Abstract
Insertion of transposable elements (TEs) into introns can lead to their activation as alternatively spliced cassette exons, an event called exonization. Exonization can enrich the complexity of transcriptomes and proteomes. Previously, we performed a genome-wide computational analysis of Ds exonization events in the monocot Oryza sativa (rice). The insertion patterns of Ds increased the number of transcripts and subsequent protein isoforms, which were determined as interior and C-terminal variants. In this study, these variants were scanned with the PROSITE database in order to identify new functional profiles (domains) that were referred to their reference proteins. The new profiles of the variants were expected to be beneficial for a selective advantage and more than 70% variants achieved this. The new functional profiles could be contributed by an exon–intron junction, an intron alone, an intron–TE junction, or a TE alone. A Ds-inserted intron may yield 167 new profiles on average, while some cases can yield thousands of new profiles, of which C-terminal variants were in major. Additionally, more than 90% of the TE-inserted genes were found to gain novel functional profiles in each intron via exonization. Therefore, new functional profiles yielded by the exonization may occur in many local regions of the reference protein.
Keywords: Ac/Ds transposon, exonization, PROSITE, protein isoforms
Introduction
Insertion of transposable elements (TEs) within eukaryotic genes is thought to be an important contributor to evolution and speciation.1 Intuitively, the TEs may disrupt the function of a gene by inserting into the exons of the gene. Even TEs insert into intronic sequences of a gene may also alter the regular splicing pattern of a pre-mRNA by alternative splicing (AS) and/or exonization.2 With AS, the inserted TE interferes with the normal splicing of a gene’s transcribed region. With exonization, the cryptic splice site of the inserted TE is incorporated (or exonized) as an alternative exon. While the prevailing original splice variant remains functional, the additional variant due to AS or exonization may evolve a new function or eventually vanish after selection. The selection may also operate to optimize the new splice sites and consequently increase the proportion of the new variant if it is advantageous.3
AS is a widespread phenomenon in higher eukaryotes. Severing et al4 performed a detailed comparison of AS events in alternative-spliced orthologs from the dicot Arabidopsis thaliana and the monocot Oryza sativa (rice) and revealed that AS has a limited role in functional expansion of the plant proteome. In the other hand, recent studies of exonization are mostly in silico analyses on mammalian TEs.5–8 For instance, the analyses for 5′ and 3′ splice sites (ie, splice donor/acceptor) formation in Alu exons have provided mechanistic insights into the process of exonization.9–12 In plants, we have previously assessed the ability of a TE to provide splice/acceptor sites in vitro by inserting a mini Ds transposon into each intron of the modified tobacco marker gene epsps.13 The results suggested that exonization may introduce a portion(s) of the TE into the resulting transcripts and alter the reading frames to enrich the complexity of proteomes.
Ds is a non-autonomous (transposase-defective) transposon, which is composed of 11 bp of terminal-inverted repeats and about 250 bp of both ends (terminal regions) of its full form transposon, Activator (Ac). We found that, firstly, Ds favored providing splice donor sites from the beginning of the inserted Ds sequence in exonization while inserting into epsps.13 Secondly, Ds inserting into an intron in a reverse pattern could offer 4 donor sites and result in different transcript isoforms having different reading frames. We further conducted a genome-wide survey of all TE-exonized transcripts in each intron of each gene in the rice genome by simulations, and yielded 58,016,056 exonized transcripts.14 About 70% of the exonized transcripts may undergo nonsense-mediated mRNA decay (NMD)15 and then yield no protein product. The remaining transcripts were translated into proteins and characterized as C-terminal or interior variants. The former, whose output peptides replaced the C terminus of the reference protein, resulted from a shift of the reading frame. The later had the same termination codon as the reference transcript but had additional peptides inserted in the middle.14 Although TE exonization can yield many different protein isoforms, the possibility of the exonized protein isoforms being used for selective advantage (eg, sources of functional isoforms) needs further examination.
In this study, we assessed the impact of exonization in terms of protein function changes by performing a detailed analysis of rice protein variants generated in our previous work. Specifically, exonized protein variants yielding new functional domains may have selective advantages. We scanned the functional profiles in the PROSITE database16 for all protein variants, together with their reference proteins, for newly added domains. The PROSITE database contains more than 2000 patterns or profiles obtained by scanning the SWISS-PROT protein database. All protein variants yielding at least one new functional profile were extracted and analyzed for content. The newly added profiles in the interior or C-terminal were classified into several categories, according to sequence types offering messages: skipped exon, intron, Ds and upcoming exon, either alone or joined with the flanking ones. The composition of the profiles in the exonized protein variants may reveal an enrichment of the proteome caused by TE exonization.
