Abstract
Infections caused by bacterial biofilms are a significant global health problem, causing considerable patient morbidity and mortality and contributing to the economic burden of infectious disease. This review describes diverse strategies to combat bacterial biofilms, focusing firstly on small molecule interference with bacterial communication and signaling pathways, including quorum sensing and two-component signal transduction systems. Secondly we discuss enzymatic approaches to the degradation of extracellular matrix components to effect biofilm dispersal. Both these approaches are based upon non-microbicidal mechanisms of action, and thereby do not place a direct evolutionary pressure on the bacteria to develop resistance. Such approaches have the potential to, in combination with conventional antibiotics, play an important role in the eradication of biofilm based bacterial infections.
Introduction
In the last twenty years, bacterial infections have reemerged as a major health threat. Hospital-acquired infections are now responsible for more deaths annually in the United States than emphysema, AIDS, Parkinson’s disease, and homicide combined [1] and cost the U.S. health care system over $20 billion annually [2]. An estimated 80% of bacterial infections in humans are caused by biofilms, according to the National Institutes of Health [3], leading the Centers for Disease Control to declare biofilms among the most pressing clinical impediments of the century [4]. Despite the increased virulence of biofilms and their obvious threat to human health, there are no clinically available drugs to inhibit or disperse biofilms in vivo [**5].
Biofilms are formed by multiple bacterial cells attached to a surface that arrange themselves into a complex tertiary structure encased in an extracellular matrix comprised of carbohydrates, proteins, and other macromolecules [6, 7]. Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, Acinetobacter baumannii and other medically relevant bacterial strains colonize clinical surfaces and medical devices via biofilms and resist common eradication methods including desiccation, antibiotic treatment, and nutrient deprivation [8]. Bacteria associated with a biofilm are up to 1,000 times more resistant to antibiotic therapies in comparison to their planktonic counterparts and are insensitive to the host immune response, allowing them to persist and promote continued infection despite aggressive antibiotic therapy [8, 9].[8, 9]. Of particular concern are biofilms that form on indwelling medical devices (IMDs), creating a continuous source of infection that often necessitates removal of the device [7].
Biofilm formation is a complex process involving multiple bacterial signaling systems including quorum sensing, nutrient and chemical signal response, and extracellular matrix formation (Figure 1). As such, very few chemical scaffolds have been identified that can inhibit or disperse bacterial biofilms. Numerous approaches have been investigated to both inhibit and disperse bacterial biofilms[**10]. This review describes approaches that involve inhibition of intercellular communication and signaling pathways with small molecules, in addition to approaches that center on degrading the integrity of the extracellular matrix. We provide an overview of important quorum sensing pathways and two component systems involved in biofilm formation and the effects of their inhibition by novel antibiofilm compounds. We also describe methods for disrupting the extracellular matrix required for the formation of robust biofilms. Finally, we conclude with future perspectives for the discovery and development of biofilm inhibitors as important and necessary therapeutic agents. This is not meant to be an exhaustive review of every anti-biofilm approach, which be beyond the scope of this document, and instead provides the reader with an overview of several of the most important anti-biofilm strategies, giving select examples in each case.
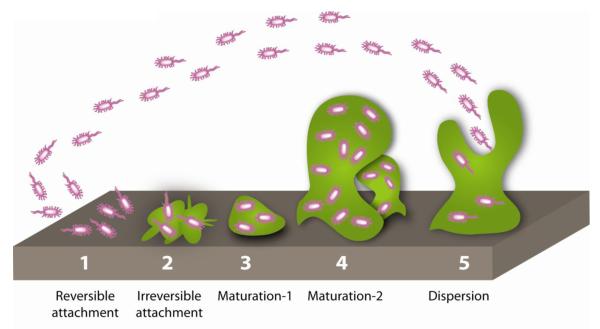
Figure 1.
Stages of the Biofilm Lifecycle. In stage 1, planktonic bacteria initiate attachment to an abiotic surface, which becomes irreversible in stage 2. Stages 3 and 4 feature biofilm maturation and growth of the three dimensional community. Dispersion occurs in stage 5 and releases planktonic bacteria from the biofilm to colonize additional sites.
Disruption of Intercellular Communication and Signaling Pathways
Quorum Sensing
Quorum sensing (QS) describes the intercellular communication required for bacterial communities to act in coordinated ways to alter gene expression based on population density [11]. QS can be reduced to interplay between two proteins; the first produces a signaling molecule known as an autoinducer (AI), and a second protein that responds to the AI. Autoinducers encompass several classes of structurally related molecules including acyl homoserine lactones (AHLs), autoinducing peptides (AIPs) and autoinducer-2 (AI-2) [12].
