Abstract
Objective
Notch receptors determine cell fate by regulating transcription, an event mediated by the Notch intracellular domain (NICD), which is generated by proteolysis brought about by Notch-ligand interactions. Since Notch activation or exposure to interleukin (Il)6 have similar effects in chondrocytes, we explored whether Il6 contributes to the mechanisms of Notch action in these cells.
Method
NICD was overexpressed in primary chondrocytes from RosaNotch mice, where the Rosa26 promoter precedes a loxP-flanked STOP cassette followed by the NICD coding sequence. Cells were infected with adenoviral vectors expressing Cre to induce NICD or green fluorescent protein as control. Gene expression was determined by quantitative reverse-transcription polymerase chain reaction. Il6 protein concentration in the culture media was determined by enzyme-linked immunosorbent assay. To test the mechanisms of Notch action on Il6 expression, cells were transfected with a fragment of the Il6 promoter or control vector pGL3, or transcriptionally arrested with 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole. Il6 was inhibited with a neutralizing antibody, whereas a normal immunoglobulin G was used as control.
Results
NICD induced Il6 mRNA and protein, and transactivated the Il6 promoter without affecting Il6 mRNA stability. Il6 neutralization had no impact on gene expression under basal conditions, and did not modify the effects of NICD on sex determining region-Y-related high mobility group-box gene 9, collagen type II α1 and collagen type X α1 expression. Conversely, Il6 neutralization opposed aggrecan (Acan) suppression and prevented matrix metalloprotease (Mmp)13 induction by NICD.
Conclusion
Il6 mediates suppression of Acan and induction of Mmp13 expression by Notch in chondrocytes.
Keywords: Notch, Mmp13, Il6, Col10a1, Acan, chondrocytes
INTRODUCTION
The Notch1 to Notch4 receptors, and the jagged (Jag)1 and 2, and delta-like (Dll)1, 3 and 4 ligands are transmembrane proteins that relay signals between adjacent cells, thereby determining cell fate and function 1. Interactions of Notch with Jag or Dll result in sequential proteolytic cleavages by extracellular metalloproteases and by the γ-secretase protein complex, leading to the release of the intracellular domain of Notch (NICD) to the cytoplasm 2. NICD translocates to the nucleus and associates with the DNA binding protein Epstein-Barr virus latency C-promoter binding factor 1, suppressor of hairless, and lag1 (CSL), also known as Rbpjκ in mice, and with mastermind-like proteins. Subsequently, the transcriptional repressors associated with Rbpjκ are displaced by transcriptional activators, resulting in the induction of Notch target genes, such as those encoding for hairy enhancer of split (Hes) and Hes related with YRPW-motif (Hey) 3,4.
Chondrocytes are cartilage forming cells of mesenchymal origin, and proliferation and differentiation of growth plate chondrocytes results in linear growth of long bones 5. Chondrocytes in articular cartilage derive from mesenchymal precursors found at the articular surface, and under physiological conditions, these cells express proteoglycan (Prg)4, sex determining region-Y-related high mobility group-box gene (Sox)9, aggrecan (Acan) and collagen type II α1 (Col2a1) and preserve the function of the extracellular matrix 6,7. Changes in the synovial fluid concentration of Prg4, and suppressed Sox9, Acan and Col2a1 transcript levels associated to increased expression of collagen type X α1 (Col10a1) by articular chondrocytes, are observed when cartilage integrity is compromised 8,9,10,11. These events are coupled to the induction of gene markers of matrix degradation, such as a disintegrin-like and metallopeptidase with thrombospondin type 1 motif (Adamts)4 and matrix metalloprotease (Mmp)13, and to the expression of mediators of inflammation, such as interleukin (Il)6 12,13,14.
Although Notch signaling suppresses chondrogenesis and differentiation of hypertrophic chondrocytes, under selected conditions, overexpression of NICD in chondrocytes induces Col10a1 and Mmp13 transcripts 15,16,17,18,19,20,21,22. Increased expression of components of the Notch signaling pathway and nuclear localization of the intracellular domains of NOTCH1 and NOTCH2 are observed in human osteoarthritic chondrocytes, indicating that activation of Notch signaling is associated with cartilage degeneration 23,24,25. Accordingly, inactivation of Rbpjκ in chondrocytes or suppression of Notch by a γ-secretase inhibitor prevents surgically induced cartilage degeneration, suggesting that activation of Notch signaling is detrimental to the integrity of articular cartilage 25. However, the mechanisms that mediate the effects of Notch on chondrocyte function are understood partially.
Notch induces Il6 expression in cells of mesenchymal origin, and exposure of bovine, rabbit, and human chondrocytes to Il6 phenocopies selected effects of Notch in murine chondrocytes 26,27,28,29. In this study, we investigated whether Notch regulates Il6 expression in chondrocytes and whether Il6 mediates the effects of Notch in these cells. To this end, primary chondrocyte-enriched cells were harvested from RosaNotch mice where Cre recombination induces the expression of NICD following excision of a loxP-flanked STOP cassette, placed between the Rosa26 promoter and the NICD coding sequence 30.
METHODS
Primary chondrocyte-enriched cell cultures
RosaNotch mice, generated by D.A. Melton (Harvard University, Cambridge, MA), were obtained from Jackson Laboratories (Bar Harbor, ME) 30. Chondrocyte-enriched cells were obtained from 3 to 4 day old male and female littermate RosaNotch or wild-type C57BL/6 mice and cultured independently in order to retain the individual identity of the donor. The distal epiphysis of the femur, tibia, humerus, ulna and radius, and the proximal epiphysis of the tibia and ulna were dissected under a Unitron Z850 stereo microscope (Unitron, Commack, NY), and trabecular bone removed to limit contamination from osteoblastic cells. Cartilage was transferred to high glucose Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies, Grand Island, New York) and digested with 0.25% trypsin in 0.9 mM EDTA (Life Technologies) for 40 min at 37 C with continuous mixing. Subsequently, cartilage was exposed to DMEM containing 10% fetal bovine serum (FBS, Atlanta Biologicals, Norcross, GA) and 200 U/ml of type II collagenase from Clostridium histolyticum (Worthington Biochemical Corporation, Lakewood, NJ) for 2 to 4 h at 37 C with continuous mixing. Following digestion, tissue debris was removed by straining through a 70 μm membrane and chondrocyte-enriched cells were collected by centrifugation at 500 g for 5 min. Cells were seeded at a density of 50,000 cells/cm2 and cultured in DMEM supplemented with 10% FBS at 37 C in a humidified 5% CO2 incubator 21,31. Experimental protocols were approved by the Animal Care and Use Committee of Saint Francis Hospital and Medical Center.
