard to aggregate formation. (1) Perhaps not unexpectedly, the solvent-exposed regions appear to precede the solvent-shielded regions in thermally induced unfolding [Fig. 1(E)] and their involvement in aggregate formation [Figs. 2(D) and 3(C,D,J-L)]. In the IgG1 and IgG2 used in this study, this process appears to occur over the temperature range (50–60°C) where the tertiary structure of the protein was significantly perturbed but the secondary structure elements primarily remained unchanged, (2) The dynamics of solvent-shielded regions in key intermediates may modulate unfolding and the aggregation propensity of these proteins. In this study, the greater dynamic nature of solvent-shielded regions in the temperature induced IgG1 versus IgG2 intermediate structures [e.g., Fig. 5(F,G)] may explain the higher Tonset and Taggregation values for aggregate formation of the IgG1 compared to the IgG2 molecule. The increased dynamic behavior with temperature for the solvent-shielded regions of the intermediates was observed with corresponding alterations in secondary structure of IgG1 and IgG2 as measured by CD.
The results presented in this manuscript both confirm and contrast with some of the findings reported earlier using a different IgG1 antibody.19 Firstly, the conformational stability of the solvent-exposed regions was lower in comparison to the solvent-shielded regions with the IgG1 and IgG2 mAbs described in this study and a different IgG1 reported previously.19 Secondly, increases in the dynamics of solvent-shielded regions, presumably in the interior of the protein, appear to predispose the protein to thermal unfolding and subsequent aggregation. These findings are consistent for both the IgG (1 and 2) molecules used in this study and the IgG1 reported previously.19 In contrast, within the pre-unfolding temperature range, the solvent-shielded regions (predominantly involved in apolar interactions in the interior of the protein) in multi-domain immunoglobulins appear to respond differently to increases in temperature, as observed in IgG1 and IgG2 in this study when compared to the previously reported IgG1.19
Acknowledgments
We acknowledge Amgen for the financial support and antibody molecules for this study. The authors would like to thank the following people (all at Amgen) for their review and discussion: Dr. Gerald Becker, Yijia Jiang, Linda Narhi, Michael Treuheit and Ping Yeh.
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.Dill KA. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 2.Pace CN, Shirley BA, McNutt M, Gajiwala K. Forces contributing to the conformational stability of proteins. FASEB J. 1996;10:75–83. doi: 10.1096/fasebj.10.1.8566551. [DOI] [PubMed] [Google Scholar]
- 3.Vlasov AP, Kravchuk ZI, Martsev SP. Non-native conformational states of immunoglobulins: thermodynamic and functional analysis of rabbit IgG. Biokhimiia. 1996;61:212–235. [PubMed] [Google Scholar]
- 4.Kameoka D, Masuzaki E, Ueda T, Imoto T. Effect of buffer species on the unfolding and the aggregation of humanized IgG. J Biochem. 2007;142:383–391. doi: 10.1093/jb/mvm145. [DOI] [PubMed] [Google Scholar]
- 5.Maity H, Maity M, Krishna MM, Mayne L, Englander SW. Protein folding: the stepwise assembly of foldon units. Proc Natl Acad Sci U S A. 2005;102:4741–4746. doi: 10.1073/pnas.0501043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindberg MO, Oliveberg M. Malleability of protein folding pathways: a simple reason for complex behaviour. Curr Opin Struct Biol. 2007;17:21–29. doi: 10.1016/j.sbi.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Shank EA, Cecconi C, Dill JW, Marqusee S, Bustamante C. The folding cooperativity of a protein is controlled by its chain topology. Nature. 2010;465:637–640. doi: 10.1038/nature09021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobson CM. The structural basis of protein folding and its links with human disease. Philos Trans R Soc Lond B Biol Sci. 2001;356:133–145. doi: 10.1098/rstb.2000.0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eichner T, Radford SE. Understanding the complex mechanisms of beta2-microglobulin amyloid assembly. FEBS J. 2011;278:3868–3883. doi: 10.1111/j.1742-4658.2011.08186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker D, Agard DA. Kinetics versus thermodynamics in protein folding. Biochemistry. 