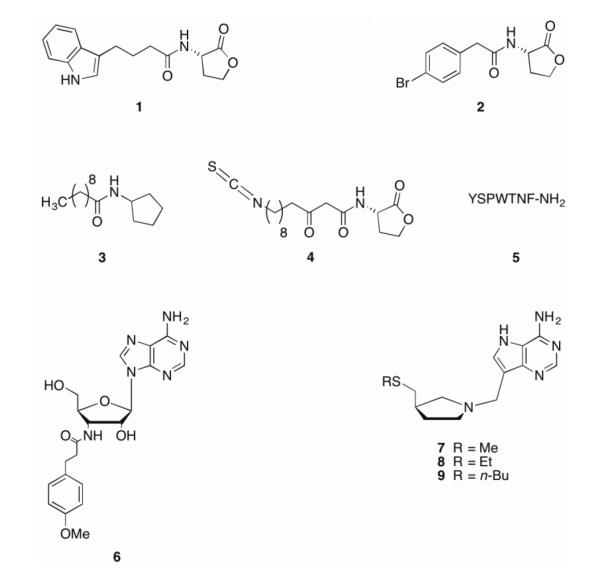
Over 70 species of Gram-negative bacteria use AHLs for intraspecies communication, with specificity imparted by variation in the oxidation state and length of the acyl side chain [12]. AHLs can freely diffuse through the bacterial membrane such that AHL concentration correlates to bacterial concentration and enables population-mediated control of gene expression. Ultimately, this gene expression results in various phenotypes including the production of virulence factors and biofilm formation [13]. As a result of their extensive study over the last three decades, AHLs have provided a scaffold for many potential biofilm inhibitors [14, 15]. The Blackwell group has reported the synthesis and activity of several unnatural AHLs, two of which (1 and 2) (Figure 2) significantly reduced biofilm formation in P. aeruginosa PA01 at 50 μM [16]. Spring et al. also investigated analogs of P. aeruginosa AHLs by replacing the lactone functionality with an N-acylated cyclopentylamide. One compound, known as C10-CPA (3), was able to abrogate biofilm formation in P. aeruginosa PA01:GFP after seven days under flow conditions at a concentration of 250 μM [17]. Using the crystal structure of the P. aeruginosa transcriptional regulator LasR [12, 18], Bottomley and coworkers designed and synthesized covalent LasR inhibitors that inhibited quorum sensing and, in the case of the lead compound, 4, could inhibit wild type PA01 biofilm formation by close to 50% at 50 μM.
Figure 2.
Quorum sensing inhibitors.
In Gram-positive bacteria, the predominant molecules used for QS are autoinducing peptides (AIPs). Many AIPs contain hydrophobic domains crucial for activity, which are hypothesized to help promote hydrophobic interactions that lead to receptor activation [19]. In S. aureus, threshold levels of AIP bind AgrC, resulting in expression of RNA-III, a small noncoding RNA that ultimately downregulates genes controlling adhesins required for biofilm formation [20]. Phosphorylation of RNA-III activating protein (RAP) activates target of RNA-III activating peptide (TRAP), thereby increasing cell adhesion and biofilm formation [21]. Conversely, RNA-III inhibiting peptide 5 (RIP), prevents phosphorylation of TRAP and thus reduces biofilm formation and cell adhesion. RIP has been evaluated extensively and has been shown to prevent infections, including those by antibiotic resistant strains, in several animal models [22] without any signs of toxicity or induction of RIP resistance [23]. Other analogs of S. aureus AIP-1 have been shown to be potent agonists of AgrC-1 QS [24] or promoters of S. aureus biofilm formation [25]. Recently, Blackwell and colleagues reported a novel class of AIP-III mimetics designed to inhibit AgrC receptors in S. aureus [26]. Although their effects on biofilm formation were not reported, the lead compounds were shown to inhibit all four AgrC receptors and block hemolysis, which is under the control of QS, at picomolar levels. Given the role of AgrC in biofilm formation, it would be very interesting to evaluate the effects of these inhibitors on S. aureus biofilm formation and maintenance.
An alternative approach to disrupting AHL based QS is to exploit the lability of the AHL scaffold and degrading AHLs enzymatically, thereby inhibiting QS. This phenomenon is known as quorum quenching, and unsurprisingly, given the role of AHL based QS in bacterial virulence, is utilized by numerous organisms, both bacterial and eukaryotic.[27] This approach to inhibiting quorum sensing was first exploited by Dong et al. in which a gene encoding a lactonase from Bacillus sp. was shown to inhibit AHL activity by hydrolyzing the lactone bond of several AHLs from P. aeruginosa and the plant pathogens Erwinia carotovora and Agrobacterium tumefaciens.[28, 29] Applying this approach as an anti-biofilm strategy, the hydrolase BpiB05, derived from the soil metagenome, was shown to affect biofilm formation, in addition to motility and pyocyanin synthesis, in P. aeruginosa. When transformed with the bpiB05 gene, P. aeruginosa PAO1 formed poorly developed biofilms that did not progress beyond initial surface attachment.[30] This work gives some indication the potential of the quorum quenching approach as an anti-biofilm strategy, though much more investigation is required.
Both Gram-positive and Gram-negative bacteria utilize autoinducer-2 (AI-2) in quorum sensing mechanisms, thus providing a basis for a universal language across bacterial species [31]. Nucleoside analogs (6-9) have been shown to interfere with AI-2 mediated quorum sensing and, in some cases, affect biofilm formation [32, 33]. Recently, Bentley and coworkers have described novel AI-2 analogs that were capable of inhibiting maturation of E. coli biofilms in vitro and that could, when combined with antibiotics near MIC levels, could almost completely clear pre-formed E. coli biofilms in a microfluidic device [*34].