Adenoviral infection
Primary chondrocyte-enriched cells from RosaNotch mice were transferred to DMEM containing 2% FBS for 1 h and exposed overnight to 100 multiplicity of infection of replication defective recombinant adenoviruses. An adenoviral vector expressing Cre recombinase under the control of the cytomegalovirus (CMV) promoter (Ad-CMV-Cre, Vector Biolabs, Philadelphia, PA) was used to excise the STOP cassette and allow NICD expression. An adenoviral vector where the CMV promoter directs expression of green fluorescent protein (GFP; Ad-CMV-GFP, Vector Biolabs) was used as control. Following infection, primary chondrocyte-enriched cells were allowed to recover for 24 to 48 h and cultured in the presence of DMEM containing 10% FBS 21. At confluence, cultures were exposed to 100 μg/ml ascorbic acid (Sigma-Aldrich, Saint Louis, MO) to prevent loss of chondrocyte phenotype and promote acquisition of a mature chondrocyte phenotype 32. Activation of Notch signaling in RosaNotch cells expressing Cre recombinase, and the effects of Notch on the chondrocyte phenotype, were confirmed in independent cultures from male or female mice and the effects of Notch were not sexually dimorphic (data not shown) 21,33,34,35,36. Therefore, observations from cell cultures obtained from mice of either sex were pooled for analysis of results.
Cytochemical staining, enzyme-linked immunosorbent assay (ELISA) and Il6 neutralization
To determine formation of chondrogenic and mineralized nodules, cultures from RosaNotch mice were fixed for 10 min at room temperature in 3.7% formaldehyde in phosphate buffered saline and subsequently stained with 1% alcian blue in 3% acetic acid, or 2% alizarin red in H2O (all from Sigma-Aldrich). Images were acquired with a Coolpix 995 digital camera mounted on an Eclipse TS100 inverted microscope equipped with contrast phase lenses (all from Nikon Inc., Melville, NY).
To explore whether NICD induces Il6 protein levels, confluent RosaNotch chondrocyte-enriched cells were cultured for 3 days before exposure for 24 h to DMEM containing 100 μg/ml ascorbic acid. Il6 concentration in the medium was measured with a mouse Il6 ELISA kit, in accordance to the manufacturer’s instruction (BD Biosciences, San Jose, California).
In selected experiments, RosaNotch chondrocyte-enriched cells were exposed to DMEM for 6 h and subsequently exposed DMEM containing 1 μg/ml neutralizing murine monoclonal immunoglobulin G (IgG) against Il6 (clone B-E8; EMD Millipore, Billerica, MA) or control murine normal IgG (Santa Cruz Biotechnology, Santa Cruz, CA).
Transient transfections
To assess whether NICD regulates Il6 transcription in chondrocytes, chondrocyte-enriched cells from RosaNotch mice were transiently transfected with a 1.3 kilobase (kb) fragment of the murine Il6 promoter cloned into pGL3 basic, upstream of the luciferase coding sequence (Il6-Luc; D.L. Allen, University of Colorado Boulder, Boulder, CO) 37. To induce Notch signaling in wild-type chondrocyte-enriched cells, a 2.4 kb DNA fragment containing the murine NICD coding sequence (J.S. Nye, Columbia University, New York City, NY) was cloned in the pcDNA 3.1 expression vector (Life Technologies, Carlsbad, CA) downstream of the CMV promoter to generate pcDNA-NICD 38. To determine the effects of NICD on Notch transactivation in wild-type chondrocytes, cells were transiently transfected with pcDNA-NICD or with pcDNA3.1, as control, and co-transfected with a construct where six multimerized dimeric CSL binding sites linked to the β-globin basal promoter govern luciferase expression (12xCSL-Luc; L.J. Strobl, Munich, Germany)39. To confirm the effects of NICD on the transactivation of the Il6 promoter, wild-type chondrocytes transiently transfected with pcDNA-NICD or with pcDNA3.1 were co-transfected with Il6-Luc. To correct for transfection efficiency, cells were co-transfected with a construct where the CMV promoter directs β-galactosidase expression (Clontech, Mountain View, CA). RosaNotch chondrocytes were exposed to a 3 μl/2 μg mix of FuGene6 (Roche, Indianapolis, IN) and DNA for 16 h before adenoviral infection with Ad-CMV-Cre or Ad-CMV-GFP. Wild-type chondrocyte cultures at 70% confluence were exposed to a 3 μl/2 μg mix of X-tremeGENE 9 (Roche) and DNA for 16 h. Cells were harvested after 48 h in luciferase extraction buffer (Roche), and luciferase and β-galactosidase activities were measured on an Optocomp luminometer (MGM Instruments, Hamden, CT), according to manufacturer’s instructions (Roche).
Quantitative reverse transcription-PCR (qRT-PCR)
Total RNA from chondrocyte-enriched cells from RosaNotch mice was extracted with the RNeasy mini kit, according to manufacturer’s instructions (Qiagen, Valencia, CA) and changes in gene expression determined by qRT-PCR. Half to one μg of total RNA were reverse-transcribed using the iScript cDNA synthesis kit (BioRad, Hercules, CA), according to manufacturer’s instructions, and amplified in the presence of specific primers (Table 1; all from IDT), and iQ SYBR Green Supermix (BioRad) at 60 C for 35 cycles. mRNA copy number was estimated by comparison to a 10-fold serial dilution of cDNA for Acan, Adamts4, Col10a1, Il6, Mmp13 and Sox9 (all from Thermo Scientific, Rockford, IL), Col2a1, Hes1, ribosomal protein l (Rpl)38 (all from American Type Culture Collection, Manassas, VA), Prg4 (Source BioScience, Nottingham, United Kingdom), Hey1 and Hey2 (both from T. Iso, University of Southern California, Los Angeles, CA) and Hey-like (HeyL; from D. Srivastava, University of Texas Southwestern Medical Center, Dallas, TX) 4,40,41,42. Reactions were conducted in a CFX96 real time PCR detection system (BioRad), fluorescence monitored during every PCR cycle at the annealing step and specificity of the reaction assessed by analysis of melting curves.
Table 1. Primers used for qRT-PCR.
