Abstract
Background
Cotton (Gossypium hirsutum L.) is one of the world’s most economically-important crops. However, its entire genome has not been sequenced, and limited resources are available in GenBank for understanding the molecular mechanisms underlying leaf development and senescence.
Methodology/Principal Findings
In this study, 9,874 high-quality ESTs were generated from a normalized, full-length cDNA library derived from pooled RNA isolated from throughout leaf development during the plant blooming stage. After clustering and assembly of these ESTs, 5,191 unique sequences, representative 1,652 contigs and 3,539 singletons, were obtained. The average unique sequence length was 682 bp. Annotation of these unique sequences revealed that 84.4% showed significant homology to sequences in the NCBI non-redundant protein database, and 57.3% had significant hits to known proteins in the Swiss-Prot database. Comparative analysis indicated that our library added 2,400 ESTs and 991 unique sequences to those known for cotton. The unigenes were functionally characterized by gene ontology annotation. We identified 1,339 and 200 unigenes as potential leaf senescence-related genes and transcription factors, respectively. Moreover, nine genes related to leaf senescence and eleven MYB transcription factors were randomly selected for quantitative real-time PCR (qRT-PCR), which revealed that these genes were regulated differentially during senescence. The qRT-PCR for three GhYLSs revealed that these genes express express preferentially in senescent leaves.
Conclusions/Significance
These EST resources will provide valuable sequence information for gene expression profiling analyses and functional genomics studies to elucidate their roles, as well as for studying the mechanisms of leaf development and senescence in cotton and discovering candidate genes related to important agronomic traits of cotton. These data will also facilitate future whole-genome sequence assembly and annotation in G. hirsutum and comparative genomics among Gossypium species.
Introduction
Cotton (Gossypium spp.) is the world’s most important agronomic fiber, as well as a significant oilseed crop. The seed is an important source of feed, foodstuff, and oil. The crop is widely cultivated in more than 80 countries, with China, India, the United States of America, and Pakistan the top four cotton producers (http://www.cotton.org/econ/cropinfo/cropdata/rankings.cfm). China is the largest producer and consumer of raw cotton. Gossypium hirsutum L., or upland cotton, is a primary cultivated species and has an allotetraploid genome (AD; 2n = 4x = 52). Gossypium hirsutum produces over 90% of the world’s fibers because of its higher yield and wider environmental adaptability [1], [2].
The advent of new molecular genetic technologies and the dramatic increase in plant gene sequence data have provided opportunities to understand the molecular basis of traits important for plant breeding, such as improved yield and plant quality. The entire genomic sequence is not available for G. hirsutum, but a large number of genomic resources have been developed for this species. These include bacterial artificial chromosomes (BACs) [3], polymorphic markers [4], and genome-wide cDNA-based or unigene expressed sequence tag (EST)–based microarrays [5]. A rapid and cost-efficient way to acquire transcriptome data for an organism with a large, complex, and unknown genome is EST sequencing; analysis of ESTs can also complement whole-genome sequencing [6]. ESTs are short, single-pass sequence reads from mRNA (cDNA). Large scale EST data represent a snapshot of genes expressed in a given tissue and/or at a given developmental stage. They are tags of expression for a given cDNA library [7]. Large EST datasets can be used to discover novel genes and carry out functional genetic studies [8], [9]. So far, a large number of G. hirsutum ESTs have been produced from cDNA libraries constructed from fibers, ovules, bolls, roots, and stems. The overwhelming majority of these EST resources have focused on fiber-development organs and have been used to explore the key genes involved in fiber development and its mechanism [10], [11]. However, large-scale EST data related to leaf development are lacking.
Leaves are specialized photosynthetic organs, and plants harvest energy and nutrients in their production. Leaf development encompasses many distinct stages, from leaf primordium formation to expansion, maturation, and abscission. The onset and progression of leaf senescence, the last phase, is accompanied by changes in expression of a large number of senescence-associated genes (SAGs). Some genes must be newly activated in leaves for the onset of senescence [12], [13]. Premature senescence, when the plant drops its leaves too early, has been occurring at an increasing frequency since the introduction of modern, high-yielding cotton cultivars like Bacillus thuringiensis (Bt) cotton. Premature leaf senescence results in reduced lint yield and poor fiber properties in cotton [14]. Understanding the molecular mechanisms of leaf senescence could greatly enhance yield and quality by guiding appropriate management to avoid premature leaf loss.
In recent decades, many advances in the understanding of leaf senescence at the molecular level have been achieved in several species, such as Arabidopsis thaliana and rice, by different experimental methods. Nine yellow-leaf-specific genes (YLS) were isolated, and RNA gel blot analysis revealed that most of them were senescence-up-regulated; the expression characteristics of YLS genes will be useful as potential molecular markers [15]. Transcript abundance in leaves of Populus tremula was studied by microarrays obtained from seven cDNA libraries, and 677 significantly up-regulated genes were identified during leaf senescence. The evidence for increased transcriptional activity before the appearance of visible signs of senescence was also found [16]. In Medicago truncatula leaves, 545 differentially-expressed genes, including 346 senescence-enhanced and 199 repressed genes, were identified by cDNA amplified fragment length polymorphism (AFLP) techniques; comparison with Arabidopsis datasets revealed common physiological events but differences in nitrogen metabolism and transcriptional regulation [17]. In rice, 533 differentially expressed genes were isolated by suppression subtractive hybridization (SSH) from early-senescent flag leaves, 183 had gene ontology (GO) annotations indicating involvement in macromolecule metabolism, protein biosynthesis regulation, energy metabolism, detoxification, pathogenicity and stress, and cytoskeleton organization [18]. A total of 140 annotated up-regulated genes in wheat flag leaves were analyzed using an in-house fabricated cDNA microarray. The results supported a protective role of mitochondria towards oxidative cell damage via the up-regulation of an alternative oxidase and possibly also succinate dehydrogenase [19]. During natural leaf senescence in Arabidopsis, 827 SAGs were identified. Comparison of these genes with artificially-induced senescence suggested that alternative pathways for essential metabolic processes such as nitrogen mobilization were used in different senescent systems [20]. Recently, a high-resolution time-course profile of gene expression during leaf senescence was obtained by microarray analysis. The dynamic changes in transcript levels were identified globally as senescence progresses, and the involvement of metabolic processes, signaling pathways, and specific transcription factors (TFs) were explicitly clarified [21]. Among the SAGs, many TFs, receptors, signaling components for hormones and stress responses, and regulators of metabolism were involved in regulating leaf senescence, indicating that senescence is governed by complex transcriptional regulatory networks.
In this study, a normalized and full-length cDNA library from different developmental stages of G. hirsutum leaves was constructed. Random sequencing of clones from the cDNA library generated a total of 9,874 high-quality ESTs, which were assembled into 5,191 unique sequences, consisting of 1,652 contigs and 3,539 singletons. Several SAGs and TFs were identified. This work will benefit the study of leaf senescence mechanisms of G. hirsutum, form a foundation for cloning the full-length sequences of these genes for genetic engineering, and also provide important resources for comparative genomic studies among closely-related species.
Results
Characterization of cDNA Library and EST Sequencing and Assembly
A normalized full-length cDNA library was constructed using leaves during the plant flowering stage. To evaluate the fullness ratios of the cDNA inserts of the library, 50 clones were randomly selected and fully sequenced; 44 (88%) contained putative full-length sequences. To assess the normalization efficiency, the relative concentration of 18S ribosomal RNA (18S) and actin in both the non-normalized and normalized cDNA populations were estimated by quantitative real-time PCR (qRT-PCR). The differences in cycle number (ΔCt) increased by 7.18 and 7.65 after library normalization, respectively. The results showed that the copies of these two genes decreased 145 and 200 fold, respectively, and suggested that the normalization quality of this library was good.
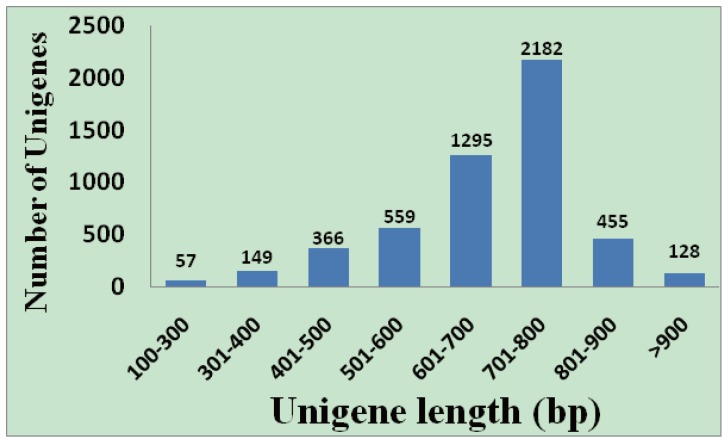
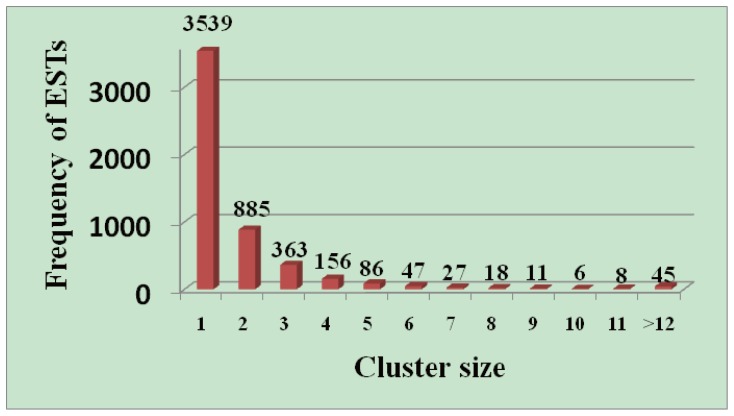
Approximately 11,623 clones were successfully single-pass sequenced from their 3′ ends. The insert sizes ranged from 900–3,000 bp, with an estimated average size of 1,200 bp. After removal of vector, poly(A) tails, contaminating microbial sequences, and those shorter than100 bp, 9,874 ESTs were considered high confidence (Q20) and were deposited in the GenBank dbEST database (JZ110066–JZ119939). Clustering and assembly of the ESTs were carried out under stringent conditions to obtain 5,191 putative unigenes, including 1,652 (31.8%) contigs that consisted of two or more ESTs and 3,539 (68.2%) singletons (Table 1). The EST redundancy of this library was 47.4%, and the unigenes had an average length of 682 bp. The distribution of high-quality EST sequences in the clusters is shown in Figure 1. Of the 1,652 contigs, 885 (53.6%) contained two ESTs, 363 (22.0%) contained three ESTs, 156 (9.4%) contained four ESTs, 86 (5.2%) contained five ESTs, 47 (2.8%) contained six ESTs, and relatively few (7.0%) contained more than six ESTs (Figure 2). The unigene mean size was only 1.9 sequences, and each contig averaged 3.8 sequences. These results also suggested that the redundancy rate of this normalized library was relatively low.