In addition to AI-2, bis-(3′5′)-cyclic di-guanylic acid (c-di-GMP) is believed to be a ubiquitous second messenger signal molecule. c-di-GMP regulates exopolysaccharide synthesis, and thereby influences exopolysaccharide-dependent biofilm formation, in proteobacteria including Vibrio cholera, P. aeruginosa, and E. coli [35], and has been implicated in biofilm dispersion in P. aeruginosa where elevated c-di-GMP levels inhibited effective surface detachment [36]. Treatment of S. aureus with exogenous c-di-GMP inhibited in vitro biofilm formation and adherence to HeLa cells [37]. While many analogs to c-di-GMP have been synthesized and evaluated [37-40], cell permeability and inferior receptor binding and affinity as compared to c-di-GMP have hampered their utility [41].
Two Component Systems
In addition to quorum sensing, bacteria use other pathways to recognize and respond to various external signals and stimuli. In many instances, the signal receptions and responses are mediated by two-component regulatory systems (TCS), which couple a membrane sensor kinase (histidine kinase, HK) to a response regulator (RR) [42, 43]. TCS have been extensively reviewed, with several recent articles highlighting the role of TCS in both virulence and antibiotic resistance [5, 44-46]. In a prototypical TCS, the HK receives extracellular signals and phosphorylates the N-terminal domain of the RR on a highly conserved aspartate residue [43, 47-49]. The phosphorylated RR typically undergoes a conformational change and dimerization leading to DNA binding and activation or repression of gene transcription via the C-terminal domain [47-50]. In this way, the bacteria can produce appropriate gene products based on extracellular signals. TCS are not found in vertebrates, thus representing a novel target for therapeutics [45]. Furthermore, there are very few essential TCSs, such that targeting them could disrupt the bacteria’s ability to respond appropriately to external stimuli without accompanying bactericidal effects, thus side-stepping direct evolutionary pressure for resistance to the TCS inhibitor [45].
Genetic screens of bacterial mutants incapable of producing biofilms has led to an extensive list of TCS involved in biofilm formation and maintenance [5]. However, despite this vast availability of protein targets, few compounds are known to target TCS responsible for biofilm formation and maintenance [5]. Walkmycin C 10 (Figure 3) is a member of the walkmycin family of natural products produced by Streptomyces sp. strain MK632-100F11 [51, 52]. Originally identified as an inhibitor of autophosphorylation of YycG in S. aureus and Bacillus subtilis, it was later found to interact with the conserved catalytic domain of the histidine kinase and has activity against several HKs [51]. In S. mutans, walkmycin C inhibits VicK, a non-essential orthologue of YycG involved in cellular growth, surface adhesion, sucrose-dependent biofilm formation, and competence development, with an IC50 of 2.87 μM (2.53 μg/mL). Walkmycin C also inhibits CiaH, an HK with roles in sucrose-dependent biofilm formation, competence development, and stress tolerance, with an IC50 of 4.87 μM (4.29 μg/mL) [51, 53]. At levels below the minimum inhibitory concentration (MIC), walkmycin C reduces biofilm mass, induces abnormal biofilm formation, and represses acid tolerance and competence in S. aureus [51].
Figure 3.
Two-component system inhibitors.
The 2-aminoimidazole (2-AI) class of small molecules, derived from marine sponge alkaloids, has shown broad spectrum biofilm inhibition and dispersion activity [10, 54-59]. Compound 11, representing the reverse amide class of 2-AIs, has been shown to inhibit and disperse biofilms in both P. aeruginosa and A. baumannii [60]. Recently, a biotinylated analogue of this compound was employed as a chemical probe to identify the A. baumannii response regulator BfmR as the molecular target of this class of compounds [**61]. BfmR belongs to the OmpR family of response regulators and has been implicated in biofilm formation [42]. A bfmR mutant showed significantly lowered ability to form biofilms and altered planktonic cellular morphology,[42, 62] thus, BfmR represents an attractive target for anti-biofilm compounds. Using a BfmR homology model, docking studies suggested that compound 11 targets the interface between the N and C-terminal domains of BfmR. The docking studies were validated with pull-down assays using truncated protein containing either the N- or C-domains only, showing that both domains make contact with the compound. Further studies indicated that 11 interacts with several other response regulators, but displays no binding to any of the tested non-response regulator proteins [**61].
