SUMMARY
JAK/STAT signaling is localized to the wing hinge, but its function is not known. Here we show that the Drosophila STAT Stat92E is downstream of Homothorax and is required for hinge development by cell-autonomously regulating hinge-specific factors. Within the hinge, Stat92E activity becomes restricted to gap domain cells that lack Nubbin and Teashirt. While gap domain cells lacking Stat92E have significantly reduced proliferation, increased JAK/STAT signaling there does not expand this domain. Thus, this pathway is necessary but not sufficient for gap domain growth. We show that reduced Wingless (Wg) signaling dominantly inhibits Stat92E activity in the hinge. However, ectopic JAK/STAT signaling does not perturbs Wg expression in the hinge. We report negative interactions between Stat92E and the notum factor Araucan, resulting in restriction of JAK/STAT signaling from the notum. In addition, we find that the distal factor Nub represses the ligand unpaired as well as Stat92E activity. These data suggest that distal expansion of JAK/STAT signaling is deleterious to wing blade development. Indeed, mis-expression of Unpaired within the presumptive wing blade causes small, stunted adult wings. We conclude that JAK/STAT signaling is critical for hinge fate specification and growth of the gap domain and that its restriction to the hinge is required for proper wing development.
Keywords: Stat92E, Upd, Hth, hinge, Nub, Wg, gap domain, Msh, Ara
INTRODUCTION
The formation of a proximal-distal (P/D) axis that runs perpendicular to the body wall is essential for the development of vertebrate limbs and invertebrate appendages. During larval development the Drosophila wing imaginal disc is progressively divided into concentric regions along the P/D axis, and these domains are specified by a distinct set of genetic factors (Klein, 2001). The wing disc is divided into pouch, hinge and notum subdomains, which give rise to distinct structures in the adult (Fig. 1A, B).
Figure 1. JAK/STAT signaling is dynamically regulated in the wing imaginal disc.
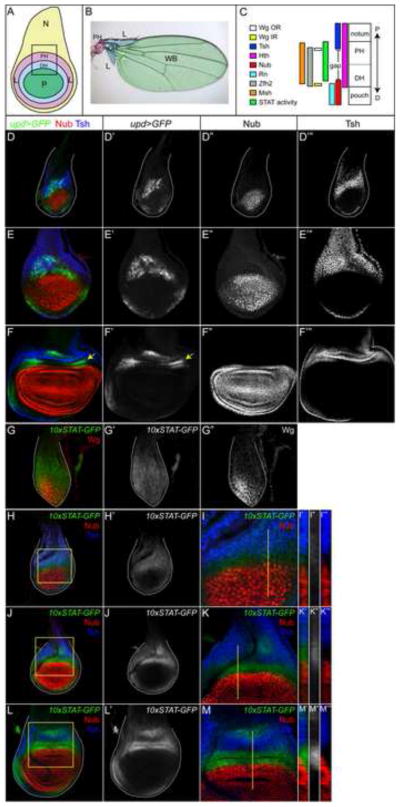
In all panels dorsal is up and anterior is to the left.
(A) Schematic of the P/D axis in a third instar wing imaginal disc, showing notum (N), proximal hinge (PH), distal hinge (DH), lateral hinge (L) and wing pouch (P).
(B) A wild type adult wing showing P/D subdivisions. Labels are the same as in A, except for the wing blade (WB), to which pouch cells fate map.
(C) Schematic representation of gene expression domains along the P/D axis of the mature wing disc.
(D–F) upd-GAL4, UAS-GFP (upd>GFP, green) expression in mid-second instar (D), early third instar (E), and late third instar (F). upd>GFP is expressed in the medial hinge within Tsh-expressing cells but it is excluded from the Nub-expressing cells in the pouch. During mid-third instar, upd appears to be expressed within the gap domain, as well as in Tsh-positive cells. D′, E′, F′ are single channels for upd>GFP and D″, E″, F″ for Nub and D‴, E‴, F‴ for Tsh.
(G–M) JAK/STAT pathway activity, as monitored by the expression of the 10xSTAT-GFP reporter, in wing discs at various stages of development. Wg is red in G. JAK/STAT signaling as shown by the expression of the 10xSTAT-GFP reporter is initially detected throughout the wing primordium (G, G′). Subsequently, it becomes restricted to the proximal regions that give rise to the hinge. An early third instar (H), a mid-third instar (J) and late third instar disc (L). Regions outlined by a yellow box in H, J and L are shown at higher resolution in I, K and M, respectively. Robust activation of the JAK/STAT pathway takes place in the gap domain (I, K, M). Locations of z-sections are marked by a yellow line in I, K and M and shown in I′-I‴, K′-K‴ and M′-M‴. I′, K′ and M′ show all three channels. I″, K″ and M″ show single channels for 10xSTAT-GFP and I‴, K‴ and M‴ show Tsh (blue) and Nub (red) channels. Nub is red and Tsh is blue in D–F, H–M.
The initial subdivision of the young wing disc into notum and wing territories is mediated by Epidermal Growth Factor Receptor (Egfr) and Wingless (Wg) signaling pathways, respectively. wg represses vein (vn) from ventro-anterior cells that will give rise to the pouch. During the second larval instar, Vn activates Egfr signaling in proximal cells, which induces Iroquois-complex (Iro-C) genes like araucan (ara). Iro-C genes encode conserved homeodomain proteins that have a selector-like function in specifying cells in the notum, and clones lacking Iro-C genes acquire dorsal hinge identity (Cavodeassi et al., 1999; Cavodeassi et al., 2000; Gomez-Skarmeta et al., 1996; McNeill et al., 1997; Wang et al., 2000; Zecca and Struhl, 2002a, b, 2007). Cells that express Iro-C genes have distinct affinities and sort out from cells that do not express Iro-C (Diez del Corral et al., 1999; Zecca and Struhl, 2002b). The distal border of Iro-C is established by the hinge-restricted factor Muscle segment homeodomain (Msh) (Dr – Flybase) and Decapentaplegic (Dpp) which repress Iro-C expression (Cavodeassi et al., 2002; Villa-Cuesta and Modolell, 2005).
Initially all cells in the wing disc express Teashirt (Tsh), a Zn-finger transcription factor, and Homothorax (Hth), a homeodomain protein (Fasano et al., 1991; Pai et al., 1998; Rieckhof et al., 1997). In second instar, tsh and hth are repressed from the presumptive pouch by wg and dpp (Azpiazu and Morata, 2000; Wu and Cohen, 2002). The nuclear protein Vestigial (Vg) indirectly induces in distal wing disc cells the genes required for wing development (del Alamo Rodriguez et al., 2002; Liu et al., 2000). These include nubbin (nub), which encodes a POU domain transcription factor (Ng et al., 1995). The nub expression domain encompasses both the wing pouch and a region of the distal hinge as well as that of rotund (rn), which encodes a Zinc finger transcription factor (Fig. 1C and (del Alamo Rodriguez et al., 2002; St Pierre et al., 2002)). Nub is required for wing patterning and growth, as wing size is severely reduced in strong nub mutations (Cifuentes and Garcia-Bellido, 1997; Ng et al., 1995). In weaker nub alleles, or in rn, tsh or hth clones, the wing hinge is deleted (Azpiazu and Morata, 2000; Casares and Mann, 2000; del Alamo Rodriguez et al., 2002). The hinge is divided into proximal and distal regions (Fig. 1C). Cells in the proximal domain express Tsh, while those in the distal hinge express Nub and Rn. Within the wing hinge, wg is expressed in two concentric rings, the inner ring (IR) in the distal domain and the outer ring (OR) in the proximal domain (Fig. 1C and (Couso et al., 1993)). The Wg IR is induced in early third instar in cells that express Nub and Rn, and it is maintained by an autoregulatory loop that requires hth and rn (del Alamo Rodriguez et al., 2002; Liu et al., 2000). The Wg IR is controlled by the spade-flag (spd-fg) enhancer, a ~1.2 kb regulatory cis-element located 9 kb upstream of the wg gene start site. The spd-fg enhancer is negatively regulated by SoxF, and the hinge is reduced in wgspd-fg mutants (del Alamo Rodriguez et al., 2002; Dichtel-Danjoy et al., 2009; Neumann and Cohen, 1996). The Wg OR is induced during mid-third instar in cells immediately distal to the Tsh expression domain through the action of a hinge-specific gene zfh2, which is required for hinge formation, and the OR is maintained by hth (Perea et al., 2009; Terriente et al., 2008; Whitworth and Russell, 2003; Wu and Cohen, 2002).
The wing hinge is divided into mutually-exclusive growth domains: distal (Nub-expressing) and proximal (Tsh-expressing) and an intermediate gap domain of Nub-negative and Tsh-negative cells (Fig. 1C and (Zirin and Mann, 2007)). In early second instar, Nub-expressing cells share an interface with Tsh-expressing cells. Ultimately, Tsh is expressed in cells that give rise to the proximal hinge, the gap domain corresponds to the region that will develop into the distal hinge, and Nub marks the wing pouch and part of the distal hinge (Ng et al., 1995; Zirin and Mann, 2007). The gap domain is established by cells in which tsh has been repressed by Wg signaling. Thereafter, this domain increases in size until the end of larval development as a result of proliferation of the Tsh-negative, Nub-negative cells (Zirin and Mann, 2007). wg is required for the proliferation of hinge cells and can induce hinge overgrowths when mis-expressed (Neumann and Cohen, 1996; Whitworth and Russell, 2003; Zirin and Mann, 2007). While wg expression is normally repressed in gap domain cells by the action of SoxF (Dichtel-Danjoy et al., 2009), it could promote growth of the gap domain by means of its sources in the Wg IR and OR. One important question to address is whether Wg is the sole or most important factor that regulates growth of the hinge or whether other signaling pathway active there have an effect on this independent growth domain.