Table 1. Summary statistics of EST data generated from 11,623Gossypium hirsutum leaves.
| Feature | Number |
| Total ESTs | 11,623 |
| High-quality ESTs | 9,874 |
| ContigsESTs in contigs | 1,6526,335 |
| Singletons | 3,539 |
| Unigenes | 5,191 |
| Average unigene sequence length (bp) | 682.5 |
Figure 1. Sequence length distribution of upland cotton ESTs after assembly.
Figure 2. Frequency and distribution of ESTs among assembled contigs.
The most abundant ESTs are shown in Table 2 (each contig contained ≥10 EST copies). Some of these genes have important roles in leaf senescence. For example, lipid transfer protein precursors (Contig1797, Contig1794, Contig1792, and Contig1778; 97 ESTs) were involved in nutrient recycling for lipid transfer. Cysteine protease (Contig1795), polyubiquitin (Contig1784), putative serine carboxypeptidase precursor (Contig1749), and aspartyl protease (Contig1748) play roles in protein degradation. Peroxidase (Contig1766) is an antioxidant important for redox regulation, and metallothionein-like protein (Contig1756) is a low-molecular-weight Cys-rich protein that functions in heavy metal detoxification to remobilize valuable metal ions. Among these unigenes, 4,876 (93.9%) had open reading frames (ORFs) that were longer than 100 bp. The average ORF was 342 bp. The mean G/C content of unigenes was approximately 42%, which was approximately equivalent to that of Arabidopsis (43.2%) and much lower than that of rice (55.2%) [22], [23].
Table 2. The most abundant ESTs detected in the Gossypium hirsutum leaf library.
| Unigene name | Number of unigenes | Putative function | Source organism | E-value |
| Contig1802 | 51 | alpha-tubulin-1 | Pisum sativum | 1.00E-105 |
| Contig1801 | 41 | protodermal factor 1.1 | Gossypium barbadense | 3.00E-48 |
| Contig1800 | 36 | alpha-expansin 1 | Gossypium hirsutum | 1.00E-103 |
| Contig1799 | 35 | alpha-expansin 1 | Gossypium hirsutum | 5.00E-81 |
| Contig1798 | 32 | E6-2 protein kinase | Gossypium barbadense | 3.00E-82 |
| Contig1797 | 32 | lipid transfer protein 4 precursor | Gossypium hirsutum | 5.00E-48 |
| Contig1796 | 31 | membrane protein f16 | Gossypium hirsutum | 7.00E-98 |
| Contig1795 | 30 | cysteine protease Cp5 | Vitis vinifera | 2.00E-68 |
| Contig1794 | 28 | lipid transfer protein 3 precursor | Gossypium hirsutum | 7.00E-54 |
| Contig1793 | 22 | chalcone isomerase | Gossypium hirsutum | 7.00E-82 |
| Contig1792 | 22 | lipid transfer protein 3 precursor | Gossypium hirsutum | 4.00E-52 |
| Contig1791 | 20 | anthocyanidin reductase | Gossypium hirsutum | 1.00E-128 |
| Contig1790 | 19 | alpha-tubulin-1 | Pisum sativum | 1.00E-99 |
| Contig1789 | 19 | proline-rich protein | Gossypium hirsutum | 5.00E-33 |
| Contig1788 | 19 | tubulin alpha chain | Heterocapsa rotundata | 2.00E-83 |
| Contig1787 | 19 | Non-specific lipid-transfer protein 3 | Prunus dulcis | 5.00E-34 |
| Contig1786 | 17 | 60S ribosomal protein L29 | Ricinus communis | 1.00E-27 |
| Contig1785 | 17 | glyceraldehyde-3-phosphate dehydrogenase C subunit | Gossypium hirsutum | 1.00E-114 |
| Contig1784 | 17 | polyubiquitin | Vitis vinifera | 1.00E-106 |
| Contig1783 | 16 | phosphoglycerate kinase | Gossypium hirsutum | 1.00E-93 |
| Contig1782 | 16 | Hypothetical protein SELMODRAFT_178975 | Selaginella moellendorffii | 1.00E-103 |
| Contig1781 | 16 | 3-ketoacyl-CoA synthase | Gossypium hirsutum | 2.00E-86 |
| Contig1780 | 16 | S28 ribosomal protein | Triticum aestivum | 3.00E-20 |
| Contig1779 | 15 | 40S ribosomal protein S2 | Ricinus communis | 3.00E-94 |
| Contig1778 | 15 | lipid transfer protein precursor | Gossypium hirsutum | 1.00E-52 |
| Contig1777 | 15 | hypothetical protein | Vitis vinifera | 1.00E-105 |
| Contig1776 | 15 | arabinogalactan protein | Gossypium hirsutum | 4.00E-77 |
| Contig1775 | 14 | tubulin beta-1 | Gossypium hirsutum | 9.00E-96 |
| Contig1774 | 14 | translation elongation factor 1A-2 | Gossypium hirsutum | 1.00E-103 |
| Contig1773 | 14 | no hit | – | – |
| Contig1772 | 14 | high-glycine tyrosine keratin-like protein | Gossypium hirsutum | 3.00E-48 |
| Contig1771 | 14 | Patellin-3 | Ricinus communis | 4.00E-77 |
| Contig1770 | 14 | histone H2B.2 | Camellia sinensis | 7.00E-43 |
| Contig1769 | 14 | protodermal factor 1.3 | Gossypium hirsutum | 9.00E-50 |
| Contig1768 | 14 | MELLADRAFT_87680 | Melampsora larici-populina | 1.00E-102 |
| Contig1767 | 13 | unnamed protein product | Vitis vinifera | 2.00E-99 |
| Contig1766 | 13 | peroxidase | Gossypium hirsutum | 3.00E-71 |
| Contig1765 | 13 | flavonoid 3′5′-hydroxylase | Gossypium hirsutum | 1.00E-119 |
| Contig1764 | 13 | conserved hypothetical protein | Ricinus communis | 2.00E-50 |
| Contig1763 | 13 | S-adenosyl-L-homocystein hydrolase | Gossypium hirsutum | 1.00E-83 |
| Contig1762 | 13 | ARALYDRAFT_890328 | Arabidopsis lyrata subsp. lyrata | 2.00E-35 |
| Contig1761 | 13 | predicted protein | Populus trichocarpa | 5.00E-77 |
| Contig1760 | 12 | S-adenosyl-L-homocystein hydrolase | Gossypium hirsutum | 3.00E-74 |
| Contig1759 | 11 | intercellular adhesion molecule 2 precursor variant | Homo sapiens | 4.00E-06 |
| Contig1758 | 11 | unknown | Medicago truncatula | 6.00E-93 |
| Contig1757 | 11 | putative SAH7 protein | Gossypium raimondii | 8.00E-63 |
| Contig1756 | 11 | metallothionein-like protein | Gossypium hirsutum | 8.00E-27 |
| Contig1755 | 11 | cydophilin | Gossypium hirsutum | 1.00E-86 |
| Contig1754 | 11 | gibberellin 20-oxidase 1 | Gossypium hirsutum | 2.00E-83 |
| Contig1753 | 11 | no hit | - | - |
| Contig1752 | 11 | unnamed protein product | Vitis vinifera | 1.00E-37 |
| Contig1751 | 10 | calreticulin | Carica papaya | 8.00E-47 |
| Contig1750 | 10 | Superoxide dismutase [Mn] | Prunus persica | 8.00E-76 |
| Contig1749 | 10 | putative serine carboxypeptidase precursor | Gossypium hirsutum | 1.00E-122 |
| Contig1748 | 10 | aspartyl protease family protein | Arabidopsis lyrata subsp. lyrata | 6.00E-66 |
| Contig1747 | 10 | heat shock protein 70 | Gossypium hirsutum | 1.00E-103 |
| Contig1746 | 10 | chalcone synthase 1 | Gossypium hirsutum | 3.00E-93 |
Unigene Functional Annotation and Functional Categorization
To annotate the unigenes, all unigenes were used in a blastx search against the NCBI non-redundant (nr) protein database with a cut-off E-value of 10−5. The nr database is commonly used as the principal target database to search for homologous proteins. Using this approach, most unique sequences (84.4%) had matches in the nr database. However, 808 sequences had no hits. Of the best matches, 1,094 (25%) were to Ricinus, 1011 (23.1%) to Vitis, 975 (22.2%) to Populus, 158 (3.6%) to Arabidopsis, 142 (3.2%) to Glycine, 49 (1.1%) to Medicago, and 27 (0.6%) to Oryza, whereas only 490 (11.2%) of the best matches were to cotton (Table 3). Comparison of our unigene data set with the NCBI nucleotide database using blastn demonstrated that 4,138 unigenes (79.7%) had significant matches. All unigenes were also blastx searched against the Swiss-Prot database, in which 2,973 (57.3%) unigenes matched. The best hits were mainly to Arabidopsis (1,514 hits, 50.9%) and rice (151 hits, 5.1%).