Forward (Fwd) and reverse (Rev) primers used to determine changes in gene expression by qRT-PCR. GenBank accession numbers for the transcript variants recognized by primer pairs are indicated.
| Gene | Strand | Primer Sequence | GenBank Accession Number |
|---|---|---|---|
| Acan | Fwd | 5′-ATGGTCCTTCTATGACATACACTCCCCG-3′ | NM_007424 |
| Rev | 5′-TTGTTACAGCGCCACCAAGG-3′ | ||
| Adamts4 | Fwd | 5′-GATGTGTGCAAGCTTACCT-3′ | NM_172845 |
| Rev | 5′-CATCCGTAACCTTTGGAGA-3′ | ||
| Col2a1 | Fwd | 5′-GACCCAAACACTTTCCAACCGCAGT-3′ | NM_031163; NM_003396 |
| Rev | 5′-TCATCAGGTCAGGTCAGCCATT-3′ | ||
| Col10a1 | Fwd | 5′-CAGGCTTTCTGGGATGCCGCTTGT-3′ | NM_009925 |
| Rev | 5′-GGGCACCTACTGCTGGGTAA-3′ | ||
| Hes1 | Fwd | 5′-ACCAAAGACGGCCTCTGAGCACAGAAAGT-3′ | NM_008235 |
| Rev | 5′-ATTCTTGCCCTTCGCCTCTT-3′ | ||
| Hey1 | Fwd | 5′-ATCTCAACAACTACGCATCCCAGC-3′ | NM_010423 |
| Rev | 5′-GTGTGGGTGATGTCCGAAGG-3′ | ||
| Hey2 | Fwd | 5′-AGCGAGAACAATTACCCTGGGCAC-3′ | NM_013904 |
| Rev | 5′-GAGGTAGTTGTCGGTGAATTGG-3′ | ||
| HeyL | Fwd | 5′-CAGTAGCCTTTCTGAATTGCGAC-3′ | NM_013905 |
| Rev | 5′-GCTTGGAGGAGCCCTGTTTCT-3′ | ||
| Il6 | Fwd | 5′-CGGCCTTCCCTACTTCACAAGTCCG-3′ | NM_031168 |
| Rev | 5′-CAGGTCTGTTGGGAGTGGTATCC-3′ | ||
| Mmp13 | Fwd | 5′-GGAAGACCTTGTGTTTGCAGAGC-3′ | NM_008607 |
| Rev | 5′-CACTGTAGACTTCTTCAGGATTCCCG-3′ | ||
| Prg4 | Fwd | 5′-CGCCTTTTCCAAAGATCAATACTA-3′ | NM_021400; NM_001110146 |
| Rev | 5′-GTGGTAATTGCTCTTGCTGTT-3′ | ||
| Rpl38 | Fwd | 5′-AGAACAAGGATAATGTGAAGTTCAAGGTTC-3′ | NM_001048057; NM_001048058; NM_023372 |
| Rev | 5′-CTGCTTCAGCTTCTCTGCCTTT-3′ | ||
| Sox9 | Fwd | 5′-CCTACTACAGTCACGCAGCCG-3′ | NM_011448 |
| Rev | 5′-GGGTTCATGTAAGTGAAGGTGGA-3′ |
RNA decay experiments
To establish the effects of NICD on the stability of Il6 mRNA, RNA polymerase 2 was inhibited in confluent chondrocyte-enriched cell cultures from RosaNotch mice by exposure to 300 μM 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB, BioMol, Plymouth Meeting, PA). Il6 mRNA levels at various times following the exposure to DRB were determined in total RNA extracts by qRT-PCR analysis. The slopes of mRNA decay were established by fitting the log10 of the percentage of Il6 transcript levels corrected for Rpl38 expression over corrected Il6 transcripts before exposure to DRB against time by linear regression.
Statistical analysis
Estimates and associated uncertainty are expressed as means and 95% confidence interval, respectively. Each observation represents an independent culture from individual mice and is the average of 2 technical replicates. Normality of data distribution was determined with 95% confidence by the Shapiro-Wilk test. Significance of statistical differences was tested for pairwise comparisons by the Student’s t-test with a p < 0.05 and for multiple comparisons by applying the Bonferroni correction with a p < 0.0250 43. Statistical differences between slopes of regression lines of corrected Il6 mRNA levels against time were determined by analysis of covariance 44. Statistical analyses were performed using Microsoft Office Excel 2003 SP2 (Microsoft, Redmond, WA) or PASW software version 18.0.0 (IBM, Armonk, NY).
RESULTS
NICD induces Il6 by a transcriptional mechanism in chondrocyte-enriched cells
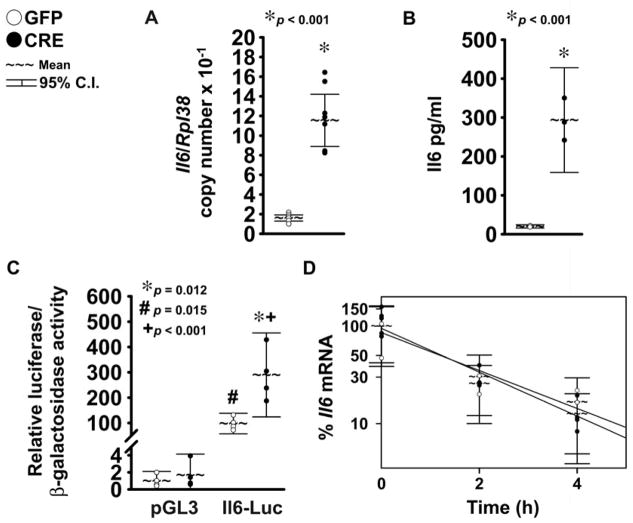
Exposure to Il6 recapitulates select effects of Notch in chondrocytes, and we tested whether Notch signaling regulates Il6 expression in chondrocyte-enriched cells from RosaNotch mice infected with Ad-CMV-Cre to induce NICD or with control Ad-CMV-GFP 26,27,28,29. Analysis of mRNA levels by qRT-PCR revealed that activation of Notch signaling induced a 7 fold increase in Il6 expression (Fig. 1A). To establish whether induction of Il6 transcripts by Notch translates into increased Il6 production, levels of Il6 protein were measured by ELISA in the supernatant of RosaNotch chondrocytes. Il6 concentration in the culture medium of controls and of cells overexpressing NICD was about 20 pg/ml and 290 pg/ml, respectively (Fig. 1B), demonstrating that Notch caused a 15 fold increase of Il6 protein levels and confirming that activation of Notch signaling in chondrocytes induces Il6.
Figure 1. NICD induces Il6 expression by transcriptional mechanisms.
