Abstract
The emergence of resistance to epidermal growth factor receptor (EGFR) inhibitor therapy is a major clinical problem for patients with non-small cell lung cancer (NSCLC). The mechanisms underlying tumor resistance to inhibitors of the kinase activity of EGFR are not fully understood. Here we found that inhibition of EGFR by erlotinib induces STAT3 phosphorylation at Tyr705 in association with increased Bcl2/Bcl-XL at both mRNA and protein levels in various human lung cancer cells. PTPMeg2 is a physiologic STAT3 phosphatase that can directly dephosphorylate STAT3 at the Tyr705 site. Intriguingly, treatment of cells with erlotinib results in downregulation of PTPMeg2 without activation of STAT3 kinases (i.e. JAK2 or c-Src), suggesting that erlotinib enhanced phosphorylation of STAT3 may occur, at least in part, from suppression of PTPMeg2 expression. Since elevated levels of phosphorylated STAT3 (pSTAT3), Bcl2 and Bcl-XL were observed in erlotinib-resistant lung cancer (HCC827/ER) cells as compared to erlotinib-sensitive parental HCC827 cells, we postulate that erlotinib-activated STAT3/Bcl2/Bcl-XL survival pathway may contribute to acquired resistance to erlotinib. Both blockage of Tyr705 phosphorylation of STAT3 by niclosamide and depletion of STAT3 by RNA interference in HCC827/ER cells reverses erlotinib resistance. Niclosamide in combination with erlotinib potently represses erlotinib-resistant lung cancer xenografts in association with increased apoptosis in tumor tissues, suggesting that niclosamide can restore sensitivity to erlotinib. These findings uncover a novel mechanism of erlotinib resistance and provide a novel approach to overcome resistance by blocking the STAT3/Bcl2/Bcl-XL survival signaling pathway in human lung cancer.
Keywords: EGFR, Erlotinib, Acquired resistance, STAT3, Bcl2, Bcl-XL, Niclosamide, NSCLC
Introduction
Lung cancer is the leading cause of cancer-related death in the United States and accounts for more than a million deaths worldwide each year (1). The overall 5-year survival rates for non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) are 16% and 6% (2), respectively, in the United States. The epidermal growth factor receptor (EGFR) has been established as a critical therapeutic target for the treatment of NSCLC because a significant proportion, up to 20% of NSCLC patients, harbor somatic mutations in the tyrosine kinase domain of the EGFR gene that promote cancer cell growth (3, 4). EGFR tyrosine kinase inhibitors (TKIs) such as erlotinib or gefitinib have been well established as one of therapeutic options for a well-defined subset of lung cancer patients (3, 4). Acquired tumor resistance to these agents typically emerges after 6-12 months of therapy and constitutes the major limitation of this otherwise effective therapeutic approach (3). The mechanisms of this resistance are not fully understood but may be associated with activation of EGFR-independent pathways, occurrence of additional EGFR gene mutations or loss of the target (4-6). For example, the EGFR T790M kinase mutation may account for approximately 50% of acquired TKI resistant cases (5, 6), and MET genomic amplification is correlated with about 5-11% of acquired TKI resistance (7-9).
Additionally, erlotinib has been found to induce MET-independent activation of STAT3-Bcl2/Bcl-XL survival signaling in early adaptive TKI resistant lung cancer tumor cells (10). STAT3 is a physiological transcription factor for Bcl2 and Bcl-XL (11-13), and erlotinib activation of STAT3 has been previously shown to result in upregulation of Bcl2/Bcl-XL thereby contributing to resistance of lung cancer cells to erlotinib (10). Inhibition of Bcl2/Bcl-XL using BH3 mimetic agent such as ABT-737, or by dual RNAi-mediated knockdown of Bcl2/Bcl-XL, was sufficient to eradicate early-resistant lung tumor cells (10). Importantly, erlotinib in combination with Bcl2/Bcl-XL inhibitor ABT-737 synergistically represses lung cancer in vivo(10).
STAT3 has been shown to be an important molecule in a variety of malignancies and has recently been validated as an attractive therapeutic target in cancer therapy, including lung cancer (14-18). The latent cytoplasmic STAT3 becomes activated through phosphorylation at residue Tyr705 by Janus associated kinase (JAK) or growth factor receptor-associated tyrosine kinase (Src) (19). Phosphorylated STAT3 dimerizes through a reciprocal Src homology 2-phospho-tyrosine interaction and accumulates in the nucleus, where it activates the transcription of a wide array of genes, including Bcl2/Bcl-XL, cyclin D1, c-Myc, etc. (20). In addition to STAT3 kinases (i.e. JAK and Src), STAT3 phosphorylation is also tightly regulated by a process of dephosphorylation, which is mediated by the protein tyrosine phosphatase, PTPMeg2 (21). PTPs are a large and structurally diverse family of enzymes that catalyze the dephosphorylation of phosphorylated proteins. PTPMeg2 has recently been identified as a new physiologic STAT3 phosphatase that directly dephosphorylates STAT3 at the Tyr705 residue (21).
Niclosamide has recently identified as a potent STAT3 inhibitor that disrupts STAT3 transcriptional activity by blocking its phosphorylation and nuclear translocation (22). In this report, we discovered that erlotinib enhances STAT3 phosphorylation by downregulation of its phosphatase PTPMeg2, which leads to elevated levels of Bcl2/Bcl-XL and consequent loss of erlotinib sensitivity. Niclosamide blocks erlotinib-induced activation of the STAT3/Bcl2/Bcl-XL survival pathway in lung cancer cells, leading to the reversal of erlotinib resistance in vitro and in vivo.
Materials and Methods
Materials
Erlotinib was purchased from LC Laboratories (Woburn, MA). Niclosamide was obtained from Sigma-Aldrich (St. Louis, MO, USA). Phospho-EGFR (Tyr1068), phospho-STAT3 (Tyr705), phospho-Akt (S473), Akt, ERK1/2, pMET (Tyr1234/1235) and cleaved caspase 3 antibodies were purchased from Cell Signaling Technology (Beverly, MA). MET, pERK1/2 and β-actin antibodies as well as PTPMeg2 shRNA and its control shRNA were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). PTPMeg2 antibody was obtained from R&D systems (R&D systems, MN). Bcl2 was obtained from Calbiochem (Darmstadt, Germany). Bcl-XL was purchased from Epitomics, Inc. (Burlingame, CA). QD605 goat anti-rabbit IgG conjugate (red), QD705 goat anti-mouse IgG conjugate (green) and ProLong® Gold antifade reagent with 4′, 6-diamidino-2-phenylindole (DAPI) were purchased from Invitrogen Life Technologies Inc (Carlsbad, CA). All other reagents used were obtained from commercial sources unless otherwise stated.
Cell lines and cell culture
HCC827, H1650, and H1975 were purchased directly from the American Type Culture Collection (ATCC, Manassas, VA) in 2010 and maintained in RPMI 1640 with 10% fetal bovine serum. These cell lines were employed for the described experiments without further authentication.
Establishment of acquired erlotinib-resistant NSCLC cell line
Erlotinib-resistant HCC827 cell line (HCC827/ER) was established by exposing parental HCC827 cells to 3.5 μM erlotinib for 2 months followed with one more month of exposure to 7.5 μM erlotinib using an alternating 5-day drug on and 5-day drug off schedule as described (23). The resistant cell population was then routinely cultured with medium containing 1 μM erlotinib. The resistance phenotype was preserved even with prolonged culture in erlotinib-free culture medium for 6 months, suggesting an irreversible phenotype (23). We did not isolate pure monoclones. The pooled populations of HCC827/ER cells were used for further studies.