Materials and Methods
The rice chromosome Genbank data was downloaded from the NCBI database ( http://www.ncbi.nlm.nih.gov/genomes/PLANTS/PlantList.html). The whole genome sequences were downloaded from MSU Rice Genome Annotation Annotation Project (Release 6.0). We used only the first CDS record for each gene to avoid redundancy. Exonization was defined as an event in which a transcript variant was created with insertion of a Ds in the intronic sequence of a gene. Therefore, we considered only genes that were completely sequenced and had at least 1 intron. A 3-step procedure to construct the exonized transcripts was performed previously.14 First, the sequence of the Ds was inserted in a forward or reverse direction to the target gene. Biologically, the insertion of Ds causes the duplication of 8 bp of G right after the insertion position, and the sequence of the Ds starts at the 9th nt after the insertion position. Second, we obtained all exonized sequences by recognizing appropriate splice donor/acceptor sites. With the Ds inserted in a forward direction in the target, the splice donor junction occurs at position 91 bp; with the Ds inserted in a reverse direction, the splice donor junction may position at 14, 18, 24 and 28 bp. Thus, one Ds may result in 1 and 4 exonized transcript variants for forward and reverse insertions, respectively. Finally, the exonized transcripts were constructed by joining the sequences of the skipped exons, the inserted intron, the Ds, and the flanking exons.
All exonized transcripts were assigned for open reading frame (ORF) analysis starting at the original start codon and terminating at the first in-frame stop codon. All transcripts were designated type I, II, III, or IV. Numbering depended on whether the in-frame stop codon occurred at the conserved region in the original splice junction, the intron was inserted by Ds, the Ds, or any exon after Ds insertion, respectively. If no in-frame stop codon was found during ORF analysis, the corresponding transcript was designated type V, and the incomplete transcript without a stop codon was outputted directly. The transcripts were further classified into 2 subtypes: an interior one if the termination codon was the same as the reference transcript (the transcript without Ds insertion); otherwise, a C-terminal transcript.
All transcripts containing a termination codon more than 55 nt upstream of the last exon/exon junction were considered putative targets for the NMD pathway15,22 and were excluded from isoform prediction. The proteins for transcripts not targeted to the NMD pathway were further translated to protein sequences. The original protein sequences and protein sequences from type III, IV, and V variants were subject to protein profile analysis, in which we scanned the sequences to search for domains (profiles) previously reported by PROSITE database (version 20.83).16 The PROSITE database consists of entries describing the protein families, domains and functional sites as well as amino acid patterns, signatures, and profiles in them. The adopted version contains 2442 functional profiles.
The functional profiles of each protein variant were compared with the ones of its reference protein. Only those variants yielding additional functional profile(s) were collected. As described in the Results section, the new functional profiles in the protein isoforms were classified as types A to F, according to their components (Fig. 1B). Further analyses of the resulting protein variants in different types were conducted using R (version 2.15.1).23
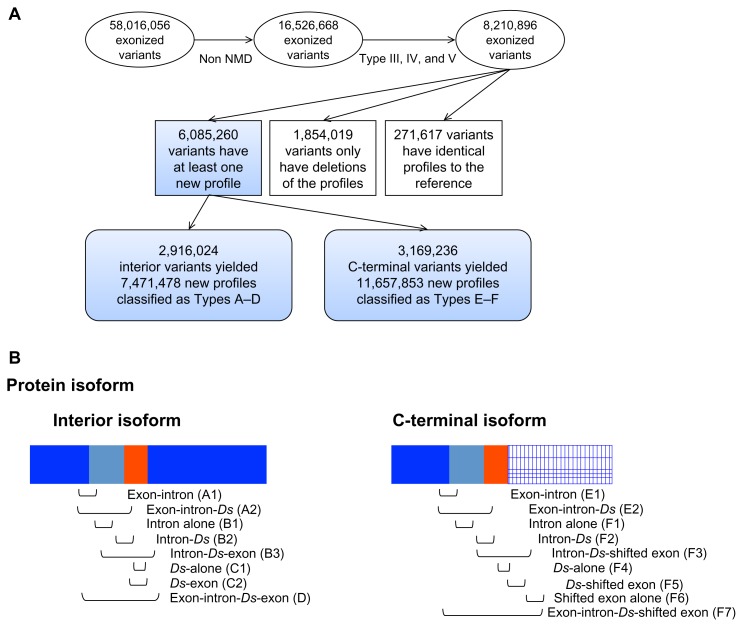
Figure 1.
(A) Flow chart of the steps for analyzing the exonized transcripts. (B) From the messages, 17 sub-types of the functional profiles were classified in interior variants or C-terminal variants.