Primary RosaNotch chondrocyte-enriched cells were infected with Ad-CMV-Cre (CRE, full circles) to induce NICD or with control Ad-CMV-GFP (GFP, empty circles). In panel A, total RNA was extracted 3 days after adenoviral infection, and changes in gene expression analyzed by qRT-PCR. Data are expressed as ratio of Il6 copy number over Rpl38 copy number. Observations represent cultures from individual mice, n = 8. * indicates differences between CRE and GFP. In panel B, Il6 concentration in the culture medium was determined by ELISA. Observations represent cultures from individual mice, n = 3. * indicates a difference between CRE and GFP. In panel C, cells were transfected with the pGL3 luciferase expression vector or with a fragment of the Il6 promoter cloned in pGL3 (Il6-Luc), and a β-galactosidase expression vector 24 h before adenoviral infection and harvested after 48 h. Data are expressed as luciferase activity corrected for β-galactosidase activity, relative to corrected luciferase activity in control cells transfected with pGL3. Observations represent cultures from individual mice n = 4. * indicates differences between CRE and GFP. # indicates a difference between Il6-Luc and pGL3 activity in cells expressing GFP. + indicates a difference between Il6-Luc and pGL3 activity in cells expressing CRE. In panel D, osteoblasts were transcriptionally arrested with DRB (time 0), and total RNA harvested at the indicated times was amplified by qRT-PCR. Data are percentage of Il6 mRNA copy number corrected for Rpl38 copy number, relative to corrected expression of Il6 at the time of exposure to DRB, plotted versus time (h). Intercept and slope of regression lines were calculated by linear regression of corrected Il6 expression over time. Circles represent means ± SEM for 4 cultures from individual mice.
To study the mechanisms that mediate induction of Il6 by Notch, the effects of Notch activation on the transactivation of a transiently transfected 1.3 kb fragment of the Il6 promoter (Il6-Luc) or on control pGL3 vector were tested in chondrocyte-enriched cells from RosaNotch littermate mice. Activity of pGL3 was not induced by Notch in comparison to controls, indicating that pGL3 does not contain regulatory DNA elements responsive to Notch signaling in chondrocytes. In control cells, activity of Il6-Luc was about 100-fold greater than the activity of pGL3, suggesting that under control conditions the Il6 promoter is transactivated in chondrocytes (Fig. 1C). In accordance with the stimulatory effects of Notch on Il6 mRNA levels, Il6-Luc was transactivated in the context of Notch induction. The extent of this effect was less pronounced than the induction of Il6 mRNA and protein levels by Notch, indicating that additional regulatory regions present in the intact Il6 locus determine the magnitude of Il6 induction by Notch. To assess whether Notch regulates the stability of Il6 transcripts, Il6 expression was determined by qRT-PCR in RosaNotch chondrocyte-enriched cells exposed to DRB, an inhibitor of mRNA transcription. The estimated half-life of the Il6 transcript was 1.2 h, and Notch did not affect the rate of Il6 mRNA decay (Fig. 1D), indicating that post-transcriptional mechanisms do not mediate the effects of Notch on Il6 expression.
To induce Notch activity in chondrocytes by alternate mechanisms, chondrocyte-enriched cells from wild-type C57BL/6 mice were transiently transfected with pcDNA-NICD or with control pcDNA3.1. To determine whether pcDNA-NICD activates the Notch signaling pathway, cells were co-transfected with pGL3 or with the 12xCSL-Luc reporter. Transient NICD overexpression had no impact on pGL3 activity whereas it induced 12xCSL-Luc transactivation, confirming activation of Notch signaling (Fig. 2). Under basal conditions, Il6-Luc activity was about 8 fold higher than pGL3 activity, whereas in the context of Notch induction Il6-Luc transactivation was about 4 fold higher than in the presence of pcDNA. These results suggest that the Il6 promoter has basal activity in chondrocytes and confirm that Notch induces Il6 expression by transcriptional mechanisms.
Figure 2. Transient overexpression of NICD transactivates the Il6 promoter.
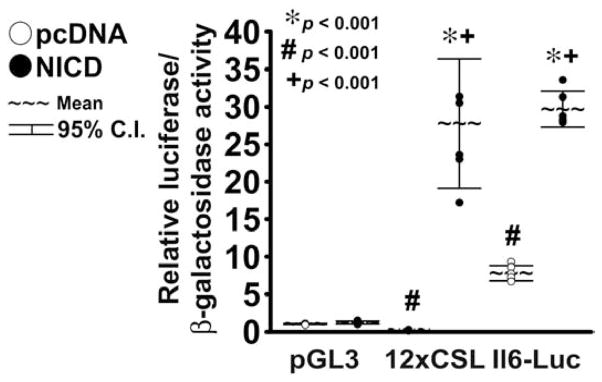
Primary chondrocyte-enriched cells from littermate wild-type C57BL/6mice were transfected with pcDNA-NICD (NICD, full circles) to induce NICD or with control pcDNA3.1 (pcDNA, empty circles). Cells were co-transfected with the pGL3 luciferase expression vector, the 12-CSL-Luc reporter (12xCSL) or a fragment of the Il6 promoter cloned in pGL3 (Il6-Luc), and a β-galactosidase expression vector. Chondrocytes were harvested after 48 h. Data are expressed as luciferase activity corrected for β-galactosidase activity, relative to corrected luciferase activity in control cells transfected with pGL3. Observations represent cultures from individual mice n = 6. * indicates differences between NICD and pcDNA. # indicates a difference between 12xCSL, or Il6-Luc, and pGL3 activity in cells transfected with pcDNA. + indicates a difference between 12xCSL, or Il6-Luc, and pGL3 activity in cells transfected with NICD.
Il6 neutralization opposes the effects of NICD on Mmp13 and Acan expression
Exposure of chondrocytes to Il6 phenocopies selected effects of Notch signaling, and we asked whether Il6 mediates the effects of Notch on the expression of chondrocyte gene markers 27,28,29. To investigate this possibility, chondrocyte-enriched cells from RosaNotch mice were exposed to a neutralizing murine monoclonal IgG raised against Il6 or murine IgG, either in the context of Notch activation or under basal conditions.