Table 3. Comparison of the Gossypium hirsutum leaf EST library with those of other species.
| Species | Number of unigenes | Percentage |
| Ricinus | 1094 | 25.0% |
| Vitis | 1011 | 23.1% |
| Populus | 975 | 22.2% |
| Gossypium | 490 | 11.2% |
| Arabidopsis | 158 | 3.6% |
| Glycine | 142 | 3.2% |
| Medicago | 49 | 1.1% |
| Homo | 37 | 0.8% |
| Jatropha | 34 | 0.8% |
| Oryza | 27 | 0.6% |
| Citrus | 19 | 0.4% |
| Cucumis | 16 | 0.4% |
| Malus | 14 | 0.3% |
| Nicotiana | 14 | 0.3% |
| Picea | 14 | 0.3% |
| Prunus | 14 | 0.3% |
| Solanum | 14 | 0.3% |
| Pisum | 12 | 0.3% |
| Sorghum | 12 | 0.3% |
| Zea | 12 | 0.3% |
| Others | 104 | 5% |
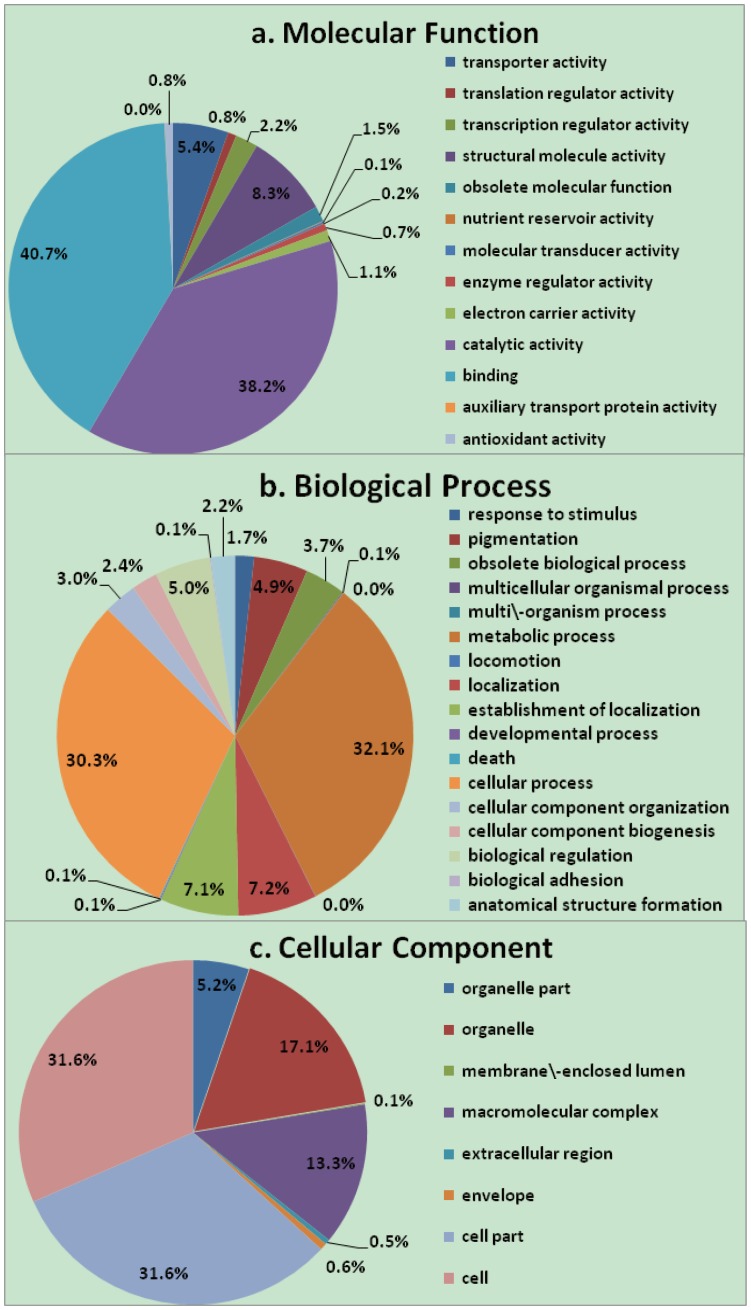
GO analysis has been widely used to classify gene functions [24]. In total, 2,416 (46.6%) unigenes fell into one or more of these categories: molecular function (2,147; 41.4%), cellular component (953; 18.4%), and biological process (1,757; 33.8%) (Figure 3). Within the molecular function category, most unigenes were assigned to molecular transducer activity (40.7%), catalytic activity (38.2%), and structural molecular activity (8.3%). The largest proportion of functionally-assigned contigs in the biological process category was categorized as metabolic process (32.1%), cellular process (30.3%), localization (7.2%), establishment of localization (7.1%), and biological regulation (5.0%). In the cellular component category, the most highly-represented groups were cell part (31.6%), cell (31.6%), and organelle (17.1%). Protein families, domains, and functional sites for the G. hirsutum unigenes were obtained through InterProScan. The most common InterPro families are presented in Table 4. A total of 3,199 unigenes fell into 1,150 InterPro families. The most frequent family was protein kinase, core (IPR000719), with 89 unigenes, followed by zinc finger, RING-type (IPR001841, 41 unigenes), WD40 repeat (IPR001680, 36 unigenes), beta tubulin (IPR000217, 30 unigenes), cytochrome P450 (IPR001128, 29 unigenes), and RNA recognition motif, RNP-1 (IPR000504, 29 unigenes).
Figure 3. Functional classifications of upland cotton 2,416 unigenes that were assigned with GO terms.
Three GO categories are presented: (a) molecular function, (b) biological process, and (c) cellular component.
Table 4. Fifty most frequent InterPro families found in the Gossypium hirsutum leaf EST library.
| No. | Interpro no. | Description | Number of unigenes |
| 1 | IPR000719 | Protein kinase, core | 89 |
| 2 | IPR001841 | Zinc finger, RING-type | 41 |
| 3 | IPR001680 | WD40 repeat | 36 |
| 4 | IPR000217 | Beta tubulin | 30 |
| 5 | IPR001128 | Cytochrome P450 | 29 |
| 6 | IPR000504 | RNA recognition motif, RNP-1 | 29 |
| 7 | IPR000886 | Endoplasmic reticulum targeting sequence | 25 |
| 8 | IPR000608 | Ubiquitin-conjugating enzyme, E2 | 25 |
| 9 | IPR002048 | Calcium-binding EF-hand | 22 |
| 10 | IPR001806 | Ras GTPase | 22 |
| 11 | IPR002198 | Short-chain dehydrogenase/reductase SDR | 21 |
| 12 | IPR000637 | HMG-I and HMG-Y, DNA-binding | 21 |
| 13 | IPR000626 | Ubiquitin | 21 |
| 14 | IPR000528 | Plant lipid transfer protein/Par allergen | 19 |
| 15 | IPR001993 | Mitochondrial substrate carrier | 18 |
| 16 | IPR015706 | RNA-directed DNA polymerase (reverse transcriptase), related | 17 |
| 17 | IPR003959 | AAA ATPase, core | 17 |
| 18 | IPR001087 | Lipolytic enzyme, G-D-S-L | 17 |
| 19 | IPR004160 | Translation elongation factor EFTu/EF1A, C-terminal | 16 |
| 20 | IPR002085 | Alcohol dehydrogenase superfamily, zinc-containing | 16 |
| 21 | IPR000425 | Major intrinsic protein | 16 |
| 22 | IPR004000 | Actin/actin-like | 15 |
| 23 | IPR001464 | Annexin | 15 |
| 24 | IPR001461 | Peptidase A1 | 15 |
| 25 | IPR000727 | Target SNARE coiled-coil region | 15 |
| 26 | IPR005123 | 2OG-Fe(II) oxygenase | 14 |
| 27 | IPR001813 | Ribosomal protein 60S | 14 |
| 28 | IPR001353 | 20S proteasome, A and B subunits | 14 |
| 29 | IPR003612 | Plant lipid transfer protein/seed storage/trypsin-alpha amylase inhibitor | 13 |
| 30 | IPR001611 | Leucine-rich repeat | 13 |
| 31 | IPR000717 | Proteasome component region PCI | 13 |
| 32 | IPR000169 | Peptidase, cysteine peptidase active site | 13 |
| 33 | IPR004159 | Protein of unknown function DUF248, methyltransferase putative | 12 |
| 34 | IPR004853 | Protein of unknown function DUF250 | 11 |
| 35 | IPR003388 | Reticulon | 11 |
| 36 | IPR002423 | Chaperonin Cpn60/TCP-1 | 11 |
| 37 | IPR002213 | UDP-glucuronosyl/UDP-glucosyltransferase | 11 |
| 38 | IPR001023 | Heat shock protein Hsp70 | 11 |
| 39 | IPR001005 | SANT, DNA-binding | 11 |
| 40 | IPR007493 | Protein of unknown function DUF538 | 10 |
| 41 | IPR004240 | Nonaspanin (TM9SF) | 10 |
| 42 | IPR003311 | AUX/IAA protein | 10 |
| 43 | IPR002052 | N-6 Adenine-specific DNA methylase | 10 |
| 44 | IPR000308 | 14-3-3 protein | 10 |
| 45 | IPR015590 | Aldehyde dehydrogenase | 9 |
| 46 | IPR004045 | Glutathione S-transferase, N-terminal | 9 |
| 47 | IPR003439 | ABC transporter related | 9 |
| 48 | IPR002109 | Glutaredoxin | 9 |
| 49 | IPR002016 | Haem peroxidase | 9 |
| 50 | IPR001509 | NAD-dependent epimerase/dehydratase | 9 |
Comparison with Previous Cotton ESTs
To evaluate potential novel sequences that did not match to sequences from other cotton species in the databases, the unigenes were used as queries in a blastn search against the Dana-Farber Cancer Institute (DFCI) Cotton Gene Index database. Approximately 24.3% of the ESTs and 19.1% unique sequences generated in this study were not highly homologous to known cotton ESTs or unique sequences. Thus, our library provides a valuable new transcript resource, with 2,400 new ESTs and 991 new unique sequences for cotton.