We previously reported that JAK/STAT pathway activity is high in the wing hinge (Bach et al., 2007; Rodrigues et al., 2012). However, a role of JAK/STAT signaling in patterning the wing disc, and in particular, the P/D axis, has not been reported. In Drosophila, three related cytokines, Unpaired (Upd) (Outstretched (Os) - Flybase), Upd2 and Upd3, activate the receptor Domeless (Dome), which leads to the activation of the sole JAK kinase called Hopscotch (Hop) and the sole STAT family transcription factor called Stat92E (Arbouzova and Zeidler, 2006). In imaginal discs, activated Stat92E induces expression of target genes such as dome, Socs36E and chinmo (Flaherty et al., 2009). Viable alleles of upd - called outstretched - have held out wings phenotype, hence the official name in Flybase, implicating the JAK/STAT pathway in wing development.
Here we study the role of the JAK/STAT pathway in development of the P/D axis and formation of the hinge in the Drosophila wing. We find that upd expression and Stat92E activity become restricted to the hinge during wing development and adult hinge structures are absent in Stat92E mutant tissue. Ectopic Stat92E activity induces and/or upregulates hinge-specific factors like Zfh2, dachsous (ds) and Msh, which are reduced cell-autonomously, in Stat92E loss-of-function clones. Interestingly, ectopic activation of Stat92E also cell autonomously represses the Iro-C protein Ara, Stat92E’s induction of Msh as well as an Msh-independent mechanism. We report that Hth is upstream of Stat92E: loss of hth results in loss of Stat92E activity and gain of Hth can induce ectopic Stat92E activity cell-autonomously in the pouch and notum. Like Hth, Wg is also upstream of Stat92E in the hinge, as loss of Wg signaling leads to loss of Stat92E activity. We demonstrate that the gap domain is not maintained in the absence of Stat92E as a result of decreased proliferation in hinge clones lacking Stat92E, suggesting that Upd is a growth factor for these cells. Stat92E activity, as well as expression of its target genes, is repressed by Nub but not Tsh, which is consistent with the largely independent expression domains of Nub and activated Stat92E. In addition, ectopic expression of Nub in the hinge represses the upd gene. These data suggest that ectopic STAT activity in the pouch is deleterious to wing blade development. Indeed, when Upd is mis-expressed in the wing pouch, wing blade development is perturbed. Taken together, these data lead to a model in which multiple signals restrict JAK/STAT signaling via repression of its activating ligand and its activated signal transducer (i.e., Stat92E) to induce proper wing development.
MATERIALS AND METHODS
Fly stocks
These stocks are described in FlyBase: Stat92E85C9; Stat92E397; wg-lacZ; wgspd-fg; wgspd-fg-lacZ; nub1 (a regulatory mutant that affects expression in the wing disc (Ng et al., 1995)); msh368; chinmo1; chinmok13009 (chinmo-lacZ); hthP2; UAS-hth; UAS-hop; UAS-gfp; UAS-tshi; UAS-nub; E132(upd)-gal4; UAS-rn; UAS-hth; hthP2;UAS-tsh; UAS-3HA-Stat92E (referred to as Stat92EFL; (Ekas et al., 2006)); UAS-GFP; UAS-lacZ; 10xSTAT-GFP (Bach et al., 2007); dpp-gal4; domePG14-gal4; MS1096-gal4; UAS-nab; UAS-TCFDN; ds-lacZ We also used:
FRT82B M(3)96C ubi-GFP
FRT82B M(3)96C arm-lacZ
3XGrh/4XdSTAT-LacZ (Tsai et al., 2007)
FRT40A M(2)24F arm-lacZ
FRT40A ubi-GFP
Clonal analyses
Clones were induced by hs-flp using the FLP/FRT technique (Xu and Rubin, 1993). To provide mutant cells with a growth advantage, we employed the Minute technique, which allows mutant cells to grow faster than surrounding heterozygous cells (Morata and Ripoll, 1975). For clonal analyses, we used both Stat92E85C9 and Stat92E397, which are strong loss-of-function mutations (Ekas et al., 2010), and obtained similar results. Larvae were heat shocked for 2 hours at 39°C during first or second instar, unless otherwise specified. Since the tsh gene lies in close proximity to the FRT80 site, we generated tsh loss-of-function flip-out clones using a hairpin construct (UAS-tshi) and UAS-lacZ to mark them. We induced AyGal4 flip-out clones (Ito et al., 1997) by heat shocking for 2 hours at 39°C. Ectopic expression was also achieved with the Gal4/UAS technique (Brand and Perrimon, 1993). Adult wings were dissected and mounted in a 2:l mixture of Canada balsam and methyl salicylate.
in situ hybridization, immunofluorescence and microscopy
in situ hybridization and antibody stainings were carried out as described in (Bach et al., 2003). We used the following antibodies: mouse anti-Wg (1:10); mouse anti-β-galactosidase (β-gal) (1:20); mouse anti-Discs large (Dlg, 1:50) (all from the Developmental Studies Hybridoma Bank (DHSB)); rat anti-Ara (1:10); rabbit anti-Msh (1:500); rabbit anti-Stat92E (Flaherty et al., 2010); guinea pig- and rabbit anti-Hth (1:2000 and 1:200, respectively); rabbit anti-Vg (1:2000); mouse anti-Nub (1:10); rabbit anti-Tsh (1:500); rat and mouse anti-Zfh2 (both at 1:300); rabbit anti-GFP (1:100) (Molecular Probes); rabbit anti-β-gal (1:2000) (Cappel); chicken anti-β-gal (1:500) (Immunology Consultants Lab, Inc.). We used fluorescent secondary antibodies at 1:250 (Jackson Laboratories). We collected fluorescent images (at 25× and 40×) using a Zeiss LSM 510 confocal microscope, and bright field pictures of adult wings (at 5×) using a Zeiss Axioplan microscope with a Spot camera.
For 5-ethynyl-2′-deoxyuridine (EdU, Invitrogen) labeling, samples were incubated for 30 minutes before fixation in Ringer’s medium containing 10μM EdU. Samples were fixed and processed normally for antibody labeling, then treated as per manufacturer’s instructions.
RESULTS
Upd expression and JAK/STAT pathway activity become restricted to the wing hinge
We recently reported the critical role of JAK/STAT signaling for growth of cells in the wing disc early in larval development (Rodrigues et al., 2012). Later in development, the ligand Upd and JAK/STAT signaling become restricted to the wing hinge (Bach et al., 2007; Mukherjee et al., 2005; Rodrigues et al., 2012). To more precisely localize Upd-producing cells with regional markers of proximal fate (e.g., Tsh) or distal fate (i.e., Nub), we monitored upd expression with E132-gal4, an insertion in the upd locus (Tsai and Sun, 2004) coupled with UAS-GFP (upd>GFP), which faithfully recapitulates upd expression in the wing disc (Bach et al., 2007; Rodrigues et al., 2012). We monitored Stat92E activity using the 10xSTAT-GFP transcriptional reporter, which contains multimerized copies of a Stat92E-responsive element from the Socs36E gene (Bach et al., 2007). As previously reported, 10xSTAT-GFP is cell-autonomously lost in either Stat92E397 or Stat92E85C9 clones (Fig. S1 and (Bach et al., 2007; Ekas et al., 2006)). Consistent with prior reports, upd is expressed in the medial hinge within Tsh-expressing cells but it is excluded from the Nub-expressing cells in the pouch (Fig. 1D–F‴). During mid-third instar, upd appears to be expressed within the gap domain, as well as in Tsh-positive cells (Fig. 1F, F′, arrow). Finally, from second through third instar, upd is expressed unevenly along the circumference of the hinge, with highest level of expression consistently in the dorsal hinge (Fig. S2A–C‴″).
As we previously reported, Stat92E activity is observed in all cells in an early-second instar disc (Fig. 1G′ and (Rodrigues et al., 2012)). During late second instar, JAK/STAT pathway activity is observed at low levels within Tsh-expressing cells, but it is almost entirely excluded from the Nub domain (Fig. 1H, I). From early to late third instar, high levels of Stat92E activity localize to the gap domain (Fig. 1J–M‴). We previously reported that high levels of stabilized Stat92E protein, which serves as a marker of transcriptionally active Stat92E, are present in hinge cells (Rodrigues et al., 2012). From late second through late third instar, three to four cells at the proximal edge of the Nub domain also exhibit high levels of GFP, and one to two rows of cells on the distal side of the Tsh domain express high levels of GFP (Fig. 1I′, I″, K′, K″, M′, M″). While 10xSTAT-GFP is expressed in nearly all cells of the hinge, it is expressed unevenly with highest level of expression in the dorsal hinge, like upd (Fig. S2D–F‴″). The similarities in the expression patterns of upd, the 10xSTAT-GFP reporter and activated Stat92E protein suggest that Upd is the ligand that activates the JAK/STAT pathway in the wing. Furthermore, using in situ hybridization analysis with established upd2 and upd3 ribo-probes (Hombria et al., 2005), we were unable to detect expression of either upd-like factor in second and third instar wing discs (not shown).
Stat92E functions in parallel to Hth in regulation of the hinge
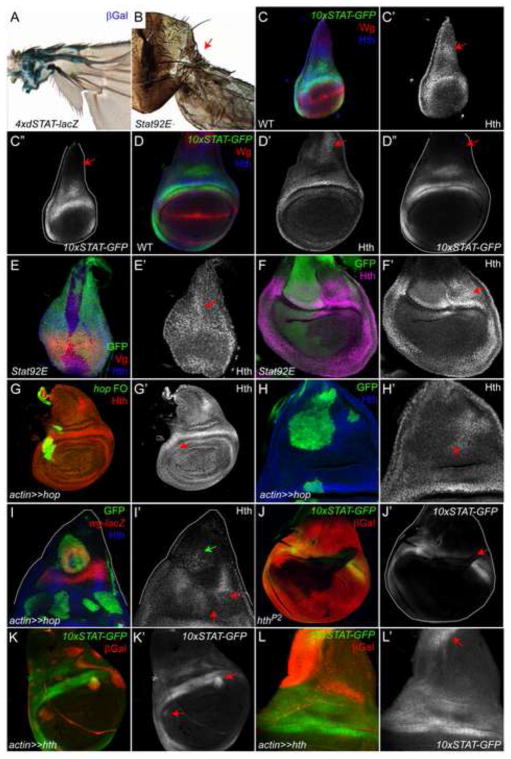
To determine the fate of cells with activated Stat92E in the adult wing, we used the 3XGrh/4XdSTAT-LacZ reporter, which contains four multimerized Stat92E binding sites regulating expression of the lacZ gene (Tsai et al., 2007). Indeed, JAK/STAT signaling is localized to the adult hinge (Fig. 2A). Furthermore, wings with Stat92E clones display a near complete lack of hinge structures and are directly attached to the thorax and locked in perpendicular alignment to the body wall hindering their ability to flutter (Fig. 2B and not shown).