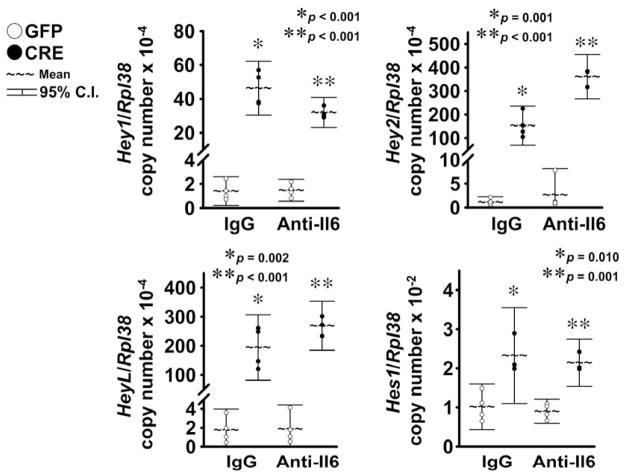
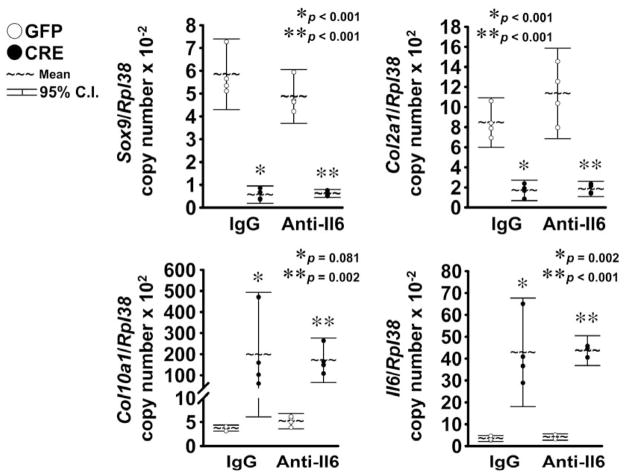
Notch induced Hey1, Hey2 and HeyL, and to a lesser extent, Hes1 transcripts, either in cells exposed to the neutralizing Il6 antibody or to normal IgG (Fig. 3), demonstrating that inhibition of Il6 does not alter Notch transactivation in chondrocyte-enriched cells. In agreement with previous results, Notch activation suppressed Sox9 and Col2a1 mRNA levels and induced Col10a1 expression in cells exposed to normal IgG 20,21,45. Similar results were obtained in the presence of the Il6 neutralizing antibody, indicating that Il6 is not required for these effects of Notch in chondrocytes. Levels of Il6 mRNA were not affected by the neutralizing Il6 antibody, either under basal conditions or in the context of NICD overexpression, excluding the possibility that the neutralizing Il6 antibody precludes induction of Il6 expression by Notch (Fig. 4). Inhibition of Il6 in the absence of Notch had no impact on Sox9, Col2a1 and Col10a1 expression, suggesting that basal Il6 levels are not required for expression of these chondrocyte gene markers.
Figure 3. Il6 neutralization does not oppose induction of Notch target genes by NICD.
Primary RosaNotch chondrocyte-enriched cells were infected with Ad-CMV-Cre (CRE, full circles) to induce NICD or with control Ad-CMV-GFP (GFP, empty circles). Cells were cultured for 3 days under conditions favoring chondrocyte maturation and serum removed for 6 h, before exposure to murine Il6 antibody (Anti-Il6) or control normal murine immunoglobulin G (IgG). Total RNA was extracted 24 h after exposure to antibodies, and changes in gene expression analyzed by qRT-PCR. Data are expressed as ratio of Hey1, Hey2, HeyL and Hes1 copy number over Rpl38 copy number. Observations represent cultures from individual mice n = 3–4. * indicates differences between CRE and GFP in the presence of IgG. ** indicates differences between CRE and GFP in the presence of Anti-Il6.
Figure 4. Il6 neutralization does not affect Sox9 and Col2a1 suppression and Col10a1 and Il6 induction by NICD.
Primary RosaNotch chondrocyte-enriched cells were infected with Ad-CMV-Cre (CRE, full circles) to induce NICD or with control Ad-CMV-GFP (GFP, empty circles). Cells were cultured for 3 days under conditions favoring chondrocyte maturation and serum removed for 6 h, before exposure to murine Il6 antibody (Anti-Il6) or control normal murine immunoglobulin G (IgG). Total RNA was extracted 24 h after exposure to antibodies, and changes in gene expression analyzed by qRT-PCR. Data are expressed as ratio of Sox9, Col2a1, Col10a1 and Il6 copy number over Rpl38 copy number. Observations represent cultures from individual mice n = 3–4. * indicates differences between CRE and GFP in the presence of IgG. ** indicates differences between CRE and GFP in the presence of Anti-Il6.
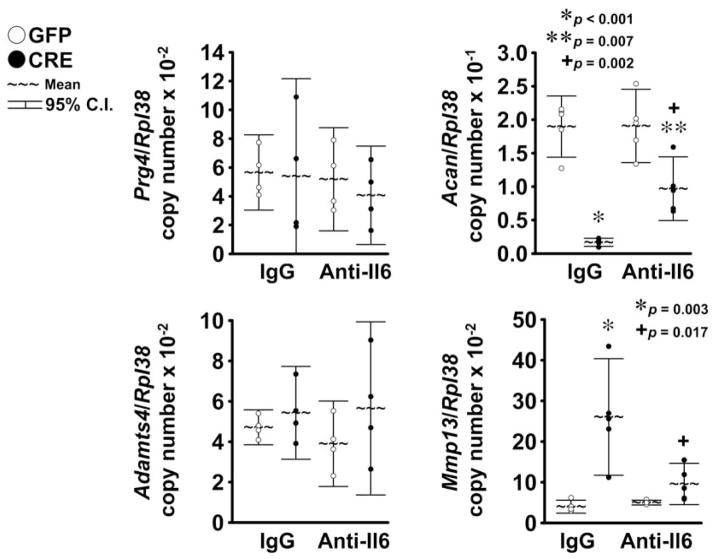
Although Notch activation had no effect on Prg4 transcripts, Notch suppressed Acan mRNA levels and this effect was opposed partially by Il6 neutralization, indicating that Notch inhibits expression of selected gene markers of chondrocyte function and this effect is mediated in part by Il6. Notch activation did not affect Ad amts4 expression, whereas it increased Mmp13 transcripts levels, confirming that under selected conditions NICD overexpression induces gene markers of cartilage matrix degradation (Fig 5) 20,21. Neutralization of Il6 opposed the stimulatory effects of Notch on Mmp13 expression, indicating that Il6 is required for this effect of Notch in chondrocytes (Fig. 5). Il6 neutralization in cells expressing GFP did not affect Prg4, Acan, Adamts4 and Mmp13 transcript levels, confirming that Il6 is dispensable for expression of selected chondrocyte gene markers.
Figure 5. Il6 neutralization opposes the effects of NICD on Acan and Mmp13 expression.
Primary RosaNotch chondrocyte-enriched cells were infected with Ad-CMV-Cre (CRE, full circles) to induce NICD or with control Ad-CMV-GFP (GFP, empty circles). Cells were cultured for 3 days under conditions favoring chondrocyte maturation and serum removed for 6 h, before exposure to murine Il6 antibody (Anti-Il6) or control normal murine immunoglobulin G (IgG). Total RNA was extracted 24 h after exposure to antibodies, and changes in gene expression analyzed by qRT-PCR. Data are expressed as ratio of Prg4, Acan, Adamts4 and Mmp13 copy number over Rpl38 copy number. Observations represent cultures from individual mice n = 3–5. * indicates differences between CRE and GFP in the presence of IgG. ** indicates differences between CRE and GFP in the presence of Anti-Il6. + indicates a difference between IgG and Anti-Il6 in the context of CRE infection.