Identification and Analysis of Leaf Senescence-related Protein Families
To identify leaf SAGs, all the unigenes were assessed by blastx against the amino acid sequences of A. thaliana genes from the leaf senescence database (LSD). Of the 5,191 unigenes, 1,339 (25.8%) had homologs matched with 455 (44.6%) SAGs of A. thaliana in the LSD, and could be classified into 29 leaf senescence-related categories (Table 5). The most abundant leaf senescence-related category was protein degradation/modification, with a total of 199 unigenes. Other highly-abundant leaf SAGs included Nutrient recycling, Lipid/Carbohydrate metabolism, Signal transduction, Transcriptional regulation, Redox regulation, Stress and detoxification, and Hormone response pathway. These functions are all closely involved with leaf senescence.
Table 5. Functional categories of Gossypium hirsutum leaf senescence-related genesa.
| Function | Totalunigenes | TotalESTs | Redundancy |
| Protein degradation/modification | 199 | 373 | 1.9 |
| Nutrient recycling | 168 | 454 | 2.7 |
| Lipid/Carbohydrate metabolism | 158 | 338 | 2.1 |
| Signal transduction | 147 | 248 | 1.7 |
| Transcriptional regulation | 133 | 210 | 1.6 |
| Redox regulation | 94 | 267 | 2.8 |
| Stress and detoxification | 51 | 124 | 2.4 |
| Hormone response pathway | 49 | 70 | 1.4 |
| Defense | 22 | 28 | 1.3 |
| Cell structure | 22 | 30 | 1.4 |
| Nucleic acid degradation | 19 | 36 | 1.9 |
| Detoxification | 9 | 23 | 2.6 |
| Metal binding | 7 | 12 | 1.7 |
| ATPases | 6 | 12 | 2.0 |
| Metabolism | 4 | 12 | 3.0 |
| Secondary metabolites | 3 | 4 | 1.3 |
| Chlorophyll degradation | 2 | 8 | 4.0 |
| Zinc finger protein | 2 | 2 | 1.0 |
| snRNP | 2 | 4 | 2.0 |
| Light signal | 2 | 4 | 2.0 |
| Dioxygenase | 2 | 4 | 2.0 |
| Others | 236 | 386 | 1.6 |
| Total | 1337 | 2649 | 2.0 |
Frequency of unigenes found in the present study withsignificant similarities to Arabidopsis thaliana genes in the leaf senescence database.
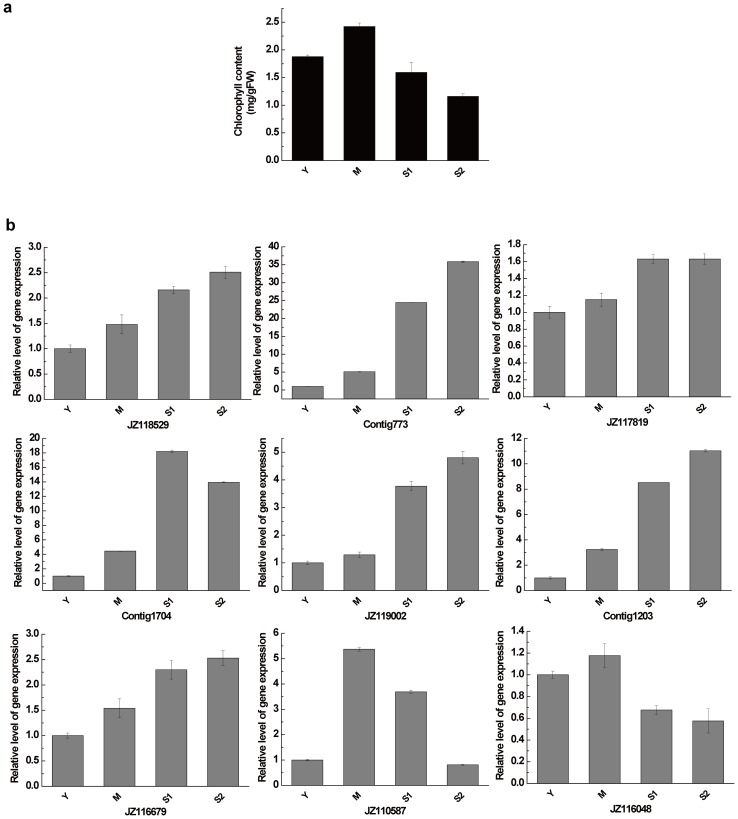
To study the expression of genes associated with leaf senescence, first, representative leaves were classified as young leaves (Y), mature leaves (M), early-senescent leaves (S1) and late-senescent leaves (S2) by their chlorophyll contents, as shown in Figure 4a. The chlorophyll content in S1 and S2 was 65% and 45% of that in M, respectively. Then, nine putative leaf senescence-related ESTs were randomly selected for qRT-PCR using RNA isolated from leaves of those four stages. Most of these ESTs were up-regulated during senescence, especially Contig773, whose expression level increased significantly in the late senescent leaves (Figure 4b). Only two ESTs, JZ110587 and JZ116048, were down-regulated. Characterization of these potential regulatory genes provided clues to the regulatory mechanism of leaf senescence.
Figure 4. Expression patterns of nine putative leaf senescence related genes from upland cotton.
(a) Chlorophyll contents per fresh weight of leaves at each of four developmental stages. (b) Changes in transcript levels of the nine putative leaf senescence-related genes at each leaf developmental stage.
Identification and Analysis of Putative Transcription Factors
Arabidopsis thaliana TFs, including 2023 TFs and 58 families, in the comprehensive PlantTFDB 2.0 database [25], were used to identify putative TFs in the cotton EST collection. Blastx searches revealed 200 (4.8% of unigenes) with matches to Arabidopsis (E-value ≤10−10) (Table 6). These TFs fell into 41 families. The most abundant family was the MYB group (22 unigenes, 11.0%), followed by bHLH (17, 8.5%), bZIP (16, 8.0%), C3H (13, 6.5%), NAC (11, 5.5%), ERF (10, 5.0%), ARF (9, 4.5%), C2H2 (9, 4.5%), and WRKY (9, 4.5%).
Table 6. The most abundant putative transcriptional factors (TFs).
| TF family | TF description | Total of unigenes | Percent (%)a |
| MYB | Myb-like DNA-binding domain | 22 | 11.0% |
| bHLH | basic/helix-loop-helix domain | 17 | 8.5% |
| bZIP | Basic leucine zipper (bZIP) motif | 16 | 8.0% |
| C3H | Zinc finger, C-x8-C-x5-C-x3-H type | 13 | 6.5% |
| NAC | No apical meristem (NAM) protein | 11 | 5.5% |
| ERF | single AP2/ERF domain | 10 | 5.0% |
| ARF | Auxin response factor | 9 | 4.5% |
| C2H2 | Zinc finger, C2H2 type | 9 | 4.5% |
| WRKY | WRKY DNA-binding domain | 9 | 4.5% |
| MIKC | MIKC-type MADS-box gene include three more domains intervening (I) domain,keratin-like coiled-coil (K) domain, and Cterminal (C) domain | 6 | 3.0% |
| TCP | TCP domain | 6 | 3.0% |
| CO-like | CONSTANS like | 5 | 2.5% |
| HB-other | Homeobox domain | 5 | 2.5% |
| HD-ZIP | HD domain with a leucine zipper motif | 5 | 2.5% |
| G2-like | Golden 2-like (GLK) | 4 | 2.0% |
| GATA | one or two highly conserved zinc finger DNA-binding domains | 4 | 2.0% |
| GRAS | three initially identified members, GAI, RGA and SCR | 4 | 2.0% |
| Trihelix | Trihelix DNA-binding domain | 4 | 2.0% |
| ARR-B | Arabidopsis response regulators(ARRs) with a Myb-like DNA binding domain(ARRM) | 3 | 1.5% |
| Dof | DNA binding with one zinc finger | 3 | 1.5% |
| SBP | SBP-domain | 3 | 1.5% |
| ZF-HD | zinc finger homeodomain | 3 | 1.5% |
| Other | – | 29 | 14.5% |
Percent = (total number of unigenes)/(total number of putative TFs). There were 200 putative TFs.
