Abstract
TNF, signaling through TNFR2, has been implicated in tissue repair, a process that in the heart may be mediated by activated resident cardiac stem cells (CSCs). The objective of our study is to determine whether ligation of TNFR2 can induce activation of resident CSCs in the setting of ischemic cardiac injury. We show that in human cardiac tissue affected by ischemia (IHD), TNFR2 is expressed on intrinsic CSCs, identified as c-kit+/CD45−/VEGFR2− interstitial round cells, which are activated as determined by entry to cell cycle and expression of Lin-28. Wild-type mouse heart organ cultures subjected to hypoxic conditions both increase cardiac TNF expression and show induced TNFR2 and Lin-28 expression in c-kit+ CSCs that have entered cell cycle. These CSC responses are enhanced by exogenous TNF. TNFR2−/− mouse heart organ cultures subjected to hypoxia increase cardiac TNF but fail to induce CSC activation. Similarly, c-kit+ CSCs isolated from mouse hearts exposed to hypoxia or TNF show induction of Lin-28, TNFR2, cell cycle entry and cardiogenic marker, α-sarcomeric actin (α-SA), responses more pronounced by hypoxia in combination with TNF. Knockdown of Lin-28 by siRNA results in reduced levels of TNFR2 expression, cell cycle entry and diminished expression of α-SA. We conclude that hypoxia-induced c-kit+ CSC activation is mediated by TNF/TNFR2/Lin-28 signaling. These observations suggest that TNFR2 signaling in resident c-kit+ CSCs induces cardiac repair, findings which provide further understanding of the unanticipated harmful effects of TNF blockade in human IHD.
Keywords: c-kit+CSCs, TNFR2, TNF, Lin-28, hypoxia
INTRODUCTION
Tumor necrosis factor (TNF, also known as TNF-α) is an inflammatory mediator produced during ischemia and implicated in ischemic myocardial damage and the development of heart failure (1, 2). Circulating levels of TNF are higher in acute myocardial infarction patients who developed heart failure compared to patients who do not (3), with a direct correlation between functional capacity, survival, and circulating TNF levels (3, 4). Experiments in mice had suggested that TNF neutralization could be beneficial (4), but clinical trials using TNF antagonists have been unsuccessful or even harmful (5). This discrepancy is unresolved.
TNF exerts its effects by binding to two distinct cell surface receptors, TNFR1 (p55 or CD120a) and TNFR2 (p75 or CD120b) (6). TNFR1 is the main receptor subtype in most cell types in resting tissues, including the heart, and its downstream signaling pathways have been studied extensively. Engagement of TNFR1 exacerbates, whereas engagement of TNFR2 ameliorates myocardial damage (7). We have previously reported that expression of TNFR1 is decreased while that of TNFR2 is increased on both human kidney and heart cells in the setting of transplant rejection (8-12). The precise signals that cause these changes are not known, but TNF and other pro-inflammatory cytokines can increase the expression of TNFR2 through new transcription, whereas TNFR1 is more commonly downregulated by these same stimuli (13, 14). These observations have led us to hypothesize that an altered expression and activation of TNFRs in human myocardium could contribute to the failure of anti-TNF therapeutics in ischemic heart disease (IHD). However, the cell types and mechanisms activated by TNF signaling through TNFR2 that improve cardiac function are unknown.
Recent compelling evidence has accumulated suggesting that the adult heart has regenerative potential (15, 16), mediated by resident cardiac stem cells (CSCs) with the ability to self-renew and differentiate (17). CSCs expressing c-kit have been isolated from human and murine heart tissue and are significantly increased in number in myocardial infarction and in acute ischemia-induced cardiac injury (18-21). Results from recent Phase 1 trials of autologous c-kit+ CSCs in patients with ischemic cardiomyopathy are encouraging (22).
Lin-28 is a RNA-binding protein essential for maintaining the pluripotency, growth and survival of embryonic stem cells (23, 24). Lin-28 has been cited as a marker of “stemness” (25). Importantly, Lin-28 has been shown to mediate mouse skeletal myogenesis (26). While barely detectable in mouse resting muscle (27), Lin-28 expression was strongly upregulated during regeneration of skeletal muscle fibers, followed by down-regulation upon completion of regeneration. This biphasic expression pattern of Lin-28 was recapitulated in cultured adult mouse primary myoblasts where Lin-28 involved in post-transcriptional regulation of insulin-like growth factor 2 (IGF-2), was extremely low during proliferation and was dramatically induced as early as 24 h following induction of differentiation. The high level of Lin-28 expression lasted for several days until completion of differentiation when it returned to the basal level suggesting a role in differentiation of muscle tissue (26). Likewise, in adult mouse small intestine, Lin-28 protein was not detected in the basal stem cells, nor terminally differentiated villous cells, but it was detected in a patch of cells between the basal stem cells and the mature villous cells (27). Thus, during regeneration of mouse skeletal muscle and small intestinal villi, induction of Lin-28 expression marks activation and differentiation of resident stem cells. Indeed, recent studies have shown significant function of baseline levels of Lin-28 in lentiviral-infected rat cardiomyoblasts in regulation of pre-miR-1 (28). However, while it is not known definitively whether induction of Lin-28 also occurs in human myocardial regeneration, increased expression of Lin-28 in c-kit expressing CSCs is likely to indicate early activation and possible commitment of these cells to the myocyte lineage.
Here we report that in human heart tissue affected by IHD there is both increased TNF expression by various cell populations and induction of TNFR2 expression on intrinsic c-kit+CD45−VEGFR2− CSCs. By multi-color immunofluorescence microscopy increased expression of TNFR2 on these cells correlates both with increased expression of Lin-28, marker of activation and potentially of commitment to the cardiac lineage, and with evidence of cell cycle entry. Similar changes in c-kit+ CSCs can be recapitulated in wild-type (but not TNFR2 null) mouse cardiac organ cultures and in c-kit+ CSCs isolated from mouse hearts subjected to hypoxia. These observations are consistent with the idea that TNFR2 signaling in resident c-kit+ CSCs induces cardiac repair and may help to explain the unanticipated harmful effects of TNF blockade in human IHD.
MATERIALS AND METHODS
Human Heart Tissue
12 human heart samples were obtained under protocols approved by the Institutional Review Boards of Yale University and of the New England Organ Bank. These samples comprised of 2 groups of 6 specimens each: (1). normal myocardium (NM) from patients who died for reasons other than cardiovascular diseases with no history of coronary artery disease; and (2). myocardium affected by ischemic heart disease (IHD) from patients with clinical and pathological evidence of prior myocardial infarctions with advanced heart failure sufficient to be listed for cardiac transplantation. These samples did not differ significantly by age, i.e. they were age-matched with mean ± SEM for NM of 47.67 ± 6.43 years old and IHD of 51.33 ± 3.35 years old; two-tailed t-test with P value = 0.06244, i.e. > 0.05. The characteristic of patients’ samples is presented in supplementary Table 1. All samples were processed for histology and analysed by immunohistochemistry, immunoblotting, in situ hybridization (ISH) and, quantitative real-time PCR (qRT-PCR).
Murine Heart Organ Cultures
Sacrifice of mice was performed under a protocol approved by the Yale Institutional Animal Care and Use Committee. Pieces of heart tissue from wild type (WT) C57BL/6 and TNFR2−/− (B6.129-tnfrsflb) mice, purchased from Jackson Laboratory (Bar Harbor, ME) were obtained immediately from surgically excised specimens. Duplicate <1mm3 fragments of tissue were placed in flat-bottomed 96-well tissue culture plate (Appleton Woods Limited, Birmingham, UK) in complete culture medium M199 and incubated at room air plus 5% CO2 or in hypoxic condition in 1% O2 and 5% CO2 in a controlled environment chamber (MACS-MG-1000 Anaerobic workstation, Don Whitley Scientific, UK) maintained at a humidified temperature of 36°C ± 1° with or without recombinant murine TNF (rmTNF) (AMS Biotechnology, Abingdon) for 0, 3, 6 or 18 h. A dose-response curve showed that both TNFRs were activated in the same concentration range. 10ng/ml, an optimal concentration, was used in all reported experiments. Multiple randomized samples from each patient were used to obtain parallel group comparisons and to assess the reliability and reproducibility of these assays. Some cultures were incubated in media alone (untreated) or pre-treated with 10 ng/ml recombinant murine TNF with or without various concentrations (150, 300 and 600 μM) of pimonidazole hydrochloride (hypoxyprobe-1) (HPI, Burlington, USA) to monitor low oxygen condition. Cultures were then harvested and either snap-frozen in isopentane-cooled in liquid-nitrogen or fixed in 4% formaldehyde for paraffin-wax embedding. 5μm-thick paraffin sections of all the samples were stained with hematoxylin and eosin (H&E) for morphological analysis and the diagnosis in all cases was verified independently by two experienced pathologists and was based entirely on examination of routinely stained slides.
Immunofluorescence (IF)
Paraffin-wax sections of NM, IHDM and murine heart organ cultures were immunostained for TNF, TNFR1 or TNFR2 and α-sarcomeric actin (α-SA, marker for cardiomyocytes (CMs)) as previously described (8, 9, 12). To assess the presence of cardiac precursor cells in human and mouse heart we have used anti-c-kit (CD117) (18) and anti-α-SA or −CD45 (pan-leukocyte marker) or −VEGFR2 (also known as flk-1 in mice or KDR in humans) (17). Parallel sections were co-immunostained for c-kit and Lin-28 or TNFR2 or phospho-Histone H3S10 (pH3S10) (nuclear protein involved in the cell cycle), followed by fluorochrome-conjugated secondary antibodies and Hoechst 33342 for nuclei detection before viewing on a Leica TCS-SPE confocal microscopy. Mouse neural stem cells were used as positive controls for c-kit and Lin-28 (29, 30) and negative controls included replacement of the primary antibodies with isotype-matched antisera. See supporting information data for detailed method and antibodies/reagents used.