Results
New functional profiles can be yielded by exon-intron junction, intron alone, intron-TE junction, TE alone, or TE-upcoming exon junction
In a previous study, genome-wide Ds exonized transcripts that did not undergo NMD were translated into proteins and characterized as C-terminal or interior variants.14 In this study, we performed a functional profile analyses with all previously determined Type III, IV, and V rice protein isoforms and their reference proteins (Fig. 1A). The number of isoforms per gene ranges between 5 and 10,030 with an average of 462.7. The functional profiles of all proteins, including the references, were analyzed in silico according to prior identification in PROSITE,16 which identifies well-characterized protein domains. However, whether the “new functional profiles” in the protein variants act as functional protein domains needed further determination. The functional profiles of each protein variant were compared with those of its reference protein. For interior variants, the exonized messages act as “peptide insertions” to the reference protein. New functional profiles may be provided by the inserted messages alone or in combination with the flanking components. For C-terminal variants, peptides from the new reading frame replace the C terminus of the reference protein. Although the C-terminal variants may behave defectively, several reports indicated that intron-exonized C-terminal variants code for functional isoforms, which were determined as new members of the reference protein.17–19 Thus, we considered both C-terminal and interior variants for further analysis.
From a total number of 8,210,896 variants, about 3% yielded functional profiles identical to their reference proteins, and only 23% variants exhibit deletions of profiles as compared to their reference proteins. We selected only those variants with additional functional profile(s). These new profiles were expected to bestow selective advantage over their reference proteins. The number of newly added profiles per gene ranges between 2 and 273,800 with average 1,078. To study the expansion of the proteome caused by TE exonization, we classified the new functional profiles in the protein isoforms as types A to F, according to the messages of the profile (Fig. 1B) from interior variants (A to D) or C-terminal variants (E and F). For type A, the functional profiles were built by exon-intron messages (A1) or exon-intron-Ds messages (A2), where the “exon” indicates the skipped exon in the exonization event. Without the messages of skipped exons, type B indicates functional profiles built by intron-alone messages (B1), intron-Ds messages (B2) or intron-Ds-exon messages (B3), where the “exon” in B3 indicates the upcoming exon of TE exonization. Type C indicates the functional profiles built by the Ds messages alone (C1) or with the upcoming exons messages (C2). Finally, type D indicates the functional profiles from messages covering the skipped exons to the upcoming exons. By the same rationale, types E1 and E2 were designated for the functional profiles of C-terminal variants built by exon-intron and exon-intron-Ds messages, respectively. Types F1 to F7 indicate the functional profiles built by messages for introns, Ds, and shifted exons (alone or joined with the flanking ones). Several protein variants yielding corresponding types described above are shown as examples in Supplemental Figure 1. Of note, for the interior variants, the amino acid sequences translated by the upcoming exons are identical to the corresponding regions in the reference protein. In contrast to the interior variants, in the C-terminal variants, genetic messages of the upcoming exons were frame-shifted.
About 74% of the TE-exonized protein isoforms yielded new functional profiles as compared with the reference proteins
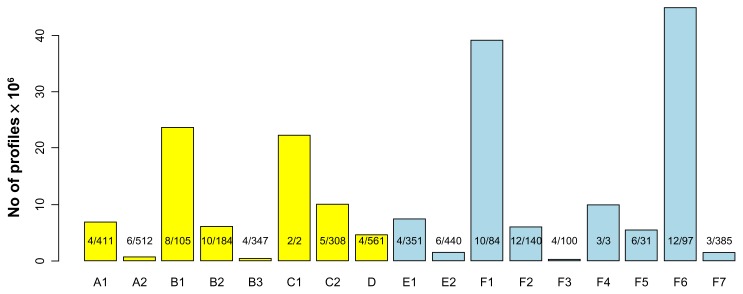
In our previous report, the number of interior variants was about 2-fold to that of C-terminal variants. Here, when we scanned the profiles in the PROSITE database for all protein variants, the number of “functional” C-terminal variants was more than that of interior variants. In all, 74% of the Ds-exonized protein variants (35% interior and 39% C-terminal) yielded new functional profiles as compared with their reference proteins. Figure 2 shows the number of profiles for types A to D (interior) and E to F (C-terminal). A variant yielded 3.14 new profiles, on average. From a total number of 19,129,331 profiles, only 876 unique profiles were determined from 2,456 profiles, indicating multiple appearances of a profile in a type (also a variant or an intron, see below). To this, we analyze the exonized variants by presenting the “total number of profiles” as well as the number of “unique profile”. The “unique profile” refers to a profile that is newly added in the variant at least once in a given categorizing criterion such as the type of peptide construction (Fig. 2), the number (per gene)/ length of the intron (Fig. 4), or all introns in each gene of rice (Fig. 5). In other words, multiple appearances of the same profile are merely counted once for the determination of the number of unique profiles. The total number of profiles serves as an indicator for the numeral limitation on evolving capacity of the peptide sequence, while the number of unique profiles is a relatively appropriate indicator for characterization of the potential protein function. For all Ds exonized variants, about 23% were type F6, with the functional profiles built by shifted exon messages. Although type F6 was the most frequent (4,493,463) among all variants, 97 unique profiles were characterized in all functional profiles. The frequency of type E2 was less than 1% (150,421), but 440 unique functional profiles were identified. Similarly, type B1 was the most frequent among the interior variants, but only 105 unique profiles were characterized. Type B3 was the least frequent among interior variants, but 347 unique profiles were obtained. Although the number of “functional” C-terminal variants was more than that of interior variants, most subtypes of the functional interior variants contained higher numbers of “unique” profiles than those of functional C-terminal variants. We found 819 unique profiles for functional interior variants and 595 for C-terminal variants.