Preparation of cell lysates and Western blot
Cells were washed with cold PBS and resuspended in ice-cold EBC buffer (0.5% Nonidet P-40, 50mM Tris, pH 7.6, 120 mM NaCl, 1 mM EDTA, and 1 mM- β-mercaptoethanol) containing protease inhibitor mixture set I. Following cell lysis by sonication and centrifugation at 14,000 × g for 15 min at 4 °C, the resulting supernatant was collected as the total cell lysate. Protein expression was analyzed by Western blot as previously described (24).
Quantitative reverse transcription-PCR (RT-PCR)
For quantitative reverse transcription-PCR, total RNA was purified and reverse transcribed with random hexamers and SuperScript III (Invitrogen). Amplification was carried out using 2 × SYBR green PCR mix (Bio-Rad, CA) by ABI 7500 real-time PCR system (Applied Biosystems) and specific primers: for human Bcl2: forward, 5′-GGT GGA GGA GCT CTT CAG G-3′ and reverse, 5′- ATA GTT CCA CAA AGG CAT CC-3′; for human Bcl-XL: forward, 5′- ATA GTT CCA CAA AGG CAT CC -3′ and reverse, 5′- TGG GAT GTC AGG TCA CTG AA-3′; for human MCL-1: forward, 5′- TAA GGA CAA AAC GGG ACT GG-3′ and reverse, 5′-ACC AGC TCC TAC TCC AGC AA-3′ and for β-actin: forward, 5′- TCA GGA TCC ACG TGC TTG TCA -3′; reverse, 5′- TAC CCT TGG ACC CAG AGG TTC TTT GA -3′. Relative gene expression quantifications were performed according to the comparative Ct method using β-actin as an internal standard. Quantification of gene expression was analyzed with the 7500 v2.0.5 software program and quantified by the 2-ΔΔCt method. Data represent the mean ± SD of three independent experiments.
RNA Interference
Lentiviral pSIH1-puro-STAT3 shRNA and pSIH1-puro-control shRNA were obtained from Addgene (Cambridge, MA). The control shRNA plasmid-A encodes a scrambled shRNA sequence that will not lead to the specific degradation of any cellular message. Control shRNA hairpin sequence: CCT AAG GTT AAG TCG CCC TCG CTC GAG CGA GGG CGA CTT AAC CTT AGG. STAT3 shRNA hairpin sequence: GAT CCG CAT CTG CCT AGA TCG GCT ATT CAA GAG ATA GCC GAT CTA GGC AGA TGT TTT TTG. PTPMeg2 shRNA (sc-44670-SH) and its control shRNA (sc-108060) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Hairpin sequence: GAT CCC TAG AGT GAA GCT AAC AAT TCA AGA GAT TGT TAG CTT CAC TCT AGT TTA. For pseudovirus production, STAT3 shRNA or PTPMeg2 shRNA was cotransfected into 293FT cells with lentivector packaging plasmid mixture (System Biosciences, CA) using nanojuice transfection kit (EMD Chemical, Inc.) as described (25). After 48h, the virus-containing media were harvested by centrifugation at 20,000 × g. Cells were infected with the virus-containing media in the presence of polybrene (8ug/ml) for 24h following which stable positive clones were selected using 1ug/ml puromycin.
Sulforhodamine B (SRB) assay
The inhibitory effects of erlotinib and niclosamide on cell growth were assessed using the sulforhodamine B (SRB) assay as described (26, 27). Cells (6 × 103 - 8 × 103) were cultured in complete RPMI 1640 medium in triplicate wells using 96-well plates. After overnight growth, cells were treated with erlotinib, niclosamide or the combination for 72h. The degree of growth inhibition was calculated by using the equation: % growth inhibition = (1 - At/Ac) × 100, where At and Ac represent the absorbance in treated and control cultures, respectively, as described previously (28).
Colony formation assay
Colony formation assay on plastic surface was conducted in 6-well plates as described (23). HCC827 and HCC827/ER cells were trypsinized (single-cell suspension) and plated in 6-well-plates (1000 cells/well). Cells were treated with erlotinib (0.1 μM) by continuous exposure for 10 days (erlotinib-containing medium was replenished every 3 days). Following treatment, culture plates were stained with crystal violet (0.1% in 20% methanol). Digital images of the plates were obtained as a permanent record prior to colony counting as described previously (29).
Lung cancer xenografts and treatments
The Institutional Animal Care and Use Committee of Emory University approved the animal experiments. Six-week-old Nu/Nu nude mice were purchased from Harlan and housed under pathogen-free conditions in microisolator cages. Xenografts were raised by injecting 5 × 106 of HCC827 and HCC827/ER cells in a balanced salt solution into subcutaneous tissue over the flank region of nude mice. Tumors were allowed to grow to an average volume of 250 mm3 prior to initiation of therapy as described (30). Tumor-bearing mice were randomized into four treatment groups (n = 8 per group) as follows: (1) vehicle control (0.5%DMSO, 100 μl/d, i.p.); (2) erlotinib (40mg/kg/d, i.p.); (3) niclosamide (20mg/kg/d, i.p.); (4) erlotinib (40mg/kg/d, i.p.) + niclosamide (20mg/kg/d, i.p.). Tumor volume was assessed by caliper measurements once every two days and calculated with the formula: V=(L×W2)/2 (L: length; W: width) as described (31). After 32 consecutive days of treatment, mice were sacrificed by inhaled CO2. Harvested tumors were used for Western blot, immunohistochemistry (IHC) or QD-IHF analysis.
Immunohistochemistry (IHC) analysis
IHC staining of tumor tissue using rabbit anti-human active caspase 3 antibodies was performed as described (30). Briefly, harvested tumors were embedded in paraffin and cut into 4-μm sections. Staining was performed using the R.T.U. Vectastain kit following the manufacturer's standard protocol (Vector Laboratories, Burlingame, CA). The tissue slides were blocked with 2.5% normal horse serum for 10 min. Samples were then incubated with rabbit anti-human active caspase 3 antibody (dilution 1:50), overnight at 4°C. After washing, the tissue slides were incubated with anti-rabbit IgG HRP secondary antibody for 10 minutes. The slides were stained with 3,3′-diaminobenzidine (DAB) (Vector Laboratories) and counterstained with hematoxylin (Vector Laboratories), dehydrated, treated with xylene, and mounted. All slides were examined and representative pictures were taken using an Olympus BX41 microscope (Olympus America, Melville, NY). Semiquantitative determination of IHC staining for active caspase 3 was performed using immunoscore based on both percentage of stained cells and staining intensity as described (32).
Quantum dot-based immunohistoflorescence (QD-IHF) and its signal quantification
QD-IHF for measurement of pSTAT3 and pEGFR in tumor tissues was performed as described previously (33-35). Briefly, harvested tumors were embedded in paraffin and cut into 4-μm sections. After deparaffinization and rehydration, antigen retrieval was performed by heating with citric acid (10 mmol/L, pH 6.0) in a microwave for 10 min. The tissue slides were blocked with 2.5% normal horse serum for 10 min before the primary antibody incubation. Rabbit anti-human pEGFR and mouse anti-human p-STAT3 antibodies were mixed at 1:50 dilution in 1×PBS containing 2.5% horse serum. Normal rabbit IgG was used as a negative control. Tissue sections were incubated with a mixed solution of p-EGFR and p-STAT3 antibodies overnight at 4°C. After washing with 1×PBS three times, QD705 goat anti-mouse IgG conjugate (green) and QD605 goat anti-rabbit IgG conjugate (red) secondary antibodies were added to the slides with further incubation for 1h at 37°C. The slides were washed three times with 1×PBS, counterstained with DAPI, mounted and stored at 4°C under dark conditions. QD imaging and quantification procedures were performed as described previously (35). The Nuance™ fluorescence microscope system (CRi consolidated with Caliper, a PerkinElmer company, Hopkinton, MA) was used for quantification of the QD-IHF signals. All cubed image files were collected from tumor tissue slides at 10 nm wavelength intervals from 420-720 nm, with an auto exposure time per wavelength interval at 200∼400× magnification. Taking the cube with a long wavelength band pass filter allowed transmission of all emission wavelengths above 420 nm. Both separated and combined QD images were obtained after establishing the QD spectral library and unmixing the image cube. For each tissue slide, 10 cubes were taken. The background signal was removed for accurate quantification of the QD signals. The average of each QD signal was obtained by selecting tumor areas on each cube for quantification by Nuance imaging software (Caliper/PerkinElmer). An average reading from the 10 cubes was obtained as a total average signal count of each tissue slide for both QD signals.