Non-Small Molecule Approaches
Enzymatic Degradation of Matrix Components
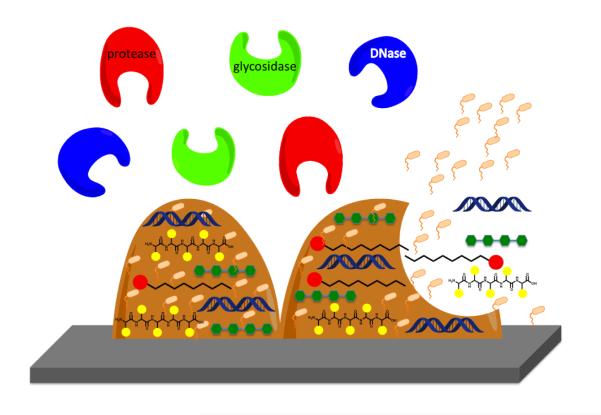
The biofilm matrix typically accounts for over 90% of the dry mass of a biofilm and consists predominantly of biopolymers produced by the bacteria themselves, these biopolymers are known as extracellular polymeric substances (EPS) and, as mentioned above, include polysaccharides, proteins, lipids and nucleic acids. The matrix forms the basis of the three-dimensional structure of the biofilm and immobilizes the cells, keeping them in close proximity to each other and allowing for cell-cell communication. The matrix contains the contents of lysed bacterial cells, serving as a nutrient source and acting as a reservoir of genes for horizontal gene transfer [*63] (Figure 4). The matrix also contributes to the tolerance of bacterial cells to antibiotics, acting as a diffusion barrier to prevent access to the cells [64]. One approach to the eradication of preformed biofilms is therefore to destroy the integrity of the biofilm matrix, typically by enzymatic degradation of components of the EPS, leading to subsequent detachment of cells from the biofilm. This mechanism of biofilm dispersal is an innate phenomenon employed by several diverse bacterial species. Bacteria secrete enzymes such as glycosidases, proteases, and DNases that degrade various components of the EPS [65]. Examples of endogenously produced matrix degrading enzymes include the DNase thermonuclease, which is produced by S. aureus, the glycoside hydrolase dispersin B, which is produced by Aggregatibacter actinomycetemcomitans, and alginate lyase, which is produced by P. aeruginosa. These enzymes, and many others, are used by the bacteria to initiate active dispersion of the biofilm, which then allows for the release of cells into the surrounding environment, contributing to bacterial survival and disease transmission [65]. Several of these matrix-degrading enzymes have been investigated as potential therapeutic agents.
Figure 4.
Schematic of the various matrix-degrading enzymes used to initiate biofilm inhibition and dispersal.
Dispersin B has been shown to inhibit the formation of biofilms by several medically relevant bacterial species including S. aureus [66], Staphylococcus epidermidis [66], E. coli [67], and Yersinia pestis [67], in addition to dispersing preformed S. epidermidis [66, 68] and E. coli [67] biofilms and sensitizing S. epidermidis biofilm cells to the action of antimicrobials [66, 69]. Dispersin B has also demonstrated activity in vivo, lowering the rate of catheter colonization by S. aureus in combination with triclosan in a rabbit model of infection [70]. Alginate lyase degrades a polysaccharide known as alginate and has been shown to enhance the microbicidal activity of aminoglycosides against P. aeruginosa biofilms in vitro [71, 72], and has also demonstrated in vivo efficacy, enhancing the clearance of mucoid P. aeruginosa when coadministered with amikacin in a rabbit model of endocarditis [73].
Extracellular DNA is an important component of the biofilm matrix [74] and the use of nucleases as an anti-biofilm strategy has been explored against a number of bacterial strains. Biofilms formed in the presence of DNase exhibit reduced biomass resulting from a reduced number and size of microcolonies within the biofilm, bacterial and decreased antibiotic tolerance [75]. The fact that degradation of extracellular DNA in the biofilm matrix by DNase has been shown to result in a biofilm that displays decreased tolerance to environmental factors makes the use of DNase an attractive anti-biofilm strategy [75]. An extracellular DNase (NucB), produced by Bacillus licheniformis induces rapid biofilm dispersal activity against preformed biofilm of several species of both Gram-positive and Gram-negative bacteria including B. subtilis, E. coli, and Micrococcus luteus [73]. S. aureus also produces a nuclease, known as Nuc, when the sigma factor B (sigB) gene is absent that has been shown to inhibit biofilm formation [76], while a nuc mutant was shown to form a thicker biofilm that contained increased levels of extracellular DNA [77]. Recombinant human DNase I (rhDNase) has been shown to inhibit biofilm formation by both S. aureus and S. epidermidis, disperse preformed S. aureus biofilms, and increase the susceptibility of S. aureus biofilm cells to killing by chlorhexidine gluconate and povidone iodine. rhDNase displayed activity in vivo, increasing the survival of S. aureus-infected Caenorhabditis elegans in when administered in combination with tobramycin [**78]. rhDNase has also been shown to enhance the microbicidal activity of aminoglycosides against P. aeruginosa biofilms in vitro [72], and to effect a significant loss of cells and biomass from biofilms of several strains of Streptococcus pneumoniae [79]. rhDNase I (also known as dornase alfa and marketed as Pulmozyme by Genentech) is used in the clinic for the treatment of pulmonary disease in cystic fibrosis (CF) patients [80], in which biofilm mediated P. aeruginosa infections are a major contributing factor to lung tissue damage [81]. Administration of Pulmozyme has been shown to lead to reduced demand for antibiotics and improved lung function in CF patients [82].
Other enzymatic anti-biofilm approaches include the use of proteases to modulate biofilms by degradation of the protein component of the biofilm matrix. It is known that endogenous proteases play a role in biofilm dispersal [83] and it has also been shown that exogenously added proteases can exhibit dispersal activity against established biofilms. For example, the serine protease Esp, which is produced by S. epidermidis, inhibits S. aureus biofilm formation and eradicates preformed biofilms of this bacterium. Esp has also been shown to enhance the susceptibility of S. aureus biofilms to the antimicrobial peptide human beta-defensin 2 (hBD2), while the activity of Exp in vivo has been demonstrated by the ability of both Esp secreting S. epidermidis and purified Esp to eliminate human nasal colonization by S. aureus [84]. The elastase LasB and proteinase K have also demonstrated anti-biofilm activity against S. aureus, while the supernatant of LasB producing P. aeruginosa was shown to induce the expression of several endogenous protease genes. Proteinase K was been shown to increase proteolytic activity, which is believed to be the mechanism by which these enzymes effect S. aureus biofilm dispersal [85]. Finally, the metalloprotease serratopeptidase (SPEP) is produced by Serratia marcescens and is widely used as an anti-inflammatory therapeutic. SPEP has been shown inhibit biofilm formation and enhance the activity of ofloxacin against biofilms of both P. aeruginosa and S. epidermidis [86] and to inhibit biofilm formation by Listeria monocytogenes [87].