1994;33:7505–7509. doi: 10.1021/bi00190a002. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez-Ruiz JM. Protein kinetic stability. Biophys Chem. 2010;148:1–15. doi: 10.1016/j.bpc.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Kozlowski S, Swann P. Current and future issues in the manufacturing and development of monoclonal antibodies. Adv Drug Deliv Rev. 2006;58:707–722. doi: 10.1016/j.addr.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Wang W, Singh S, Zeng DL, King K, Nema S. Antibody structure, instability, and formulation. J Pharm Sci. 2007;96:1–26. doi: 10.1002/jps.20727. [DOI] [PubMed] [Google Scholar]
- 14.Shire SJ. Formulation and manufacturability of biologics. Curr Opin Biotechnol. 2009;20:708–714. doi: 10.1016/j.copbio.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 15.Kamerzell TJ, Middaugh CR. The complex inter-relationships between protein flexibility and stability. J Pharm Sci. 2008;97:3494–3517. doi: 10.1002/jps.21269. [DOI] [PubMed] [Google Scholar]
- 16.Ramsey JD, Gill ML, Kamerzell TJ, Price ES, Joshi SB, Bishop SM, Oliver CN, Middaugh CR. Using empirical phase diagrams to understand the role of intramolecular dynamics in immunoglobulin G stability. J Pharm Sci. 2009;98:2432–2447. doi: 10.1002/jps.21619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lilyestrom WG, Shire SJ, Scherer TM. Influence of the cosolute environment on IgG solution structure analyzed by small-angle X-ray scattering. J Phys Chem B. 2012;116:9611–9618. doi: 10.1021/jp303839t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thakkar SV, Joshi SB, Jones ME, Sathish HA, Bishop SM, Volkin DB, Middaugh CR. Excipients differentially influence the conformational stability and pretransition dynamics of two IgG1 monoclonal antibodies. J Pharm Sci. 2012;101:3062–3077. doi: 10.1002/jps.23187. [DOI] [PubMed] [Google Scholar]
- 19.Thakkar SV, Kim JH, Samra HS, Sathish HA, Bishop SM, Joshi SB, Volkin DB, Middaugh CR. Local dynamics and their alteration by excipients modulate the global conformational stability of an lgG1 monoclonal antibody. J Pharm Sci. 2012;101:4444–4457. doi: 10.1002/jps.23332. [DOI] [PubMed] [Google Scholar]
- 20.Herbst R, Gast K, Seckler R. Folding of firefly (Photinus pyralis) luciferase: aggregation and reactivation of unfolding intermediates. Biochemistry. 1998;37:6586–6597. doi: 10.1021/bi972928i. [DOI] [PubMed] [Google Scholar]
- 21.Zerovnik E, Turk V, Waltho JP. Amyloid fibril formation by human stefin B: influence of the initial pH-induced intermediate state. Biochem Soc Trans. 2002;30:543–547. doi: 10.1042/bst0300543. [DOI] [PubMed] [Google Scholar]
- 22.Sambashivan S, Liu Y, Sawaya MR, Gingery M, Eisenberg D. Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure. Nature. 2005;437:266–269. doi: 10.1038/nature03916. [DOI] [PubMed] [Google Scholar]
- 23.Rousseau F, Wilkinson H, Villanueva J, Serrano L, Schymkowitz JW, Itzhaki LS. Domain swapping in p13suc1 results in formation of native-like, cytotoxic aggregates. J Mol Biol. 2006;363:496–505. doi: 10.1016/j.jmb.2006.07.061. [DOI] [PubMed] [Google Scholar]
- 24.Wahlbom M, Wang X, Lindstrom V, Carlemalm E, Jaskolski M, Grubb A. Fibrillogenic oligomers of human cystatin C are formed by propagated domain swapping. J Biol Chem. 2007;282:18318–18326. doi: 10.1074/jbc.M611368200. [DOI] [PubMed] [Google Scholar]