Statistical analysis
Significant differences between two groups were analyzed using two-sided unpaired Student's t-test and p value < 0.05 was considered statistically significant. Statistical analysis was performed with Graphpad InStat 3 software (San Diego, CA) (36).
Results
Erlotinib induces activation of the STAT3/Bcl2/Bcl-XL survival pathway in association with downregulation of PTPMeg2
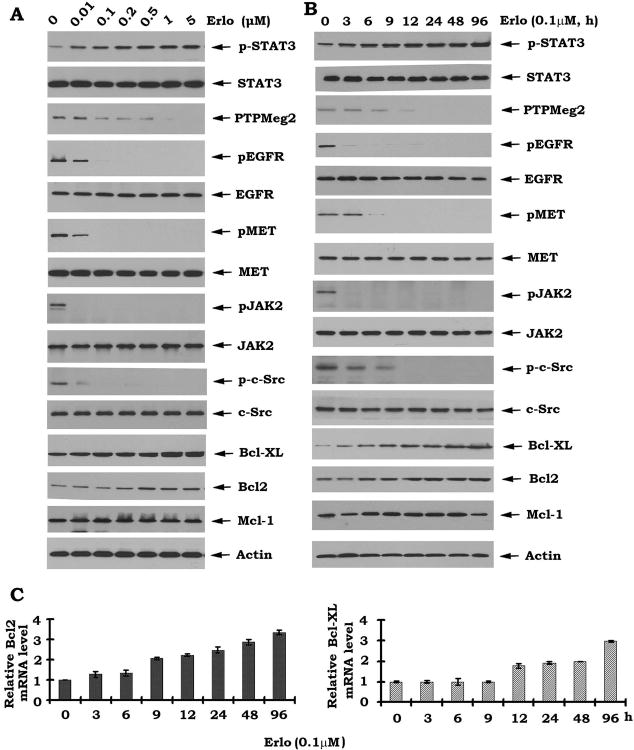
To test whether inhibition of EGFR by erlotinib activates the STAT3/Bcl2/Bcl-XL survival signaling pathway, human lung cancer HCC827 cells were treated with increasing concentrations of erlotinib for 48 hours. Inhibition of EGFR by erlotinib resulted in increased phosphorylation of STAT3 at Tyr705 (i.e. activation) and elevated Bcl-2/Bcl-XL in dose- and time-dependent manners (Fig.1A, B). Since decreased phosphorylation of EGFR and MET was observed following erlotinib treatment in HCC827 cells (Fig.1A, B), this is consistent with the expected activity of erlotinib as an EGFR inhibitor. It is known that STAT3 functions as a physiologic transcriptional factor of Bcl2 and Bcl-XL (11-13). The mRNA levels for Bcl-2 and Bcl-XL were increased in HCC827 following cell exposure to erlotinib (Fig. 1C), suggesting that erlotinib-induced activation of STAT3 may activate Bcl2/Bcl-XL at the transcriptional level. Similar studies were also performed in other cell lines (i.e. H1650, H1975 etc.). Results indicate that erlotinib can also inhibit pEGFR and enhance pSTAT3 in both H1650 and H1975 cells. Interestingly, erlotinib is more sensitive to H1650 cells in inhibition of pEGFR than H1975 cells containing T790M mutation. The basic levels of pSTAT3 in H1975 cells are higher than H1650 cells (Fig. S1). This confirms that erlotinib-induced activation of STAT3 is not limited to a specific cell type.
Figure 1.
Inhibition of EGFR by erlotinib downregulates PTPMeg2 in association with activation of the STAT3/Bcl2/Bcl-XL pathway in lung cancer HCC827 cells. A, HCC827 cells were treated with increasing concentrations of erlotinib (Erlo) for 48h. pEGFR, PTPMeg2, pSTAT3, Bcl2, Bcl-XL, etc., were analyzed by Western blot. B, HCC827 cells were treated with erlotinib (0.1 μM) for various times. Levels of Bcl2 or Bcl-XL mRNA were analyzed by RT-PCR.
To uncover the mechanism(s) by which erlotinib induces STAT3 phosphorylation, the effects of erlotinib on STAT3 kinases (i.e. c-Src or JAK2) and phosphatase (i.e. PTPMeg2) were tested. Treatment of HCC827 cells with erlotinib significantly reduced PTPMeg2 without activation of c-Src or JAK2. Conversely, erlotinib reduced phosphorylation of c-Src and JAK2 (Fig. 1A and B). These findings indicate that erlotinib-induced STAT3 phosphorylation and activation may occur through downregulation of its physiologic phosphatase PTPMeg2.
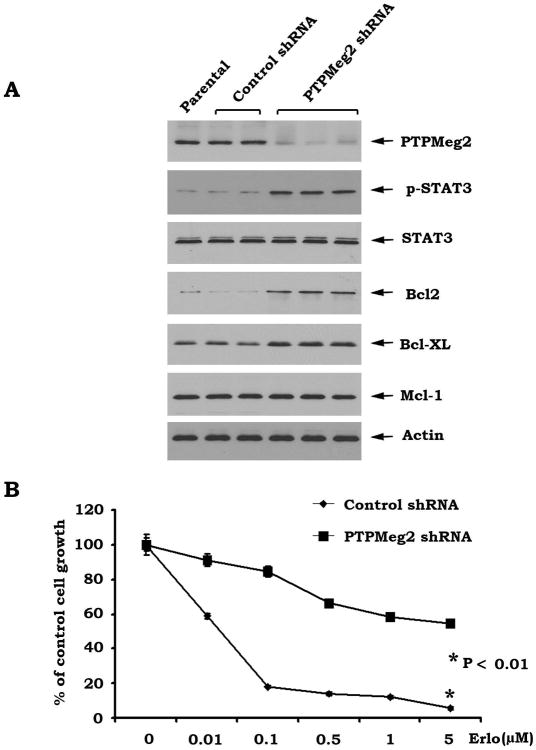
Specific knockdown of PTPMeg2 by RNA interference (RNAi) results in activation of the STAT3/Bcl2/Bcl-XL survival pathway in human lung cancer cells
To genetically demonstrate whether PTPMeg2 regulates the STAT3/Bcl2/Bcl-XL survival pathway, PTPMeg2 was depleted from HCC827 cells by RNA interference using PTPMeg2 shRNA. Cells expressing PTPMeg2 shRNA displayed more than 95% reduction of PTPMeg2 protein expression as compared to cells expressing control shRNA (Fig. 2A). The specific knockdown of PTPMeg2 results in elevated levels of pSTAT3, Bcl2 and Bcl-XL (Fig. 2A), consistent with its identification as a physiologic STAT3 phosphatase(21). Importantly, upregulation of the STAT3/Bcl2/Bcl-XL pathway by specific depletion of PTPMeg2 leads to the significant resistance of HCC827 cells to erlotinib (Fig. 2B).
Figure 2.
Depletion of PTPMeg2 from HCC827 cells leads to upregulation of pSTAT3 and Bcl2/Bcl-XL. A, PTPMeg2 shRNA or control shRNA was transfected into HCC827 cells. Expression levels of pSTAT3, Bcl-XL, Bcl2 and Mcl-1 were analyzed by Western blot. B, HCC827 cells expressing PTPMeg2 shRNA or control shRNA were treated with increasing concentrations of erlotinib for 48h. Cell growth was determined by SRB assays. Data are mean ± SD from three independent experiments.