Detection of Hypoxyprobe-1 in Murine Heart Organ Cultures
Exposure of murine heart organ cultures to low oxygen conditions was assessed using anti-hypoxyprobe-1 antibody as previously described (31). See supporting information data for detailed method.
ISH and qRT-PCR and Immunoblotting
Paraffin-wax sections of NM and IHDM were hybridized with digoxigenin-labeled anti-sense probes specific for human c-kit and Lin-28 and murine organ cultures with probes specific to mouse TNF, TNFR1 and TNFR2 (MWG-Biotech, UK) as previously described (10, 11). Gene expression was visualized using alkaline-phosphatase/BCIP/NBT substrate (Sigma-Aldrich, UK). Corresponding sense probes were used as negative controls. Probe sequences are provided in SI text. For qRT-PCR, total RNA was isolated using Taqman expression systems. TaqMan_Gene Expression Assay ID:TNFR1 (Hs00533560), TNFR2 (Hs00153550),TNFR (Hs99999043), c-kit; (Hs00174029) and Lin-28 (Hs00702808)(Applied Biosystems, Foster City, CA). RPLPO (Hs99999902_m1) and TBP (Hs99999910_m1) were used for normalization. Heart tissue homogenates from same study groups was analysed by immunoblotting as previously described (11) using antibodies to TNFR1, TNFR2, TNF, pH3S10, Lin-28, c-kit and normalised to β-actin. Immunoblots were quantified using Image J software version v1.47k. See supporting information data for detailed methods.
Processing, Isolation, and Culture of c-kit+ Cells from the Adult Murine Heart
c-kit+ CSCs were enzymatically dissociated from 30 adult mice hearts (10-13 week old) with NOD or C57BL6 background into single cell suspension using a previously described method (32). Hearts were dissected into 2 halves and rinsed thoroughly with Ca2+-Mg2+-free phosphate-buffered solution (PBS) (Invitrogen, Paisley, UK). Enzymatic dissociation of the hearts was carried out using collagenase II (~600 U/ml; Lorna Laboratories, UK), DNase I (~12 U/ml, Invitrogen, UK) and HBSS on a GentleMACs Disassociator (Miltenyi) according to the manufacturer's instructions. Tissue was then incubated on a MACsMix rotator (Miltenyi) for 30 minutes at 37°C, centrifuged at 300g for 5 min cell suspension, passed through 40 μm nylon mesh to remove cell clumps, rinsed with HBSS and centrifuged at 300g for 10 min. Single cell suspension was suspended in cardiosphere-growth medium [35% complete IMDM/65% DMEM–Ham F-12 mix containing 2% B27, 0.1 mmol/L 2-mercaptoethanol, 10 ng/mL epidermal growth factor [EGF], 20 ng/mL basic fibroblast growth factor [bFGF], 40 nmol/L cardiotrophin-1, 40 nmol/L thrombin, antibiotics, and L-Glu] (32). The cells were cultured on sterile petri dishes and incubated for 3-5 days and collected using 0.1% trypin/0.2% EDTA and enrichment of the c-kit+ cells achieved by Magnetic Cell Sorting system (MACS), Miltenyi Biotech). A total of 6×108 cells were magnetically labeled with CD117-Microbeads in PBE buffer [PBS, 2mM EDTA and 0.5% BSA] for 15 min at 4°C according to the manufacturer's instructions and passed through the MACS® column placed in the magnetic field of a MiniMACS™ Separator. The magnetically labeled CD117+ cells were retained within the column and eluted as the positively selected cell fraction. Enriched CD-117+ cells ~6×105 cells/mL were seeded in poly-L-lysine-coated 8-well slide chambers (Thermo-scientific, Essex, UK) in a differentiation medium [IMDM supplemented with 10% fetal calf serum, 100 U/mL penicillin G, L-glutamine DMEM-Ham F-12 mix containing 2% B27, 0.1 mmol/L 2-mercaptoethanol, 10 ng/mL epidermal growth factor [EGF], 20 ng/mL basic fibroblast growth factor [bFGF], 40 nmol/L cardiotrophin-1, 40 nmol/L thrombin, L-Glutamine,10 uM 5’azacytidine(33), 100 nM oxytocin(33) and 40 nM thrombin (32)]. All reagents were from Invitrogen, except 5’azacytidine and 2-mercaptoethanol (Sigma-Aldrich, Gillingham, UK), EGF and bFGF were from R&D Systems, Oxford, UK. After 2 days the medium was replaced with fresh growth medium. Once confluent, the CD117+ cells were incubated in media alone in normoxic conditions (untreated controls) or treated with 10ng/ml recombinant mouse TNF (R&D systems) and/or exposed to hypoxia for 6 h.
RNA Interference
c-kit+ CSCs from adult mouse hearts were transfected with control siRNA (siRNAControl; cat~sc-37007; Santa Cruz Biotechnology or On-TARGET plus Non-targeting siRNA cat~D-001810-01-05; Dharmacon) or with RNA targeting mouse Lin-28 (siRNALin-28-1; cat~sc-106990; Santa Cruz Biotechnology) or TARGET plus mouse Lin28siRNA-SMART-pool (siRNALin-28-2; cat~L-051530-01; Dharmacon) at 40 nM in the presence of TurboFect™ Transfection Reagent (Fermentas, Cambridge, UK) according to the manufacturer's instructions for 48h prior to treatment with recombinant murine TNF (10ng/ml) and/or hypoxia for 6 h. Cells were immunostained using PE-conjugated anti-CD-117 or rabbit anti-c-kit antibody and Biotin-conjugated anti-Lineage marker (comprising of a panel of monoclonal antibodies that recognizes all mature hematopoietic lineages) and a mouse monoclonal antibody to Lin-28, pH3S10, anti-TNFR2 or anti-α-SA. Followed by secondary antibody with anti-rabbit or anti-mouse-Northern Light-498 (NL−498) or NL−557 (R&D Systems) or anti-Streptavidin-FITC or Texas Red (Vector Laboratories, Peterborough, UK) and examined on a Leica SPE confocal microscope (Leica Microsystems, Knowlhill, Milton Keynes, UK). Image for each fluorophore was acquired sequentially using the same constant acquisition time and settings rather than simultaneously to avoid crosstalk between channels. We have examined the specificity of the Lin-28siRNAs using the mouse teratocarcoma cell line P19 (29).
Data Analysis
All results are expressed as mean ± SEM unless otherwise stated. Positive cells in NM and IHDM human samples were counted in 10 randomly chosen fields of view at x40 Mag (with each field of view containing at least 280 cells, most of which are CMs ~84% and the remaining are ICs and vascular structures). Sections were viewed using a Nikon OPTIPHOT-2 (Nikon, Surrey, UK) in a blind-folded manner. Statistical differences analyses were performed using Student's t test or ANOVA followed by Bonferroni's correction in GraphPad Prism version 5.02. Isolated c-kit+CSCs from murine hearts were scored in a similar manner (with each field of view containing at least 30 cells). The total number of positive cells from 10 random fields was divided by the total cell numbers to generate the % of positive cells. Each mouse heart experiment was repeated three times and the same statistically significant differences between experimental groups were observed in all three independent experiments although the absolute values varied.
RESULTS
TNFR2 and pH3S10 are Increased in IHDM
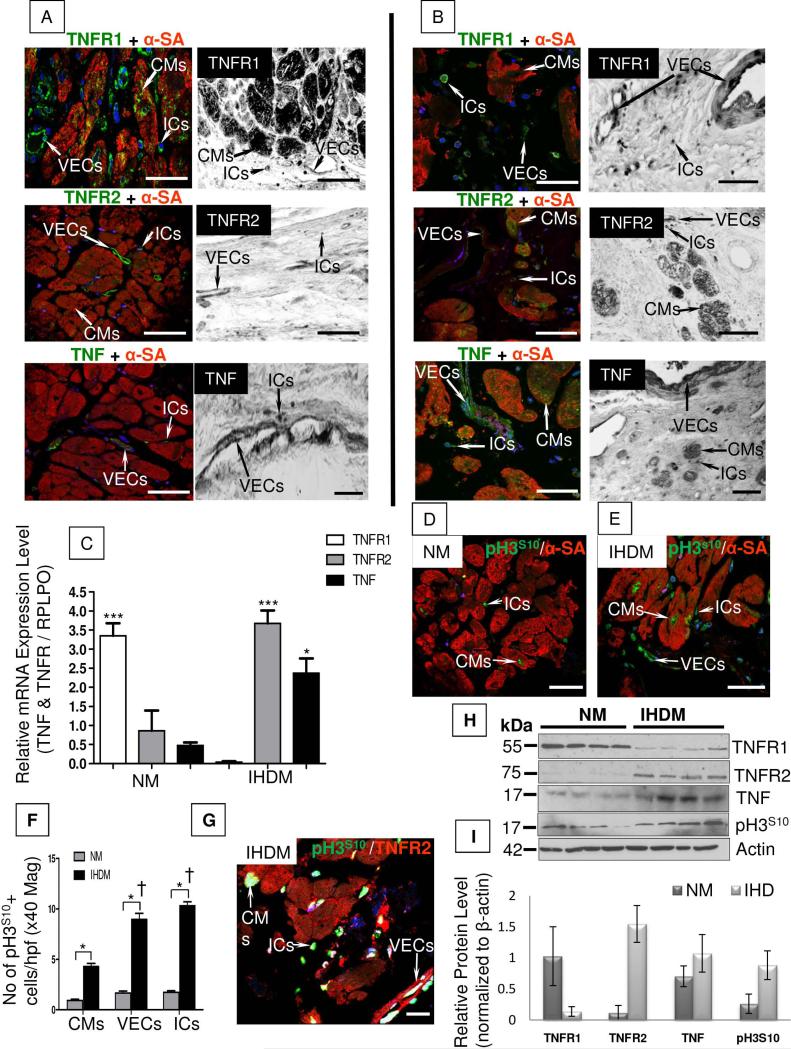
Specimens of NM showed no abnormalities on H&E stained sections (supplemental online Fig.1A), whereas sections of IHDM displayed CM damage and/or loss, areas of fibrosis containing cellular debris, and disrupted vasculature with numerous small, round ICs present within the fibrotic regions (supplemental online Fig. 1B). The expression of TNF and TNFRs in NM and IHDM were examined by IF, ISH and qRT-PCR (Figs. 1A and 1B, quantified in 1C and supplementary online Fig 2). Consistent with our previous findings (8), TNFR1 protein and mRNA were constitutively expressed in CMs, identified by expression of α-SA, in NM; TNFR1 was also present in some microvessels (possibly VECs and/or perivascular cells), occasional ICs, and, in rarefibroblasts (sections not shown). In contrast, TNFR2 and TNF protein and transcripts were mainly confined to ICs and microvessels. Compared to NM, sections of IHDM demonstrated reduced expression of TNFR1 in CMs, but a strong signal in VECs and ICs, and increased TNFR2 expression in CMs, VECs and ICs. Similar to NM, TNF expression in IHDM specimens was mainly confined to VECs and ICs (supplemental online Table 2). By qRT-PCR (Fig.1C and supplementary online Fig. 2), tissue extracts demonstrated a statistically significant increase in TNFR1 transcripts in NM compared to IHDM (***p<0.0001). In contrast, TNF (*p<0.05) and TNFR2 (***p<0.0001) transcripts were both markedly increased in IHDM compared to NM. Collectively, these data indicate that TNF and TNFRs are differentially expressed in NM and IHDM and, more specifically, that TNFR2 expression increases with ischemic injury.