Antibodies as an Anti Biofilm Strategy
A recently reported non-small molecule approach to the discovery of anti-biofilm agents is the use of antibodies for the eradiation of bacterial biofilms. Monoclonal antibodies (mAbs) that bind the P. aeruginosa Psl, a ubiquitous cell surface anchored exopolysaccharide that plays a role in the formation and maintenance of biofilms by acting as a scaffold for other biofilm initiating components [88, **89], were identified from a screen of an M13 phage-based human antibody library. Lead mAbs were shown to inhibit host cell attachment by P. aeruginosa and impart significant protection in multiple animal models of P. aeruginosa infection including a mouse acute lethal pneumonia model and a thermal injury model [**89]. Antibodies to the partially de-N-acetylated form of the Staphylococcal surface polymer poly-N-acetylglucosamine (PNAG), which promotes biofilm formation, increased killing of S. aureus by human neutrophils, while passive immunization of mice with anti-dPNAG-DT rabbit sera resulted in increased clearance of S. aureus from the blood compared to mice treated with normal rabbit sera [90].
Conclusion and Future Perspectives
Bacterial biofilms are a major threat to human health as they are inherently resistant to clearance by both the host immune system and antibiotics. This review highlights recent strategies to combat biofilms both through small molecules and protein-based methods focusing on strategies that do not rely on toxic mechanisms to inhibit or disperse biofilms. As the scientific community learns more about bacterial biofilms and searches for methods to combat them, we must be mindful of the lessons learned from antibiotic development over the last century. Finding treatments that can alter the phenotype of the bacteria without inducing and selecting for genetic modifications that can lead to resistance is key in winning the battle against these pathogens. Antibiofilm strategies that focus on interrupting complex regulatory systems involved in biofilm formation and maintenance without killing the bacteria should disrupt the biofilm without also selecting for a resistant population. Such strategies have the potential, when paired with conventional antibiotics, to prevent or destroy biofilms, thereby greatly impacting human health and medicine. Furthermore, the identification of treatments that can disrupt bacterial regulation and communication may have great implications as antibiotic adjuvants as well as antibiofilm therapies.
Biologically-Inspired Strategies for Combating Bacterial Biofilms Highlights.
Biofilms are inherently resistant to antibiotics and are a major health threat
Quorum sensing antagonists inhibit biofilm formation
Inhibition of two-component systems disrupt biofilm formation
Approaches to target the extracellular matrix to disrupt biofilms are discussed
Acknowledgements
We thank the NIH (R01GM055769) and the DOD DMRDP program (W81XWH-11-2-0115) for their support. The DMRDP program is administered by the Department of Army; The U.S. Army Medical Research Acquisition Activity, 820 Chandler Street, Fort Detrick, MD 21702-5014 is the awarding and administering office. The content of this manuscript does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Klevens RM, Morrison MA, Nadle J, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. Jama-J Am Med Assoc. 2007;298(15):1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 2.Roberts RR, Hota B, Ahmad I, et al. Hospital and societal costs of antimicrobial-resistant infections in a Chicago teaching hospital: implications for antibiotic stewardship. Clin Infect Dis. 2009;49(8):1175–1184. doi: 10.1086/605630. [DOI] [PubMed] [Google Scholar]
- 3.Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2003;2(2):114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 4.Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8(9):881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **5.Worthington RJ, Blackledge MS, Melander C. Small Molecule Inhibition of Bacterial Two-Component Systems to Combat Antibiotic Resistance and Virulence. Future Medicinal Chemistry. 2013 doi: 10.4155/fmc.13.58. This review describes two-component systems and outlines their role in bacterial virulence and biofilm formation. Small molecule inhibitors of these systems are also discussed in depth.
- 6.Yang L, Liu Y, Wu H, et al. Combating biofilms. FEMS Immunol Med Microbiol. 2011 doi: 10.1111/j.1574-695X.2011.00858.x. [DOI] [PubMed] [Google Scholar]
- 7.Richards JJ, Melander C. Controlling bacterial biofilms. Chembiochem. 2009;10(14):2287–2294. doi: 10.1002/cbic.200900317. [DOI] [PubMed] [Google Scholar]
- 8.Gaddy JA, Actis LA. Regulation of Acinetobacter baumannii biofilm formation. Future Microbiol. 2009;4(3):273–278. doi: 10.2217/fmb.09.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogers SA, Huigens RW, Cavanagh J, Melander C. Synergistic Effects between Conventional Antibiotics and 2-Aminoimidazole-Derived Antibiofilm Agents. Antimicrob Agents Ch. 2010;54(5):2112–2118. doi: 10.1128/AAC.01418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **10.Worthington RJ, Richards JJ, Melander C. Small molecule control of bacterial biofilms. Org Biomol Chem. 2012;10(37):7457–7474. doi: 10.1039/c2ob25835h. This is an in-depth review of bacterial biofilm formation and the small molecules that both inhibit and promote this process.