DISCUSSION
In this study, we explored the mechanisms that mediate the regulation of gene expression by the Notch signaling pathway in chondrocytes. Several studies have established Notch as a suppressor of chondrogenesis in vivo, and this effect is consistent with the role of Notch as an inhibitor of mesenchymal cell differentiation 1. However, the role of Notch signaling in differentiated cells of the chondrocyte lineage is controversial, since both suppression of hypertrophic differentiation and elongation of the hypertrophic zone of the growth plate have been described as effects of NICD overexpression under the control of the Col2a1 promoter 18,20,22. The validity of the RosaNotch system to study the effects of Notch signaling in primary chondrocyte-enriched cell cultures was confirmed by the induction of Notch target genes following infection with Ad-CMV-Cre (data not shown). In agreement with previous findings, Notch induced expression of Hey1, 2 and L to a greater extent than Hes1 (data not shown) 21.
We confirmed that induction of Notch signaling in primary chondrocytes, irrespective of donor sex, inhibits progression to a mature chondrocyte phenotype while inducing expression of Col10a1 and Mmp13 20,21. In previous work we reported that chondrocyte-enriched cell cultures present minimal expression of osteoblast gene markers, suggesting presence of osteoblastic cells 21. Under selected conditions, osteoblasts express Mmp13, raising the possibility that increased Mmp13 transcripts levels exhibited by chondrocyte-enriched cell cultures overexpressing NICD is due to presence of contaminating osteoblasts 46. However, Notch activation did not increase Mmp13 expression in primary osteoblast cultures (data not shown), indicating that the effect is selective to cells of the chondrocyte lineage. NICD overexpression led to the induction of Il6 and suppression of Acan mRNA levels, but did not affect Prg4 or Adamts4 expression. These findings indicate that Notch activation in murine chondrocytes affects selected gene markers of chondrocyte function and cartilage matrix degradation. It is important to mention that the model culture used contains both articular and growth plate chondrocytes, limiting our ability to discern the cell population that is responding to the Notch signal.
Notch transactivated the Il6 promoter and induced release of Il6 without affecting the stability of the Il6 mRNA, demonstrating that Notch induces Il6 expression by transcriptional mechanisms. Activity of the Il6 promoter and expression of Il6 were observed in control cultures, and this is consistent with work from other laboratories reporting expression of Il6 by chondrocytes under basal conditions 47. However, concentration of Il6 in the supernatant of control cells was minimal and the neutralizing Il6 antibody had no impact on chondrocyte gene markers expression in the absence of Notch, suggesting that basal levels of Il6 are dispensable for chondrocyte function. Induction of Il6 by Notch is in agreement with previous studies carried out in bone marrow stromal cells, macrophages and breast cancer cells, and confirms that Il6 is a direct target gene of Notch signaling 26,48,49.
Activation of the Notch signaling pathway was not altered by Il6 neutralization, excluding the possibility that the effects of Il6 inhibition in the context of Notch induction are secondary to suppression of Notch signaling. Although Il6 inhibited Sox9 and Col2a1 expression, Il6 neutralization did not prevent suppression of both genes by Notch in RosaNotch chondrocyte-enriched cells, and this is in agreement with previous studies demonstrating that HEY1 and HES1 mediate the inhibitory effects of NICD on COL2A1 expression in human chondrocytes 27,28,29,45. The effects of Il6 on Col10a1 expression were not reported, and induction of Col10a1 by Notch was not affected by Il6 neutralization, raising the possibility that Col10a1, similarly to Hey genes and Hes1, is a direct target of Notch signaling. In agreement with the effects of Il6 on Mmp13 in chondrocytes, Il6 neutralization precluded Mmp13 induction by Notch, indicating that Il6 mediates this effect of Notch. Conversely, Il6 neutralization partially opposed suppression of Acan transcripts by Notch in chondrocytes 27,28,29. HEY1 overexpression inhibits ACAN expression in human bone marrow-derived chondrocytes, suggesting that suppression of Acan mRNA levels by Notch in murine cells is mediated by induction of both Hey1 and Il6 45.
In conclusion, Il6 mediates induction of Mmp13 expression by Notch and contributes to the inhibitory effect of Notch on Acan mRNA levels in cells of the chondrocyte lineage.
Acknowledgments
The authors thank Drs. D.A. Melton for RosaNotch mice, D.L. Allen for Il6 promoter fragment, J.S. Nye for NICD cDNA and L.J. Strobl for 12xCSL-Luc. The authors also thank D. Bridgewater for technical assistance and M. Yurczak for secretarial assistance.
ROLE OF THE FUNDING SOURCE
This work was supported by Grant DK045227 from the National Institute of Diabetes and Digestive and Kidney Diseases (E.C.), and Research Fellowship 5371 from the Arthritis Foundation (S.Z.). Sponsors of this study had no role in the study design, collection, analysis and interpretation of data; in the writing of the manuscript; and in the decision to submit the manuscript for publication.
Abbreviation list
- Acan
aggrecan
- Ad
adenovirus
- Adamts
a disintegrin-like and metallopeptidase with thrombospondin type 1 motif
- CMV
cytomegalovirus
- Col2a1
collagen type II α1
- Col10a1
collagen type X α1
- CSL
Epstein-Barr virus latency C promoter binding factor 1/suppressor of hairless/lag 1
- Dll
delta-like
- DMEM
Dulbecco’s modified Eagle’s medium
- ELISA
enzyme-linked immunosorbent assay
- FBS
fetal bovine serum
- Fwd
forward
- GFP
green fluorescent protein
- Hes
hairy enhancer of split
- Hey
hes related with YRPW motif
- HeyL
Hey-like
- IDT
Integrated DNA Technologies
- Il6
interleukin 6
- IgG
immunoglobulin G
- Jag
jagged
- kb
kilobase
- luc
luciferase
- Mmp
matrix metalloprotease
- NICD
Notch intracellular domain
- PCR
polymerase chain reaction
- Prg
proteoglycan
- qRT-PCR
quantitative reverse transcription-PCR
- Rev
reverse
- Rpl38
ribosomal protein l38
- SEM
standard error of the mean
- Sox
sex determining region-Y-related high mobility group-box gene
Footnotes
COMPETING INTERESTS STATEMENT
We have nothing to disclose.