Erlotinib resistance is associated with activation of the STAT3/Bcl2/Bcl-XL survival pathway
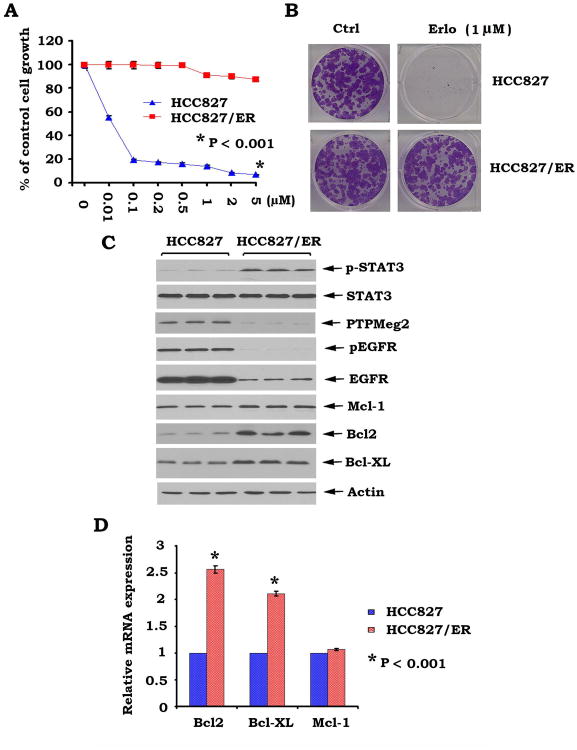
The HCC827 cell line harbors the erlotinib sensitive mutation 1 (i.e. Del E746-A750) without the resistant mutation 2 (i.e.T790M) (37). This renders HCC827 cells highly sensitive to treatment with erlotinib and other EGFR-TKI such as gefitinib (23, 37). To demonstrate whether erlotinib-induced activation of the STAT3/Bcl2/Bcl-XL signaling pathway is associated with erlotinib resistance, a lung cancer cell line with acquired erlotinib resistance (i.e.HCC827/ER) was established as described in “Methods” (23). SRB and colony formation assays confirmed that HCC827 parental cells are sensitive, but HCC827/ER cells are resistant to erlotinib (Fig. 3A, B). Intriguingly, increased levels of pSTAT3, Bcl2 and Bcl-XL were observed in HCC827/ER cells as compared to HCC827 cells (Fig. 3C). Real time PCR confirmed that increased protein levels of Bcl2 and Bcl-XL in HCC827/ER cells resulted from increased mRNA levels of Bcl-2 and Bcl-XL (Fig. 3D), suggesting that increased Bcl2/Bcl-XL levels result from STAT3 transcriptional regulation. Importantly, decreased levels of STAT3 phosphatase PTPMeg2 were also observed in HCC827/ER cells (Fig. 3C), which may explain why elevated levels of pSTAT3 were observed in HCC827/ER cells as compared to HCC827 parental cells. These results strongly indicate that erlotinib resistance may occur, at least in part, from altered PTPMeg2/STAT3/Bcl2/Bcl-XL signaling pathway in human lung cancer cells. It is known that secondary mutations of EGFR are associated with erlotinib resistance (6, 38). DNA sequencing analysis of genomic DNA isolated from HCC728 and HCC827/ER cells revealed that no secondary erlotinib resistance mutations (e.g. T790M, T854A, L747S and D761Y) were observed in either cell line, suggesting that the acquisition of erlotinib resistance by HCC827/ER cells did not result from these secondary genetic mutations in this system. However, we cannot rule out other possible genetic changes that may occur in HCC829/ER cells.
Figure 3.
Decreased PTPMeg2 and increased levels of pSTAT3 and Bcl2/Bcl-XL are associated with erlotinib resistance in human lung cancer cells. A, HCC827 cells and acquired erlotinib resistant HCC827/ER cells were treated with increasing concentrations of erlotinib (Erlo) for 48h. Cell growth was analyzed by SRB assay. B, HCC827 and HCC827/ER cells were treated with erlotinib (Erlo, 1 μM) and colony formation assay was performed as described in “Methods”. C, pSTAT3, PTPMeg2, Bcl2, Bcl-XL, etc., were analyzed by Western Blot. D, mRNA levels of Bcl2, Bcl-XL and Mcl-1 were analyzed by RT-PCR.
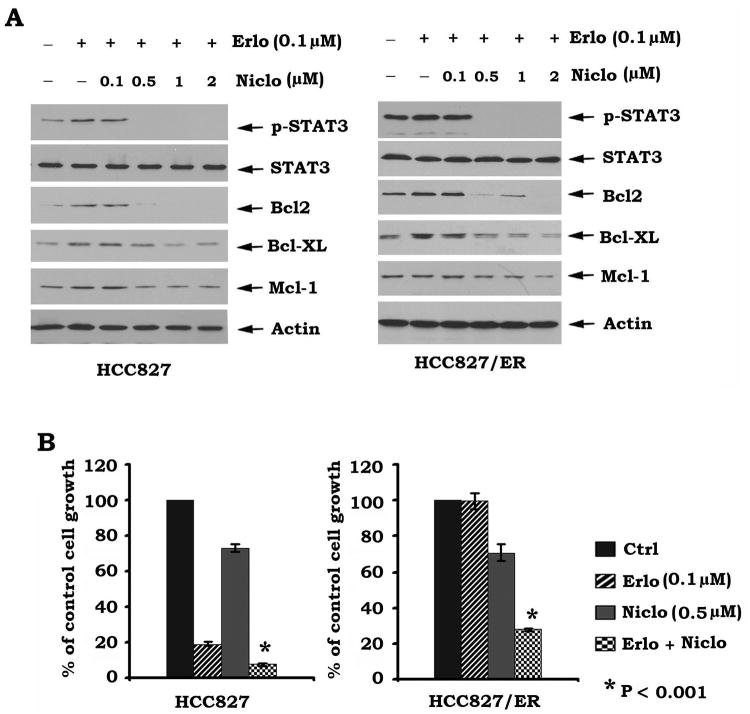
Inhibition of STAT3 by niclosamide blocks erlotinib-activated STAT3/Bcl2/Bcl-XL pathway and reverses erlotinib resistance of human lung cancer cells
Our findings reveal that activation of the STAT3/Bcl2/Bcl-XL pathway by erlotinib contributes to the development of resistance against erlotinib in human lung cancer cells (Figs. 1 and 3). To determine whether inhibition of the STAT3/Bcl2/Bcl-XL pathway overcomes erlotinib resistance, HCC827 and HCC827/ER cells were treated with erlotinib (0.1 μM) in the absence or presence of increasing concentrations of niclosamide (a STAT3 inhibitor, 0.1∼2 μM) for 72 h (Fig. S2). Results indicate that niclosamide blocks erlotinib-induced STAT3 phosphorylation and subsequently reduces Bcl2/Bcl-XL in both HCC827 or HCC827/ER cells (Figs. 4A). Functionally, niclosamide not only sensitized HCC827 cells to erlotinib but also reversed erlotinib resistance of HCC827/ER cells (Fig. 4B).
Figure 4.
Treatment of cells with niclosamide blocks erlotinib-induced activation of STAT3/Bcl2/Bcl-XL and reverses erlotinib resistance. A, HCC827 and HCC827/ER cells were treated with erlotinib (Erlo, 0.1 μM) in the absence or presence of increasing concentrations of niclosamide (Niclo) for 48h. pSTAT3, Bcl2, Bcl-XL and Mcl-1 were analyzed by Western Blot. B, HCC827 and HCC827/ER cells were treated with various concentrations of erlotinib (Erlo), niclosamide (Niclo, 0.5 μM) or their combination for 48 h. Cell growth was evaluated by SRB assay.