Figure 2.
The number of newly added profiles in 17 subtypes (yellow for 8 types of interior variants and blue for 9 types of C-terminal variants). The maximum number of unique profiles per intron to total number of unique profiles in each type is provided.
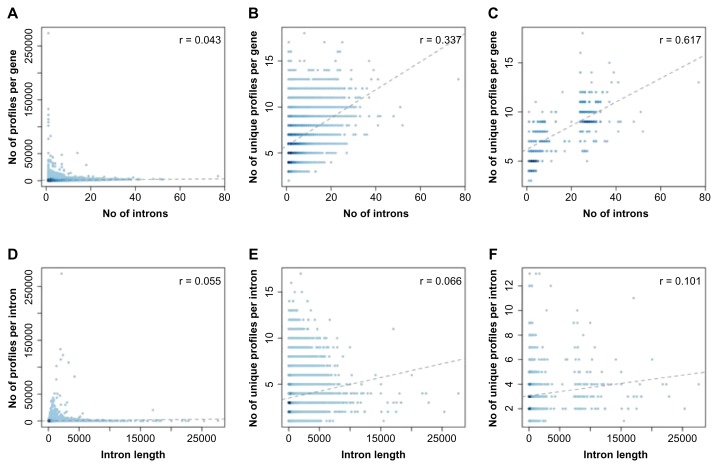
Figure 4.
The (unique) number of profiles per gene according to number of introns (upper panel) and the (unique) number of profiles per intron according to introns length (lower panel). (A), (B), (D), and (E) The results for all genes considered. (C) and (F) The results for the top 100 genes containing the largest number of introns and the top 100 genes containing the longest introns, respectively.
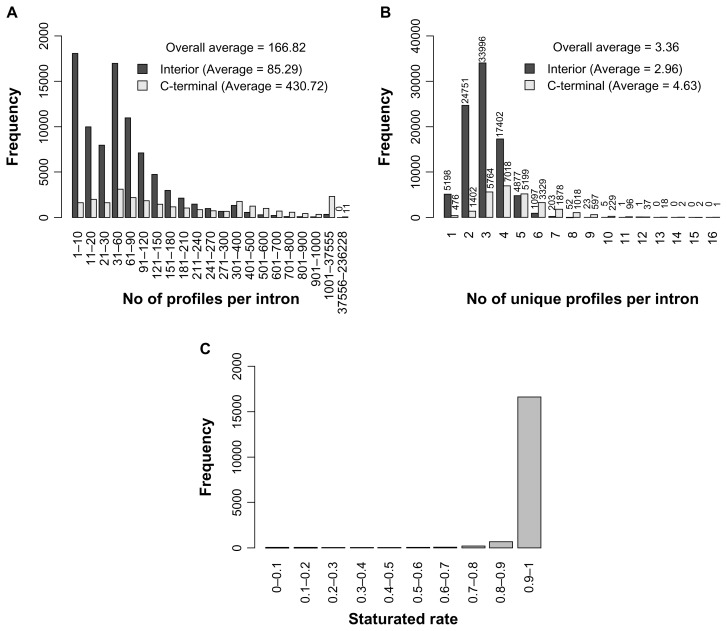
Figure 5.
The distributions of (A) the numbers and (B) the unique numbers of profiles per intron. (C) The distribution of the saturated rate for all interior isoforms.