Figure 2. Stat92E is critical for hinge development.
(A) Expression of the 4xdSTAT-lacZ reporter in the adult wing hinge (blue) shows that JAK/STAT pathway activity remains active in the hinge until adulthood.
(B) Stat92E clones lead to loss of hinge structures (arrow) resulting in a wing blade that connects directly to the thorax. Clones are unmarked in the adult.
(C, D) Stat92E activity (10xSTAT-GFP, green) is largely overlapping with Hth (blue) in an early (C) and middle third instar wing disc (D) in the hinge. However, Hth is expressed in the notum (C′, D′, arrows) whereas 10xSTAT-GFP is not (C″, D″, arrows). Wg is red is C and D. C′, D′ are single channels for Hth; C″, D″ for 10xSTAT-GFP.
(E, F) Hth protein (blue in E and magenta in F) is slightly increased in Stat92E that lack GFP in early (E) and late third instar (F) discs. Vg is red is E. Arrows in E′ and F′ indicate regions of elevated Hth protein. E′, F′ are single channels for Hth.
(G) Hth (red, arrow in G′) is not altered by ectopic activation of the JAK/STAT pathway in Hop flip-out clones residing in the notum (labeled actin≫hop, green). G′ is single channel for Hth.
(H, I) Hth (blue) is repressed in Hop flip-out clones (green) residing in the notum (H′, I′, red arrows). Hth can be induced in an ectopic growth domain induced by a Hop flip-out clone residing next to the wg-lacZ (red) domain in the notum (I′, green arrow). H′, I′ are single channels for Hth.
(J) Stat92E activation (green, arrow in I′) is lost in a hth clone (indicated by the loss of LacZ (red)). J′ is single channel for 10xSTAT-GFP.
(K, L) Stat92E activity (green) can be upregulated by Hth flip-out clones (red, labeled actin≫hth) that reside in within the pouch (K′, arrows) and the notum (L′, arrow). K′, J′ are single channels for 10xSTAT-GFP.
Hth is a key regulator of hinge fate (Azpiazu and Morata, 2000; Casares and Mann, 2000). Stat92E activity shares considerable overlap with Hth expression during third instar in the presumptive hinge (Fig. 2C–D″). However, Hth is expressed in the medial notum in third instar (Fig. 2C′, D′, arrows and (Azpiazu and Morata, 2000; Casares and Mann, 2000)), whereas 10xSTAT-GFP is not (Fig. 2C″, D‴, arrows). To determine if Stat92E impacts Hth expression, we examined Hth protein in Stat92E clones. Hth is slightly but consistently elevated in Stat92E clones at early and late stages of wing development (Fig. 2E′, F′, arrows), suggesting that Stat92E may negatively regulate Hth. We addressed whether ectopic activation of JAK/STAT signaling could alter Hth expression. We generated Hop-expressing clones, which activate Stat92E in a ligand-independent and cell-autonomous manner (Rodrigues et al., 2012). When Hop-expressing clones reside within the hinge, Hth expression is not changed (Fig. 2G′, arrow). By contrast, cell-autonomous reductions in Hth levels were observed in Hop expressing clones residing within the notum (Fig. 2H′, I′, red arrows). Finally, ectopic Hth is observed in Hop flip-out clones residing in the dorsal notum next to the wg stripe that induce re-patterning and an ectopic outgrowth (Fig. 2I′, green arrow). We next investigated how loss- of gain-of-function mutations in Hth affected JAK/STAT signaling. 10xSTAT-GFP and epithelial folds corresponding to hinge fate are lost in hth clones (Fig. 2J′, arrow), indicating that Stat92E is downstream of Hth in the hinge. Consistent with this observation, Stat92E activity is frequently increased in a cell-autonomous manner in Hth-expressing clones residing at the proximal edge of the pouch (Fig. 2K′, arrows). The upregulation of Stat92E activity in these clones never extends to the distal part of the clone, presumably because pouch factors like Nub and Rn (see below) repress Stat92E activity there. Lastly, ectopic 10xSTAT-GFP expression is observed within Hth-expressing clones encompassing the dorsal notum (Fig. 2L′, arrow). Similar to the pouch examples, the ectopic Stat92E activity in Hth-expressing clones within the notum is cell-autonomous but does not fill the clone boundary, presumably because notum factors repress it (see below). Taken together, we conclude that JAK/STAT signaling may exert a negative effect on Hth protein levels and that Hth - with its ability to induce Stat92E activity - resides at the top of the hinge regulatory network.
Stat92E induces hinge-specific genes and represses notum-specific genes
To determine the effect of Stat92E activity on hinge-specific genes, we monitored expression of Msh, ds and Zfh2 in clones with increased or decreased JAK/STAT signaling. In wild-type third instar discs, Msh exhibits considerable overlap with 10xSTAT-GFP in the hinge (Fig. 3A–A″). Zfh2 and ds have been previously reported to be localized to the hinge and Zfh2 is required for its development (Terriente et al., 2008; Whitworth and Russell, 2003; Zecca and Struhl, 2010). Consistent with Stat92E residing near the top of the hierarchy of genes controlling hinge development, Msh, ds and Zfh2 are reduced in a cell-autonomous manner in Stat92E clones residing in the presumptive dorsal or ventral hinge or pleura (Fig. 3B′, C′, E′, arrows). In addition, Msh is autonomously induced in Hop-expressing clones residing in the notum clearly away from the Msh endogenous expression domain (Fig. 3F–F″). Zfh2 seems to be induced by ectopic Stat92E activation in a more restricted manner, weakly and only in a limited part of Hop-expressing clones residing outside of the hinge (Fig. 3D′, arrows). One important function of Msh is to counteract the Iro-C protein Ara and they are expressed in complementary patterns (Fig. 3G and (Villa-Cuesta and Modolell, 2005)). Ara is ectopically expressed in the hinge region in Stat92E clones (Fig. 3H′, red arrow). This appears to be cell-autonomous because Ara is not ectopically expressed in neighboring heterozygous tissue that contains one functional Stat92E allele (Fig. 3H′, green arrow). We tested whether ectopic activation of JAK/STAT signaling could cell-autonomously repress Ara within its endogenous domain. Ara is autonomously repressed in Hop-expressing clones that activate JAK/STAT signaling (Fig. 3I′, arrows). In most instances, the loss of Ara within the clone is associated with upregulation of Msh (Fig. 3I′, I″, green arrow). However, occasionally we observed repression of Ara in Hop-expressing clones that do not have ectopic Msh (Fig. 3I′, I″, red arrow). We conclude that JAK/STAT signaling can repress Ara expression through Msh-dependent and Msh-independent means.
Figure 3. Activated Stat92E induces hinge-specific genes and represses notum genes.
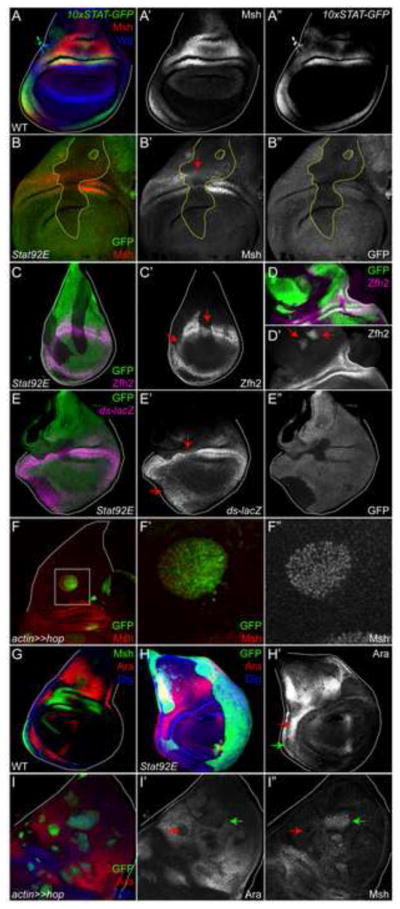
(A) In a wild type mid- to late third instar disc, Stat92E activity (green) largely overlaps with that of Msh (red) in the proximal hinge. Wg is blue. A′ is single channel for Msh; A″ for 10xSTAT-GFP.
(B) Msh (red) is lost in a cell-autonomous manner in Stat92E clones (B′, arrow), which lacks GFP. Clone boundaries are outlined by dashed yellow lines. B′ is single channel for Msh; B″ for GFP.
(C) Zfh2 (magenta) is reduced in a cell-autonomous manner (C′, arrows) in Stat92E clones, which lack GFP. C′ is single channel for Zfh2.
(D) Zfh2 (magenta) is ectopically induced in a cell-autonomous manner within some regions of Hop-expressing clones, for example at the edges of clones residing in the notum (D′, arrows). D′ is single channel for Zfh2.
(E) ds-lacZ (magenta) is reduced in a cell-autonomous manner (E′, arrows) in Stat92E clones, which lack GFP. E′ is single channel for ds-lacZ; E″ for GFP.
(F) Msh (red) can be induced in a cell autonomous manner (F′, arrow) in Hop-expressing clones residing in the notum. Dlg is blue. F′ is a close-up of the Hop flip-out clone. F″ is the single for Msh.
(G) Msh (green) and Ara (red) have mutually exclusive expression patterns. Dlg is blue.