Figure 1. Combined-immunofluorescence and in situ hybridization for TNF, TNFRs and phospho-histone H3S10 (pH3S10) in normal myocardium (NM) and ischemic heart disease myocardium (IHDM).
(A). NM show TNFR1 protein and mRNA expression in cardiomyocytes (CMs), also positive for α-sarcomeric actin (α-SA; marker for CMs), in vascular endothelial cells (VECs) and in interstitial cells (ICs) while TNFR2 and TNF protein and mRNA are confined to VECs and ICs but absent in CMs. (B). In contrast in IHDM, TNFR1 protein and mRNA is down-regulated in CMs and mainly present in VECs and ICs. In comparison, TNFR2 and TNF protein and mRNA expression is up-regulated in CMs, VECs and ICs. (C). qRT-PCR of tissue extracts from same study groups show strong signal for TNFR1 mRNA in NM versus IHDM, with increased TNF and TNFR2 mRNA in IHDM versus NM. *p<0.05 and ***p<0.0001. Expression (D and E) and quantification (F) of pH3S10 show a rare signal in NM, more pronounced in IHDM with a higher level of expression in VECs and ICs versus CMs.*p<0.05 and †p<0.05. (G). Co-localization of TNFR2 and pH3S10 is seen in CMs, VECs and ICs in IHDM. (H). Immunoblot of the same study groups show up-regulation of TNF, TNFR2 and pH3S10 and down-regulation of TNFR1 in IHDM with β-actin as loading protein. (I). Densitometric analysis, normalized to β-actin levels, with one sample for each protein set to 1. Results expressed as mean ± SD; n=4/study group. x40 Magnification, Scale bars; 50μm; 1E-zoomed 1.22x, Scale bar 50μm; ID-Scale bar; 75μm; 1G-zoomed 2.39x, Scale bar; 75μm; confocal images; n=6/study group with similar findings in two other independent experiments; immunoblot; n=4/study group.
We recently demonstrated an association between increased TNFR2 expression and cell cycle entry of CMs and VECs in injured myocardium of cardiac allografts (8). We therefore compared the expression of pH3S10, an indicator of cell cycle entry, in NM and IHDM. A negligible level of staining was detected in CMs in NM (Fig. 1D). In contrast, IHDM showed a marked increase in pH3S10 in α-SA-positive CMs and in VECs and ICs with the highest level of pH3S10 detected in VECs and in ICs (†p<0.001) (Fig. 1E, quantified in 1F and supplemental online Table 2). Co-localization of pH3S10 and TNFR2 is detected in CMs, VECs and in ICs (Fig. 1G). A similar pattern and frequency of co-localization for TNFR2 and Ki67 or PCNA was seen in IHDM (supplementary online Fig 3). Immunoblotting confirmed increased levels of expression of TNF, TNFR2 and pH3S10 in extracts of myocardium from IHD specimens compared to NM (Fig. 1H, quantified in Fig. 1I).
Cardiac Stem Cells Positive for c-kit are Increased and Co-express Lin-28 in IHDM
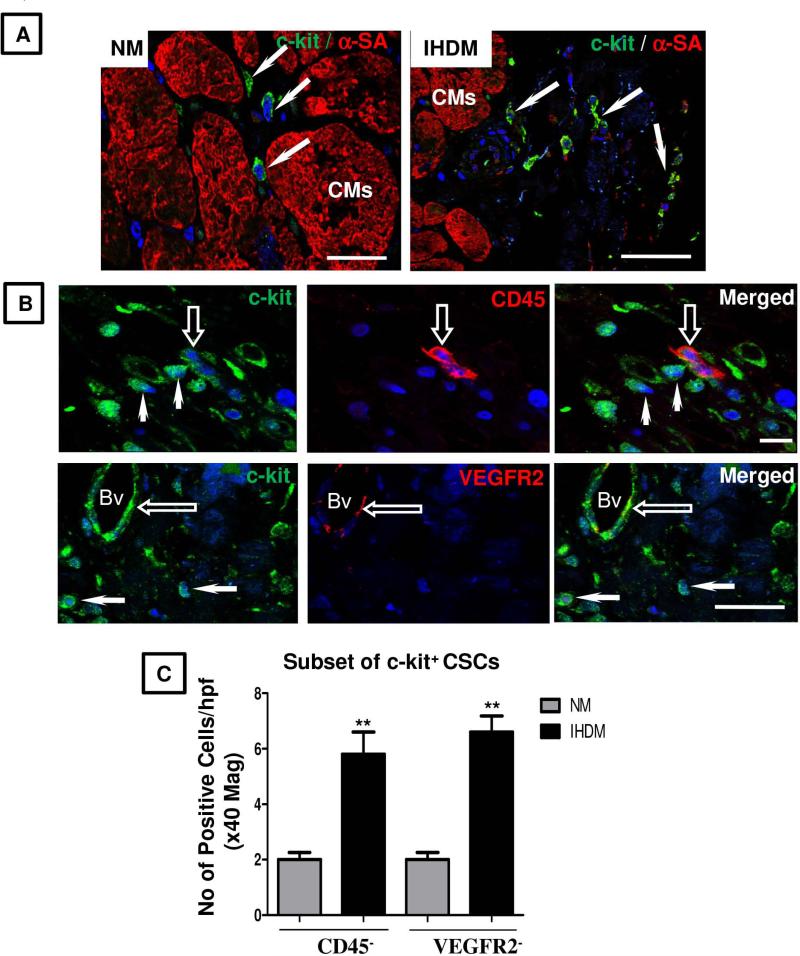
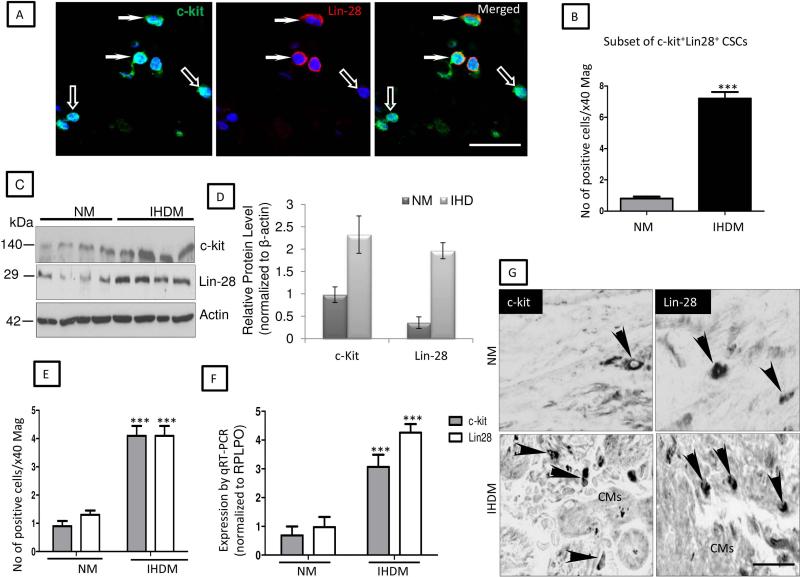
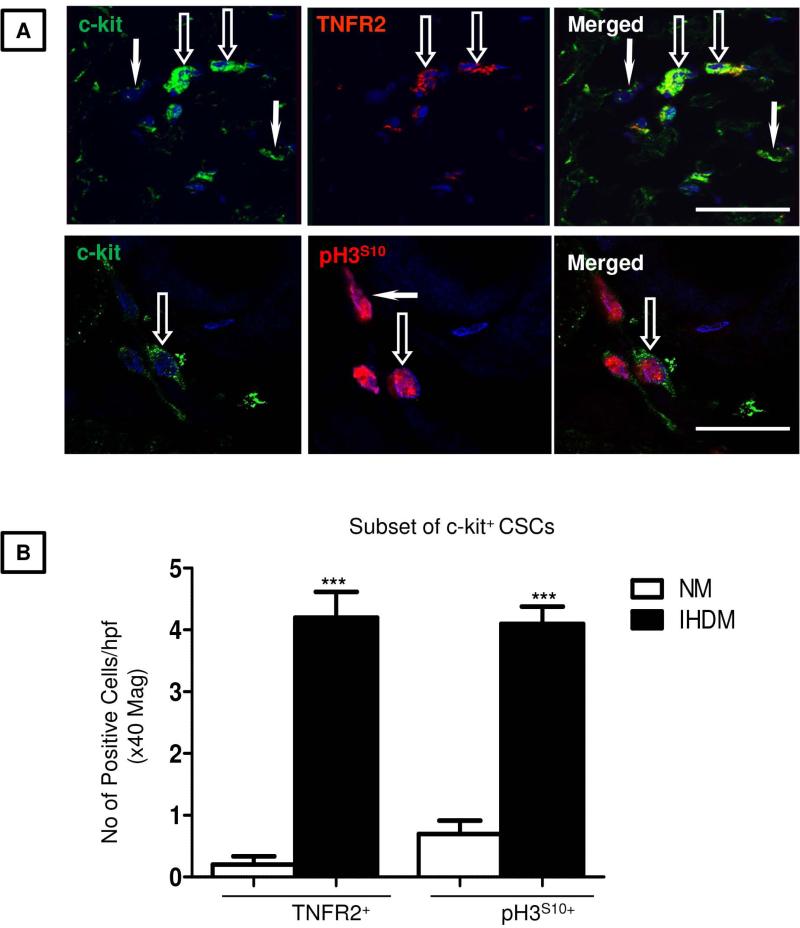
Experimental and clinical studies in humans and animals indicate the presence of precursor cells within the heart that have the capacity to reconstitute and restore cardiac function in damaged myocardium (34, 15, 16). To determine whether the small round ICs observed in our heart samples are potential cardiogenic precursors, sections of NM or IHDM were immunostained for expression of c-kit and α-SA (Fig. 2A). In NM, only occasional ICs showed strong signal for c-kit while negative for α-SA (≤ 3 cells in high power field at x40 magnification). In contrast, a striking increase in c-kit+ cells (≥12 cells/hpf) was observed in sections of IHDM mainly confined to ischemic zones. No signal for c-kit was detected in negative controls when anti-c-kit antibody was replaced with an isotype-matched nonspecific antibody (supplemental online Fig. 4A) and a strong signal present in mouse neural stem cells (positive controls) (supplemental online Fig. 4B). The majority of these c-kit+ cells are small round ICs that lack CD45 or VEGFR2 (17) (Fig. 2B, quantified in Fig. 2C) and are putative CSCs. In skeletal muscle and small intestinal epithelium, activated stem cells express Lin-28 (26, 27). We therefore determined whether Lin-28 is expressed in IHDM and whether it co-localizes with c-kit by IF. Only a few Lin-28+ ICs cells were present in NM compared to IHDM, which showed an increased number of ICs positive for Lin-28, negative for α-SA or CD45 or VEGFR2 (supplemental online Figs. 5A-5E, quantified in supplemental online Table 2). No signal for Lin-28 was detected in IHDM when anti-Lin-28 antibody was replaced with an isotype-matched nonspecific antibody but a strong signal was seen in mouse neural stem cells (positive control) (supplemental online Figs. 5D and 5E). Of particular interest, Lin-28 is detected in c-kit+ CSCs (≤ 8 cells/hpf, Fig. 3A, quantified in Fig. 3B). We interpret c-kit+CD45−VEGFR2− small round ICs that express Lin-28 as being activated resident CSCs. Immunoblotting confirmed increased expression of c-kit and Lin-28 in IHDM compared to NM (Fig. 3C, quantified in Fig. 3D). Gene expression was concordant with protein expression; transcripts of both markers