- 11.Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311(5764):1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amara N, Mashiach R, Amar D, et al. Covalent inhibition of bacterial quorum sensing. J Am Chem Soc. 2009;131(30):10610–10619. doi: 10.1021/ja903292v. [DOI] [PubMed] [Google Scholar]
- 13.Finch RG, Pritchard DI, Bycroft BW, Williams P, Stewart GS. Quorum sensing: a novel target for anti-infective therapy. J Antimicrob Chemother. 1998;42(5):569–571. doi: 10.1093/jac/42.5.569. [DOI] [PubMed] [Google Scholar]
- 14.Hentzer M, Wu H, Andersen JB, et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. Embo J. 2003;22(15):3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geske GD, O’neill JC, Blackwell HE. Expanding dialogues: from natural autoinducers to non-natural analogues that modulate quorum sensing in Gram-negative bacteria. Chem Soc Rev. 2008;37(7):1432–1447. doi: 10.1039/b703021p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geske GD, Wezeman RJ, Siegel a P, Blackwell HE. Small molecule inhibitors of bacterial quorum sensing and biofilm formation. J Am Chem Soc. 2005;127(37):12762–12763. doi: 10.1021/ja0530321. [DOI] [PubMed] [Google Scholar]
- 17.Ishida T, Ikeda T, Takiguchi N, Kuroda A, Ohtake H, Kato J. Inhibition of quorum sensing in Pseudomonas aeruginosa by N-acyl cyclopentylamides. Appl Environ Microbiol. 2007;73(10):3183–3188. doi: 10.1128/AEM.02233-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bottomley MJ, Muraglia E, Bazzo R, Carfi A. Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J Biol Chem. 2007;282(18):13592–13600. doi: 10.1074/jbc.M700556200. [DOI] [PubMed] [Google Scholar]
- 19.Lyon GJ, Muir TW. Chemical signaling among bacteria and its inhibition. Chem Biol. 2003;10(11):1007–1021. doi: 10.1016/j.chembiol.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Bordi C, De Bentzmann S. Hacking into bacterial biofilms: a new therapeutic challenge. Ann Intensive Care. 2011;1(1):19. doi: 10.1186/2110-5820-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fux CA, Stoodley P, Hall-Stoodley L, Costerton JW. Bacterial biofilms: a diagnostic and therapeutic challenge. Expert Rev Anti Infect Ther. 2003;1(4):667–683. doi: 10.1586/14787210.1.4.667. [DOI] [PubMed] [Google Scholar]
- 22.Giacometti A, Cirioni O, Gov Y, et al. RNA III inhibiting peptide inhibits in vivo biofilm formation by drug-resistant Staphylococcus aureus. Antimicrob Agents Ch. 2003;47(6):1979–1983. doi: 10.1128/AAC.47.6.1979-1983.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balaban N, Cirioni O, Giacometti A, et al. Treatment of Staphylococcus aureus biofilm infection by the quorum-sensing inhibitor RIP. Antimicrob Agents Ch. 2007;51(6):2226–2229. doi: 10.1128/AAC.01097-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan WC, Coyle BJ, Williams P. Virulence regulation and quorum sensing in staphylococcal infections: competitive AgrC antagonists as quorum sensing inhibitors. J Med Chem. 2004;47(19):4633–4641. doi: 10.1021/jm0400754. [DOI] [PubMed] [Google Scholar]
- 25.Fowler SA, Stacy DM, Blackwell HE. Design and synthesis of macrocyclic peptomers as mimics of a quorum sensing signal from Staphylococcus aureus. Org Lett. 2008;10(12):2329–2332. doi: 10.1021/ol800908h. [DOI] [PubMed] [Google Scholar]
- 26.Tal-Gan Y, Stacy DM, Foegen MK, Koenig DW, Blackwell HE. Highly Potent Inhibitors of Quorum Sensing in Staphylococcus aureus Revealed Through a Systematic Synthetic Study of the Group-III Autoinducing Peptide. J Am Chem Soc. 2013 doi: 10.1021/ja3112115. 130517075635007. [DOI] [PubMed] [Google Scholar]
- 27.Dong YH, Zhang LH. Quorum sensing and quorum-quenching enzymes. J Microbiol. 2005;43(Spec No):101–109. [PubMed] [Google Scholar]
- 28.Dong YH, Xu JL, Li XZ, Zhang LH. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc Natl Acad Sci U S A. 2000;97(7):3526–3531. doi: 10.1073/pnas.060023897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong YH, Wang LH, Xu JL, Zhang HB, Zhang XF, Zhang LH. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature. 2001;411(6839):813–817. doi: 10.1038/35081101. [DOI] [PubMed] [Google Scholar]
- 30.Bijtenhoorn P, Schipper C, Hornung C, et al. BpiB05, a novel metagenome-derived hydrolase acting on N-acylhomoserine lactones. J Biotechnol. 2011;155(1):86–94. doi: 10.1016/j.jbiotec.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 31.Hardie KR, Heurlier K. Establishing bacterial communities by ‘word of mouth’: LuxS and autoinducer 2 in biofilm development. Nat Rev Microbiol. 2008;6(8):635–643. doi: 10.1038/nrmicro1916. [DOI] [PubMed] [Google Scholar]
- 32.Brackman G, Celen S, Baruah K, et al. AI-2 quorum-sensing inhibitors affect the starvation response and reduce virulence in several Vibrio species, most likely by interfering with LuxPQ. Microbiology. 2009;155(Pt 12):4114–4122. doi: 10.1099/mic.0.032474-0. [DOI] [PubMed] [Google Scholar]
- 33.Gutierrez JA, Crowder T, Rinaldo-Matthis A, Ho MC, Almo SC, Schramm VL. Transition state analogs of 5′-methylthioadenosine nucleosidase disrupt quorum sensing. Nat Chem Biol. 2009;5(4):251–257. doi: 10.1038/nchembio.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *34.Roy V, Meyer MT, Smith JA, et al. AI-2 analogs and antibiotics: a synergistic approach to reduce bacterial biofilms. Appl Microbiol Biotechnol. 2013;97(6):2627–2638. doi: 10.1007/s00253-012-4404-6. The authors report the use of AI-2 analogs to reduce the formation of E. coli biofilms in vitro. They also use a novel flow cell device to show that the same molecules, when paired with the antibiotic gentimicin, are capable of clearing pre-formed E. coli biofilms.