Specific knockdown of STAT3 restores sensitivity of lung cancer cells to erlotinib
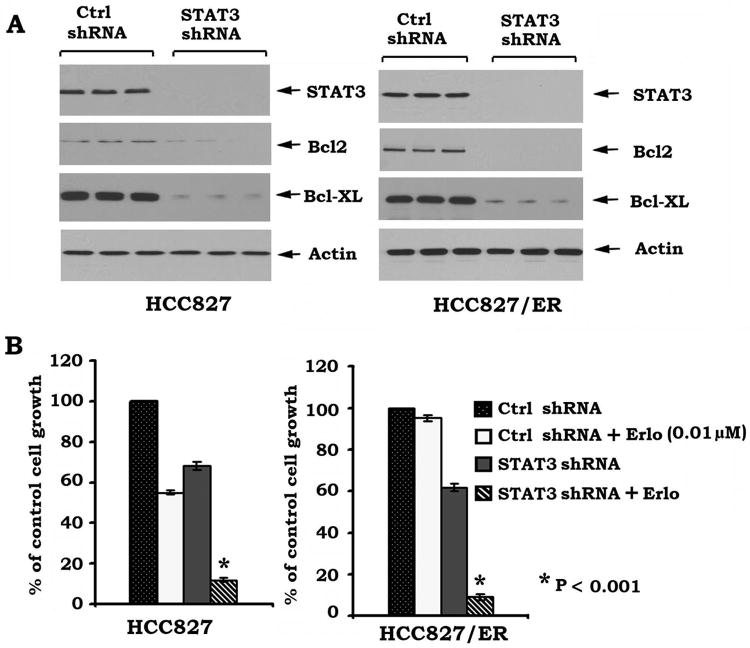
To test whether the STAT3 survival pathway is required for erlotinib resistance, STAT3 was depleted from HCC827 and HCC827/ER cells by RNA interference using STAT3 shRNA. Transfection of STAT3 shRNA significantly reduced the expression level of endogenous STAT3 by more than 99% (Fig. 5A). The effect of STAT3 shRNA on STAT3 expression was highly specific, as shown by the control shRNA having no effect (Fig. 5A). Importantly, silencing of STAT3 downregulated Bcl2 and Bcl-XL (Fig. 5A). SRB experiments indicated that depletion of STAT3 by RNA interference not only reverses erlotinib resistance of HCC827/ER cells but also sensitizes HCC827 cells to erlotinib (Fig. 5B). These findings suggest that the STAT3/Bcl2/Bcl-XL survival pathway may be involved in de novo resistance and is required for development of acquired resistance to erlotinib in human lung cancer cells.
Figure 5.
Specific depletion of STAT3 reverses erlotinib resistance. A, STAT3 shRNA or control shRNA was transfected into HCC827 cells. Expression levels of STAT3, Bcl-XL and Bcl2 were analyzed by Western blot. B, HCC827 cells expressing STAT3 shRNA or control shRNA were treated with erlotinib (Erlo) for 48h. Cell growth was determined by SRB assays.
Niclosamide overcomes acquired erlotinib resistance in vivo leading to long-term tumor-free survival
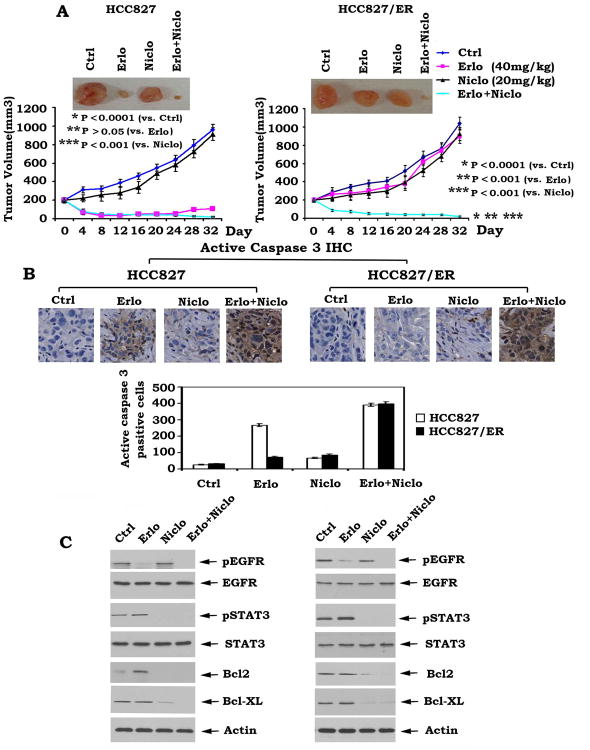
To test whether niclosamide overcomes the acquired erlotinib resistance of lung cancer in vivo, xenografts derived from HCC827 cells or HCC827/ER cell lines were treated in groups of eight mice with erlotinib (40mg/kg/d) (10), niclosamide (20mg/kg/d) (39) or their combination for 32 days as described in “Methods”. The control group was treated with 0.5% DMSO vehicle. We observed that lung cancer xenografts derived from HCC827 cells were very sensitive to erlotinib treatment with significant tumor shrinkage, whereas xenografts derived from HCC827/ER cells were resistant to erlotinib treatment with tumor volumes comparable to those in the vehicle-treated control group (Fig. 6A). By contrast, combination of niclosamide and erlotinib significantly repressed lung tumors derived from erlotinib resistant HCC827/ER cells, indicating that niclosamide can reverse acquired erlotinib resistance and restore sensitivity of HCC827/ER xenografts to erlotinib treatment in vivo (Fig. 6A). Intriguingly, two of the eight HCC827/ER xenograft-bearing mice treated with combined erlotinib and niclosamide showed complete tumor regression (Fig. S3), which is persisting as of the time of this publication (more than 8 months from the end of treatment on day 32).To determine whether erlotinib alone or in combination with niclosamide represses lung cancer via apoptosis in vivo, representative samples from harvested tumor tissues were analyzed by IHC for active caspase 3 as previously described (30). Results revealed that erlotinib enhances active caspase 3 positive cells only in HCC827 tumor tissues but not in HCC827/ER tumor tissues (Fig. 6B). In contrast, the combination of the two drugs increased active caspase 3 positive cells in both HCC827 and HCC827/ER tumor tissues (Fig. 6B). Western blot analysis of tumor lysates revealed that treatment of HCC827 and HCC827/ER mice with niclosamide also blocks STAT3 phosphorylation in association with reduced Bcl2 and Bcl-XL (Fig. 6C).
Figure 6.
Combination of erlotinib and niclosamide overcomes acquired erlotinib resistance in vivo. A, Mice bearing HCC827 or HCC827/ER lung cancer xenografts were treated with vehicle control, erlotinib (Erlo, 40mg/kg/d), niclosamide (Niclo, 20mg/kg/d) or their combination for 32 days. Each group includes 8 mice. Tumor volume was measured once every 2 days. After 32 days, the mice were sacrificed and the tumors were removed and analyzed. Representative tumor pictures were taken. B, Active caspase 3 was analyzed in tumor tissues at the end of experiments by IHC staining and quantified as described in “Methods”. C, Expression levels of pEGFR, pSTAT3, Bcl2 and Bcl-XL from tumor tissues in various treatment groups were analyzed by Western blot.