(H) In a Stat92E clone, Ara (red) expression is expanded into the into the lateral hinge (H′, red arrow). By contrast, Ara is not increased in neighboring heterozygous cells that contain one functional copy of Stat92E (H′, green arrow). H′ is single channel for Ara.
(I) In Hop-expressing clones (green), Ara (red) is repressed in a cell-autonomous manner (arrows in I′). Msh is blue. Most often Ara repression is associated with upregulation of Msh within the Hop flip-out clone (I″, I″, green arrows). However, less frequently, we observe repression of Ara in the Hop flip-out clone without increases in Msh (I′, I″, red arrows). I′ is single channel for Ara; I″ for Msh.
JAK/STAT pathway activity is required for growth of the gap domain
The gap domain is an independent growth domain formed in late second instar after Wg represses Tsh from the pouch (Zirin and Mann, 2007). Cells in the gap domain lack Nub and Tsh, and this domain expands from late second instar until the end of larval development. Stat92E activity is detected at high levels in cells within the gap domain (Fig. 1M′), raising the possibility that JAK/STAT signaling is required for its formation and/or maintenance. Consistent with the loss of the hinge domain in adults with Stat92E clones, we observed a cell-autonomous loss of the gap domain in Stat92E clones induced 48 hours AED (compare yellow line to white line in Fig. 4A–A‴ ″). This domain is also reduced in Stat92E clones induced later in development at 96 hours AED (compare yellow line to white line in Fig. 4B–B‴). These data suggest a continuous requirement for JAK/STAT activity in growth of the gap domain. We addressed whether factors expressed in the hinge and at least partly regulated by JAK/STAT signaling are also required for formation or maintenance of the gap domain. We found that the gap domain in msh mutant clones is comparable to control tissue from the same disc (Fig. S3A–C). In addition, chinmo, which is regulated by JAK/STAT signaling in the eye disc (Flaherty et al., 2010), is not also required for maintenance of the gap domain, despite being expressed in the hinge (Fig. S3D–G). Thus, two factors that are expressed in the hinge and can be regulated by JAK/STAT signaling are not required for formation or maintenance of the gap domain.
Figure 4. JAK/STAT signaling is required for growth of the gap domain.
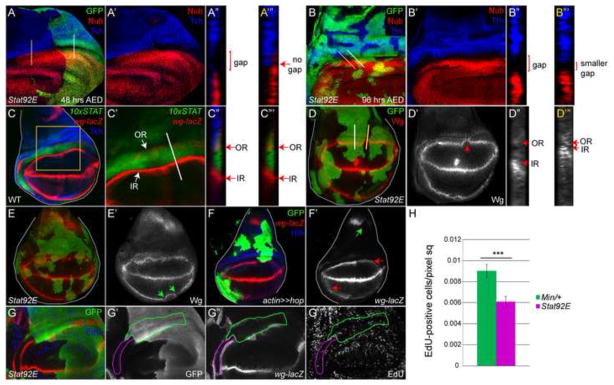
(A, B) Stat92E clones induced at 48 (A) or 96 (B) hours AED, marked by the absence of GFP, result in loss (A) or reduction (B) of the gap domain and juxtaposition of the Tsh (blue) and Nub (red) domains. xz sections are marked in (A, B) by white and yellow lines crossing heterozygous (A″, B″) and mutant (A‴, B‴) tissue, respectively. Nub and Tsh expression are shown together in A′ and B′.
(C) In wild type, the Wg OR (red) lies within the proximal region in cells that express high levels of JAK/STAT signaling (green). By contrast the Wg IR (red) abuts the distal end of JAK/STAT activity in the gap domain. C′ shows a close up of the area outlined by the yellow box in C. The region that was scanned as an xz section is marked by a white line in C′. C″ shows 10xSTAT-GFP, wg and Tsh (blue). C‴ shows 10xSTAT-GFP and wg. Note the overlap of 10xSTAT-GFP with the Wg OR and mutual exclusion of 10xSTAT-GFP with the Wg IR.
(D, E) Stat92E clones in the hinge marked by the absence of GFP result in a proximal shift of the Wg IR (red) moving it closer to the Wg OR (also red) (D′, E′, arrow) compared to surrounding heterozygous tissue. xz sections of Wg (white) through heterozygous Stat92E tissue (white line) is shown in D″ and through Stat92E homozygous mutant tissue (yellow line) is shown in D‴. Note the collapse of the Wg IR onto the Wg OR in D‴. D′, E′ are single channels for Wg.
(F) The region between the Wg IR and OR (marked by wg-lacZ, red) is not increased in Hop flip-out clones (green) (F′, arrows). By contrast, wg in the notum is repressed by ectopic activation of the JAK/STAT pathway (F, green arrow). F′ is single channel for wg-lacZ.
(G, H) Within the gap domain, Stat92E mutant tissue, marked by the absence of GFP, contains fewer EdU-positive (blue) cells than heterozygous tissue, marked by the presence of GFP. wg-lacZ (red) marks the Wg IR and OR. A magenta line marks the Stat92E mutant tissue and a green line marks the heterozygous tissue located between the Wg IR and Wg OR (G′-G‴). G′ is single channel for GFP; G″ for wg-lacZ; G‴ for EdU. Graphical representation of EdU-positive cells in Stat92E mutant (magenta bar) or heterozygous (labeled Min/+) (green bar) tissue in 10 wing discs. The mean of the number of EdU positive cells/pixels sq +/− the standard deviation of the mean is 0.0090 +/ 0.0006 in Min/+ tissue and 0.0061 +/− 0.00051 in Stat92E mutant tissue. Asterisks indicate a statistically significant difference between these genotypes (P<0.002).
The gap domain resides largely between the Wg IR and OR (Fig. 4C–C‴ and (Zirin and Mann, 2007)). In wild type, Stat92E activity abuts the Wg IR and encompasses the OR (Fig. 4C‴). In Stat92E clones, the Wg IR shifts proximally and collapses into a single ring with the Wg OR, while the position of the OR is unaffected (Fig. 4D‴, arrows). By contrast, the distance between Wg IR and OR is unaffected in adjacent Minute/+ tissue (Fig. 4D″, arrows). The reduction in the distance between in the Wg IR and OR is not limited to the dorsal hinge but rather can be observed throughout the hinge (Fig. 4E′, green arrows). We next addressed if we could expand the gap domain or affect the Wg rings by ectopic activation of the JAK/STAT signaling pathway. We and others previously showed that ectopic Stat92E can repress wg in the eye, antennal and leg discs and in the stripe of wg in the notum of the wing disc (Ayala-Camargo et al., 2007; Ekas et al., 2006; Tsai et al., 2007). We find that ectopic JAK/STAT signaling in clones does not alter the Wg IR or OR and does not increase the size of the gap domain (Fig. 4F′, red arrows), consistent with our prior report (Rodrigues et al., 2012). We verified that our Hop flip-out clones had ectopic Stat92E activity because a Hop-expressing clone in the notum autonomously represses wg expression there (Fig. 4F′, green arrow). Therefore, we conclude that JAK/STAT signaling is necessary but not sufficient for the growth of the domain between the Wg IR and OR.
To assay directly if JAK/STAT signaling could regulate growth of the gap domain, we generated Stat92E clones after 96 hours of development in order to reduce (but not eliminate) the gap domain. We labeled the discs with EdU to mark cells that were replicating their DNA. We restricted our analysis to tissue located between Wg IR and OR in the dorsal hinge. We counted the number of EdU-positive cells between the Wg IR and OR in Stat92E tissue as compared to EdU-positive cells in adjacent Minute/+ control tissue. Fig. 4G is a representative example of the 10 discs we analyzed that had Stat92E mutant and control tissue in the dorsal hinge. The number of EdU-positive cells in each genotype was then divided by the area (pixels sq) examined (Fig. 4G–H). We reasoned in that if Stat92E mutant cells and Minute/+ cells were undergoing S phase at the same rate, there would be a similar number of EdU positive cells/pixels sq in each genotype. However, we observed a statistically significant decrease (P<0.002) in the number of EdU-positive cells/pixels sq in Stat92E clones (Fig. 4H, magenta bar) as compared to adjacent heterozygous tissue (Fig. 4H, green bar). These data indicate that dorsal hinge cells lacking Stat92E cycle more slowly than control cells.
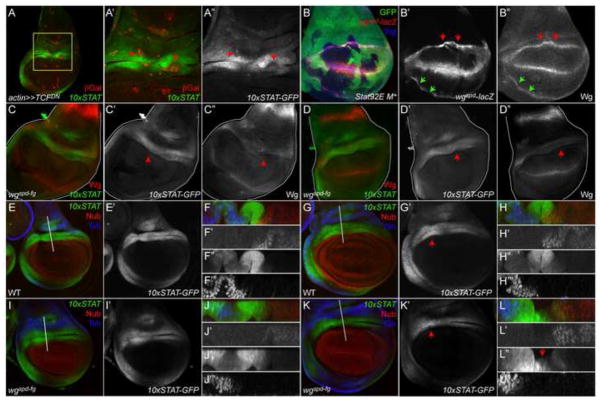
Reducing Wg signaling dominantly inhibits Stat92E activity in the wing hinge
Prior studies have shown that inhibition of Wg signaling results in reduction of the gap domain and collapse of the two Wg rings onto each other (Zirin and Mann, 2007). Since this phenotype is also observed in Stat92E clones, we wanted to assess if alterations in Wg signaling could affect Stat92E activity. We asked if reduction in Wg signaling - by mis-expression of a dominant-negative TCF (TCFDN)(van de Wetering et al., 1997) - affected JAK/STAT signaling. Clones over-expressing TCFDN were small due to reduced growth and survival, consistent with a previous report (Johnston and Sanders, 2003). When these clones resided in the 10xSTAT-GFP endogenous domain in the dorsal hinge, the expression of the Stat92E transcriptional reporter was cell autonomously reduced and the epithelial fold was perturbed within the clone (Fig. 5A–A″), presumably as a result of the decreased survival of TCFDN-expressing clones. These data suggest that JAK/STAT signaling is genetically downstream of Wg signaling in the wing hinge.