were detected in NM, with a striking increase expression in IHDM (***p<0.0001) (Figs.3E-3G and supplemental online Figs. 5F). Most c-kit+ CSCs in IHDM were also TNFR2+, with only a rare CSC TNFR2−, with some c-kit+ CSCs also positive for pH3S10 (Fig. 4A, quantified in Fig. 4B), providing evidence of cell cycle entry. These findings by IF are supported by changes in protein expression detected by immunoblotting, by ISH analysis of tissues and by qRT-PCR measurements of transcripts in tissue extracts. Collectively, these observations are consistent with an association between TNFR2 expression, cell cycle entry and activation of resident CSCs found in cardiac tissue from patients with IHD and suggest the hypothesis that TNF signaling through induced TNFR2 on c-kit+ CSCs results in stem cell activation and cell proliferation.
Figure 2. Co-localization of c-kit and α-sarcomeric actin or CD45 or VEGFR2 in normal myocardium (NM) and ischemic heart disease myocardium (IHDM).
(A). NM show a few c-kit+ CSCs, negative for α-sarcomeric actin (α-SA), noticeably increased in IHDM (arrows). (B). c-kit+ CSCs are CD45− and VEGFR2− (arrows), with an occasional c-kit+ CSC, CD45+ and VEGFR2+, with the latter also detected in some blood vessels (Bv, open-arrows). (C). Quantification of c-kit+/CD45- and c-kit+/VEGFR2− CSCs. **p<0.001; Bars=mean + SEM; Scale bars; A-50μm, B-upper panel-10μm; lower panel-50μm; n=6/study group with similar findings in two other independent experiments.
Figure 3. Protein and gene expression for c-kit+ and Lin-28 in normal myocardium (NM) and ischemic heart disease myocardium (IHDM).
(A). Sections of IHDM show some CSCs strongly ckit+/Lin-28+ (arrows), with a few cells c-kit+/Lin-28− (open-arrows). (B). Quantification of immunostaining show increased number of c-kit+/Lin-28+ CSCs in IHDM versus NM (***p<0.0001). (C). Immunoblot show increased c-kit and Lin-28 in IHDM with β-actin used as loading protein. (D). Densitometric analysis, normalized to β-actin levels, with one of the samples for each protein set to 1, n=4/study group. (E and F). Quantification of ISH and qRT-PCR, show an increased level of c-kit and Lin-28 transcripts in IHDM versus NM (***p<0.0001). (G). ISH of NM and IHDM show CSCs positive for c-kit and Lin-28 mRNA (arrowheads); Scale bars; A, 25μm; G, 75μm; n=6/study group with similar findings in two other independent experiments.
Figure 4. Co-localization for c-kit and TNFR2 or pH3S10 in normal myocardium (NM) and ischemic heart disease myocardium (IHDM).
(A). TNFR2 or pH3S10 is seen in some c-kit+ CSCs in IHDM (open-arrows), with a few c-kit+/TNFR2− or c-kit−/pH3S10+CSCs (arrows). (B). Quantification of immunostaining show increased c-kit+/TNFR2+and c-kit+/pH3S10+ CSCs in IHDM versus NM (***p<0.0001), Scale bars; 25μm; n=6/study group with similar findings in two other independent experiments.
Hypoxia and/or TNF Induce Up-regulation of TNF and TNFR2 in CMs in Murine Heart Organ Cultures
We next analyzed the effects of stimuli associated with ischemia on expression of TNF and TNFRs in murine heart organ cultures. Cultures were incubated either in normoxic or hypoxic conditions at various time points (0, 3, 6 or 18 h). Low oxygen conditions were confirmed by immunostaining for hypoxyprobe-1 (supplemental online Fig. 6). Normoxic cultures showed a constitutive expression of TNFR1 in CMs, VECs, and ICs, with TNF or TNFR2 expression mainly confined to VECs and ICs (sections not shown). In contrast, exposure to hypoxia for up to 18 h resulted in a diminished expression of TNFR1 in CMs but induced an increase in TNF expression in VECs and ICs and increased TNFR2 expression in VECs, ICs and CMs (supplemental online Table 3). To assess whether the effects of hypoxia can be induced by TNF, we added exogenous TNF to normoxic or hypoxic cultures. Addition of exogenous TNF in normoxic cultures induced TNF expression in VECs and ICs and markedly increased TNFR2 expression in CMs, VECs and ICs at all the times points, with an even greater increase in the intensity of staining observed in cultures incubated in hypoxia in combination with TNF (supplemental online Table 3 and supplemental online Fig. 7A). Gene expression analysis by ISH (supplemental online Fig. 7B) correlated with protein expression showing increased levels of TNF mRNA in VECs and ICs following TNF treatment, indicating that exogenous TNF was inducing endogenous TNF production in these cells. TNF and/or hypoxia resulted in a similar induction of TNF expression in WT and TNFR2−/− mice (supplemental online Fig. 7C). Hypoxic conditions caused a time-dependent expression of TNFR2 mRNA in VECs, ICs and CMs and TNF in combination with hypoxia induced a more marked increase in TNFR2 mRNA expression in all three cell types. As expected, no TNFR2 mRNA was detected in TNFR2−/−cultures. These data suggest that hypoxia-induced TNF could be responsible for the up-regulation of TNFR2 and the down-regulation of TNFR1 observed in heart cells in the setting of ischemia.