Quantum dot (QD)-based immunohistofluorescence (QD-IHF) analysis of pEGFR and pSTAT3 in tumor tissues
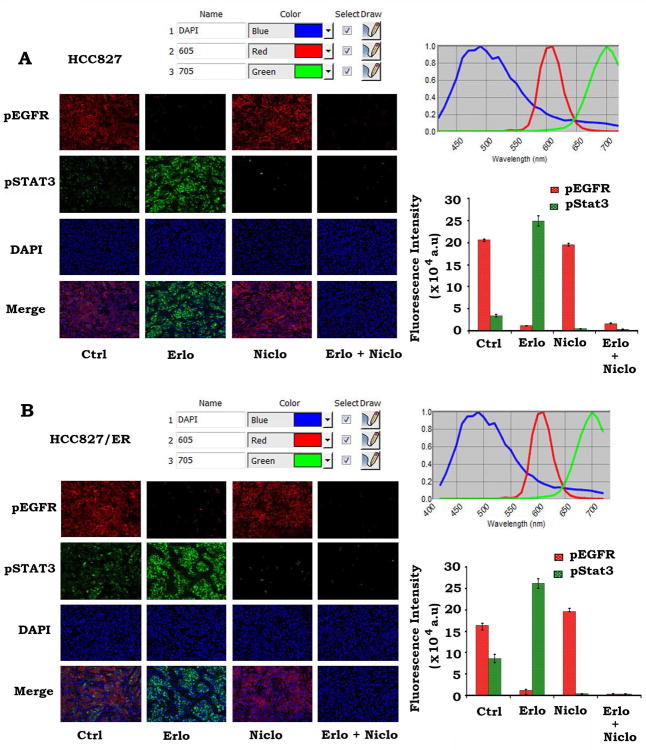
The QD-IHF technique is a valuable tool for simultaneous and concurrent immunostaining of multiple biomarkers in formalin-fixed paraffin-embedded (FFPE) tissues, thereby allowing for quantification of several biomarkers simultaneously on the same tissue slide (33-35, 40). To confirm whether inhibition of EGFR by erlotinib enhances pSTAT3 and whether erlotinib-induced phosphorylation of STAT3 can be inhibited by niclosamide in tumor tissues, pEGFR and pSTAT3 were simultaneously analyzed by QD-IHF on the same tissue slide using primary antibodies and QD-conjugated secondary antibodies with two different emission wavelengths (i.e. QD705 (green) and QD605 (red)). Both separated and combined QD images were obtained after determining the QD spectral library and unmixing the cubed image. QD images from HCC827 and HCC827/ER xenografts showed that pEGFR is localized at the cell membrane and pSTAT3 is located in the nucleus (Fig. 7A, B). Erlotinib treatment downregulated pEGFR in association with increased pSTAT3 in both HCC827 and HCC827/ER xenografts (Fig. 7A, B). Importantly, erlotinib enhanced pSTAT3 could be suppressed by niclosamide in both HCC827 and HCC827/ER tumor tissues (Fig. 7A, B).
Figure 7.
QD-IHF analysis of pEGFR and pSTAT3 in tumor tissues. A and B, HCC827 or HCC827/ER xenografts were treated with vehicle control, erlotinib (Erlo, 40mg/kg/d), niclosamide (Niclo, 20mg/kg/d) or their combination for 32 days. pEGFR and pSTAT3 were analyzed in tumor tissues at the end of experiments by QD-IHF and quantified as described in “Methods”.
Toxicity analysis in vivo
Erlotinib (40mg/kg/d) in combination with niclosamide (20mg/kg/d) potently suppressed tumor growth and was well tolerated as demonstrated by the absence of any significant weight loss in the treated animals compared to other treatment conditions (Figs.S4A and S5A). Also, there were no observable alterations in vital organ functions as reflected by the results of liver, kidney and bone marrow function tests (ALT, AST and BUN, WBC, RBC, Hb and platelets; Figs.S4B and S5B). Histopathology of harvested normal tissues (heart, liver, lung, brain, spleen, kidney, intestine, etc.) revealed no evidence of normal tissue toxicities after treatment with erlotinib or niclosamide alone or in combination for 32 days (Figs.S4C and S5C).
Discussion
The EGFR tyrosine kinase inhibitors (TKIs), erlotinib and gefitinib, are effective agents for the treatment of NSCLC patients whose tumors bear activating somatic mutations in EGFR gene (exon 19 deletions or exon 21 L858R substitution) (23). The vast majority of patients with EGFR-mutant advanced NSCLCs will show an initial impressive tumor response to EGFR-TKIs. However, over a median period of approximately 12 months, resistance develops leading to a loss of treatment efficacy of the EGFR-TKIs (6, 38, 41). Two major mechanisms of resistance to EGFR-TKIs have been identified: (1) secondary resistance mutations and (2) “oncogene kinase switch” (i.e. amplification of the MET oncogene). These two mechanisms (T790M mutation and MET amplification) account for 70% of all cases of acquired resistance to EGFR-TKIs (42). Other secondary resistance mutations (i.e. D761Y, L747S and T854A) seem to be rare (<5%). Since “multiple” molecular mechanisms at play in “late resistance” besides T790M-EGFR and MET-amplification(7), identification of these mechanisms that mediate the remaining instances of otherwise unexplained acquired EGFR-TKI resistance is critical for lung cancer therapy (38). In the present report, we discovered that inhibition of EGFR by erlotinib induces activation of STAT3 by suppressing its physiological phosphatase PP™eg2 leading to increased Bcl2/Bcl-XL, which contributes to the acquired erlotinib resistance (Figs 1-3). Interestingly, Mcl-1 did not seem to be altered or impacted in the erlotinib resistance in this model system. It is currently unclear why erlotinib- activated STAT3 only up-regulated Bcl-2/Bcl-XL but did not affect Mcl-1 expression. Further work may be required to uncover the mechanism.
Our findings support and have now extended those of a previous report (10). It is possible that early p-STAT3 activation induced by erlotinib in the early adaptive drug resistant tumor cells may ultimately become late resistance by irreversibly maintaining activation of the p-STAT3/Bcl2/Bcl-xL survival pathway. Our studies are focused on irreversible “late resistance” rather than “early adaptive TKI resistance”. It is currently unclear how erlotinib stimulates STAT3 phosphorylation. Most published studies have attributed increased STAT3 phosphorylation to over-activation of JAK or Src kinase (19). As shown in Fig. 1, erlotinib inhibited JAK2 and c-Src phosphorylation. Thus, erlotinib-induced STAT3 phosphorylation does not likely result from JAK2 or c-Src in this case. It is well known that STAT3 phosphorylation status is also tightly regulated by dephosphorylation through its physiologic phosphatase PTPMeg2 (21). Since erlotinib inhibited the physiological STAT3 phosphatase PTPMeg2 (Fig. 1A, B), which could reduce its ability to dephosphorylate STAT3. Because reduced dephosphorylation could lead to elevated phosphorylation, erlotinib-induced STAT3 phosphorylation may occur, at least in part, through suppression of its phosphatase PTPMeg2. It is currently unclear whether other kinase(s) may be involved in erlotinib-stimulated STAT3 phosphorylation.
Selective depletion of PTPMeg2 activates the STAT3/Bcl2/Bcl-XL survival pathway that also contributes to decreased sensitivity to erlotinib (Fig. 2). Since decreased PTPMeg2 and increased pSTAT3/Bcl2/Bcl-XL were also observed in HCC827/ER cells with acquired erlotinib resistance (vs. parental HCC827), this indicates that erlotinib activation of STAT3/Bcl2/Bcl-XL by suppression PTPMeg2 contributes to acquired erlotinib resistance. It is important that other mechanisms of resistance such as acquired genetic mutations (i.e. T790M, T854A, L747S and D761Y) were excluded by DNA sequencing in our models.
Niclosamide (C13H8Cl2N2O4, MW: 327.119) is an FDA-approved small molecule drug of the teniacide anthelmintic family that is effective against human tapeworms (43). It exerts its anthelminthic effects by uncoupling oxidative phosphorylation in the tapeworm (44). Niclosamide is safe, well tolerated, inexpensive and readily available (45, 46). Interestingly, niclosamide has recently been identified as a new small molecule STAT3 inhibitor because it can inhibit Tyr705 site phosphorylation as well as transcriptional activity of STAT3, but has no obvious inhibitory effect on the activation of the upstream proteins JAK2 and Src (22). Our results reveal that niclosamide not only blocks erlotinib-induced STAT3 phosphorylation but also suppresses its downstream effectors, Bcl2 and Bcl-XL, in both HCC827 and HCC827/ER cells (Fig. 4A), suggesting that niclosamide functions as a STAT3 inhibitor to block erlotinib-activated STAT3/Bcl2/Bcl-XL survival pathway in human lung cancer cells. Importantly, niclosamide can reverse erlotinib resistance and restore the sensitivity of HCC827/ER cells to erlotinib (Fig. 4B). Since specific depletion of STAT3 using RNAi sensitizes HCC827 cells to erlotinib and also overcomes erlotinib resistance in HCC827/ER cells through reduced Bcl2/Bcl-XL (Fig. 5), this suggests that the STAT3/Bcl2/Bcl-XL survival pathway is essential for optimal efficacy and the development of resistance to erlotinib therapy.