Figure 5. Wg signaling is dominant to JAK/STAT signaling in the wing hinge.
(A) TCFDN-expressing clones, which inhibit Wg signaling, (marked by βGal (red)) exhibit cell-autonomous loss of 10xSTAT-GFP (green) and the hinge epithelial fold (B′, B″, arrows). Wg is blue. B′ is a close of the area outlined in yellow in B. B″ is single channel for 10xSTAT-GFP.
(B) The wgspd-fg-LacZ reporter (magenta), corresponding to wg expression in the Wg IR (and at the dorso-ventral boundary), shifts proximally in Stat92E clones, which lack GFP. Red and green arrows in C′, C″ indicate in proximal movement of the wgspd-fg-LacZ reporter within Stat92E clone boundaries.
(C, D) 10xSTAT-GFP (green) activity is reduced in a third instar wgspd-fg mutant disc, particularly the distal most fold of reporter expression (C′, D′, arrow). The Wg IR (red) is also lost in this mutant background (C″ D″). C′, D′ is single channel for 10xSTAT-GFP; C″, D″ for Wg.
(E–L) In a second instar (E) and third instar (G) disc from +/SM5~TM6B disc (labeled WT), 10xSTAT-GFP (green) is expressed in two folds in the hinge, one proximal and one distal. Nub is Red and Tsh is blue. F, H show xz scans through the region indicated by the white lines in E and G. Stat92E activity (F″, H″) is concentrated in two domain between the Nub (F″, H″) and Tsh (F‴, H‴) domains. In a second instar wgspd-fg mutant disc, Stat92E activity (green) (I, I′) appears similar to sibling controls (E, E′). However, by mid-third instar, the distal domain of 10xSTAT-GFP expression is greatly reduced in wgspd-fg (K, K′) compared to sibling controls (G, G′). J, L show xz scans through the region indicated by the white lines in I and K. In a second instar wgspd-fg disc, Stat92E activity (J″) is concentrated in two domain between the Nub (F″) and Tsh (F‴) domains similar to sibling controls (F″). However, in a third instar wgspd-fg disc, Stat92E activity (L″) is greatly reduced in the distal fold (L″, arrow), in stark contrast to a sibling control (H″). F, H, J, L is the merge where 10xSTAT-GFP is green, Nub is red and Tsh is blue. F′, H′, J′, L′ are single channels for 10xSTAT-GFP; F″, H″, J″, L″ for Nub and F‴, H‴, J‴, L‴ for Tsh.
To further investigate the relationship between the Wg and JAK/STAT pathways in the hinge, we examined expression of a wg regulatory enhancer driving expression of lacZ that recapitulates the pattern of the Wg IR wgspd-fg-lacZ (Neumann and Cohen, 1996). We observed that wgspd-fg-lacZ moves proximally in Stat92E clones (Fig. 5B′, B″, arrows) in a manner reminiscent of wg-lacZ (Fig. 4G″) or Wg protein (Fig. 4D′, E′). The effect of loss of Stat92E on the wgspd-fg-lacZ reporter was observed throughout the entire circumference of the Wg IR, as with Wg protein (Fig. 5B″, green arrows). Finally, we examined the expression of 10xSTAT-GFP in the wgspd-fg mutant. These mutants have reduced expression of the Wg IR, impaired growth of the presumptive hinge and lack distal hinge structures in the adult (del Alamo Rodriguez et al., 2002; Dichtel-Danjoy et al., 2009; Neumann and Cohen, 1996). We carried out these experiments using a stock in which the wgspd-fg allele was in trans to the SM5~TM6B compound chromosome, which facilitates the identification of homozygous larvae by the lack of Tb. We confirmed that the Wg IR is absent in these discs (Fig. 5C″, D″, arrows). In early to mid-third instar, we find that 10xSTAT-GFP is expressed normally in the dorsal hinge of wgspd-fg mutants in the two epithelial folds, one proximal and one distal (Fig. 5I–J‴), as it is in sibling controls (Fig. 5E–F‴). By late third instar, we find a reduction in the dorsal hinge folds in wgspd-fg mutants as well as in 10xSTAT-GFP activity in the dorsal and lateral hinge (Fig. 5C′, D′, K′, arrows, Fig. 5K–L‴). The reduction in the gap domain is particular apparent for the distal fold of 10xSTAT-GFP expression (Fig. 5L″, arrow). By contrast, a third instar disc from a sibling control retained both the proximal and distal folds of 10xSTAT-GFP activity (Fig. 5G–H‴). These data indicate that the reduction in wg expression in the hinge reduces growth of cells in the gap domain even though they are capable of activating JAK/STAT signaling. Taken together, our results reveal that Wg signaling is dominant to Stat92E activity in these cells. Our results also reveal that JAK/STAT signaling likely does not directly regulate wg expression in the hinge but keeps the Wg IR and OR separated by promoting growth of the gap domain.
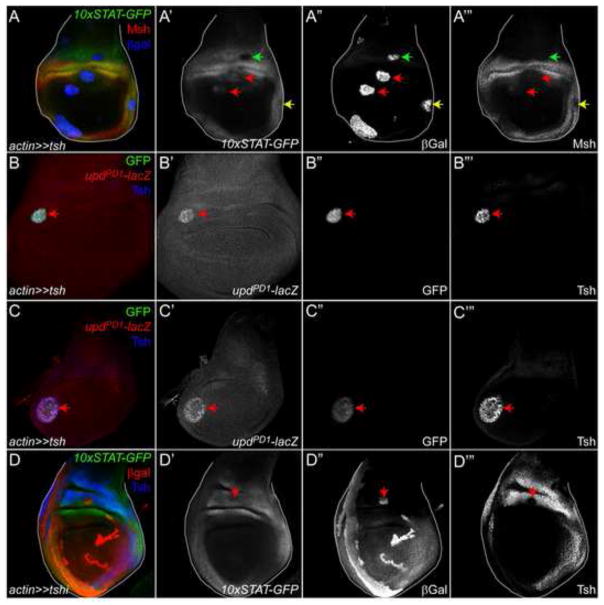
No mutual inhibition between Tsh and Stat92E
We analyzed the expression of 10xSTAT-GFP in loss- and gain-of-function clones for Tsh. Since Stat92E activity and Tsh are co-localized in some cells in the proximal hinge, we predicted that Stat92E activity would not be substantially altered when Tsh levels were experimentally manipulated. However, we found that 10xSTAT-GFP is repressed in a cell-autonomous manner when the Tsh-expressing clone arose within the proximal hinge (Fig. 6A′-A‴, green arrow) but not those that arose in the ventral hinge (Fig. 6A′-A‴, yellow arrow), despite the fact that Msh is repressed in both clones (Fig. 6A‴, green and yellow arrow). By contrast, when Tsh-expressing clones arose in the presumptive pouch, we observed induction of this reporter (Fig. 6A′-A‴, red arrows), presumably as a result of the re-patterning caused by ectopic Tsh (Azpiazu and Morata, 2000; Wu and Cohen, 2002; Zirin and Mann, 2007). We also observed cell-autonomous induction of an upd-lacZ enhancer trap updPD1 (Sun et al., 1995) in Tsh-expressing clones in the hinge and in the pouch (Fig. 6B–C‴, arrows). We reasoned that the ectopic induction of upd by Tsh likely led to autocrine and paracrine activation of Stat92E observed in some Tsh-expressing clones.
Figure 6. Stat92E and Tsh do not exhibit mutual antagonism.
(A) 10xSTAT-GFP is repressed in Tsh-clones residing in the proximal hinge (green arrow, A′, A″) but not in the lateral hinge (A′, A″, yellow arrow), despite Msh being repressed in both clones (A‴, green and yellow arrow). By contrast, 10xSTAT-GFP is induced in Tsh-expressing clones residing in the pouch (arrows in A′, A‴). Msh (red) is weakly induced in some Tsh-expressing clones in the pouch (red arrows in A‴). A′ is single channel for 10xSTAT-GFP; A″ for βGal; A‴ for Msh.
(B, C) An upd enhancer trap (updPD1, marked by βGal (red)) is cell-autonomously induced in Tsh-expressing clones (arrows in B–B‴ and C–C‴), which express both GFP (green) and Tsh (blue). The upregulation of upd occurs in both the hinge (B) and the pouch (B). Note that the endogenous domain of Tsh is not visible in B‴ because the gain of the confocal was decreased to not over-saturate the signal. B′ and C′ are single channel for βGal; B″ and C″ for GFP; B‴ and C″ for Tsh.
(D) Endogenous 10xSTAT-GFP is not altered in a Tsh RNAi (tshi)-expressing clone (marked by βGal (red)) residing in the proximal hinge (arrow in D′-D‴). The lack of Tsh protein within the clone (arrow in D‴) indicates the effective of the knockdown. Tsh is blue in D. D′ is single channel for 10xSTAT-GFP ; D′ for βGal; D‴ for Tsh.
We hypothesized that if Tsh represses JAK/STAT signaling to the gap domain, the lack of Tsh should result in increased activity of the pathway, particularly if the clone arose in the proximal hinge. We were unable to examine the effects of tsh loss-of-function clones on Stat92E activity due to the close proximity of the tsh gene to FRT80. Instead, we reduced Tsh expression by ectopically expressing a tsh RNAi construct (tshi) (Zirin and Mann, 2007). Although Tsh protein is undetectable in a tshi clone (Fig. 6D‴, arrow), JAK/STAT signaling is unaltered in it (Fig. D″, arrow). We acknowledge that it is possible that the lack of repression of 10xSTAT-GFP in tshi clones could be explained by a redundant repression by tip-top, a Tsh paralogue that is expressed in an overlapping pattern to that of tsh (Bessa et al., 2009). We also monitored the expression of Tsh in clones that had ectopic JAK/STAT signaling. Tsh expression is not altered in Hop-expressing cells (Fig 7A, A‴, green arrows). These data suggest that activated Stat92E does not repress Tsh. These data, taken together with the continuous overlap of Tsh with basal JAK/STAT signaling in the dorsal wing disc throughout larval development (Fig. 1H–M‴), suggest that Tsh and activated Stat92E do not exhibit strong repressive interactions.