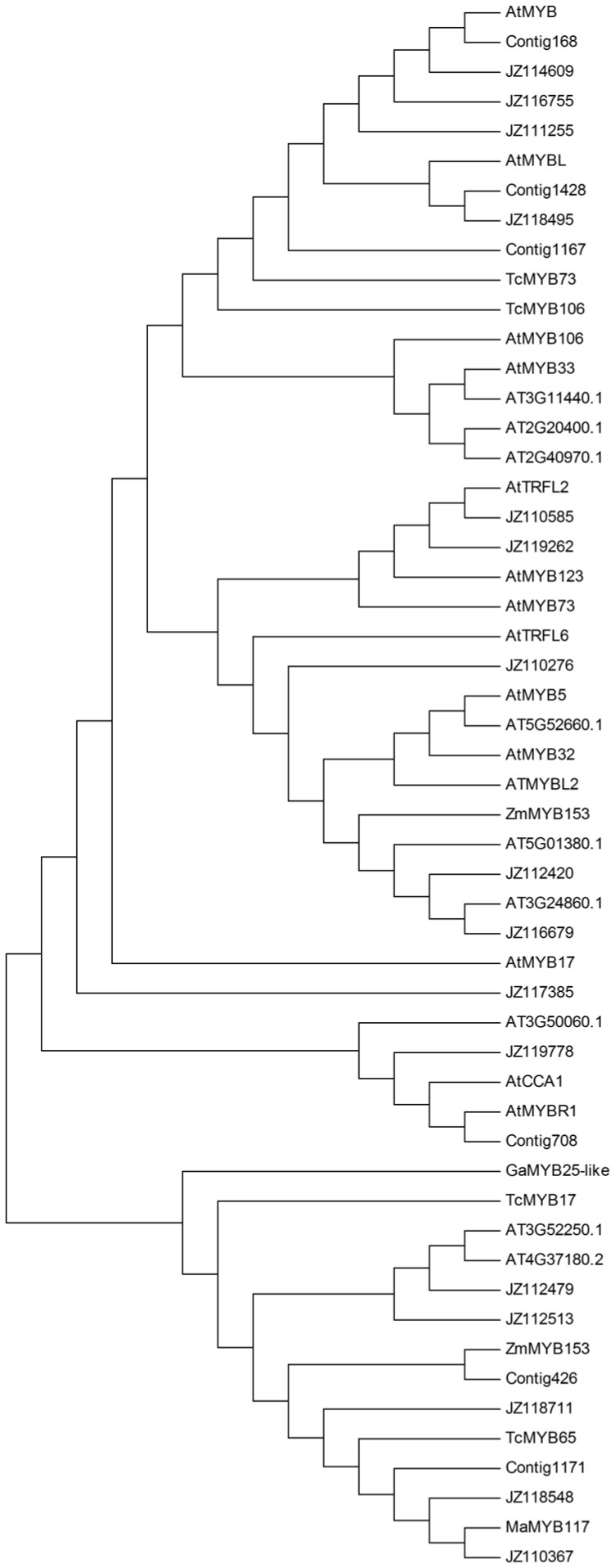
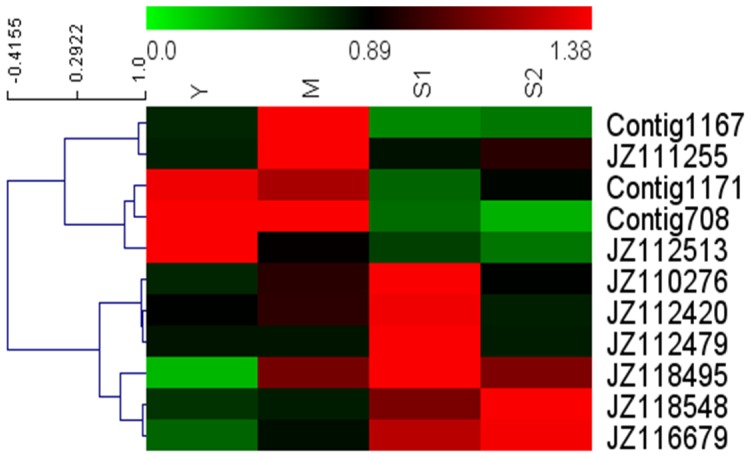
To identify the potential roles of these TFs during leaf senescence, the most abundant MYB family in this normalized library was selected and its expression pattern was analyzed. As shown in Figure 5, several putative cotton MYB orthologs matched AtMYBL (AT1G49010), ZmMYB153 (GRMZM2G050550), AtMYB (AT4G01280), AT3G24860.1, AtMYBR1 (AT5G67300), AT3G52250.1, and AT4G37180.2, which play roles in leaf senescence [26]–[28]. Using qRT-PCR, we confirmed the transcript abundance of selected ESTs encoding putative MYB TFs in leaves at different developmental stages (Figure 6). JZ118495, JZ116679 and JZ112479 were putative cotton orthologs of AtMYBL (AT1G49010), AT3G24860.1 and AT4G37180.2, respectively, from A. thaliana (Figure 5). The expression of JZ118495 increased in M stage and reached a maximum in S1 stage, while that of JZ116679 increased gradually during leaf senescence and peaked in S2 stage, and JZ112479 was expressed at high levels in the S1 stage but at reduced levels in the S2 stage (Figure 6). Six of 11 ESTs were highly expressed in senescent leaves; most increased in the expression level in the S1 stage, including JZ110276, JZ112420, JZ112479, and JZ118495. Other transcripts, such as Contig1167, JZ111255, Contig 1171, Contig708 and JZ112513 were down-regulated during leaf senescence. The results indicated that these MYB TFs may be involved in controlling leaf senescence in cotton.
Figure 5. Phylogeny analysis of putative MYB transcription factors.
Twenty-two putative cotton MYB transcription factors and thirty-one putative MYB transcription factors from other plant species were aligned and analyzed by neighbor-joining in MEGA4.
Figure 6. Expression patterns of 11.
qRT-PCR was used to evaluate the relative levels of these ESTs at each leaf development stage. The patterns were clustered and viewed using software MeV4.7.4.
Cloning of Upland Cotton YLS Homologous Genes: Sequence, Phylogenetic, and Expression Analyses
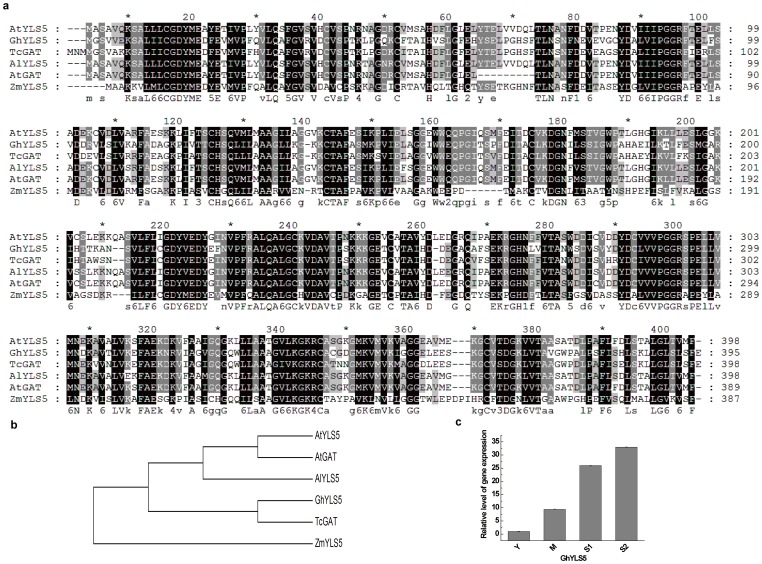
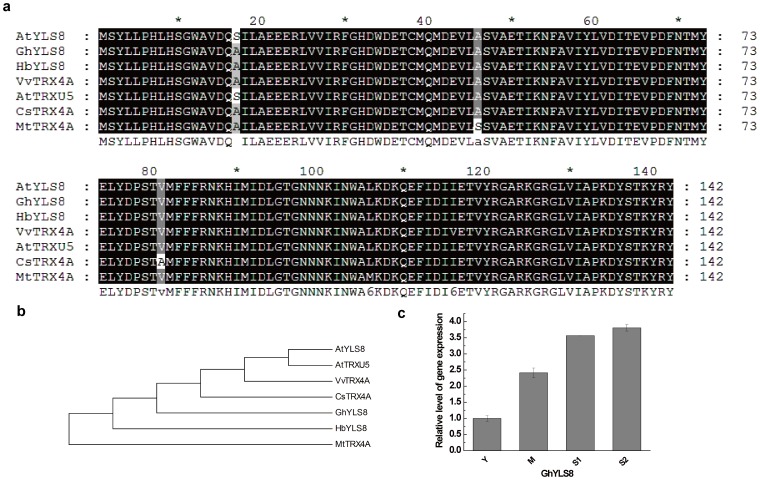
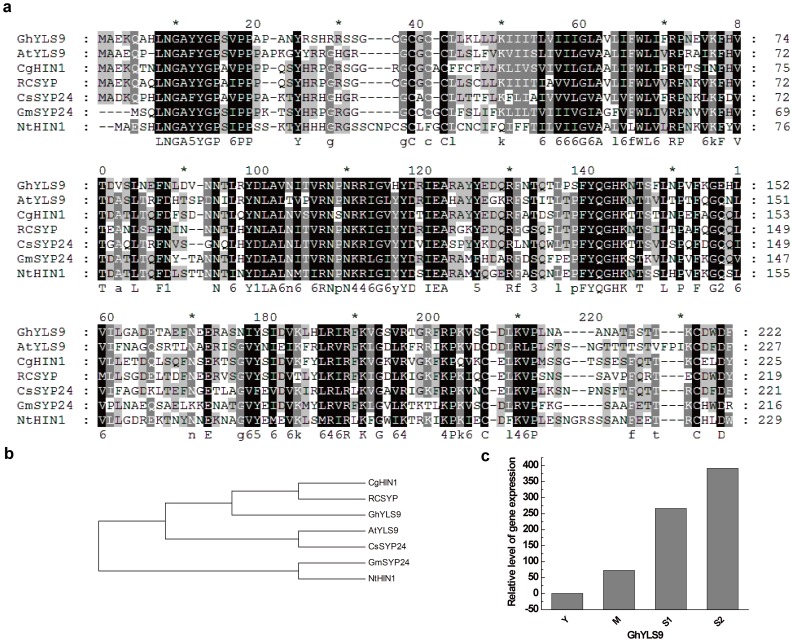
To confirm that our full-length library was an efficient method for rapid functional gene discovery in upland cotton, three A. thaliana homologs of yellow-leaf-specific genes (YLS) were cloned and analyzed. The Arabidopsis YLS proteins were used as queries to search our EST database with tBLASTn. Three unique full-length sequences were found in upland cotton and named GhYLS5 (JX163920), GhYLS8 (JX163921), and GhYLS9 (JX163922). In A. thaliana, the YLS5 gene encoded a proteaseI (pfpI)-like protein of 398 amino acid residues that was expressed weakly in young leaves and strongly in senescent leaves. This gene can be induced by artificial senescence processes such as darkness, ethylene, and ABA treatment [15]. The GhYLS5 gene had an ORF of 1,188 bp and encoded a protein of 395 amino acid residues. Multiple sequence alignment showed that GhYLS5 proteins were homologous to the glutamine amidotransferase (GAT) of A. thaliana and Theobroma cacao and with YLS5 of A. thaliana, Arabidopsis lyrata and Zea mays with identities of 51–84% (Figure 7a, b). Arabidopsis YLS8 contained an ORF encoding a Dim1 homolog of 142 amino acid residues that had high expression in senescent and virus-infected leaves [15], [29]. The GhYLS8 gene had an ORF of 429 bp, encoding a protein of 142 amino acid residues. The protein of GhYLS8 was highly conserved, with very high sequence homology to YLS8 from A. thaliana, Hevea brasiliensis, Matthiola longipetala, Iberis amara, and Lepidium sativum and to thioredoxin-like protein 4A (TRX4A) from A. thaliana, Cucumis sativus, Vitis vinifera and Medicago truncatula (Figure 8a, b). The YLS9 gene (also called NHL10) of Arabidopsis contained an ORF encoding a polypeptide of 227 amino acid residues, whose sequence was similar to tobacco hairpin-induced gene (HIN1) and Arabidopsis non-race specific disease resistance gene (NDR1). Expression of this gene is induced by Cucumber mosaic virus, spermine, and senescence [15], [29], [30].GhYLS9 gene had an ORF of 669 bp, encoding a protein of 222 amino acid residues. GhYLS9 proteins were homologous to syntaxin (SYP) from Ricinus communis, Cucumis sativus and Glycine max, HIN1 from Casuarina glauca and Nicotiana tabacum, and YLS9 from A. thaliana, with identities of 51–62% (Figure 9a, b). The expression of three GhYLS transcripts were also analyzed using qRT-PCR at different leaf developmental stages (Figures 7c, 8c, 9c). The three genes were all up-regulated in senescent leaves. In particular, expression of GhYLS9 was nearly 400-fold higher than in young leaves. These results suggested that leaf senescence related-genes could be identified from our library using -homologous sequence searches.
Figure 7. Analysis of GhYLS5 relationships.
(a) Multiple sequence alignment of GhYLS5 and other homologous proteins in plants: Theobroma cacao GAT (EOX94596), Arabidopsis thaliana GAT (NP_850303), A. thaliana a YLS5 (AB047808), Arabidopsis lyrata YLS5 (XP_002881620), and Zea mays YLS5 (NP_001146927). (b) Phylogenetic tree of these plant proteins constructed with MEGA 4 (c) Changes in transcript levels of GhYLS5 genes at each leaf development stage.
Figure 8. Analysis of GhYLS8 relationships.
(a) Multiple sequence alignment of GhYLS8 and other homologous proteins in plants: Arabidopsis thaliana YLS8 (AB047811), Hevea brasiliensis YLS8 (XP_004148041), Cucumis sativus TRX4A (XP_004163626), Medicago truncatula TRX4A (XP_003590204), A. thaliana TRXU5(AED91278) and Vitis vinifera TRX4A (XP_002310072). (b)Phylogenetic tree of these plant proteins constructed with MEGA 4 (c) Changes in the transcript levels of GhYLS8 genes at each leaf development stage.
Figure 9. Analysis of GhYLS9 relationships.