To further evaluate whether blockage of the STAT3/Bcl2/Bcl-XL survival pathway by niclosamide overcomes erlotinib resistance in vivo, erlotinib-sensitive (i.e. HCC827) and erlotinib- resistant (HCC827/ER) xenografts were compared with various treatments as shown in Fig. 6. Low dose niclosamide (20mg/kg/d) in combination with a standard dose of erlotinib (40mg/kg/d) effectively overcame erlotinib resistance in HCC827/ER xenografts (Figs. 6, S4 and S5). The long term tumor regression seen in some of the mice treated with the combination of erlotiinb and niclosamide (Fig. S3) suggests that this combination may be worthy of clinical translation in human subjects with lung cancer.
Quantum dots (QDs) are nanoscale particles made from inorganic semiconductors that can produce different fluorescence signals depending on their size and components (35). Compared with organic dyes, QDs have superior signal brightness and photo-stability, relatively long excited-state lifetime, and optimized signal-to-background ratios (47). Due to their long excitation time and narrow emission spectra, QDs can be excited simultaneously through one appropriate excitation source. These properties render QDs ideal for multiplexed biological imaging and they have been used for both molecular and cellular labeling (40, 47-49). Since immunoblotting can only show the total level of each protein, QDs combined with an interrelated imaging and quantification system were used to quantify the localization of multiple proteins in the same sample. QD-based images showed that pEGFR mainly localizes on membranes, and pSTAT3 is located in the nucleus, as expected (Fig. 7). QD-quantification also confirmed that treatment of mice with erlotinib significantly inhibits EGFR phosphorylation at Tyr1068 in association with upregulation of STAT3 phosphorylation at Tyr705 in tumor tissues (Fig. 7). Niclosamide specifically blocked erlotinib-induced STAT3 phosphorylation without affecting pEGFR (Fig. 7). The results from this QD-based analysis not only provide additional evidence that erlotinib activates STAT3, but also help explain how niclosamide can reverse erlotinib resistance in vivo (Fig. 6).
In summary, we have identified a previously unrecognized mechanism of acquired resistance to EGFR targeted therapy in lung cancer cell lines. This mechanism employs the PTPMeg2/STAT3/Bcl2/Bcl-XL survival signaling pathway and is independent of the secondary mutation resistance mechanism in NSCLC cells. Erlotinib-induced STAT3 phosphorylation occurs through suppression of its physiologic phosphatase PTPMeg2 without activation of STAT3 upstream kinases (i.e. JAK or c-Src). Inhibition of the STAT3/Bcl2/Bcl-XL survival pathway by niclosamide can overcome acquired erlotinib resistance in vitro and in vivo. Based on our findings, combined treatment with niclosamide and erlotinib may represent a novel and effective strategy for treatment of NSCLC, including for those patients who have already developed resistance to standard EGFR-TKI therapy.
Supplementary Material
Acknowledgments
We thank Anthea Hammond for professional editing of the manuscript.
Grant Support: This work was supported by NCI, National Institutes of Health Grants R01CA112183 (X. Deng) and R01CA136534 (X. Deng), and by Flight Attendant Medical Research Institute Clinical Innovator Award (X. Deng).
Footnotes
Disclosure of Potential Conflicts of Interest: The authors disclose no potential conflicts of interest
Authors ' Contributions: Conception and design: X. Deng, R. Li
Development of methodology: X. Deng, R. Li, Z. Hu
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): R. Li, Z. Hu, G. L. Sica, Z. Chen
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): X. Deng, R. Li, Z. Hu
Writing, review, and/or revision of the manuscript: X. Deng, R. Li, S.Y. Sun, Z. Chen, T.K. Owonikoko, G. L. Sica, S. S. Ramalingam, W. J. Curran, F. R. Khuri
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): R. Li, X. Deng, W. J. Curran, F. R. Khuri, S.Y. Sun
Study supervision: X. Deng
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 3.da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol. 2011;6:49–69. doi: 10.1146/annurev-pathol-011110-130206. [DOI] [PubMed] [Google Scholar]
- 4.Gridelli C, Maione P, Bareschino MA, Schettino C, Sacco PC, Ambrosio R, et al. Erlotinib in the treatment of non-small cell lung cancer: current status and future developments. Anticancer Res. 2010;30:1301–10. [PubMed] [Google Scholar]
- 5.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–92. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 7.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75r. doi: 10.1126/scitranslmed.3002003. a26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, et al. Analysis of Tumor Specimens at the Time of Acquired Resistance to EGFR-TKI Therapy in 155 Patients with EGFR-Mutant Lung Cancers. Clin Cancer Res. 2013;19:2240–7. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arcila ME, Oxnard GR, Nafa K, Riely GJ, Solomon SB, Zakowski MF, et al. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res. 2011;17:1169–80. doi: 10.1158/1078-0432.CCR-10-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan W, Tang Z, Yin L, Morrison B, Hafez-Khayyata S, Fu P, et al. MET-independent lung cancer cells evading EGFR kinase inhibitors are therapeutically susceptible to BH3 mimetic agents. Cancer Res. 2011;71:4494–505. doi: 10.1158/0008-5472.CAN-10-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Catlett-Falcone R, Landowski TH, Oshiro MM, Turkson J, Levitzki A, Savino R, et al. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10:105–15. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- 12.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao YX, Pestell RG, Albanese C, et al. Stat3 as an oncogene (vol 98, pg 295, 1998) Cell. 1999;99 doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 13.Alas S, Bonavida B. Rituximab inactivates signal transducer and activation of transcription 3 (STAT3) activity in B-non-Hodgkin's lymphoma through inhibition of the interleukin 10 autocrine/paracrine loop and results in down-regulation of Bcl-2 and sensitization to cytotoxic drugs. Cancer Res. 2001;61:5137–44. [PubMed] [Google Scholar]
- 14.Bollrath J, Phesse TJ, von Burstin VA, Putoczki T, Bennecke M, Bateman T, et al. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 2009;15:91–102. doi: 10.1016/j.ccr.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Lee H, Deng J, Kujawski M, Yang C, Liu Y, Herrmann A, et al. STAT3-induced S1PR1 expression is crucial for persistent STAT3 activation in tumors. Nat Med. 2010;16:1421–8. doi: 10.1038/nm.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lesina M, Kurkowski MU, Ludes K, Rose-John S, Treiber M, Kloppel G, et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19:456–69. doi: 10.1016/j.ccr.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 17.Gao SP, Mark KG, Leslie K, Pao W, Motoi N, Gerald WL, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846–56. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haura EB, Zheng Z, Song L, Cantor A, Bepler G. Activated epidermal growth factor receptor-Stat-3 signaling promotes tumor survival in vivo in non-small cell lung cancer. Clin Cancer Res. 2005;11:8288–94. doi: 10.1158/1078-0432.CCR-05-0827. [DOI] [PubMed] [Google Scholar]