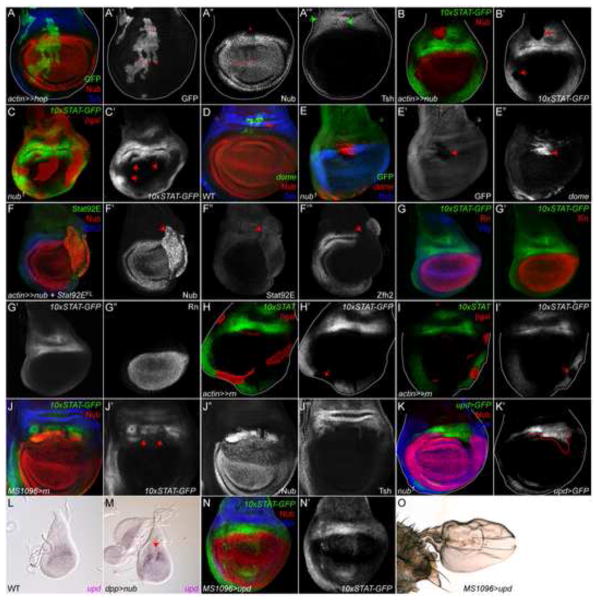
Figure 7. Nub represses Stat92E activity and upd production.
(A) Hop-expressing clones (green) moderately repress Nub (red) (small red arrows in A′ and A″) but do not alter Tsh (blue) (green arrows in A‴). A′ is single channel for GFP; A″ for Nub; A‴ for Tsh.
(B) Nub-expressing clones (red) residing within the hinge autonomously (B′, arrow) repress 10xSTAT-GFP (green). B′ is single channel for 10xSTAT-GFP.
(C) 10xSTAT-GFP extends distally into the pouch in nub1 mutant clones, which lack βGal (red). Nub is blue. B″ is single channel for 10xSTAT-GFP and the expansion into the pouch is indicated by the arrows.
(D, E) dome-gal4, UAS-GFP (labeled “dome” (green)) is observed only in the dorsal medial hinge (arrow in C′). dome-gal4, UAS-lacZ (marked by βGal (red) in mosaic nub1 mutant clones (marked by the absence of GFP) is ectopically expressed in the pouch (E′, E″, arrows). Nub is blue. E′ is single channel for GFP; E″ for βGal.
(F) Clonal mis-expression of Stat92EFL (green) together with Nub (red) does not restore expression of Zfh2 (blue) and hinge folds to the clone, suggesting that Nub represses Stat92E activity and not Stat92E expression. The clone is most easily visualized by ectopic Nub expression. F′ is single channel for Nub; F″ for Stat92E; F‴ for Zfh2.
(G) 10xSTAT-GFP expression domain (green) is separated from the Rn domain (red) by the Wg IR (blue). G′ shows merge of 10xSTAT-GFP and Rn channels. G″ is single channel for 10xSTAT-GFP; G‴ for Rn.
(H, I) Rn-expressing clones that are marked with βgal (red) residing within the hinge autonomously (H′, I′, arrows) repress 10xSTAT-GFP (green). H′ and I′ are single channels for 10xSTAT-GFP.
(J) 10xSTAT-GFP (green) is downregulated when Rn is mis-expressed by MS1096-Gal4. Nub is red and Tsh is blue. Arrows in J′ mark the perturbations to 10xSTAT-GFP in the dorsal hinge. J′ is single channel for 10xSTAT-GFP; J″ for Nub; J‴ for Tsh.
(K) upd-gal4, UAS-GFP (upd>GFP) is ectopically expressed in pouch cells in nub1 mutant clones marked by the absence of Nub in red and βgal in blue. K′ is the single channel for upd>gfp with the clone boundary outlined by red line.
(L, M) in situ hybridization reveals the expression of upd within the medial hinge in a wild type second instar wing disc (L). In a dpp>nub second instar disc, upd is repressed within the dpp domain (arrow, M).
(N, O) MS1096-driven over-expression of upd (MS1096>upd) in the pouch and dorsal wing results in ectopic activation of the JAK/STAT pathway in the pouch (N′), and small wings in the adult (O).
Nub and Rn restrict JAK/STAT activity to the presumptive hinge
Given the progressive exclusion of Stat92E activity from the presumptive pouch during wing development and the fact that Nub can act as a direct transcriptional repressor (Neumann and Cohen, 1998), we tested the hypothesis that Nub represses of one or more of the components of the JAK/STAT pathway. Cells that ectopically express Nub always exhibit autonomous loss of JAK/STAT pathway activity regardless of their position along the hinge circumference (n ≥300 Nub-expressing clones examined) (Fig 7B′, arrows and data not shown). Furthermore, in nub1 clones, Stat92E activity is ectopically observed in the pouch along the entire circumference of the hinge-pouch interface (Fig. 7C′, arrows). To rule out the trivial possibility that ectopic expression of Stat92E activity in nub1 clones was due to spurious results of the 10xSTAT-GFP reporter, we examined expression of dome, another well-characterized Stat92E target gene that encodes the Upd receptor. In the embryo and eye disc the expression of the dome gene is substantially increased by activation of the JAK/STAT pathway in a positive feedback loop (Bach et al., 2003; Flaherty et al., 2009; Lovegrove et al., 2006). In wild type third instar wing discs, the dome>gfp transcriptional reporter is faintly detected in the dorsal hinge (Fig. 7D, arrow). In nub1 mutant clones, dome is ectopically expressed in the proximal wing pouch (Fig. 7E–E″, arrow). These results indicate that JAK/STAT signaling is increased within the hinge when Nub levels are reduced. Consistent with the model that Nub inhibits Stat92E activity (and not Stat92E expression), we are unable to restore hinge folds and Zfh2 expression in clones that mis-express both Nub and full-length Stat92E transgenes (Fig. 7F–F‴, arrow).
We note that the ectopic expression of 10xSTAT-GFP and dome do not extend fully into the presumptive pouch within nub clones (Fig. 7C′, E″, arrows). We reasoned that additional pouch factors like Rn might repress JAK/STAT signaling in clones with reduced Nub levels. Indeed, co-labeling for 10xSTAT-GFP, Wg and Rn revealed that Rn and activated Stat92E are separated by the Wg IR (Fig. 7G–G‴). Consistent with our hypothesis that pouch factors other than Nub can restrict JAK/STAT signaling, we observe cell autonomous repression of 10xSTAT-GFP in Rn-expressing clones in the lateral and ventral hinge (Fig. 7H′, I′, arrows). 10xSTAT-GFP is downregulated in the dorsal hinge when Rn is mis-expressed using MS1096-Gal4 (Fig. 7J′, arrows). The MS1096-Gal4 driver is initially expressed in the dorsal wing disc, and at later larval stages in the ventral pouch (Capdevila and Guerrero, 1994; Neumann and Cohen, 1996).
We also addressed if Nub could inhibit expression the ligand Upd, which is expressed in a characteristic five-spot pattern in a wild-type wing disc (Fig. 1F′ and (Bach et al., 2007; Mukherjee et al., 2005)). Indeed, upd>GFP reporter is ectopically activated in nub1 mutant clones throughout the dorsal hinge (where it is usually restricted to two dots) and in the proximal wing pouch in a cell-autonomous manner (Fig. 7K, K′). We observe a similar result in nub1 clones straddling in the lateral and ventral hinge (data not shown). We then tested the hypothesis that Nub represses upd production. We expressed Nub outside of its endogenous domain and monitored upd mRNA by in situ hybridization. In wild type second instar wing discs, upd is expressed within the central hinge (Fig. 7L), consistent with a previous report (Mukherjee et al., 2005). By contrast, upd is strongly repressed within its endogenous domain by mis-expression of UAS-nub along the A/P axis with dpp-gal4 (Fig. 7M, arrow). These results suggest that Nub represses the upd gene. Taken together with results indicating that Nub also inhibits Stat92E activity, we favor the interpretation that Upd synthesis and Stat92E activity are incompatible with cells that have acquired pouch identity.
Finally, we addressed the issue of whether ectopic JAK/STAT signaling in clones could affect expression of Nub. We observed that Nub protein levels are moderately reduced in a cell-autonomous manner in Hop flip-out clones residing in the pouch (Fig. 7A″, arrows). These data suggest that activated Stat92E can moderately repress Nub.
Down-regulation of JAK/STAT activity from the pouch is necessary for wing blade development
The data we have presented to this point indicate that defective distal signals, such as loss of Nub, fail to restrict JAK/STAT pathway signaling to the hinge, resulting in ectopic activation of Stat92E distally. Previous studies have shown that ectopic expression of hinge factors obstruct wing blade development and result in proximalization of the wing, exemplified by the formation of small wings (Azpiazu and Morata, 2000; Casares and Mann, 2000). To determine the physiological relevance of restriction of JAK/STAT pathway activation to the hinge, we ectopically expressed Upd in the pouch using the MS1096-Gal4 driver. Indeed, mis-expression of upd by this driver results in ectopic activation of the JAK/STAT pathway in the developing wing pouch (Fig. 7N, N′) and small/stunted adult wing blades (Fig. 7O). These data indicate that JAK/STAT pathway activity needs to be constrained to the hinge for proper wing blade development to occur.