(a) Multiple sequence alignment of GhYLS9 and other homologous proteins in plants: Arabidopsis thaliana YLS9 (AB047812), Casuarina glauca HIN1 (ABZ80409), Nicotiana tabacum HIN1 (BAD22533), Ricinus communis SYP(XP_002532540), Cucumis sativus SYP24 (XP_004136508) and Glycine max SYP24 (XP_003554459). (b) Phylogenetic tree of these plant proteins constructed with MEGA 4 (c) Changes in transcript levels of GhYLS9 genes at each leaf development stage.
Discussion
Gossypium hirsutum is one of the most economically-important species in its genus. Unfortunately, to date, its genome has not been completely sequenced. Recent efforts have demonstrated that EST sequencing is an efficient and relatively low-cost approach for large-scale gene discovery, annotation, and comparative genomics research [31]. In G. hirsutum, although many ESTs are available, the total number is less than that of some field crops and model plants, and most ESTs in GenBank are from fibers or fiber-bearing ovules [11], [32]–[34] and provide little or no information regarding leaf development. Therefore, G. hirsutum leaf ESTs must be sequenced to examine the functional genomics of cotton leaf development. In this study, we produced 9,874 high-quality ESTs that assembled into 5,191 unigenes from a normalized leaf cDNA library. The leaf samples spanned all development stages, including unexpanded young leaves, fully-expanded mature leaves, and senescent leaves, at the plant blooming stage. This is the first such database and largest number of unique sequences from G. hirsutum leaf tissues to include all developmental periods. This EST resource provides a foundation for molecular control of G. hirsutum leaf growth and development and for future whole-genome sequencing and analysis of the functional genome and gene expression patterns.
Normalized cDNA libraries overcome problems caused by differential expression of genes and are an efficient and cost-effective tool for obtaining large-scale unique EST sequences and for gene identification [35]. Our cDNA library was normalized by saturation hybridization with genomic DNA, assuming relatively uniform copy numbers of most of genes in the genome. EST assembly revealed a novelty rate of 52.6%, a redundancy rate of 47.4%, and 68.2% of unigenes that contained only one EST. Thus, there remains considerable potential to discover additional novel sequences by sequencing randomly-selected cDNAs from this library. Alpha-tubulin 10 (TUA10) and ubiquitin (UBI1), the most redundant transcripts in cotton leaves, were represented by only 19 and 17 clones in our ESTs, respectively. Furthermore, the copies of two highly abundant genes actin and 18S, decreased 145 and 200 fold after cDNA library was normalized, respectively. These results reflect the quality of the normalized library and also showed that this approach was an efficient tool for gene identification because it reduced variation among abundant clones and increased the probability of sequencing rare transcripts.
The majority of annotated sequences with BLAST hits were transcripts from the rosid clade, to which cotton (Malvales; eurosids II clade) also belongs. Ricinus (25% of the best matches) and Populus (22.2%) belong to the eurosids I clade, while Vitis (23.1%) is a basal rosid [36]. Although A. thaliana and O. sativa are well-studied model systems with completely-sequenced genomes, these organisms were best matches to only 3.6% and 0.6% of our unique sequences, respectively. Yu [37] investigated the conservation of colinearity between cotton BAC sequences and other model plant genomes; on a phylogeny of single-copy orthologous genes from cotton, Arabidopsis, poplar, grape, rice, and maize, poplar was the closest relative to cotton. Arabidopsis thaliana, P. trichocarpa, and G. hirsutum are dicots, while O. sativa is a monocot, which may account for the differences in similarity among their sequences. Only 11.2% of the hits were to cotton sequences already available in GenBank, highlighting the lack of sequence information for this genus and the value of our EST sequences. Clearly, genome sequencing of G. hirsutum represents a vital and urgent need. Furthermore, we discovered 2,400 new cotton ESTs and 991 unique cotton sequences when comparing our data to the DFCI Cotton Gene Index database. Our data will contribute to the enrichment of cotton genetic and physical maps.
In previous studies, much attention was focused on leaf senescence, especially in Arabidopsis and rice [18], [38]–[42]. Leaf senescence constitutes the last stage of leaf development and strongly affects cotton yield. Currently, however, the dynamic regulatory mechanisms of leaf senescence in cotton remain unclear. A large number of SAGs have been identified in various plants through microarray analyses [21], [43]. Some of them have been found to be TFs belonging to several different families, especially NAC, WRKY, C2H2-type zinc finger, AP2/EREBP, and MYB protein families [44], [45]. Characterization of these potential regulatory genes led to discovery of a few important senescence regulatory genes and provided some insight into the regulatory mechanism of leaf senescence. Using data from the PlantTFDB 2.0 database, we found 200 unigenes from our library that had high similarity with 163 TFs from 41 families. The most well-represented TF family in our library was the MYB group, followed by the bHLH, bZIP, C3H, NAC, ERF, ARF, C2H2, and WRKY families. Analysis of the expression patterns of several putative MYB transcript families showed that the expression level of some transcripts changed significantly during leaf senescence. This result indicated that some MYB TFs may play roles during leaf senescence. These results also were in accordance with those of previous studies. In addition to these TF families, several others known to be involved in plant development were also present in our data.