- 35.Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. Journal of bacteriology. 2005;187(5):1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karaolis DK, Rashid MH, Chythanya R, Luo W, Hyodo M, Hayakawa Y. c-di-GMP (3′-5′-cyclic diguanylic acid) inhibits Staphylococcus aureus cell-cell interactions and biofilm formation. Antimicrob Agents Ch. 2005;49(3):1029–1038. doi: 10.1128/AAC.49.3.1029-1038.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brouillette E, Hyodo M, Hayakawa Y, Karaolis DK, Malouin F. 3′,5′-cyclic diguanylic acid reduces the virulence of biofilm-forming Staphylococcus aureus strains in a mouse model of mastitis infection. Antimicrob Agents Ch. 2005;49(8):3109–3113. doi: 10.1128/AAC.49.8.3109-3113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kline T, Jackson SR, Deng W, Verlinde CL, Miller SI. Design and synthesis of bis-carbamate analogs of cyclic bis-(3′-5′)-diguanylic acid (c-di-GMP) and the acyclic dimer PGPG. Nucleosides Nucleotides Nucleic Acids. 2008;27(12):1282–1300. doi: 10.1080/15257770802554150. [DOI] [PubMed] [Google Scholar]
- 40.Ching SM, Tan WJ, Chua KL, Lam Y. Synthesis of cyclic di-nucleotidic acids as potential inhibitors targeting diguanylate cyclase. Bioorg Med Chem. 2010;18(18):6657–6665. doi: 10.1016/j.bmc.2010.07.068. [DOI] [PubMed] [Google Scholar]
- 41.Kalia D, Merey G, Nakayama S, et al. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem Soc Rev. 2013;42(1):305–341. doi: 10.1039/c2cs35206k. [DOI] [PubMed] [Google Scholar]
- 42.Tomaras a P, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiol-Sgm. 2008;154:3398–3409. doi: 10.1099/mic.0.2008/019471-0. [DOI] [PubMed] [Google Scholar]
- 43.Kenney LJ. Structure/function relationships in OmpR and other winged-helix transcription factors. Curr Opin Microbiol. 2002;5(2):135–141. doi: 10.1016/s1369-5274(02)00310-7. [DOI] [PubMed] [Google Scholar]
- 44.Gooderham WJ, Hancock RE. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol Rev. 2009;33(2):279–294. doi: 10.1111/j.1574-6976.2008.00135.x. [DOI] [PubMed] [Google Scholar]
- 45.Gotoh Y, Eguchi Y, Watanabe T, Okamoto S, Doi A, Utsumi R. Two-component signal transduction as potential drug targets in pathogenic bacteria. Curr Opin Microbiol. 2010;13(2):232–239. doi: 10.1016/j.mib.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 46.Schaller GE, Shiu SH, Armitage JP. Two-component systems and their co-option for eukaryotic signal transduction. Curr Biol. 2011;21(9):R320–330. doi: 10.1016/j.cub.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 47.Bourret RB, Hess JF, Simon MI. Conserved Aspartate Residues and Phosphorylation in Signal Transduction by the Chemotaxis Protein Chey. P Natl Acad Sci USA. 1990;87(1):41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stock JB, Stock a M, Mottonen JM. Signal Transduction in Bacteria. Nature. 1990;344(6265):395–400. doi: 10.1038/344395a0. [DOI] [PubMed] [Google Scholar]
- 49.Martinezhackert E, Stock a M. Structural relationships in the OmpR family of winged-helix transcription factors. J Mol Biol. 1997;269(3):301–312. doi: 10.1006/jmbi.1997.1065. [DOI] [PubMed] [Google Scholar]
- 50.Barbieri CM, Mack TR, Robinson VL, Miller MT, Stock a M. Regulation of Response Regulator Autophosphorylation through Interdomain Contacts. Journal of Biological Chemistry. 2010;285(42):32325–32335. doi: 10.1074/jbc.M110.157164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eguchi Y, Kubo N, Matsunaga H, Igarashi M, Utsumi R. Development of an antivirulence drug against Streptococcus mutans: repression of biofilm formation, acid tolerance, and competence by a histidine kinase inhibitor, walkmycin C. Antimicrob Agents Chemother. 2011;55(4):1475–1484. doi: 10.1128/AAC.01646-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okada A, Igarashi M, Okajima T, et al. Walkmycin B targets WalK (YycG), a histidine kinase essential for bacterial cell growth. J Antibiot (Tokyo) 2010;63(2):89–94. doi: 10.1038/ja.2009.128. [DOI] [PubMed] [Google Scholar]