DISCUSSION
Here we show the critical role of the JAK/STAT pathway in formation and growth of the wing hinge. We find that Stat92E activity, which becomes progressively localized to the wing hinge during larval stages, is critical for the development of hinge structures at larval and adult stages. Promotion of hinge fate by JAK/STAT signaling occurs through cell-autonomous upregulation of hinge-specific factors like Ds, Msh and Zfh2 (Fig. 8). The fact that Ds and Zfh2 are reduced but not lost in Stat92E clones and that Zfh2 is only upregulated in Hop flip-out clones residing in specific locations and in certain parts of the clone suggests that Stat92E augments Ds and Zfh2 levels but is probably not their primarily regulator. By contrast, Msh appears to be more robustly regulated by Stat92E in a cell-autonomous manner, suggesting it may be a direct target. Stat92E activity also antagonizes Ara expression, through Msh, a known Ara antagonist (Villa-Cuesta and Modolell, 2005), as well as through Msh-independent means. The repressive actions of Ara on cells that have Msh and activated Stat92E would restrict cells with activated JAK/STAT signaling out of the notum towards the hinge. In addition, the pouch factor Nub through repression of upd and Stat92E activity would restrict cells with JAK/STAT signaling out of the pouch towards the hinge. The relatively modest extent of the ectopic expression of dome>gfp, 10xSTAT-GFP and upd>gfp into the pouch in nub clones indicates that there are additional repressors of the pathway in cells there. The fact that ectopic clonal mis-expression of Rn causes autonomous repression of 10xSTAT-GFP strongly suggests that Rn is one of these other factors. Our results suggest that Stat92E activity must be excluded from the pouch. Indeed forced mis-expression of upd within the pouch perturbs wing blade development.
Figure 8. Models.
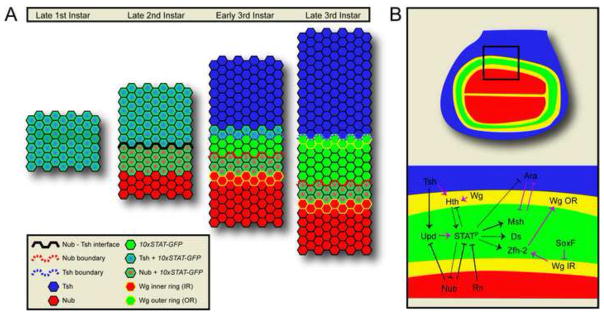
(A) Model representing the relative location of activated Stat92E throughout development with respect to Nub, Tsh and Wg IR and Wg OR.
(B) Model of factors investigated here. Black line indicate interactions reported in this study. Purple lines indicate previously known interactions. We showed that Tsh can induce Upd, which activates Stat92E. We find that JAK/STAT signaling, which is progressively refined to the hinge, upregulates factors Msh, ds and Zfh2. Our data suggest that Ara is repressed by activated Stat9E in Msh-dependent and Msh-independent manners. Within the hinge, Hth and Wg are both upstream of and dominant to activated Stat92E. Stat92E has little effect on Wg signaling but it can repress Hth. Pouch factors like Nub and Rn restrict Upd/JAK/STAT signaling proximally. Dashed lines represent weaker interactions.
Given the importance of Hth in the development of the hinge, we sought to determine its relationship to Stat92E. We find that Stat92E activity is lost in hth clones, suggesting that hth is upstream to Stat92E. Consistent with this, Stat92E activity is induced within Hth-expressing clones residing in the pouch and in the dorsal notum. The ectopic activation of Stat92E within Hth clones does not fill the entire clone boundary but only occurs in a restricted domain, suggesting that factors in the pouch (like Nub and Rn) and the notum (like Ara) repress Stat92E expression and/or activity. We also note that Hth protein is slightly but consistently increased in Stat92E loss-of-function clones, suggesting that Stat92E may normally restrain Hth to some degree. Indeed, Hop-expressing clones residing with the notum exhibit cell-autonomous reduction in Hth expression. In sum, we propose a model in which Hth is the dominant player in hinge formation but Stat92E also has important roles in this region.
We find that JAK/STAT signaling is critical for growth of the gap domain. Stat92E mutant cells in the gap domain proliferate less than cells from adjacent control tissue. This results is a shorter distance between Wg IR and OR and a smaller gap domain (Zirin and Mann, 2007). Our results support a model in which Stat92E is not required for formation of the gap domain but rather for its growth, particularly since we are unable to find any inhibitory action of Stat92E on Wg, which is a potent regulator of growth of the hinge. However, unlike the hinge over-growth observed with gain of wg or loss of its repressor SoxF (Dichtel-Danjoy et al., 2009; Neumann and Cohen, 1996; Whitworth and Russell, 2003), ectopic activation of JAK/STAT signaling cannot expand the gap domain. Thus, JAK/STAT signaling is necessary but not sufficient for gap domain proliferation. Our results indicate that Wg is upstream of JAK/STAT signaling in the hinge since Stat92E activity is lost cell-autonomously in TCFDN-expressing clones and progressively from the gap domain in wgspd-fg mutant wing disc.
In sum, we propose that notum and pouch signals confine JAK/STAT signaling to the proximal domain, where activated Stat92E is required to confer proximal fates by regulation of hinge markers, as well as to induce growth of the gap domain. Our data is consistent with a model in which Hth and Wg act upstream of JAK/STAT signaling in hinge fate acquisition and with JAK/STAT signaling being permissive in this process. Since the factors that modulate Stat92E activity or that are controlled by JAK/STAT signaling are evolutionarily conserved, this raises the intriguing possibility that similar regulatory networks exist in higher organisms.
Supplementary Material
STAT upregulates hinge factors Msh, Ds and Zfh2 and represses notum factors Iro-C.
STAT acts downstream of Wg and Hth and is required for growth of the gap domain.
Nubbin represses Unpaired and STAT activity from the pouch.
Mis-expression of Unpaired in the pouch causes small, stunted adult wings.
Hinge restriction of STAT activity is essential for normal wing growth and patterning.
Acknowledgments
We thank our colleagues, the Bloomington stock center and the DHSB for flies and antibodies. We are particularly grateful to Sonsoles Campuzano for sending the msh allele and Ara antibody to us so quickly. We are very grateful to the members of the NYU Biology Department and Esteban Mazzoni for their generosity in the aftermath of Superstorm Sandy. We also thank two anonymous reviewers whose comments greatly improved this study. This work was supported by March of Dimes Basil O’Connor Starter Award (5-FY06-579), Breast Cancer Alliance, Inc. Young Investigator Award, and NIH grant R01-GM085075 (all to EAB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arbouzova NI, Zeidler MP. JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development (Cambridge, England) 2006;133:2605–2616. doi: 10.1242/dev.02411. [DOI] [PubMed] [Google Scholar]
- Ayala-Camargo A, Ekas LA, Flaherty MS, Baeg GH, Bach EA. The JAK/STAT pathway regulates proximo-distal patterning in Drosophila. Dev Dyn. 2007;236:2721–2730. doi: 10.1002/dvdy.21230. [DOI] [PubMed] [Google Scholar]
- Azpiazu N, Morata G. Function and regulation of homothorax in the wing imaginal disc of Drosophila. Development (Cambridge, England) 2000;127:2685–2693. doi: 10.1242/dev.127.12.2685. [DOI] [PubMed] [Google Scholar]
- Bach EA, Ekas LA, Ayala-Camargo A, Flaherty MS, Lee H, Perrimon N, Baeg GH. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr Patterns. 2007;7:323–331. doi: 10.1016/j.modgep.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Bach EA, Vincent S, Zeidler MP, Perrimon N. A sensitized genetic screen to identify novel regulators and components of the Drosophila janus kinase/signal transducer and activator of transcription pathway. Genetics. 2003;165:1149–1166. doi: 10.1093/genetics/165.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessa J, Carmona L, Casares F. Zinc-finger paralogues tsh and tio are functionally equivalent during imaginal development in Drosophila and maintain their expression levels through auto- and cross-negative feedback loops. Dev Dyn. 2009;238:19–28. doi: 10.1002/dvdy.21808. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development (Cambridge, England) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Capdevila J, Guerrero I. Targeted expression of the signaling molecule decapentaplegic induces pattern duplications and growth alterations in Drosophila wings. The EMBO journal. 1994;13:4459–4468. doi: 10.1002/j.1460-2075.1994.tb06768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casares F, Mann RS. A dual role for homothorax in inhibiting wing blade development and specifying proximal wing identities in Drosophila. Development (Cambridge, England) 2000;127:1499–1508. doi: 10.1242/dev.127.7.1499. [DOI] [PubMed] [Google Scholar]
- Cavodeassi F, Diez Del Corral R, Campuzano S, Dominguez M. Compartments and organising boundaries in the Drosophila eye: the role of the homeodomain Iroquois proteins. Development (Cambridge, England) 1999;126:4933–4942. doi: 10.1242/dev.126.22.4933. [DOI] [PubMed] [Google Scholar]
- Cavodeassi F, Modolell J, Campuzano S. The Iroquois homeobox genes function as dorsal selectors in the Drosophila head. Development (Cambridge, England) 2000;127:1921–1929. doi: 10.1242/dev.127.9.1921. [DOI] [PubMed] [Google Scholar]
- Cavodeassi F, Rodriguez I, Modolell J. Dpp signalling is a key effector of the wing-body wall subdivision of the Drosophila mesothorax. Development (Cambridge, England) 2002;129:3815–3823. doi: 10.1242/dev.129.16.3815. [DOI] [PubMed] [Google Scholar]
- Cifuentes FJ, Garcia-Bellido A. Proximo-distal specification in the wing disc of Drosophila by the nubbin gene. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:11405–11410. doi: 10.1073/pnas.94.21.11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couso JP, Bate M, Martinez-Arias A. A wingless-dependent polar coordinate system in Drosophila imaginal discs. Science (New York, N Y. 1993;259:484–489. doi: 10.1126/science.8424170. [DOI] [PubMed] [Google Scholar]