Hypoxia and/or TNF Induce Expression and Activation of c-kit+ CSCs and Expression of TNFR2 and Cell Cycle Entry in c-kit+ CSCs in Organ Cultures of Cardiac Tissue from WT but not TNFR2−/− Mice
We next assessed the presence of resident CSCs in mouse hearts and whether hypoxia and TNF effect their activation. Analysis of murine heart tissue confirmed the presence of a population of interstitial c-kit+ CSCs that lacked CD45 and VEGFR2, which accounted for ~ 45% of the c-kit+ population (Fig. 5A). Hypoxia induced an increase in the percentage of c-kit+ cells expressing Lin-28 in a time-dependent manner with the most marked increase in expression occurred after 18 h of treatment with hypoxia and TNF in combination (Fig. 5B and Table 1). Following 18 h of culture under control conditions 17% of c-kit+ cells expressed Lin-28 and after 18 h of treatment under hypoxic conditions with TNF 69% of c-kit+ cells expressed Lin-28. The number of CSCs expressing Lin-28 was negligible in hearts from TNFR2−/− mice compared WT cultures, and TNF and hypoxia did not alter expression of this activation marker (Table 1). Hypoxia and/or TNF also increased the percentage of c-kit+ cells expressing TNFR2 and entering cell cycle. Following 18 h of culture under control conditions 1% of c-kit+ cells expressed TNFR2, and 3% stained positive for pH3S10. After 18 h of treatment under hypoxic conditions with TNF, 26% of c-kit+ cells expressed TNFR2, and 28% stained positive for pH3S10 (Table 1 and Fig. 5C). In contrast to WT, pH3S10expression was negligible in TNFR2−/− hearts (Table 1). These data indicate that both TNF and hypoxia induce activation of c-kit+ CSCs and that the presence of TNFR2 is important for TNF-mediated activation of CSCs and subsequent cell cycle entry.
Figure 5. Hypoxia and/or TNF induce activation of c-kit+CSCs in murine heart organ cultures.
(A). Representative confocal images show c-kit+/CD45−/VEGFR2− CSCs, (open-arrows), with a few c-kit+/CD45+ (arrows) and VEGFR2+blood vessel (Bv). (B).TNF plus hypoxia result in a sequential induction of Lin-28 in c-kit+ CSCs, with the highest level of expression in cultures incubated for 18 h vs0, 3 or 6 h. (C). Cultures incubated in TNF plus hypoxia for 18 h show induction of TNFR2 and pH3S10 in c-kit+ CSCs (open-arrows), with a few c-kit+/TNFR2− CSCs (arrows). Scale bars; 50μm; n=3/study groupwith similar findings in two other independent experiments.
Table 1.
Quantitative analysis of two-color immunofluorescence for c-kit and Lin-28 or phospho-Histone H3S10 (pH3S10) or TNFR2 in murine heart organ cultures from wild Type (WT) or TNFR2−/− mice incubated either in normoxic or hypoxic conditions, with or without TNF for 18 h. Percentages represent the % of c-kit+ cells that express Lin-28, pH3S10 or TNFR2. In comparison to expression in WT, TNFR2−/− mice show a diminished level of all the 3 markers. Data analyzed by ANOVA and Bonferroni's correction.
| Normoxia | Hypoxia | ||||
|---|---|---|---|---|---|
| % of c-kit cells expressing cells | −TNF | +TNF | −TNF | +TNF | |
| WT | Lin-28+ | 17 ± 2% | 48 ± 2.7% | 62 ± 1.6%‡ | 69 ± 1.7%† |
| pH3S10+ | 3 ± 0.2% | 18 ± 1.5% | 22 ± 2.1%‡ | 28 ± 1.5%† | |
| TNFR2+ | 1 ± 0.1% | 13 ± 1.3% | 19 ± 1.2%‡ | 26 ± 2.0%† | |
| TNFR2−/− | Lin-28+ | <1% | <1% | <1% | <1% |
| pH3S10+ | <1% | <1% | <1% | <1% | |
| TNFR2+ | <1% | <1% | <1% | <1% | |
Values are mean ± SEM; n=3/genotype. Bold=p<0.05-vs normoxia (−TNF)
p<0.001-vs TNF or hypoxia alone
p<0.01-vs normoxia (+TNF).
Hypoxia and/or TNF-induced Expression of Lin-28 and TNFR2 result in Activation and Differentiation of isolated Mouse c-kit+ CSCs
To investigate the roles of Lin-28 and TNFR2 in activation and differentiation of CSCs, c-kit+ cells were isolated from adult mouse heart and cultured under control conditions. These cells were negative for lineage marker (Lin−), pan leukocyte marker CD45, cardiomyocyte marker α-SA and hematopoietic progenitor marker CD133. Very few c-kit+ cells incubated in media alone (untreated) expressed Lin-28 (1%), TNFR2 (2.2%) or pH3S10 (1.5%) (supplementary online Fig. 8A). Treatment with TNF or hypoxia alone for 6 h resulted in a noticeable induction of Lin-28 (4.2%), TNFR2 (8.0%) and pH3S10 (6%), which were more pronounced with hypoxia in combination with TNF (c-kit+/Lin-28+ ~10%, c-kit+/TNFR2+ ~13% and c-kit+/pH3S10+ ~14%) (supplementary online Figs. 8B & 8C). Hypoxia and TNF also induced expression of α-SA in some cells (3.5%), which then showed weak or no signal for c-kit. Addition of differentiation agents (5-azacytidine (32) and oxytoxin (33)) to culture medium prior to exposure to hypoxia or to TNF resulted in a more pronounced expression of Lin-28, TNFR2 or pH3S10 (Lin-28 ~14%, TNFR2 ~17% and pH3S10 ~20%), with a slight increase in α-SA expression ~7% in some cells, negative for c-kit. Exposure to hypoxia in combination with TNF resulted in an even higher expression for all the 4 proteins (Lin-28 ~40%, TNFR2 ~36% and, pH3S10 ~46%, α-SA ~16%), with identifiable sarcomeres detected in the α-SA+ cells. Expression of α-SA strongly suggests that c-kit+ cells represent myogenic precursors, and their differentiation is associated with loss of c-kit and expression of Lin-28 and TNFR2. A functional role for Lin-28 was demonstrated by siRNA knockdown using two siRNALin-28s. A reduction in Lin-28 in siRNALin-28-1 transfected cells versus siRNAControl was accompanied by a reduction in TNFR2 (13%), pH3S10 (6.6%) and α-SA expression (1.5%) versus TNFR2; (32%); pH3S10 (41%); α-SA expression (15%) in cells transfected with siRNAControl (Fig. 6). Similar findings were observed using siRNALin-28-2 (supplementary online Fig. 9) and knockdown efficiency of about 80% was observed with both the siRNALin-28s in P19 cells (supplementary online Fig. 10). Collectively these data support the role of TNF, Lin-28 and TNFR2 in activation and differentiation of c-kit+ CSCs to a cardiogenic lineage.
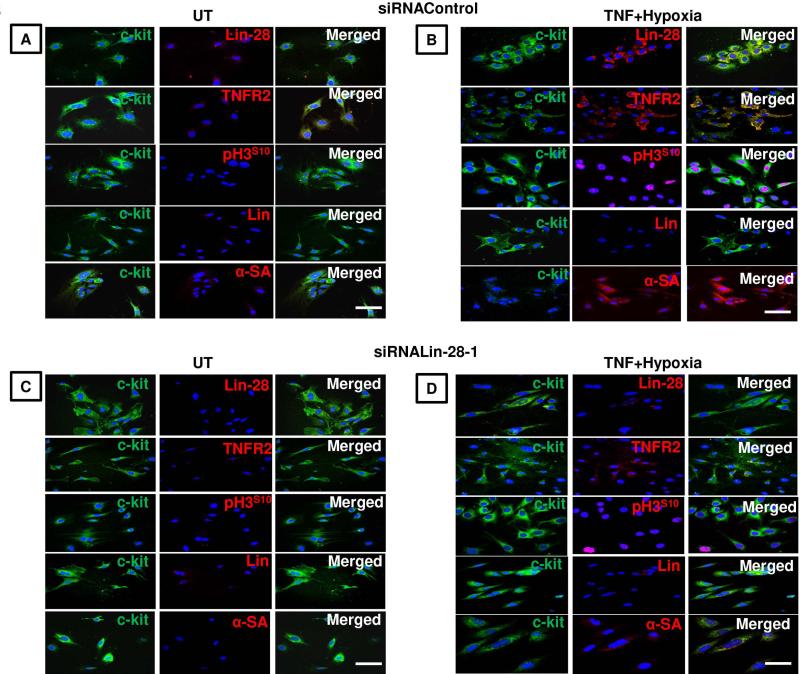
Figure 6. c-kit+CSCs isolated from murine hearts transfected with either siRNAControl or siRNALin28-1 and left in media alone in normoxia (untreated; UT) or treated with TNF plus hypoxia for 6 h.
(A). UT siRNAControl-transfected cells, Lin− and α-SA−, show negligible levels of Lin-28, TNFR2 and pH3S10. (B). In contrast, cells exposed to TNF plus hypoxia shows an increased expression of c-kit, Lin-28, TNFR2, pH3S10 and α-SA. (C). UT siRNALin28-1 transfected cells show comparable levels of c-kit, TNFR2 and pH3S10expression to siRNAControl-transfected cells. (D). In contrast, exposure to TNF plus hypoxia result in a significant diminished level of Lin-28, accompanied by a reduction in TNFR2, pH3S10and, α-SA compared to siRNAControl-transfected cells. Blue nuclei with Hoechst 33342; Scale bars; 50μm; n=3 with similar findings in two other independent experiments.