AUTHOR CONTRIBUTIONS
E.C. obtained funding, provided substantial contributions to the design of the study, revised the manuscript critically for important intellectual content and approved the final version to be submitted. S.Z. obtained funding, designed the study, acquired and analyzed data, drafted and revised the article and approved the final version to be submitted. S.Z. takes responsibility for the integrity of the work of the work as a whole, from inception to finished article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zanotti S, Canalis E. Notch and the Skeleton. Mol Cell Biol. 2010;30:886–896. doi: 10.1128/MCB.01285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kovall RA. More complicated than it looks: assembly of Notch pathway transcription complexes. Oncogene. 2008;27:5099–5109. doi: 10.1038/onc.2008.223. [DOI] [PubMed] [Google Scholar]
- 4.Iso T, Sartorelli V, Poizat C, Iezzi S, Wu HY, Chung G, et al. HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Mol Cell Biol. 2001;21:6080–6089. doi: 10.1128/MCB.21.17.6080-6089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olsen BR, Reginato AM, Wang W. Bone development. Annu Rev Cell Dev Biol. 2000;16:191–220. doi: 10.1146/annurev.cellbio.16.1.191. [DOI] [PubMed] [Google Scholar]
- 6.Marcelino J, Carpten JD, Suwairi WM, Gutierrez OM, Schwartz S, Robbins C, et al. CACP, encoding a secreted proteoglycan, is mutated in camptodactyly-arthropathy-coxa vara-pericarditis syndrome. Nat Genet. 1999;23:319–322. doi: 10.1038/15496. [DOI] [PubMed] [Google Scholar]
- 7.Pacifici M, Koyama E, Iwamoto M. Mechanisms of synovial joint and articular cartilage formation: recent advances, but many lingering mysteries. Birth Defects Res C Embryo Today. 2005;75:237–248. doi: 10.1002/bdrc.20050. [DOI] [PubMed] [Google Scholar]
- 8.Elsaid KA, Fleming BC, Oksendahl HL, Machan JT, Fadale PD, Hulstyn MJ, et al. Decreased lubricin concentrations and markers of joint inflammation in the synovial fluid of patients with anterior cruciate ligament injury. Arthritis Rheum. 2008;58:1707–1715. doi: 10.1002/art.23495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ballard BL, Antonacci JM, Temple-Wong MM, Hui AY, Schumacher BL, Bugbee WD, et al. Effect of tibial plateau fracture on lubrication function and composition of synovial fluid. J Bone Joint Surg Am. 2012;94:e64. doi: 10.2106/JBJS.K.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldring MB, Marcu KB. Cartilage homeostasis in health and rheumatic diseases. Arthritis Res Ther. 2009;11:224. doi: 10.1186/ar2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Kraan PM, van den Berg WB. Chondrocyte hypertrophy and osteoarthritis: role in initiation and progression of cartilage degeneration? Osteoarthritis Cartilage. 2012;20:223–232. doi: 10.1016/j.joca.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Fosang AJ, Rogerson FM. Identifying the human aggrecanase. Osteoarthritis Cartilage. 2010;18:1109–1116. doi: 10.1016/j.joca.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 13.Roach HI, Yamada N, Cheung KS, Tilley S, Clarke NM, Oreffo RO, et al. Association between the abnormal expression of matrix-degrading enzymes by human osteoarthritic chondrocytes and demethylation of specific CpG sites in the promoter regions. Arthritis Rheum. 2005;52:3110–3124. doi: 10.1002/art.21300. [DOI] [PubMed] [Google Scholar]
- 14.Kapoor M, Martel-Pelletier J, Lajeunesse D, Pelletier JP, Fahmi H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat Rev Rheumatol. 2011;7:33–42. doi: 10.1038/nrrheum.2010.196. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe N, Tezuka Y, Matsuno K, Miyatani S, Morimura N, Yasuda M, et al. Suppression of differentiation and proliferation of early chondrogenic cells by Notch. J Bone Miner Metab. 2003;21:344–352. doi: 10.1007/s00774-003-0428-4. [DOI] [PubMed] [Google Scholar]
- 16.Fujimaki R, Toyama Y, Hozumi N, Tezuka K. Involvement of Notch signaling in initiation of prechondrogenic condensation and nodule formation in limb bud micromass cultures. J Bone Miner Metab. 2006;24:191–198. doi: 10.1007/s00774-005-0671-y. [DOI] [PubMed] [Google Scholar]
- 17.Hilton MJ, Tu X, Wu X, Bai S, Zhao H, Kobayashi T, et al. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med. 2008;14:306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mead TJ, Yutzey KE. Notch pathway regulation of chondrocyte differentiation and proliferation during appendicular and axial skeleton development. Proc Natl Acad Sci U S A. 2009;106:14420–14425. doi: 10.1073/pnas.0902306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong Y, Jesse AM, Kohn A, Gunnell LM, Honjo T, Zuscik MJ, et al. RBPjkappa-dependent Notch signaling regulates mesenchymal progenitor cell proliferation and differentiation during skeletal development. Development. 2010;137:1461–1471. doi: 10.1242/dev.042911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohn A, Dong Y, Mirando AJ, Jesse AM, Honjo T, Zuscik MJ, et al. Cartilage-specific RBPjkappa-dependent and -independent Notch signals regulate cartilage and bone development. Development. 2012;139:1198–1212. doi: 10.1242/dev.070649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zanotti S, Canalis E. Notch suppresses nuclear factor of activated T cells (NFAT) transactivation and Nfatc1 expression in chondrocytes. Endocrinology. 2013;154:762–772. doi: 10.1210/en.2012-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S, Tao J, Bae Y, Jiang MM, Bertin T, Chen Y, et al. Notch gain of function inhibits chondrocyte differentiation via Rbpj-dependent suppression of Sox9. J Bone Miner Res. 2013;28:649–659. doi: 10.1002/jbmr.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karlsson C, Brantsing C, Egell S, Lindahl A. Notch1, Jagged1, and HES5 are abundantly expressed in osteoarthritis. Cells Tissues Organs. 2008;188:287–298. doi: 10.1159/000121610. [DOI] [PubMed] [Google Scholar]