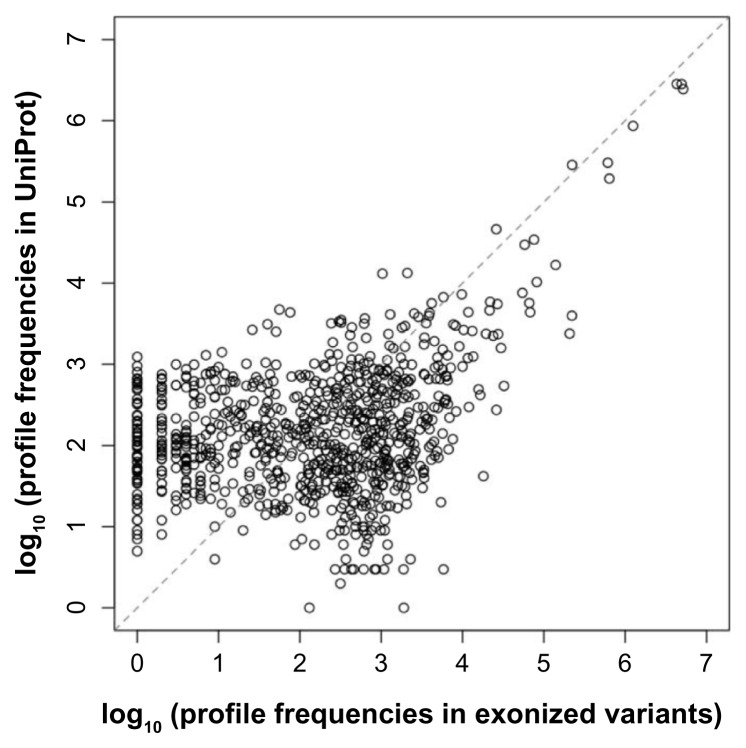
To determine whether these unique profiles were representative, we used PROSITE to search all determined 538,259 proteins in Swiss-Prot (11/28/2012 release), which contains manually annotated and reviewed proteins of the UniProt database (Supplemental Table 1). All determined proteins were supported by experimental evidence. The results were compared to those obtained from exonized variants. The frequencies of the PROSITE profiles in Swiss-Prot were correlated to those of the profiles newly added after Ds insertion (Fig. 3). This feature indicates that the new profiles of the protein variants are unbiased to limiting functions. Of note, the functional profiles for types A1, B1, E1, F1 and F6 may also occur by regular AS events, characterized as non–Ds-specific profiles. The number of these profiles (12.2 million) was much higher than that of Ds-specific profiles (6.9 million). Yet, the number of unique profiles was 1.5-fold greater for Ds-specific than non–Ds-specific profiles (833 vs. 565). Therefore, Ds-specific profiles provided more unique profiles for selective advantage (see Discussion).
Figure 3.
The frequency of the PROSITE profiles in Swiss-Prot as compared with those of profiles newly added to rice genes after Ds exonization. The dashed line is the 45-degree straight line across the origin.
Contribution of intron number and intron length of the inserted gene to functional protein variant by Ds exonization
Our previous report indicated that little of the Ds genetic message of exonized transcripts would be translated to the protein isoforms. Forward Ds contributed 20 amino acids to the new protein isoforms, and reverse Ds only a maximum of 7. The complexity of the new functional profiles in protein variants is expected to be based on the incorporated intron that Ds was inserted into. We therefore analyzed the correlation of profile number and intron number or intron length of a gene. Figure 4 shows the total and unique profiles’ numbers by the intron number (Fig. 4A and B, respectively) and those by the intron length (Fig. 4D and E). Neither intron number nor length contributed to increasing the number of novel profiles per intron, because the correlation coefficients were close to 0 (correlation = 0.043 and 0.055, respectively). However, if the number of unique profile was taken into consideration, the linear correlations of these 2 factors to the number of “unique” profiles increased (correlation = 0.337 and 0.066, respectively; Fig. 4B and E). To highlight this feature, we compared unique profile numbers in the top 100 genes containing the largest number of introns and the top 100 genes containing the longest introns (Fig. 4C and F). The results showed that intron number of a gene may contribute more to a unique functional profile after TE exonization.
More than 90% of genes can yield at least one additional functional profile in every intron via TE exonization
Since exonization occurs when a TE inserts into a gene’s intron, we studied how many total and unique profiles an exonized intron may yield. Figure 5A shows that, for interior variants, an intron can yield up to 37,555 new functional profiles (85.3, on average) via Ds insertion and subsequent exonization events. For C-terminal variants, an intron can yield up to 236,228 new profiles (430.7, on average). The extremely high number of profiles with C-terminal variants may result from the fact that a new reading frame replaces the C terminus of the reference protein. The extreme case of a intron yielding 236,228 profiles occurred when the Ds inserts into the 1st intron of Os05 g0162500.
Because a unique profile (eg, an A1 type) may present repeatedly in variants from the same intron, we analyzed the number of unique profiles that an intron may yield. The interior variants could yield up to 12 profiles (2.96, on average) and the C-terminal variants up to 16 (4.63, on average) (Fig. 5B). Furthermore, we determined the number of genes in which every intron can yield at least 1 functional variant via TE exonization, which we termed “exonized-intron saturation”. About 90% of all genes were saturated (Fig. 5C), and 4% achieved near-saturation (with all introns but one). These results indicate that a Ds-inserted gene can yield protein variants with new functional profiles located in every portion of the reference protein according to their exon–exon junction sites. Such an outcome will greatly enrich the protein complexity, leading to a selective advantage.