DISCUSSION
We have previously reported up-regulation of TNFR2 in association with CM cell cycle entry in human cardiac allograft rejection (8). In this study, we investigated the expression of TNF and TNFR2 in IHDM and gained insights into their role in ischemic injury using c-kit+ cells isolated from mouse hearts and an organ culture model (35, 8, 12) of WT and TNFR2−/− mice hearts incubated in hypoxic conditions as a model of ischemic injury. We report several new findings. In NM TNFR1 protein and transcript, but not TNFR2 or TNF, are strongly expressed in CMs, with TNFR2 and TNF expression seen only in VECs and ICs. In IHDM, TNFR1 expression is reduced, and TNFR2 is up-regulated consistent with the response to injury in other tissues (9, 12, 36). TNFR2 up-regulation in CMs, VECs and ICs in IHDM is associated with increased expression of markers of cell cycle entry, consistent with a role for TNFR2 in tissue repair. Our data reaffirm findings of Higuchi et al (7) using transgenic mice with TNF-induced cardiomyopathy who have shown that ablation of the TNFR2 gene exacerbates heart failure and reduces survival, whereas ablation of TNFR1 blunts heart failure and improve survival.
We observed increased numbers of ICs in ischemic areas in IHDM, the majority of which express the CSC marker c-kit and lack leukocyte (CD45), and VECs (VEGFR2) markers. We propose that small round ICs which express c-kit but lack CD45 and VEGFR2 are resident CSCs. Many c-kit+ CSCs in ischemic areas co-express TNFR2, Lin-28, and nuclear pH3S10, findings that potentially implicate TNFR2 in activation of c-kit+ CSCs and CSCs cell cycle entry. These observations in cardiac tissue from patients with IHD can be replicated in murine cardiac tissue and in c-kit+ cells isolated from mouse hearts cultured under hypoxic conditions with or without TNF. Most importantly, TNF and hypoxia increase expression and cell cycle entry of c-kit+/Lin28+ CSCs in WT but not TNFR2−/− mice,and these changes are more pronounced in cultures exposed to hypoxia in combination with TNF. We were unable to perform FACS and Western blot analysis on isolated c-kit+cells due to low number of cell yield. To address whether inhibition of Lin-28 plays a functional role in the activation of c-kit+ CSCs, we reduced Lin-28 expression using Lin-28-mouse specific siRNA prior to exposure to hypoxia and TNF. We observed suppression of Lin-28 led to a decrease in the population of c-kit+ cells expressing pH3S10. This was also accompanied by a noticeable reduction in TNFR2 and α-SA expression. A previous genome-wide study in human embryonic stem cells have suggested that Lin28 may regulate the expression of at least three TNFR superfamily members (TNFRSF12A, RNFRSF10C, and TNFRSF1B) at post-transcriptional level, as revealed by the significant enrichment of the respective mRNAs in Lin28-containing ribonucleoprotein complexes (see Supplementary Table S1 in Penget al, (24)). Collectively, these results imply that Lin-28, induced by TNF signalling through TNFR2, then contributes to enhanced TNFR2 expression and c-kit+ CSC proliferation and/or differentiation to the cardiogenic lineage (37).
Our identification of CSCs in the adult human heart and their increase in ischemic injury is consistent with previous studies (15, 16, 19, 20, 32, 21). CSCs can lead to CM regeneration and restoration of cardiac function (38, 34, 15, 22, 21). In particular, human c-kit+ CSCs have the ability to self-renew, are clonogenic and multipotent, and able to differentiate into CMs and, to a lesser extent, into smooth muscle cells and VECs (17). Initiation of the reparative response is thought to be associated with cardiac release of pro-inflammatory cytokines (39) and chemokines(40). Our new findings indicate that in ischemic injury local production of TNF coupled with hypoxia-mediated induction of TNFR2 on resident c-kit+ CSCs trigger factors that activate c-kit+ CSCs in their niche, causing them to undergo transient proliferation and eventually differentiation into mature CMs. Following our previous observations of the presence of a population of CMs that enter cell cycle (8); it is not unreasonable to speculate, from our data here, that these myocytes may originate from activation of c-kit+ CSCs.
The primary results in this study are from human tissue but a murine heart organ culture model and c-kit+ cells isolated from mouse hearts has provided a useful alternative tool in gaining insights in the effects of TNF and hypoxia (35). In conclusion, we have provided evidence for the first time that c-kit+ CSCs can express TNFR2 and that binding of TNF to this receptor can result in CSC activation and cell cycle entry in association with Lin-28. These findings provide a potential mechanism for cardiac repairthat could have clinical application. Although TNF inhibition has been reported to be beneficial in certain models of myocardial ischemia (4), substantial clinical evidence contradicts this notion (41, 5). Our new data suggests that a reduction in TNFR2/Lin-28-mediated c-kit+ CSCs activation could explain the harmful effects on TNF blockade in IHD and that TNFR1 blockade in a manner that spares TNFR2 signaling may be a better solution for the treatment of heart failure.
Supplementary Material
Supplementary Figure 1. Photomicrograph of hematoxylin and eosin stained section of normal human myocardium (NM) and ischemic heart disease (IHDM). (A) NM show no pathological changes while (B) IHDM show cellular and micro-anatomical features consistent with ischemic injury such as damage and/or loss of cardiomyocytes (CMs; circled), extensive areas of fibrosis containing cellular debris, isolated interstitial cells, blood vessels and collagen fibres in the ischemic zone (*) (VECs-vascular endothelial cells, Scale bars; 100μm).
Supplementary Figure 2. TNF and TNFR mRNA expression in normal myocardium (NM) and ischemic heart disease myocardium (IHDM) measured by qRT-PCR normalized to TBP housekeeping gene. TNFR1 mRNA is increased in NM versus IHDM but decreased in IHDM, where TNF and TNFR2 are both increased.*p<0.05 and ***p<0.0001; n=6/study group.
Supplementary Figure 3. Co-immunolocalization of TNFR2 and Ki67 or PCNA in ischemic heart disease myocardium (IHDM). A strong signal for ki67 or PCNA is detected in nuclear of some cardiomyocytes, positive forTNFR2 (arrows) and in a few ICs (arrowhead). Scale bar; 75μm; =6/study group with similar findings in two other independent experiments.
Supplementary Figure 4. Controls for c-kit immunostaining. (A). A negative control section of ischemic disease myocardium (IHDM) shows absence of c-kit when the primary antibody is replaced with isotype-specific antisera, with a strong staining for α-sarcomeric actin (red) on cardiomyocytes (B). In contrast, positive controls of mouse neuronal stem cells show strong signal for c-kit (green). Scale bars; 100μm.
Supplementary Figure 5. Representative confocal images of Lin-28 and α-SA or CD45 or VEGFR2 in normal human myocardium (NM) and ischemic heart disease myocardium (IHDM). (A). NM show a few interstitial cells Lin-28+/α-SA−, increased in (B) IHDM (arrows). (C). Lin-28+/CD45− or VEGFR2− CSCs (arrows), with VEGFR2+detectedin blood vessels (Bv). (D, E). Lin-28 is undetected in IHDM when primary antibody is replaced by isotype-matched antisera (negative control) and a strong signal in mouse neural stem cells (positive control). Blue nuclei stained with Hoechst 33342. (F). qRT-PCR show an increase c-kit and Lin-28 mRNA, normalized to TBP housekeeping gene, in IHDM versus NM (***p<0.0001). Scale bars; 25μm; n=6/study group with similar findings in two other independent experiments.
Supplementary Figure 6. Photomicrographs of murine heart organ cultures incubated either in normoxia or hypoxia with or without 300μM hydroxyprobe-1 (pimonidazole hydrochloride) at various time points (0, 3, 6 or 18 h). A strong signal for hydroxyprobe-1 is evident in cultures exposed to hypoxia, visualized using 3’3-diaminobenzidine; (DAB; brown color) indicative of low oxygen level. Scale bars; 100μm.
Supplementary Figure 7. Murine heart organ cultures incubated in normoxia or hypoxia with or without TNF for 6 h. (A). Exposure toTNF plus hypoxia result in induction of TNF protein in vascular endothelial cells (VECs) and interstitial cells (ICs) and, in TNFR2 in cardiomyocytes (CMs), VECs and ICs. Blue nuclei stained with Hoechst 33342. (B). In situ hybridization showinduction of TNF and TNFR2 mRNA in CMs and ICs in cultures exposed to TNF or hypoxia alone, more pronounced in cultures exposed to TNF plus hypoxia. In contrast, control cultures (normoxia without TNF) show a strong signal for TNFR1 mRNA expression in CMs, diminished in cultures exposed to hypoxia. As expected, TNFR2 mRNA is undetected in TNFR2−/−mice (right panels). (C). Quantification of TNF immunostaining in wild type (WT) and TNFR2−/− mice organ cultures. TNF is induced by exogenous TNF (T) or hypoxia (H) alone at 6 h, more pronounced byTNF plus hypoxia (T+H), with the highest expression in ICs, then VECs and lastly in CMs. ***p<0.001 and *p<0.05. Mean + SEM; Scale bars; A, 100μm, B, 200μm. n= 3/genotype with similar findings in two other independent experiments.
Supplementary Figure 8. c-kit+CSCs isolated from murine hearts by enzymatic dissociation and immune-magnetic separation incubated in media alone (untreated-UT) or treated with TNF and/or Hypoxia for 6 h. (A). UT c-kit+ CSCs, Lin− and α-SA−, show negligible levels of Lin-28, TNFR2, and pH3S10. (B). TNF result in induction of Lin-28, TNFR2 and pH3S10, more pronounced by TNF plus hypoxia, accompanied by induction of α-SA expression in some cells with identifiable sarcomeres, weakly positive or negative for c-kit. (C). Scale bar; 50μm; n=3 with similar findings in two other independent experiments.