Leaf senescence is an integrated response of leaf cells to age and other internal and environmental signals. It is an exceptionally complex and dynamic genetic process [46]. Arabidopsis thaliana is a favorite model for the molecular genetic study of leaf senescence [47]–[49]. The LSD is also a platform to study leaf senescence [50]. Of the unigenes in our library, 1,339 could be classified into 29 SAG categories by a BLAST search against A. thaliana senescence-related proteins (1,021), such as nutrient recycling, Lipid/Carbohydrate metabolism, and hormone response pathway. During leaf senescence, nutrients in the leaf are reallocated to younger leaves, growing seeds, or other growing organs in a process of nutrient salvage, e.g., hydrolysis of macromolecules and subsequent remobilization, which requires complex array of metabolic pathways [51]. Many the genes involved in lipid metabolism function in leaf senescence. Lipid-degrading enzymes, such as lytic acyl hydrolase, phosphatidic acid phosphatase, phospholipase D, and lipoxygenase appear to be involved in hydrolysis and metabolism of the membrane lipid in senescing leaves [51], [52]. Changed expression of the Arabidopsis acyl hydrolase gene in transgenic plants led to altered leaf senescence phenotypes [53]. The hormonal pathways appear to affect all stages of leaf senescence. In this work, numerous genes belonging to hormone response pathways were also identified. These results indicated that many previously-known leaf SAGs and pathways were included in this library. Three GhYLS genes were successfully cloned and analyzed. Their expression profiles revealed that their transcripts accumulated in leaves during senescence. Thus, these genes could potentially serve as molecular markers for distinguishing the complex regulatory networks of leaf senescence processes. This library provides a robust sequence resource and will be a useful tool for cloning the full-length sequences of functional genes for further leaf senescence-related analysis in G. hirsutum.
Materials and Methods
Plant Material
Upland cotton CCRI 36 (a short-season cultivar) was grown on the experimental farm of the Cotton Research Institute of Chinese Academy of Agricultural Sciences, Anyang, Henan Province. At the blooming stage, unexpanded leaves of the same size near the tops of stems were selected and marked. The day when leaves were fully expanded was considered the first day. Leaves were collected every 5 d for 70 d. Samples from each time point were pooled from at least 10 plants, frozen immediately in liquid nitrogen, and stored at –80°C.
RNA Isolation and cDNA Library Construction
Total RNA was isolated by an improved CTAB method [54], and equal amounts of total RNA sampled at different time points were mixed to construct a full-length normalized cDNA library. Purification of mRNA from total RNA was carried out using the FastTrack® 2.0 Kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. cDNAs were synthesized using the Superscript Full-length Library Construction Kit II (Invitrogen) according to the manufacturers’ protocols, cloned into a Gateway pDONR222 vector (Invitrogen) by the BP cloning process, and transformed into Escherichia coli strain DH10B competent cells (Invitrogen) through electroporation using an E. coli Pulser (BTX Harvard Apparatus, Holliston, MA, USA). After the full-length library was constructed, plasmid DNA was extracted with the PureLink™ HQ Mini Plasmid DNA Purification Kit (Invitrogen). Normalization was performed by saturation hybridization between genomic DNA and mixed plasmid DNA from the cDNA library [55]. Then, clones were randomly selected and fully sequenced to test fullness ratios of the cDNA inserts of the library. Putative full-length cDNA sequences were identified by comparison with all available ORF-complete mRNA sequences from the NCBI nr protein database [56]. Finally, qRT-PCR was used to estimate the relative concentration of a highly abundant clone in both the non-normalized and the normalized cDNA populations.
EST Sequencing, Editing, and Assembly
Clones were randomly picked and transferred into 384-well plates. Selected clones were sequenced from the 3′ end on an ABI 3730 automatic DNA sequencer (Applied Biosystems, Foster City, CA, USA) using the M13 universal primer (M13R:CAGGAAACAGCTATGACC) and the BigDye Terminator Cycle Sequencing Kit (ABI) at the Invitrogen Sequencing Center. All sequences were clustered using the Phred/Phrap/Consed software package [57], [58]. The 3′ DNA EST sequence chromatogram files were base-called and quality trimmed (low-quality bases with<Q20 and <99% accuracy were removed) using Phred. Crossmatch (http://www.phrap.org/) and Repeat-Masker (http://www.repeatmasker.org/) were used to remove vector sequences and to identify and mask repeat sequences. Contaminating microbial sequences were eliminated using VecScreen (http://www.ncbi.nlm.nih.gov/VecScreen/VecSc-reen.html), and poly(A) tails were deleted. Sequences that passed the quality control screening for high-confidence base calls (Q20) and with lengths longer than 100 bp were defined as high quality EST and deposited into the dbESTs division of GenBank. The processed EST sequence files were combined and assembled into contigs and singlets (unisequences) using Phrap with a high stringency level (95% sequence identity with 20 bp overlap).
To validate potential novel ESTs and unique sequences that did not match any sequences in related cotton species in the existing databases, all the high-quality ESTs and assembled unigenes were compared against ESTs and unigenes already available in the DFCI Cotton Gene Index (http://compbio.dfci.harvard.edu/cgi-bin/tgi/gimain.pl?gudb=cotton) database, which contains 351,954 cotton ESTs and 2,315 ETs fully assembled into 117,992 unique sequences. With such stringent criteria, an EST was considered as new if it had at least 10% of its sequence with less than 95% of identity to any other EST or unigene in the public EST database.
Prediction of ORFs, Unigene Functional Annotation, and Functional Categorization
All unique sequences were searched for putative ORFs with the Getorf program of EMBOSS-4.1.0 [59], and the longest sequences were used for functional analysis. Unigenes were compared with a variety of databases, including the NCBI non-redundant nucleotide and non-redundant protein databases, and Swiss-Prot, using either blastn (E-value≤10−5) or blastx (E-value≤10−5) [60]. To identify putative leaf SAGs and TFs, blastx (E-value≤10−5) searches against amino acid sequences of A. thaliana genes from a leaf senescence database (LSD) [55], [61] and a comprehensive plant TF database (PlantTFDB) [25] were used. Batch searches of the unigenes were performed using the local BLAST tools available at ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast/LATEST/. To assign GO terms, functional annotation was performed using Blast2GO software based on sequence similarity [62]–[64]. Furthermore, to improve annotations, results from an InterProScan search [65] (http://www.ebi.ac.uk/interpro/index.html) were merged with GO annotations and searched in the BlastProDom, FPrint-Scan, HMMPIR, HMMPfam, HMMSmart, HMMTigr, ProfileScan, ScanRegExp, and SuperFamily databases.
Leaf Senescence Related Homolog Identification and Expression Pattern Analysis
To examine gene expressions during leaf development, the leaves used for qRT-PCR were harvested from approximately 10 individual plants for each stage. Total chlorophyll of the samples was measured as described by Lichtenthaler (1987) [66].Homologs of leaf senescence-related protein sequences were identified and randomly selected according to the LSD function annotation. Total RNA was extracted by an improved CTAB method as described above. cDNA was reverse transcribed from RNA by PrimeScript® RT Reagent Kit with gDNA Eraser (Takara, Otsu, Japan) with an Oligo dT Primer and random six-mers as the RT primer according to the manufacturer’s protocol. The specific primer pairs for nine selected genes and the internal control gene actin are listed in Table S1. qRT-PCR was performed with the SYBR Green PCR Master Mix (Takara) as recommended by the manufacturer in an ABI 7500 Real-time PCR System (Applied Biosystems) with three replicates. To analyze changes in gene expression, values from triplicate real-time PCRs were normalized to the expression level of actin and to the Y sample by the 2–ΔΔCt method [67]. Arabidopsis YLS genes were used as queries to tBLASTn search against the cDNA library. The identified clones were sequenced in both directions with the internal primers. The amino-acid multiple-sequence alignment was analyzed using GeneDoc. Phylogenetic analysis was performed using the neighbor-joining method in MEGA 4 [68]. Expression patterns were detected by qRT-PCR as described above.
Supporting Information
Primers used in gene-specific qRT-PCR of leaf senescence related genes.