- 19.Bharti AC, Donato N, Aggarwal BB. Curcumin (diferuloylmethane) inhibits constitutive and IL-6-inducible STAT3 phosphorylation in human multiple myeloma cells. J Immunol. 2003;171:3863–71. doi: 10.4049/jimmunol.171.7.3863. [DOI] [PubMed] [Google Scholar]
- 20.Inghirami G, Chiarle R, Simmons WJ, Piva R, Schlessinger K, Levy DE. New and old functions of STAT3: a pivotal target for individualized treatment of cancer. Cell Cycle. 2005;4:1131–3. doi: 10.4161/cc.4.9.1985. [DOI] [PubMed] [Google Scholar]
- 21.Su F, Ren F, Rong Y, Wang Y, Geng Y, Feng M, et al. Protein tyrosine phosphatase Meg2 dephosphorylates signal transducer and activator of transcription 3 and suppresses tumor growth in breast cancer. Breast Cancer Res. 2012;14:R38. doi: 10.1186/bcr3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren XM, Duan L, He QA, Zhang Z, Zhou Y, Wu DH, et al. Identification of Niclosamide as a New Small-Molecule Inhibitor of the STAT3 Signaling Pathway. Acs Med Chem Lett. 2010;1:454–9. doi: 10.1021/ml100146z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Fan S, Koo J, Yue P, Chen ZG, Owonikoko TK, et al. Elevated expression of eukaryotic translation initiation factor 4E is associated with proliferation, invasion and acquired resistance to erlotinib in lung cancer. Cancer Biol Ther. 2012;13:272–80. doi: 10.4161/cbt.18923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito T, Deng X, Carr B, May WS. Bcl-2 phosphorylation required for anti-apoptosis function. J Biol Chem. 1997;272:11671–3. doi: 10.1074/jbc.272.18.11671. [DOI] [PubMed] [Google Scholar]
- 25.Huang S, Okumura K, Sinicrope FA. BH3 mimetic obatoclax enhances TRAIL-mediated apoptosis in human pancreatic cancer cells. Clin Cancer Res. 2009;15:150–9. doi: 10.1158/1078-0432.CCR-08-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun SY, Yue P, Dawson MI, Shroot B, Michel S, Lamph WW, et al. Differential effects of synthetic nuclear retinoid receptor-selective retinoids on the growth of human non-small cell lung carcinoma cells. Cancer Res. 1997;57:4931–9. [PubMed] [Google Scholar]
- 27.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1:1112–6. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- 28.Sun SY, Rosenberg LM, Wang X, Zhou Z, Yue P, Fu H, et al. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 2005;65:7052–8. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, Yue P, Kim YA, Fu H, Khuri FR, Sun SY. Enhancing mammalian target of rapamycin (mTOR)-targeted cancer therapy by preventing mTOR/raptor inhibition-initiated, mTOR/rictor-independent Akt activation. Cancer Res. 2008;68:7409–18. doi: 10.1158/0008-5472.CAN-08-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–81. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 31.Liu AW, Cai J, Zhao XL, Jiang TH, He TF, Fu HQ, et al. ShRNA-targeted MAP4K4 inhibits hepatocellular carcinoma growth. Clin Cancer Res. 2011;17:710–20. doi: 10.1158/1078-0432.CCR-10-0331. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Sun SY, Owonikoko TK, Sica GL, Curran WJ, Khuri FR, et al. Rapamycin induces Bad phosphorylation in association with its resistance to human lung cancer cells. Mol Cancer Ther. 2012;11:45–56. doi: 10.1158/1535-7163.MCT-11-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xing Y, Chaudry Q, Shen C, Kong KY, Zhau HE, Chung LW, et al. Bioconjugated quantum dots for multiplexed and quantitative immunohistochemistry. Nat Protoc. 2007;2:1152–65. doi: 10.1038/nprot.2007.107. [DOI] [PubMed] [Google Scholar]
- 34.Xu J, Muller S, Nannapaneni S, Pan L, Wang Y, Peng X, et al. Comparison of quantum dot technology with conventional immunohistochemistry in examining aldehyde dehydrogenase 1A1 as a potential biomarker for lymph node metastasis of head and neck cancer. Eur J Cancer. 2012 doi: 10.1016/j.ejca.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang DH, Su L, Peng XH, Zhang H, Khuri FR, Shin DM, et al. Quantum dot-based quantification revealed differences in subcellular localization of EGFR and E-cadherin between EGFR-TKI sensitive and insensitive cancer cells. Nanotechnology. 2009;20:225102. doi: 10.1088/0957-4484/20/22/225102. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, Hawk N, Yue P, Kauh J, Ramalingam SS, Fu H, et al. Overcoming mTOR inhibition-induced paradoxical activation of survival signaling pathways enhances mTOR inhibitors' anticancer efficacy. Cancer Biol Ther. 2008;7:1952–8. doi: 10.4161/cbt.7.12.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gandhi J, Zhang J, Xie Y, Soh J, Shigematsu H, Zhang W, et al. Alterations in genes of the EGFR signaling pathway and their relationship to EGFR tyrosine kinase inhibitor sensitivity in lung cancer cell lines. PLoS One. 2009;4:e4576. doi: 10.1371/journal.pone.0004576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen KS, Kobayashi S, Costa DB. Acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancers dependent on the epidermal growth factor receptor pathway. Clin Lung Cancer. 2009;10:281–9. doi: 10.3816/CLC.2009.n.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sack U, Walther W, Scudiero D, Selby M, Kobelt D, Lemm M, et al. Novel effect of antihelminthic Niclosamide on S100A4-mediated metastatic progression in colon cancer. J Natl Cancer Inst. 2011;103:1018–36. doi: 10.1093/jnci/djr190. [DOI] [PubMed] [Google Scholar]
- 40.Huang DH, Peng XH, Su L, Wang DS, Khuri FR, Shin DM, et al. Comparison and Optimization of Multiplexed Quantum Dot-Based Immunohistofluorescence. Nano Research. 2010;3:61–8. [Google Scholar]
- 41.Engelman JA, Janne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2008;14:2895–9. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 42.Yoshida T, Zhang G, Haura EB. Targeting epidermal growth factor receptor: central signaling kinase in lung cancer. Biochem Pharmacol. 2010;80:613–23. doi: 10.1016/j.bcp.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 43.Lateef M, Zargar SA, Khan AR, Nazir M, Shoukat A. Successful treatment of niclosamide- and praziquantel-resistant beef tapeworm infection with nitazoxanide. Int J Infect Dis. 2008;12:80–2. doi: 10.1016/j.ijid.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 44.Weinbach EC, Garbus J. Mechanism of action of reagents that uncouple oxidative phosphorylation. Nature. 1969;221:1016–8. doi: 10.1038/2211016a0. [DOI] [PubMed] [Google Scholar]
- 45.Merschjohann K, Steverding D. In vitro trypanocidal activity of the anti-helminthic drug niclosamide. Exp Parasitol. 2008;118:637–40. doi: 10.1016/j.exppara.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 46.Jin Y, Lu Z, Ding K, Li J, Du X, Chen C, et al. Antineoplastic mechanisms of niclosamide in acute myelogenous leukemia stem cells: inactivation of the NF-kappaB pathway and generation of reactive oxygen species. Cancer Res. 2010;70:2516–27. doi: 10.1158/0008-5472.CAN-09-3950. [DOI] [PubMed] [Google Scholar]
- 47.Smith AM, Gao X, Nie S. Quantum dot nanocrystals for in vivo molecular and cellular imaging. Photochem Photobiol. 2004;80:377–85. doi: 10.1562/0031-8655(2004)080<0377:QDNFIV>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 48.Whaley SR, English DS, Hu EL, Barbara PF, Belcher AM. Selection of peptides with semiconductor binding specificity for directed nanocrystal assembly. Nature. 2000;405:665–8. doi: 10.1038/35015043. [DOI] [PubMed] [Google Scholar]
- 49.Luo G, Long J, Zhang B, Liu C, Ji S, Xu J, et al. Quantum dots in cancer therapy. Expert Opin Drug Deliv. 2012;9:47–58. doi: 10.1517/17425247.2012.638624. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.