Supplementary Figure 9. c-kit+CSCs isolated from murine hearts and transfected with siRNAControl or siRNALin-28-2 and either left in media alone under conditions of normoxia (untreated; UT) or treated with TNF under conditions of hypoxia for 6 h. (A). UT siRNAControl-transfected cells show negligible levels of Lin-28, pH3S10 and TNFR2. (B). In contrast, TNF plus hypoxia result in induction of Lin-28, pH3S10 and TNFR2. (C). UT cells transfected with siRNALin-28-2 show a similar pattern of staining as UT siRNAControl-transfected cells. (D). In contrast, exposure to TNF plus hypoxia result in diminished Lin-28, with a noticeable reduced expression of TNFR2, pH3S10 compared to siRNAControl-transfected cells. Blue nuclei stained with Hoechst 33342. Scale bar; 50μm; n=3 with similar findings in two other independent experiments.
Supplementary Figure 10. Mouse P19 cells were transfected with siRNAControl or siRNALin-28-1 or siRNALin-28-2 for 96 h. Total RNA was isolated and analyzed with qRT-PCR. Data analyzed using the ΔΔCT method normalized to GAPDH. Both siRNALin-28s show a knockdown efficiency of about 80%; error bars represent mean ± SD; **p<0.001; n=4 with similar findings in two other independent experiments.
ACKNOWLEDGEMENTS
We are grateful to Drs Sarah Howlett, Maja Wallberg (Cambridge Institute of Medical Research, Cambridge), Jane Goodall, Tim Fitzmaurice and Alexi Crosby (Department of Medicine, Cambridge University) for kindly providing murine hearts. We also thank Mrs Suzanne Diston for her help with the references and the Tissue Bank (Department of Histopathology, Cambridge University Hospital NHS Trust) for their assistance in tissue processing.
The work was supported by the British Heart Foundation, the National Institute for Health Research Cambridge Biomedical Research Centre and the U.S. National Institutes of Health (R01-HL036003)
Footnotes
Author contributions:
RSM - Conception and Design, Collection and assembly of data, Data Analysis and Interpretation, Manuscript writing, final approval of manuscript
WLu - Collection and assembly of data
JW - Collection and assembly of data
JY - Data Analysis and Interpretation
TJS - Data Analysis and Interpretation
RW - Collection and assembly of data
CS - Collection and assembly of data
PW - Data Analysis and Interpretation
MG - Provision of study material or patients
QH - Provision of study material or patients
AHL - Provision of study material or patients
GT - Provision of study material or patients
YH - Provision of study material or patients
WM – Provision of study material or patients
JSP - Conception and Design, Data Analysis and Interpretation, editing and final approval of manuscript
JRB - Conception and Design, Data Analysis and Interpretation, editing and final approval of manuscript, financial support
DISCLOSURES
The authors declare no conflict of interest
REFERENCES
- 1.Meldrum DR. Tumor necrosis factor in the heart. Am J Physiol. 1998;274:R577–595. doi: 10.1152/ajpregu.1998.274.3.R577. [DOI] [PubMed] [Google Scholar]
- 2.Safranow K, Dziedziejko V, Rzeuski R, et al. Plasma concentrations of TNF-alpha and its soluble receptors sTNFR1 and sTNFR2 in patients with coronary artery disease. Tissue Antigens. 2009;74:386–392. doi: 10.1111/j.1399-0039.2009.01332.x. [DOI] [PubMed] [Google Scholar]
- 3.Levine B, Kalman J, Mayer L, et al. Elevated circulating levels of tumor necrosis factor in severe chronic heart failure. N Engl J Med. 1990;323:236–241. doi: 10.1056/NEJM199007263230405. [DOI] [PubMed] [Google Scholar]
- 4.Maekawa N, Wada H, Kanda T, et al. Improved myocardial ischemia/reperfusion injury in mice lacking tumor necrosis factor-alpha. J Am Coll Cardiol. 2002;39:1229–1235. doi: 10.1016/s0735-1097(02)01738-2. [DOI] [PubMed] [Google Scholar]
- 5.Mann DL, McMurray JJ, Packer M, et al. Targeted anticytokine therapy in patients with chronic heart failure: results of the Randomized Etanercept Worldwide Evaluation (RENEWAL). Circulation. 2004;109:1594–1602. doi: 10.1161/01.CIR.0000124490.27666.B2. [DOI] [PubMed] [Google Scholar]
- 6.Bradley JR. TNF-mediated inflammatory disease. J Pathol. 2008;214:149–160. doi: 10.1002/path.2287. [DOI] [PubMed] [Google Scholar]
- 7.Higuchi Y, McTiernan CF, Frye CB, et al. Tumor necrosis factor receptors 1 and 2 differentially regulate survival, cardiac dysfunction, and remodeling in transgenic mice with tumor necrosis factor-alpha-induced cardiomyopathy. Circulation. 2004;109:1892–1897. doi: 10.1161/01.CIR.0000124227.00670.AB. [DOI] [PubMed] [Google Scholar]
- 8.Al-Lamki RS, Brookes AP, Wang J, et al. TNF receptors differentially signal and are differentially expressed and regulated in the human heart. Am J Transplant. 2009;9:2679–2696. doi: 10.1111/j.1600-6143.2009.02831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Lamki RS, Sadler TJ, Wang J, et al. Tumor necrosis factor receptor expression and signaling in renal cell carcinoma. Am J Pathol. 2010;177:943–954. doi: 10.2353/ajpath.2010.091218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Lamki RS, Wang J, Skepper JN, et al. Expression of tumor necrosis factor receptors in normal kidney and rejecting renal transplants. Lab Invest. 2001;81:1503–1515. doi: 10.1038/labinvest.3780364. [DOI] [PubMed] [Google Scholar]
- 11.Al-Lamki RS, Wang J, Thiru S, et al. Expression of silencer of death domains and death-receptor-3 in normal human kidney and in rejecting renal transplants. Am J Pathol. 2003;163:401–411. doi: 10.1016/S0002-9440(10)63670-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Lamki RS, Wang J, Vandenabeele P, et al. TNFR1- and TNFR2-mediated signaling pathways in human kidney are cell type-specific and differentially contribute to renal injury. FASEB J. 2005;19:1637–1645. doi: 10.1096/fj.05-3841com. [DOI] [PubMed] [Google Scholar]
- 13.Kalthoff H, Roeder C, Brockhaus M, et al. Tumor necrosis factor (TNF) up-regulates the expression of p75 but not p55 TNF receptors, and both receptors mediate, independently of each other, up-regulation of transforming growth factor alpha and epidermal growth factor receptor mRNA. J Biol Chem. 1993;268:2762–2766. [PubMed] [Google Scholar]
- 14.Winzen R, Wallach D, Kemper O, et al. Selective up-regulation of the 75-kDa tumor necrosis factor (TNF) receptor and its mRNA by TNF and IL-1. J Immunol. 1993;150:4346–4353. [PubMed] [Google Scholar]
- 15.Beltrami AP, Barlucchi L, Torella D, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 16.Beltrami AP, Urbanek K, Kajstura J, et al. Evidence that human cardiac myocytes divide after myocardial infarction. N Engl J Med. 2001;344:1750–1757. doi: 10.1056/NEJM200106073442303. [DOI] [PubMed] [Google Scholar]
- 17.Bearzi C, Rota M, Hosoda T, et al. Human cardiac stem cells. Proc Natl Acad Sci U S A. 2007;104:14068–14073. doi: 10.1073/pnas.0706760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altarche-Xifro W, Curato C, Kaschina E, et al. Cardiac c-kit+AT2+ cell population is increased in response to ischemic injury and supports cardiomyocyte performance. Stem Cells. 2009;27:2488–2497. doi: 10.1002/stem.171. [DOI] [PubMed] [Google Scholar]
- 19.Castaldo C, Di Meglio F, Nurzynska D, et al. CD117-positive cells in adult human heart are localized in the subepicardium, and their activation is associated with laminin-1 and alpha6 integrin expression. Stem Cells. 2008;26:1723–1731. doi: 10.1634/stemcells.2007-0732. [DOI] [PubMed] [Google Scholar]
- 20.Di Meglio F, Castaldo C, Nurzynska D, et al. Localization and origin of cardiac CD117-positive cells: identification of a population of epicardially-derived cells in adult human heart. Ital J Anat Embryol. 2010;115:71–78. [PubMed] [Google Scholar]
- 21.Urbanek K, Torella D, Sheikh F, et al. Myocardial regeneration by activation of multipotent cardiac stem cells in ischemic heart failure. Proc Natl Acad Sci U S A. 2005;102:8692–8697. doi: 10.1073/pnas.0500169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolli R, Chugh AR, D'Amario D, et al. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet. 2011;387:1847–1857. doi: 10.1016/S0140-6736(11)61590-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Heo I, Joo C, Kim YK, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Peng S, Chen LL, Lei XX, et al. Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells. 2011;29:496–504. doi: 10.1002/stem.591. [DOI] [PubMed] [Google Scholar]
- 25.Richards M, Tan SP, Tan JH, et al. The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells. 2004;22:51–64. doi: 10.1634/stemcells.22-1-51. [DOI] [PubMed] [Google Scholar]
- 26.Polesskaya A, Cuvellier S, Naguibneva I, et al. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007;21:1125–1138. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang DH, Moss EG. Temporally regulated expression of Lin-28 in diverse tissues of the developing mouse. Gene Expr Patterns. 2003;3:719–726. doi: 10.1016/s1567-133x(03)00140-6. [DOI] [PubMed] [Google Scholar]