- del Alamo Rodriguez D, Terriente J, Galindo MI, Couso JP, Diaz-Benjumea FJ. Different mechanisms initiate and maintain wingless expression in the Drosophila wing hinge. Development (Cambridge, England) 2002;129:3995–4004. doi: 10.1242/dev.129.17.3995. [DOI] [PubMed] [Google Scholar]
- Dichtel-Danjoy ML, Caldeira J, Casares F. SoxF is part of a novel negative-feedback loop in the wingless pathway that controls proliferation in the Drosophila wing disc. Development (Cambridge, England) 2009;136:761–769. doi: 10.1242/dev.032854. [DOI] [PubMed] [Google Scholar]
- Diez del Corral R, Aroca P, JL Gm-S, Cavodeassi F, Modolell J. The Iroquois homeodomain proteins are required to specify body wall identity in Drosophila. Genes & development. 1999;13:1754–1761. doi: 10.1101/gad.13.13.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekas LA, Baeg GH, Flaherty MS, Ayala-Camargo A, Bach EA. JAK/STAT signaling promotes regional specification by negatively regulating wingless expression in Drosophila. Development (Cambridge, England) 2006;133:4721–4729. doi: 10.1242/dev.02675. [DOI] [PubMed] [Google Scholar]
- Ekas LA, Cardozo TJ, Flaherty MS, McMillan EA, Gonsalves FC, Bach EA. Characterization of a dominant-active STAT that promotes tumorigenesis in Drosophila. Developmental biology. 2010;344:621–636. doi: 10.1016/j.ydbio.2010.05.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano L, Roder L, Core N, Alexandre E, Vola C, Jacq B, Kerridge S. The gene teashirt is required for the development of Drosophila embryonic trunk segments and encodes a protein with widely spaced zinc finger motifs. Cell. 1991;64:63–79. doi: 10.1016/0092-8674(91)90209-h. [DOI] [PubMed] [Google Scholar]
- Flaherty MS, Salis P, Evans CJ, Ekas LA, Marouf A, Zavadil J, Banerjee U, Bach EA. chinmo is a functional effector of the JAK/STAT pathway that regulates eye development, tumor formation and stem cell self-renewal in Drosophila. Developmental Cell. 2010;18:556–568. doi: 10.1016/j.devcel.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty MS, Zavadil J, Ekas LA, Bach EA. Genome-wide expression profiling in the Drosophila eye reveals unexpected repression of notch signaling by the JAK/STAT pathway. Dev Dyn. 2009;238:2235–2253. doi: 10.1002/dvdy.21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Skarmeta JL, Diez del Corral R, de la Calle-Mustienes E, Ferre-Marco D, Modolell J. Araucan and caupolican, two members of the novel iroquois complex, encode homeoproteins that control proneural and vein-forming genes. Cell. 1996;85:95–105. doi: 10.1016/s0092-8674(00)81085-5. [DOI] [PubMed] [Google Scholar]
- Hombria JC, Brown S, Hader S, Zeidler MP. Characterisation of Upd2, a Drosophila JAK/STAT pathway ligand. Developmental biology. 2005;288:420–433. doi: 10.1016/j.ydbio.2005.09.040. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development (Cambridge, England) 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Johnston LA, Sanders AL. Wingless promotes cell survival but constrains growth during Drosophila wing development. Nature cell biology. 2003;5:827–833. doi: 10.1038/ncb1041. [DOI] [PubMed] [Google Scholar]
- Klein T. Wing disc development in the fly: the early stages. Current opinion in genetics & development. 2001;11:470–475. doi: 10.1016/s0959-437x(00)00219-7. [DOI] [PubMed] [Google Scholar]
- Liu X, Grammont M, Irvine KD. Roles for scalloped and vestigial in regulating cell affinity and interactions between the wing blade and the wing hinge. Developmental biology. 2000;228:287–303. doi: 10.1006/dbio.2000.9939. [DOI] [PubMed] [Google Scholar]
- Lovegrove B, Simoes S, Rivas ML, Sotillos S, Johnson K, Knust E, Jacinto A, Hombria JC. Coordinated control of cell adhesion, polarity, and cytoskeleton underlies Hox-induced organogenesis in Drosophila. Curr Biol. 2006;16:2206–2216. doi: 10.1016/j.cub.2006.09.029. [DOI] [PubMed] [Google Scholar]
- McNeill H, Yang CH, Brodsky M, Ungos J, Simon MA. mirror encodes a novel PBX-class homeoprotein that functions in the definition of the dorsal-ventral border in the Drosophila eye. Genes & development. 1997;11:1073–1082. doi: 10.1101/gad.11.8.1073. [DOI] [PubMed] [Google Scholar]
- Morata G, Ripoll P. Minutes: mutants of drosophila autonomously affecting cell division rate. Developmental biology. 1975;42:211–221. doi: 10.1016/0012-1606(75)90330-9. [DOI] [PubMed] [Google Scholar]
- Mukherjee T, Hombria JC, Zeidler MP. Opposing roles for Drosophila JAK/STAT signalling during cellular proliferation. Oncogene. 2005;24:2503–2511. doi: 10.1038/sj.onc.1208487. [DOI] [PubMed] [Google Scholar]
- Neumann CJ, Cohen SM. Distinct mitogenic and cell fate specification functions of wingless in different regions of the wing. Development (Cambridge, England) 1996;122:1781–1789. doi: 10.1242/dev.122.6.1781. [DOI] [PubMed] [Google Scholar]
- Neumann CJ, Cohen SM. Boundary formation in Drosophila wing: Notch activity attenuated by the POU protein Nubbin. Science (New York, NY. 1998;281:409–413. doi: 10.1126/science.281.5375.409. [DOI] [PubMed] [Google Scholar]
- Ng M, Diaz-Benjumea FJ, Cohen SM. Nubbin encodes a POU-domain protein required for proximal-distal patterning in the Drosophila wing. Development (Cambridge, England) 1995;121:589–599. doi: 10.1242/dev.121.2.589. [DOI] [PubMed] [Google Scholar]
- Pai CY, Kuo TS, Jaw TJ, Kurant E, Chen CT, Bessarab DA, Salzberg A, Sun YH. The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, extradenticle, and suppresses eye development in Drosophila. Genes & development. 1998;12:435–446. doi: 10.1101/gad.12.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea D, Terriente J, Diaz-Benjumea FJ. Temporal and spatial windows delimit activation of the outer ring of wingless in the Drosophila wing. Developmental biology. 2009;328:445–455. doi: 10.1016/j.ydbio.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, Mann RS. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- Rodrigues AB, Zoranovic T, Ayala-Camargo A, Grewal S, Reyes-Robles T, Krasny M, Wu DC, Johnston LA, Bach EA. Activated STAT regulates growth and induces competitive interactions independently of Myc, Yorkie, Wingless and ribosome biogenesis. Development (Cambridge, England) 2012;139:4051–4061. doi: 10.1242/dev.076760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Pierre SE, Galindo MI, Couso JP, Thor S. Control of Drosophila imaginal disc development by rotund and roughened eye: differentially expressed transcripts of the same gene encoding functionally distinct zinc finger proteins. Development (Cambridge, England) 2002;129:1273–1281. doi: 10.1242/dev.129.5.1273. [DOI] [PubMed] [Google Scholar]
- Sun YH, Tsai CJ, Green MM, Chao JL, Yu CT, Jaw TJ, Yeh JY, Bolshakov VN. White as a reporter gene to detect transcriptional silencers specifying position-specific gene expression during Drosophila melanogaster eye development. Genetics. 1995;141:1075–1086. doi: 10.1093/genetics/141.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terriente J, Perea D, Suzanne M, Diaz-Benjumea FJ. The Drosophila gene zfh2 is required to establish proximal-distal domains in the wing disc. Developmental biology. 2008;320:102–112. doi: 10.1016/j.ydbio.2008.04.028. [DOI] [PubMed] [Google Scholar]
- Tsai YC, JGY, PHC, Posakony JW, Barolo S, Kim J, Henry Sun Y. Upd/Jak/STAT signaling represses wg transcription to allow initiation of morphogenetic furrow in Drosophila eye development. Developmental biology. 2007;306:760–771. doi: 10.1016/j.ydbio.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Tsai YC, Sun YH. Long-range effect of upd, a ligand for Jak/STAT pathway, on cell cycle in Drosophila eye development. Genesis. 2004;39:141–153. doi: 10.1002/gene.20035. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- Villa-Cuesta E, Modolell J. Mutual repression between msh and Iro-C is an essential component of the boundary between body wall and wing in Drosophila. Development (Cambridge, England) 2005;132:4087–4096. doi: 10.1242/dev.01977. [DOI] [PubMed] [Google Scholar]
- Wang SH, Simcox A, Campbell G. Dual role for Drosophila epidermal growth factor receptor signaling in early wing disc development. Genes & development. 2000;14:2271–2276. doi: 10.1101/gad.827000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth AJ, Russell S. Temporally dynamic response to Wingless directs the sequential elaboration of the proximodistal axis of the Drosophila wing. Developmental biology. 2003;254:277–288. doi: 10.1016/s0012-1606(02)00036-2. [DOI] [PubMed] [Google Scholar]
- Wu J, Cohen SM. Repression of Teashirt marks the initiation of wing development. Development (Cambridge, England) 2002;129:2411–2418. doi: 10.1242/dev.129.10.2411. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development (Cambridge, England) 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Zecca M, Struhl G. Control of growth and patterning of the Drosophila wing imaginal disc by EGFR-mediated signaling. Development (Cambridge, England) 2002a;129:1369–1376. doi: 10.1242/dev.129.6.1369. [DOI] [PubMed] [Google Scholar]
- Zecca M, Struhl G. Subdivision of the Drosophila wing imaginal disc by EGFR-mediated signaling. Development (Cambridge, England) 2002b;129:1357–1368. doi: 10.1242/dev.129.6.1357. [DOI] [PubMed] [Google Scholar]
- Zecca M, Struhl G. Recruitment of cells into the Drosophila wing primordium by a feed-forward circuit of vestigial autoregulation. Development (Cambridge, England) 2007;134:3001–3010. doi: 10.1242/dev.006411. [DOI] [PubMed] [Google Scholar]
- Zecca M, Struhl G. A feed-forward circuit linking wingless, fat-dachsous signaling, and the warts-hippo pathway to Drosophila wing growth. PLoS biology. 2010;8:e1000386. doi: 10.1371/journal.pbio.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirin JD, Mann RS. Nubbin and Teashirt mark barriers to clonal growth along the proximal-distal axis of the Drosophila wing. Developmental biology. 2007;304:745–758. doi: 10.1016/j.ydbio.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.