- 24.Grogan SP, Miyaki S, Asahara H, D’Lima DD, Lotz MK. Mesenchymal progenitor cell markers in human articular cartilage: normal distribution and changes in osteoarthritis. Arthritis Res Ther. 2009;11:R85. doi: 10.1186/ar2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hosaka Y, Saito T, Sugita S, Hikata T, Kobayashi H, Fukai A, et al. Notch signaling in chondrocytes modulates endochondral ossification and osteoarthritis development. Proc Natl Acad Sci U S A. 2013;110:1875–1880. doi: 10.1073/pnas.1207458110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sethi N, Dai X, Winter CG, Kang Y. Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell. 2011;19:192–205. doi: 10.1016/j.ccr.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Legendre F, Dudhia J, Pujol JP, Bogdanowicz P. JAK/STAT but not ERK1/ERK2 pathway mediates interleukin (IL)-6/soluble IL-6R down-regulation of Type II collagen, aggrecan core, and link protein transcription in articular chondrocytes. Association with a down-regulation of SOX9 expression. J Biol Chem. 2003;278:2903–2912. doi: 10.1074/jbc.M110773200. [DOI] [PubMed] [Google Scholar]
- 28.Poree B, Kypriotou M, Chadjichristos C, Beauchef G, Renard E, Legendre F, et al. Interleukin-6 (IL-6) and/or soluble IL-6 receptor down-regulation of human type II collagen gene expression in articular chondrocytes requires a decrease of Sp1. Sp3 ratio and of the binding activity of both factors to the COL2A1 promoter. J Biol Chem. 2008;283:4850–4865. doi: 10.1074/jbc.M706387200. [DOI] [PubMed] [Google Scholar]
- 29.Novakofski KD, Torre CJ, Fortier LA. Interleukin-1alpha, -6, and -8 decrease Cdc42 activity resulting in loss of articular chondrocyte phenotype. J Orthop Res. 2012;30:246–251. doi: 10.1002/jor.21515. [DOI] [PubMed] [Google Scholar]
- 30.Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci U S A. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfander D, Cramer T, Schipani E, Johnson RS. HIF-1alpha controls extracellular matrix synthesis by epiphyseal chondrocytes. J Cell Sci. 2003;116:1819–1826. doi: 10.1242/jcs.00385. [DOI] [PubMed] [Google Scholar]
- 32.Lefebvre V, Garofalo S, Zhou G, Metsaranta M, Vuorio E, de Crombrugghe B. Characterization of primary cultures of chondrocytes from type II collagen/beta-galactosidase transgenic mice. Matrix Biol. 1994;14:329–335. doi: 10.1016/0945-053x(94)90199-6. [DOI] [PubMed] [Google Scholar]
- 33.Zanotti S, Smerdel-Ramoya A, Stadmeyer L, Durant D, Radtke F, Canalis E. Notch Inhibits Osteoblast Differentiation And Causes Osteopenia. Endocrinology. 2008;149:3890–3899. doi: 10.1210/en.2008-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zanotti S, Smerdel-Ramoya A, Canalis E. Reciprocal regulation of notch and nuclear factor of activated T-cells (NFAT)c1 transactivation in osteoblasts. J Biol Chem. 2011;286:4576–4588. doi: 10.1074/jbc.M110.161893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zanotti S, Smerdel-Ramoya A, Canalis E. Nuclear factor of activated T-cells (NFAT)C2 inhibits Notch receptor signaling in osteoblasts. J Biol Chem. 2013;288:624–632. doi: 10.1074/jbc.M112.340455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Canalis E, Parker K, Feng JQ, Zanotti S. Osteoblast lineage-specific effects of notch activation in the skeleton. Endocrinology. 2013;154:623–634. doi: 10.1210/en.2012-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen DL, Uyenishi JJ, Cleary AS, Mehan RS, Lindsay SF, Reed JM. Calcineurin activates interleukin-6 transcription in mouse skeletal muscle in vivo and in C2C12 myotubes in vitro. Am J Physiol Regul Integr Comp Physiol. 2010;298:R198–R210. doi: 10.1152/ajpregu.00325.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nye JS, Kopan R, Axel R. An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development. 1994;120:2421–2430. doi: 10.1242/dev.120.9.2421. [DOI] [PubMed] [Google Scholar]
- 39.Strobl LJ, Hofelmayr H, Stein C, Marschall G, Brielmeier M, Laux G, et al. Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch1 transactivate genes by interacting with the cellular protein RBP-J kappa. Immunobiology. 1997;198:299–306. doi: 10.1016/s0171-2985(97)80050-2. [DOI] [PubMed] [Google Scholar]
- 40.Tso JY, Sun XH, Kao TH, Reece KS, Wu R. Isolation and characterization of rat and human glyceraldehyde-3-phosphate dehydrogenase cDNAs: genomic complexity and molecular evolution of the gene. Nucleic Acids Res. 1985;13:2485–2502. doi: 10.1093/nar/13.7.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakagawa O, Nakagawa M, Richardson JA, Olson EN, Srivastava D. HRT1, HRT2, and HRT3: a new subclass of bHLH transcription factors marking specific cardiac, somitic, and pharyngeal arch segments. Dev Biol. 1999;216:72–84. doi: 10.1006/dbio.1999.9454. [DOI] [PubMed] [Google Scholar]
- 42.Kouadjo KE, Nishida Y, Cadrin-Girard JF, Yoshioka M, St-Amand J. Housekeeping and tissue-specific genes in mouse tissues. BMC Genomics. 2007;8:127. doi: 10.1186/1471-2164-8-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bland JM, Altman DG. Multiple significance tests: the Bonferroni method. BMJ. 1995;310:170. doi: 10.1136/bmj.310.6973.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sokal RR, Rohlf FJ. Biometry. 2. 1981. [Google Scholar]
- 45.Grogan SP, Olee T, Hiraoka K, Lotz MK. Repression of chondrogenesis through binding of notch signaling proteins HES-1 and HEY-1 to N-box domains in the COL2A1 enhancer site. Arthritis Rheum. 2008;58:2754–2763. doi: 10.1002/art.23730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Canalis E, Delany AM. Mechanisms of glucocorticoid action in bone. Ann N Y Acad Sci. 2002;966:73–81. doi: 10.1111/j.1749-6632.2002.tb04204.x. [DOI] [PubMed] [Google Scholar]
- 47.Guerne PA, Carson DA, Lotz M. IL-6 production by human articular chondrocytes. Modulation of its synthesis by cytokines, growth factors, and hormones in vitro. J Immunol. 1990;144:499–505. [PubMed] [Google Scholar]
- 48.Wongchana W, Palaga T. Direct regulation of interleukin-6 expression by Notch signaling in macrophages. Cell Mol Immunol. 2012;9:155–162. doi: 10.1038/cmi.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin S, Mutvei AP, Chivukula IV, Andersson ER, Ramskold D, Sandberg R, et al. Non-canonical Notch signaling activates IL-6/JAK/STAT signaling in breast tumor cells and is controlled by p53 and IKKalpha/IKKbeta. Oncogene. 2012 doi: 10.1038/onc.2012.517. [DOI] [PMC free article] [PubMed] [Google Scholar]