- 28.Hatzistergos KE, Quevedo H, Oskouei BN, et al. Bone marrow mesenchymal stem cells stimulate cardiac stem cell proliferation and differentiation. Circ Res. 2010;107:913–922. doi: 10.1161/CIRCRESAHA.110.222703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eda A, Tamura Y, Yoshida M, Hohjoh H. Systematic gene regulation involving miRNAs during neuronal differentiation of mouse P19 embryonic carcinoma cell. Biochem Biophys Res Commun. 2009;388:648–653. doi: 10.1016/j.bbrc.2009.08.040. [DOI] [PubMed] [Google Scholar]
- 30.Turner DL, Cepko CL. A common progenitor for neurons and glia persists in rat retina late in development. Nature. 1987;328:131–136. doi: 10.1038/328131a0. [DOI] [PubMed] [Google Scholar]
- 31.Hofer SO, Mitchell GM, Penington AJ, et al. The use of pimonidazole to characterise hypoxia in the internal environment of an in vivo tissue engineering chamber. Br J Plast Surg. 2005;58:1104–1114. doi: 10.1016/j.bjps.2005.04.033. [DOI] [PubMed] [Google Scholar]
- 32.Messina E, De Angelis L, Frati G, et al. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circ Res. 2004;95:911–921. doi: 10.1161/01.RES.0000147315.71699.51. [DOI] [PubMed] [Google Scholar]
- 33.Matsuura K, Nagai T, Nishigaki N, et al. Adult cardiac Sca-1-positive cells differentiate into beating cardiomyocytes. J Biol Chem. 2004;279:11384–11391. doi: 10.1074/jbc.M310822200. [DOI] [PubMed] [Google Scholar]
- 34.Assmus B, Schachinger V, Teupe C, et al. Transplantation of Progenitor Cells and Regeneration Enhancement in Acute Myocardial Infarction (TOPCARE-AMI). Circulation. 2002;106:3009–3017. doi: 10.1161/01.cir.0000043246.74879.cd. [DOI] [PubMed] [Google Scholar]
- 35.Al-Lamki RS, Bradley JR, Pober JS. Human organ culture as a tool for analyzing the response to tumor necrosis factor. Current Trends in Immunology. 2011;12:49–66. [Google Scholar]
- 36.Yang J, You Z, Kim HH, et al. Genetic analysis of the role of tumor necrosis factor receptors in functional outcome after traumatic brain injury in mice. J Neurotrauma. 2010;27:1037–1046. doi: 10.1089/neu.2009.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaruba MM, Soonpaa M, Reuter S, Field LJ. Cardiomyogenic potential of C-kit(+)-expressing cells derived from neonatal and adult mouse hearts. Circulation. 2010;121:1992–2000. doi: 10.1161/CIRCULATIONAHA.109.909093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Assmus B, Rolf A, Erbs S, et al. Clinical outcome 2 years after intracoronary administration of bone marrow-derived progenitor cells in acute myocardial infarction. Circ Heart Fail. 2009;3:89–96. doi: 10.1161/CIRCHEARTFAILURE.108.843243. [DOI] [PubMed] [Google Scholar]
- 39.Fazel S, Cimini M, Chen L, et al. Cardioprotective c-kit+ cells are from the bone marrow and regulate the myocardial balance of angiogenic cytokines. J Clin Invest. 2006;116:1865–1877. doi: 10.1172/JCI27019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui Y, Madeddu P. The Role of Chemokines, Cytokines and Adhesion Molecules in Stem Cell Trafficking and Homing. Curr Pharm Des. 2011 doi: 10.2174/138161211797904109. [DOI] [PubMed] [Google Scholar]
- 41.Chung ES, Packer M, Lo KH, et al. Randomized, double-blind, placebo-controlled, pilot trial of infliximab, a chimeric monoclonal antibody to tumor necrosis factor-alpha, in patients with moderate-to-severe heart failure: results of the anti-TNF Therapy Against Congestive Heart Failure (ATTACH) trial. Circulation. 2003;107:3133–3140. doi: 10.1161/01.CIR.0000077913.60364.D2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Photomicrograph of hematoxylin and eosin stained section of normal human myocardium (NM) and ischemic heart disease (IHDM). (A) NM show no pathological changes while (B) IHDM show cellular and micro-anatomical features consistent with ischemic injury such as damage and/or loss of cardiomyocytes (CMs; circled), extensive areas of fibrosis containing cellular debris, isolated interstitial cells, blood vessels and collagen fibres in the ischemic zone (*) (VECs-vascular endothelial cells, Scale bars; 100μm).
Supplementary Figure 2. TNF and TNFR mRNA expression in normal myocardium (NM) and ischemic heart disease myocardium (IHDM) measured by qRT-PCR normalized to TBP housekeeping gene. TNFR1 mRNA is increased in NM versus IHDM but decreased in IHDM, where TNF and TNFR2 are both increased.*p<0.05 and ***p<0.0001; n=6/study group.
Supplementary Figure 3. Co-immunolocalization of TNFR2 and Ki67 or PCNA in ischemic heart disease myocardium (IHDM). A strong signal for ki67 or PCNA is detected in nuclear of some cardiomyocytes, positive forTNFR2 (arrows) and in a few ICs (arrowhead). Scale bar; 75μm; =6/study group with similar findings in two other independent experiments.
Supplementary Figure 4. Controls for c-kit immunostaining. (A). A negative control section of ischemic disease myocardium (IHDM) shows absence of c-kit when the primary antibody is replaced with isotype-specific antisera, with a strong staining for α-sarcomeric actin (red) on cardiomyocytes (B). In contrast, positive controls of mouse neuronal stem cells show strong signal for c-kit (green). Scale bars; 100μm.
Supplementary Figure 5. Representative confocal images of Lin-28 and α-SA or CD45 or VEGFR2 in normal human myocardium (NM) and ischemic heart disease myocardium (IHDM). (A). NM show a few interstitial cells Lin-28+/α-SA−, increased in (B) IHDM (arrows). (C). Lin-28+/CD45− or VEGFR2− CSCs (arrows), with VEGFR2+detectedin blood vessels (Bv). (D, E). Lin-28 is undetected in IHDM when primary antibody is replaced by isotype-matched antisera (negative control) and a strong signal in mouse neural stem cells (positive control). Blue nuclei stained with Hoechst 33342. (F). qRT-PCR show an increase c-kit and Lin-28 mRNA, normalized to TBP housekeeping gene, in IHDM versus NM (***p<0.0001). Scale bars; 25μm; n=6/study group with similar findings in two other independent experiments.
Supplementary Figure 6. Photomicrographs of murine heart organ cultures incubated either in normoxia or hypoxia with or without 300μM hydroxyprobe-1 (pimonidazole hydrochloride) at various time points (0, 3, 6 or 18 h). A strong signal for hydroxyprobe-1 is evident in cultures exposed to hypoxia, visualized using 3’3-diaminobenzidine; (DAB; brown color) indicative of low oxygen level. Scale bars; 100μm.
Supplementary Figure 7. Murine heart organ cultures incubated in normoxia or hypoxia with or without TNF for 6 h. (A). Exposure toTNF plus hypoxia result in induction of TNF protein in vascular endothelial cells (VECs) and interstitial cells (ICs) and, in TNFR2 in cardiomyocytes (CMs), VECs and ICs. Blue nuclei stained with Hoechst 33342. (B). In situ hybridization showinduction of TNF and TNFR2 mRNA in CMs and ICs in cultures exposed to TNF or hypoxia alone, more pronounced in cultures exposed to TNF plus hypoxia. In contrast, control cultures (normoxia without TNF) show a strong signal for TNFR1 mRNA expression in CMs, diminished in cultures exposed to hypoxia. As expected, TNFR2 mRNA is undetected in TNFR2−/−mice (right panels). (C). Quantification of TNF immunostaining in wild type (WT) and TNFR2−/− mice organ cultures. TNF is induced by exogenous TNF (T) or hypoxia (H) alone at 6 h, more pronounced byTNF plus hypoxia (T+H), with the highest expression in ICs, then VECs and lastly in CMs. ***p<0.001 and *p<0.05. Mean + SEM; Scale bars; A, 100μm, B, 200μm. n= 3/genotype with similar findings in two other independent experiments.
Supplementary Figure 8. c-kit+CSCs isolated from murine hearts by enzymatic dissociation and immune-magnetic separation incubated in media alone (untreated-UT) or treated with TNF and/or Hypoxia for 6 h. (A). UT c-kit+ CSCs, Lin− and α-SA−, show negligible levels of Lin-28, TNFR2, and pH3S10. (B). TNF result in induction of Lin-28, TNFR2 and pH3S10, more pronounced by TNF plus hypoxia, accompanied by induction of α-SA expression in some cells with identifiable sarcomeres, weakly positive or negative for c-kit. (C). Scale bar; 50μm; n=3 with similar findings in two other independent experiments.
Supplementary Figure 9. c-kit+CSCs isolated from murine hearts and transfected with siRNAControl or siRNALin-28-2 and either left in media alone under conditions of normoxia (untreated; UT) or treated with TNF under conditions of hypoxia for 6 h. (A). UT siRNAControl-transfected cells show negligible levels of Lin-28, pH3S10 and TNFR2. (B). In contrast, TNF plus hypoxia result in induction of Lin-28, pH3S10 and TNFR2. (C). UT cells transfected with siRNALin-28-2 show a similar pattern of staining as UT siRNAControl-transfected cells. (D). In contrast, exposure to TNF plus hypoxia result in diminished Lin-28, with a noticeable reduced expression of TNFR2, pH3S10 compared to siRNAControl-transfected cells. Blue nuclei stained with Hoechst 33342. Scale bar; 50μm; n=3 with similar findings in two other independent experiments.
Supplementary Figure 10. Mouse P19 cells were transfected with siRNAControl or siRNALin-28-1 or siRNALin-28-2 for 96 h. Total RNA was isolated and analyzed with qRT-PCR. Data analyzed using the ΔΔCT method normalized to GAPDH. Both siRNALin-28s show a knockdown efficiency of about 80%; error bars represent mean ± SD; **p<0.001; n=4 with similar findings in two other independent experiments.