- 25.Hart T, Hosszu LL, Trevitt CR, Jackson GS, Waltho JP, Collinge J, Clarke AR. Folding kinetics of the human prion protein probed by temperature jump. Proc Natl Acad Sci U S A. 2009;106:5651–5656. doi: 10.1073/pnas.0811457106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hafner-Bratkovic I, Bester R, Pristovsek P, Gaedtke L, Veranic P, Gaspersic J, Mancek-Keber M, Avbelj M, Polymenidou M, Julius C, Aguzzi A, Vorberg I, Jerala R. Globular domain of the prion protein needs to be unlocked by domain swapping to support prion protein conversion. J Biol Chem. 2011;286:12149–12156. doi: 10.1074/jbc.M110.213926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu C, Sawaya MR, Eisenberg D. beta(2)-microglobulin forms three-dimensional domain-swapped amyloid fibrils with disulfide linkages. Nat Struct Mol Biol. 2011;18:49–55. doi: 10.1038/nsmb.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lang L, Kurnik M, Danielsson J, Oliveberg M. Fibrillation precursor of superoxide dismutase 1 revealed by gradual tuning of the protein-folding equilibrium. Proc Natl Acad Sci U S A. 2012;109:17868–17873. doi: 10.1073/pnas.1201795109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neudecker P, Robustelli P, Cavalli A, Walsh P, Lundstrom P, Zarrine-Afsar A, Sharpe S, Vendruscolo M, Kay LE. Structure of an intermediate state in protein folding and aggregation. Science. 2012;336:362–366. doi: 10.1126/science.1214203. [DOI] [PubMed] [Google Scholar]
- 30.Campos LA, Bueno M, Lopez-Llano J, Jimenez MA, Sancho J. Structure of stable protein folding intermediates by equilibrium phi-analysis: the apoflavodoxin thermal intermediate. J Mol Biol. 2004;344:239–255. doi: 10.1016/j.jmb.2004.08.081. [DOI] [PubMed] [Google Scholar]
- 31.Ayuso-Tejedor S, Garcia-Fandino R, Orozco M, Sancho J, Bernado P. Structural analysis of an equilibrium folding intermediate in the apoflavodoxin native ensemble by small-angle X-ray scattering. J Mol Biol. 2011;406:604–619. doi: 10.1016/j.jmb.2010.12.027. [DOI] [PubMed] [Google Scholar]
- 32.Ayuso-Tejedor S, Angarica VE, Bueno M, Campos LA, Abian O, Bernado P, Sancho J, Jimenez MA. Design and structure of an equilibrium protein folding intermediate: a hint into dynamical regions of proteins. J Mol Biol. 2010;400:922–934. doi: 10.1016/j.jmb.2010.05.050. [DOI] [PubMed] [Google Scholar]
- 33.Garcia-Fandino R, Bernado P, Ayuso-Tejedor S, Sancho J, Orozco M. Defining the nature of thermal intermediate in 3 state folding proteins: apoflavodoxin, a study case. PLoS Comput Biol. 2012;8:e1002647. doi: 10.1371/journal.pcbi.1002647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nowak M. Immunoglobulin kappa light chain and its amyloidogenic mutants: a molecular dynamics study. Proteins. 2004;55:11–21. doi: 10.1002/prot.10606. [DOI] [PubMed] [Google Scholar]
- 35.Qin Z, Hu D, Zhu M, Fink AL. Structural characterization of the partially folded intermediates of an immunoglobulin light chain leading to amyloid fibrillation and amorphous aggregation. Biochemistry. 2007;46:3521–3531. doi: 10.1021/bi061716v. [DOI] [PubMed] [Google Scholar]
- 36.Lee H, Kirchmeier M, Mach H. Monoclonal antibody aggregation intermediates visualized by atomic force microscopy. J Pharm Sci. 2011;100:416–423. doi: 10.1002/jps.22279. [DOI] [PubMed] [Google Scholar]
- 37.Kamerzell TJ, Esfandiary R, Joshi SB, Middaugh CR, Volkin DB. Protein-excipient interactions: mechanisms and biophysical characterization applied to protein formulation development. Adv Drug Deliv Rev. 2011;63:1118–1159. doi: 10.1016/j.addr.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 38.Lakowicz JR. Principles of fluorescence spectroscopy. 3rd ed. New York:: Springer; 2006. p. 954. p. xxvi. [Google Scholar]
- 39.Eftink MR, Ghiron CA. Dynamics of a protein matrix revealed by fluorescence quenching. Proc Natl Acad Sci U S A. 1975;72:3290–3294. doi: 10.1073/pnas.72.9.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eftink MR, Ghiron CA. Exposure of tryptophanyl residues in proteins. Quantitative determination by fluorescence quenching studies. Biochemistry. 1976;15:672–680. doi: 10.1021/bi00648a035. [DOI] [PubMed] [Google Scholar]
- 41.Middaugh CR, Litman GW. Investigations of the molecular basis for the temperature-dependent insolubility of cryoglobulins. VI. Quenching by acrylamide of the intrinsic tryptophan fluorescence of cryoglobulin and non-cryoglobulin IgM proteins. Biochim Biophys Acta. 1978;535:33–43. doi: 10.1016/0005-2795(78)90030-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