Discussion
A single reverse-insertion of Ds may yield 4 exonized transcript isoforms and subsequently 4 protein isoforms. Our previous study13 and unpublished results indicated that 4 splice donors were used for exonization with different efficiencies in different plant species. For simplicity, the results were previously presented under the assumption that the donors are used with the same efficiency. Although we have previously simulated abundant protein variants by TE exonization, the differences between the variants and their reference proteins need further study, specifically any new function obtained for selective advantage in the variants. We used the database PROSITE, with about 2,500 unique functional profiles (domains), to assess the function of protein variants. About 3% of the total TE-exonized protein isoforms retained functional profiles identical to their reference proteins. These variants were expected to behave nearly the same as their reference proteins. 23% of variants only showed deleted profiles as compared with their references and were expected to behave defectively. Although a protein losing a regulatory domain can sometimes behave alternatively, these 2 portions were excluded in this study. Therefore, the selective advantage of the exonized variants prior to their references were expected based on the rest 74% of variants, which yielded additional new functional profiles compared to their reference proteins. Although these variants yielded a myriad of new functional profiles, only 876 unique profiles were characterized, which account for about 1/3 of the total number of 2,456 profiles in PROSITE.
Whether the new profiles of the exonized variants may be biased to limiting functions is questionable. To assess this issue, we used PROSITE to search all 538,259 proteins determined in Swiss-Prot and found several profiles with high frequency (Supplemental Table 1, see more discussion below). About 1,000 unique profiles present in all proteins scanned less than 100 times, while only 100 unique profiles scanned more than 1000 times. The frequency of the PROSITE profiles appearing in Swiss-Prot were similar to the frequency of profiles newly added by Ds exonization events (Fig. 3). The 10 profiles with exonized variant frequencies were higher than 5 contribute the most to the high correlation. Yet, after removing the 10 profiles, the frequencies remained moderately correlated (correlation coefficient = 0.589). Therefore, the new profiles in the variants were expected to be representative for predicting possible functions for selective advantage. Additional unique profiles added to the PROSITE database will reveal more functional profiles of the exonized protein variants.
Next, we determined the original messages of the functional profiles in the exonized variants. For example, the functional profiles from type A1, B1, E1, F1, and F6 may also be found in regular AS events, without the need for a TE. Figure 2 shows that the number was higher for non–Ds-specific than Ds-specific profiles. Yet, a unique profile (eg, an A1 type) may present repeatedly in variants from the same gene whose intron was inserted by a Ds with the same reading frame. This will greatly increase non–Ds-specific profiles. Nonetheless, these interior variants containing the same A1 profile may still have different functions because of the length of the inserted peptides, which results from different Ds insertion sites in an intron. The number of unique profiles was 1.5-fold more for Ds-specific than non–Ds-specific profiles. Of the 833 unique Ds-specific profiles, only 3, namely PS00005, PS00006 and PS00008, were built by Ds alone; the former 2 were indicated as “PATTERN with a high probability of occurrence” (see also discussion below). Therefore, the Ds messages in the exonized variants may contribute in 2 ways: (1) when Ds alone carries a functional profile, the profile can increase the complexity of the exonized variants in different portions of the reference proteins (eg, interior variants harboring the same unique C1 profile) or (2) the Ds messages fuse with the flanking exon and/or intron messages to build new functional profiles for complex exonized variants. These features indicate the contribution of TE exonization for selective advantage.
TE exonization occurs only when a TE inserts into an intron. Also, TE-derived sequences play functional roles in intron size expansion.20 We attempted to reveal the contribution of the exonized intron for functional profiles. It is expected that the longer an intron, the greater number of profiles it can yield. We did not find a significant correlation between the ability to yield new profiles and increased intron number or length of a gene (Fig. 4A and D), supposedly because a few profiles (eg, an A1 type profile) present repeatedly in variants from the same intron. This feature complicates the analysis of the possible contribution of intron number and length of a gene. Indeed, when we ruled out this factor (ie, only considered the ability to increase the number of unique profiles by increasing the intron number and length of a gene), we observed a positive correlation (Fig. 4B–F). Although intron number may contribute more to protein variants than intron length, further analyses indicated that these 2 factors cannot be considered independently for the complexity of proteomes (Supplemental Fig. 2). The intron length provides a higher number of yet repeated profiles; these profiles may still have different functions influenced by the length of the inserted peptides, which result from different Ds insertion sites in an intron.