(DOC)
Funding Statement
This work was supported by the National Basic Research Program of China (No. 2010CB126006) and the earmarked fund for the China Agriculture Research System (CARS-18). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mei M, Syed NH, Gao W, Thaxton PM, Smith CW, et al. (2004) Genetic mapping and QTL analysis of fiber-related traits in cotton (Gossypium). Theor Appl Genet 108: 280–291. [DOI] [PubMed] [Google Scholar]
- 2. Han ZG, Guo WZ, Song XL, Zhang TZ (2004) Genetic mapping of EST-derived microsatellites from the diploid Gossypium arboreum in allotetraploid cotton. Mol Genet Genomics 272: 308–327. [DOI] [PubMed] [Google Scholar]
- 3. Udall JA, Swanson JM, Haller K, Rapp RA, Sparks ME, et al. (2006) A global assembly of cotton ESTs. Genome Res 16: 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang C, Ulloa M, Roberts PA (2006) Identification and mapping of microsatellite markers linked to a root-knot nematode resistance gene (rkn1) in Acala NemX cotton (Gossypium hirsutum L.). Theor Appl Genet 112: 770–777. [DOI] [PubMed] [Google Scholar]
- 5. Wu Y, Machado AC, White RG, Llewellyn DJ, Dennis ES (2006) Expression profiling identifies genes expressed early during lint fibre initiation in cotton. Plant Cell Physiol 47: 107–127. [DOI] [PubMed] [Google Scholar]
- 6. Karsi A, Cao D, Li P, Patterson A, Kocabas A, et al. (2002) Transcriptome analysis of channel catfish (Ictalurus punctatus): initial analysis of gene expression and microsatellite-containing cDNAs in the skin. Gene 285: 157–168. [DOI] [PubMed] [Google Scholar]
- 7. Boguski MS, Lowe TM, Tolstoshev CM (1993) dbEST–database for “expressed sequence tags”. Nat Genet 4: 332–333. [DOI] [PubMed] [Google Scholar]
- 8. Pashley CH, Ellis JR, McCauley DE, Burke JM (2006) EST databases as a source for molecular markers: lessons from Helianthus. J Hered 97: 381–388. [DOI] [PubMed] [Google Scholar]
- 9. Brautigam M, Lindlof A, Zakhrabekova S, Gharti-Chhetri G, Olsson B, et al. (2005) Generation and analysis of 9792 EST sequences from cold acclimated oat, Avena sativa. BMC Plant Biol 5: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li XB, Cai L, Cheng NH, Liu JW (2002) Molecular characterization of the cotton GhTUB1 gene that is preferentially expressed in fiber. Plant Physiol 130: 666–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Samuel YS, Cheung F, Lee JJ, Ha M, Wei NE, et al. (2006) Accumulation of genome-specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J 47: 761–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58: 115–136. [DOI] [PubMed] [Google Scholar]
- 13. Guo Y, Gan S (2005) Leaf senescence: signals, execution, and regulation. Curr Top Dev Biol 71: 83–112. [DOI] [PubMed] [Google Scholar]
- 14. Hezhong D, Weijiang L, Wei T, Zhenhuai L (2006) Yield,quality and leaf senescence of cotton grown at varying planting dates and plant densities in the Yellow River Valley of China. Field Crop Research 98: 106–115. [Google Scholar]
- 15. Yoshida S, Ito M, Nishida I, Watanabe A (2001) Isolation and RNA gel blot analysis of genes that could serve as potential molecular markers for leaf senescence in Arabidopsis thaliana. Plant Cell Physiol 42: 170–178. [DOI] [PubMed] [Google Scholar]
- 16. Andersson A, Keskitalo J, Sjodin A, Bhalerao R, Sterky F, et al. (2004) A transcriptional timetable of autumn senescence. Genome Biol 5: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. De Michele R, Formentin E, Todesco M, Toppo S, Carimi F, et al. (2009) Transcriptome analysis of Medicago truncatula leaf senescence: similarities and differences in metabolic and transcriptional regulations as compared with Arabidopsis, nodule senescence and nitric oxide signalling. New Phytol 181: 563–575. [DOI] [PubMed] [Google Scholar]
- 18. Liu L, Zhou Y, Zhou G, Ye R, Zhao L, et al. (2008) Identification of early senescence-associated genes in rice flag leaves. Plant Mol Biol 67: 37–55. [DOI] [PubMed] [Google Scholar]
- 19. Gregersen PL, Holm PB (2007) Transcriptome analysis of senescence in the flag leaf of wheat (Triticum aestivum L.). Plant Biotechnol J 5: 192–206. [DOI] [PubMed] [Google Scholar]
- 20. Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, et al. (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585. [DOI] [PubMed] [Google Scholar]
- 21. Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, et al. (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuhl JC, Cheung F, Yuan Q, Martin W, Zewdie Y, et al. (2004) A unique set of 11,008 onion expressed sequence tags reveals expressed sequence and genomic differences between the monocot orders Asparagales and Poales. Plant Cell 16: 114–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu J, Hu S, Wang J, Wong GK, Li S, et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92. [DOI] [PubMed] [Google Scholar]
- 24. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang H, Jin J, Tang L, Zhao Y, Gu X, et al. (2011) PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res 39: 1114–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang X, Ju HW, Chung MS, Huang P, Ahn SJ, et al. (2011) The R-R-type MYB-like transcription factor, AtMYBL, is involved in promoting leaf senescence and modulates an abiotic stress response in Arabidopsis. Plant Cell Physiol 52: 138–148. [DOI] [PubMed] [Google Scholar]
- 27. Sekhon RS, Childs KL, Santoro N, Foster CE, Buell CR, et al. (2012) Transcriptional and metabolic analysis of senescence induced by preventing pollination in maize. Plant Physiol 159: 1730–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, et al. (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ascencio-Ibanez JT, Sozzani R, Lee TJ, Chu TM, Wolfinger RD, et al. (2008) Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol 148: 436–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zheng MS, Takahashi H, Miyazaki A, Hamamoto H, Shah J, et al. (2004) Up-regulation of Arabidopsis thaliana NHL10 in the hypersensitive response to Cucumber mosaic virus infection and in senescing leaves is controlled by signalling pathways that differ in salicylate involvement. Planta 218: 740–750. [DOI] [PubMed] [Google Scholar]
- 31. Lindqvist C, Scheen AC, Yoo MJ, Grey P, Oppenheimer DG, et al. (2006) An expressed sequence tag (EST) library from developing fruits of an Hawaiian endemic mint (Stenogyne rugosa, Lamiaceae): characterization and microsatellite markers. BMC Plant Biol 6: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Taliercio E, Allen RD, Essenberg M, Klueva N, Nguyen H, et al. (2006) Analysis of ESTs from multiple Gossypium hirsutum tissues and identification of SSRs. Genome 49: 306–319. [DOI] [PubMed] [Google Scholar]
- 33. Shi YH, Zhu SW, Mao XZ, Feng JX, Qin YM, et al. (2006) Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18: 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Samuel YS, Cheung F, Lee JJ, Ha M, Wei NE, et al. (2006) Accumulation of genome-specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J 47: 761–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee BY, Howe AE, Conte MA, D’Cotta H, Pepey E, et al. (2010) An EST resource for tilapia based on 17 normalized libraries and assembly of 116,899 sequence tags. BMC Genomics 11: 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bremer B, Bremer K, Chase MW (2003) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Botanical Journal of the Linnean Society 141: 399–436. [Google Scholar]
- 37.Shuxun Y (2010) Progress in Upland cotton sequencing. International Cotton Initiative Genome(ICGI) Research Conference. Canberra, Australia. pp. 2.
- 38. Hung KT, Kao CH (2003) Nitric oxide counteracts the senescence of rice leaves induced by abscisic acid. J Plant Physiol 160: 871–879. [DOI] [PubMed] [Google Scholar]
- 39. Hung KT, Kao CH (2004) Hydrogen peroxide is necessary for abscisic acid-induced senescence of rice leaves. J Plant Physiol 161: 1347–1357. [DOI] [PubMed] [Google Scholar]
- 40. Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, et al. (2006) Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis . Proc Natl Acad Sci U S A 103: 814–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kong Z, Li M, Yang W, Xu W, Xue Y (2006) A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol 141: 1376–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van der Graaff E, Schwacke R, Schneider A, Desimone M, Flugge UI, et al. (2006) Transcription analysis of arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141: 776–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang HS, et al. (2002) Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14: 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46: 601–612. [DOI] [PubMed] [Google Scholar]
- 45. Hinderhofer K, Zentgraf U (2001) Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta 213: 469–473. [DOI] [PubMed] [Google Scholar]
- 46. Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58: 115–136. [DOI] [PubMed] [Google Scholar]
- 47. Bleecker AB, Patterson SE (1997) Last exit: senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell 9: 1169–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Buchanan-Wollaston V, Earl S, Harrison E, Mathas E, Navabpour S, et al. (2003) The molecular analysis of leaf senescence–a genomics approach. Plant Biotechnol J 1: 3–22. [DOI] [PubMed] [Google Scholar]
- 49. Lim PO, Woo HR, Nam HG (2003) Molecular genetics of leaf senescence in Arabidopsis. Trends Plant Sci 8: 272–278. [DOI] [PubMed] [Google Scholar]
- 50. Liu X, Li Z, Jiang Z, Zhao Y, Peng J, et al. (2011) LSD: a leaf senescence database. Nucleic Acids Res 39: 1103–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thompson JE, Froese CD, Madey E, Smith MD, Hong Y (1998) Lipid metabolism during plant senescence. Prog Lipid Res 37: 119–141. [DOI] [PubMed] [Google Scholar]
- 52. Thompson J, Taylor C, Wang TW (2000) Altered membrane lipase expression delays leaf senescence. Biochem Soc Trans 28: 775–777. [PubMed] [Google Scholar]
- 53. Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46: 601–612. [DOI] [PubMed] [Google Scholar]
- 54. Wan CY, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223: 7–12. [DOI] [PubMed] [Google Scholar]
- 55. Chu ZH, Peng KM, Zhang LD, Zhou B, Wei J, et al. (2003) Construction and characterization of a normalized whole-life-cycle cDNA library of rice. Chinese Sci Bull 48: 229–235. [Google Scholar]
- 56. Zhang JY, Lee YC, Torres-Jerez I, Wang M, Yin Y, et al. (2013) Development of an integrated transcript sequence database and a gene expression atlas for gene discovery and analysis in switchgrass (Panicum virgatum L.). Plant J 74: 160–173. [DOI] [PubMed] [Google Scholar]
- 57. Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8: 186–194. [PubMed] [Google Scholar]
- 58. Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8: 175–185. [DOI] [PubMed] [Google Scholar]
- 59. Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277. [DOI] [PubMed] [Google Scholar]
- 60. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 61. Liu X, Li Z, Jiang Z, Zhao Y, Peng J, et al. (2011) LSD: a leaf senescence database. Nucleic Acids Res 39: 1103–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, et al. (2004) The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res 32: 258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 64. Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, et al. (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36: 3420–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, et al. (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33: 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods enzymol 148: 350–382. [Google Scholar]
- 67. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 68. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in gene-specific qRT-PCR of leaf senescence related genes.
(DOC)