Next, we studied the number of new profiles that an intron can yield. Figure 5A shows that an intron can yield 167 new profiles, on average, in all exonized protein variants, while an extreme case (C-terminal) can yield up to 236,228 profiles. For the C-terminal variants, genetic messages of the upcoming exons were frame-shifted. This feature may result in an increasing number of total and unique profiles in C-terminal variants. Another factor that contributes to the results shown in Figure 5A may be the several profiles that PROSITE provides. A few profiles (eg, PS00005, PS00006 and PS00008) were defined as having sequences of 3 to 4 amino acids or of 6 with less stringency. These profiles are apt to match a local region in a protein variant. Indeed, this kind of profile contributed about 75% of the total new profiles of the exonized variants. For example, PS00005 and PS00006 are designated as protein kinase C phosphorylation and casein kinase II phosphorylation sites, respectively. The former requires a protein sequence containing 3 amino acids, starting with S or T and ending with R or K, and an arbitrary amino acid in the middle. The later requires a protein sequence containing 4 amino acids, starting with S or T and ending with D or E, and 2 arbitrary amino acids in the middle. Because of their high similarity, simultaneously aligning both profiles to a variant is easy, and therefore, they are recorded twice. The frequency of these 3 profiles present in all defined proteins was also greater than 75%, the same result as for the exonized variants described above (Supplemental Table 1). The intron number of a TE-inserted gene provides the possibility for changing the different regions of the reference protein. Specifically, the TE-exonized messages act as “peptide insertion” for the interior variants. Therefore, the inserted peptide can occur in many regions in the reference protein according to the exon–exon junction sites. By retaining all amino acids of the reference protein, the new functional profiles of the inserted peptide may modify (adding and/or changing) local portions in the reference protein for selective advantage. Then, we studied the proportion of genes in which functional variants can be yielded in every intron via TE exonization. Figure 5C shows that more than 90% of total genes would yield protein variants with new functional profiles in every local region according to their intron sites. About 5% of total genes achieved saturation with all introns but 1. Thus, even with the same unique profile (eg, a C1 type) in many variants for the same reference protein, the variants may still behave differently because the new functional profiles are located in different portions of the reference protein.
In conclusion, via exonization events, TE inserts in introns of genes can yield protein variants with abundant and complex functional profiles by providing splice donor and/or acceptor sites.21 Therefore, TE exonization can enrich the proteome for evolution because the TE is a medium to add and/or change the local domain, in large or small scale, of the reference protein. As a consequence, TE exonization diversifies a gene’s products for selective advantage without disrupting the gene’s function.
Supplementary Materials
Examples of the 17-type exonized protein variants described in the main text. For example, in Type A1, ‘Os10g0125300_4Ds1_5_38_1’ is the identifier indicating that a Type IV interior variant (1 or interior and 0 for c-terminal variant) was yielded after forwardly inserting (Ds1: forward insertion; Ds2-5 reverse insertion using 4 different donor sites) the Ds sequence after the nucleotide 38 in the 5th intron of gene Os10g0125300. A PROSITE profile called ‘PS00029’ is newly created (marked by the red rectangles) in ‘Os10g0125300_4Ds1_5_38_1’ variant as compared with the original protein sequence (designated as ‘Os10g0125300_NA_NA’). The definition and the PROSITE language (ie, L-x(6)-L-x(6)-L-x(6)-L) of PS00029 are also provided.
The scatter plots of (unique) number of profiles versus number of introns divided by total intron length in kilobases for all gene variants studied. As compared with the results in Figure 4B and E in the main text, the positive correlation between number of profiles and number of introns was weakened without including intron lengths.
Frequencies of all profiles of PROSITE appearing in all Ds exonized protein variants and all determined 538,259 proteins in Swiss-Prot.
A simulated example of a Ds-inserted gene, both transcripts and protein variants.
Acknowledgements
The authors would like to thank Dr. Chien-Yu Chen from Department of Bio-Industrial Mechatronics Engineering at National Taiwan University for providing valuable comments, and the National Center for High-Performance Computing in Taiwan for providing computational resources.
Footnotes
Author Contributions
YCC conceived and designed the experiments. TYC and LDL analyzed the data. YCC wrote the first draft of the manuscript. YCC and LDL contributed to the writing of the manuscript and jointly developed the structure and arguments for the paper. The final manuscript was reviewed and approved by all authors.
Competing Interests
Author(s) disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication the authors have provided signed confirmation of their compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests.
Funding
This project was supported by the National Science Council of Taiwan (Grant No. NSC 101-2313-B-002-001-MY3).
References
- 1.Sela N, Kim E, Ast G. The role of transposable elements in the evolution of non-mammalian vertebrates and invertebrates. Genome Biol. 2010;11(6):R59. doi: 10.1186/gb-2010-11-6-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9(5):397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmitz J, Brosius J. Exonization of transposed elements: A challenge and opportunity for evolution. Biochimie. 2011;93(11):1928–34. doi: 10.1016/j.biochi.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 4.Severing EI, van Dijk AD, Stiekema WJ, van Ham RC. Comparative analysis indicates that alternative splicing in plants has a limited role in functional expansion of the proteome. BMC Genomics. 2009;10:154. doi: 10.1186/1471-2164-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy A, Sela N, Ast G. TranspoGene and microTranspoGene: transposed elements influence on the transcriptome of seven vertebrates and invertebrates. Nucleic Acids Res. 2008;36(Database issue):D47–52. doi: 10.1093/nar/gkm949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mersch B, Sela N, Ast G, Suhai S, Hotz-Wagenblatt A. SERpredict: detection of tissue- or tumor-specific isoforms generated through exonization of transposable elements. BMC Genet. 2007;8:78. doi: 10.1186/1471-2156-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mola G, Vela E, Fernández-Figueras MT, Isamat M, Muñoz-Mármol AM. Exonization of Alu-generated splice variants in the survivin gene of human and non-human primates. J Mol Biol. 2007;366(4):1055–63. doi: 10.1016/j.jmb.2006.11.089. [DOI] [PubMed] [Google Scholar]
- 8.Sela N, Mersch B, Gal-Mark N, Lev-Maor G, Hotz-Wagenblatt A, Ast G. Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu’s unique role in shaping the human transcriptome. Genome Biol. 2007;8(6):R127. doi: 10.1186/gb-2007-8-6-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krull M, Petrusma M, Makalowski W, Brosius J, Schmitz J. Functional persistence of exonized mammalian-wide interspersed repeat elements (MIRs) Genome Res. 2007;17(8):1139–45. doi: 10.1101/gr.6320607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lev-Maor G, Sorek R, Levanon EY, Paz N, Eisenberg E, Ast G. RNA-editing-mediated exon evolution. Genome Biol. 2007;8(2):R29. doi: 10.1186/gb-2007-8-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ram O, Schwartz S, Ast G. Multifactorial interplay controls the splicing profile of Alu-derived exons. Mol Cell Biol. 2008;28(10):3513–25. doi: 10.1128/MCB.02279-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sorek R, Lev-Maor G, Reznik M, al et. Minimal conditions for exonization of intronic sequences: 5′ splice site formation in alu exons. Mol Cell. 2004;14(2):221–31. doi: 10.1016/s1097-2765(04)00181-9. [DOI] [PubMed] [Google Scholar]
- 13.Huang KC, Yang HC, Li KT, Liu LY, Charng YC. Ds transposon is biased towards providing splice donor sites for exonization in transgenic tobacco. Plant Mol Biol. 2012;79(4–5):509–19. doi: 10.1007/s11103-012-9927-9. [DOI] [PubMed] [Google Scholar]
- 14.Liu LY, Charng YC. Genome-wide survey of ds exonization to enrich transcriptomes and proteomes in plants. Evol B ioinform Online. 2012;8:575–87. doi: 10.4137/EBO.S10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76(1):51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 16.Hulo N, Bairoch A, Bulliard V, al et. The PROSITE database. Nucleic Acids Res. 2006;34(Database Issue):D227–30. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu JW, Chandra D, Tang SH, Chopra D, Tang DG. Identification and characterization of Bimgamma, a novel proapoptotic BH3-only splice variant of Bim. Cancer Res. 2002;62(10):2976–81. [PubMed] [Google Scholar]
- 18.Wu M, Li L, Sun Z. Transposable element fragments in protein-coding regions and their contributions to human functional proteins. Gene. 2007;401(1–2):165–71. doi: 10.1016/j.gene.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 19.Yi P, Zhang W, Zhai Z, Miao L, Wang Y, Wu M. Bcl-rambo beta, a special splicing variant with an insertion of an Alu-like cassette, promotes etoposide- and Taxol-induced cell death. FEBS Lett. 2003;534(1–3):61–8. doi: 10.1016/s0014-5793(02)03778-x. [DOI] [PubMed] [Google Scholar]
- 20.Wang D, Su Y, Wang X, Lei H, Yu J. Transposon-derived and satellite-derived repetitive sequences play distinct functional roles in Mammalian intron size expansion. Evol Bioinform Online. 2012;8:301–19. doi: 10.4137/EBO.S9758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charng YC, Liu LD. The extent of Ds1 transposon to enrich transcriptomes and proteomes by exonization. Bot Stud. 2013;54(14) doi: 10.1186/1999-3110-54-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hori K, Watanabe Y. Context analysis of termination codons in mRNA that are recognized by plant NMD. Plant Cell Physiol. 2007;48(7):1072–8. doi: 10.1093/pcp/pcm075. [DOI] [PubMed] [Google Scholar]
- 23.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Examples of the 17-type exonized protein variants described in the main text. For example, in Type A1, ‘Os10g0125300_4Ds1_5_38_1’ is the identifier indicating that a Type IV interior variant (1 or interior and 0 for c-terminal variant) was yielded after forwardly inserting (Ds1: forward insertion; Ds2-5 reverse insertion using 4 different donor sites) the Ds sequence after the nucleotide 38 in the 5th intron of gene Os10g0125300. A PROSITE profile called ‘PS00029’ is newly created (marked by the red rectangles) in ‘Os10g0125300_4Ds1_5_38_1’ variant as compared with the original protein sequence (designated as ‘Os10g0125300_NA_NA’). The definition and the PROSITE language (ie, L-x(6)-L-x(6)-L-x(6)-L) of PS00029 are also provided.
The scatter plots of (unique) number of profiles versus number of introns divided by total intron length in kilobases for all gene variants studied. As compared with the results in Figure 4B and E in the main text, the positive correlation between number of profiles and number of introns was weakened without including intron lengths.
Frequencies of all profiles of PROSITE appearing in all Ds exonized protein variants and all determined 538,259 proteins in Swiss-Prot.
A simulated example of a Ds-inserted gene, both transcripts and protein variants.