- 53.Qi F, Merritt J, Lux R, Shi W. Inactivation of the ciaH Gene in Streptococcus mutans diminishes mutacin production and competence development, alters sucrose-dependent biofilm formation, and reduces stress tolerance. Infect Immun. 2004;72(8):4895–4899. doi: 10.1128/IAI.72.8.4895-4899.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richards JJ, Reed CS, Melander C. Effects of N-pyrrole substitution on the anti-biofilm activities of oroidin derivatives against Acinetobacter baumannii. Bioorg Med Chem Lett. 2008;18(15):4325–4327. doi: 10.1016/j.bmcl.2008.06.089. [DOI] [PubMed] [Google Scholar]
- 55.Huigens RW, 3rd, Rogers SA, Steinhauer a T, Melander C. Inhibition of Acinetobacter baumannii, Staphylococcus aureus and Pseudomonas aeruginosa biofilm formation with a class of TAGE-triazole conjugates. Org Biomol Chem. 2009;7(4):794–802. doi: 10.1039/b817926c. [DOI] [PubMed] [Google Scholar]
- 56.Bunders CA, Richards JJ, Melander C. Identification of aryl 2-aminoimidazoles as biofilm inhibitors in Gram-negative bacteria. Bioorg Med Chem Lett. 2010;20(12):3797–3800. doi: 10.1016/j.bmcl.2010.04.042. [DOI] [PubMed] [Google Scholar]
- 57.Peng L, Desousa J, Su Z, et al. Inhibition of Acinetobacter baumannii biofilm formation on a methacrylate polymer containing a 2-aminoimidazole subunit. Chem Commun (Camb) 2011;47(17):4896–4898. doi: 10.1039/c1cc10691k. [DOI] [PubMed] [Google Scholar]
- *58.Stowe SD, Richards JJ, Tucker a T, Thompson R, Melander C, Cavanagh J. Anti-biofilm compounds derived from marine sponges. Mar Drugs. 2011;9(10):2010–2035. doi: 10.3390/md9102010. Compounds derived from marine sponge metabolites are discussed with a focus on those that exhibit antibiofilm properties. Both natural and synthetic analogs are discussed.
- 59.Yeagley a A, Su Z, Mccullough KD, Worthington RJ, Melander C. N-substituted 2-aminoimidazole inhibitors of MRSA biofilm formation accessed through direct 1,3-bis(tert-butoxycarbonyl)guanidine cyclization. Org Biomol Chem. 2013;11(1):130–137. doi: 10.1039/c2ob26469b. [DOI] [PubMed] [Google Scholar]
- 60.Ballard TE, Richards JJ, Wolfe a L, Melander C. Synthesis and Antibiofilm Activity of a Second-Generation Reverse-Amide Oroidin Library: A Structure-Activity Relationship Study. Chemistry-a European Journal. 2008;14(34):10745–10761. doi: 10.1002/chem.200801419. [DOI] [PubMed] [Google Scholar]
- **61.Thompson RJ, Bobay BG, Stowe SD, et al. Identification of BfmR, a response regulator involved in biofilm development, as a target for a 2-Aminoimidazole-based antibiofilm agent. Biochemistry. 2012;51(49):9776–9778. doi: 10.1021/bi3015289. A 2-aminoimidazole antibiofilm compound was used as a probe to identify BfmR as its molecular target in A. baumannii. This is the first reported small molecule scaffold shown to interact with a bacterial response regulator.
- 62.Manikal VM, Landman D, Saurina G, Oydna E, Lal H, Quale J. Endemic carbapenem-resistant Acinetobacter species in Brooklyn, New York: Citywide prevalence, interinstitutional spread, and relation to antibiotic usage. Clinical Infectious Diseases. 2000;31(1):101–106. doi: 10.1086/313902. [DOI] [PubMed] [Google Scholar]
- *63.Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–633. doi: 10.1038/nrmicro2415. This is an excellent review on the composition and formation of the biofilm matrix.
- 64.Mah TF, O’toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9(1):34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 65.Kaplan JB. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J Dent Res. 2010;89(3):205–218. doi: 10.1177/0022034509359403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Izano EA, Amarante MA, Kher WB, Kaplan JB. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microbiol. 2008;74(2):470–476. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Itoh Y, Wang X, Hinnebusch BJ, Preston JF, 3rd, Romeo T. Depolymerization of beta-1,6-N-acetyl-D-glucosamine disrupts the integrity of diverse bacterial biofilms. J Bacteriol. 2005;187(1):382–387. doi: 10.1128/JB.187.1.382-387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaplan JB, Ragunath C, Velliyagounder K, Fine DH, Ramasubbu N. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother. 2004;48(7):2633–2636. doi: 10.1128/AAC.48.7.2633-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Donelli G, Francolini I, Romoli D, et al. Synergistic activity of dispersin B and cefamandole nafate in inhibition of staphylococcal biofilm growth on polyurethanes. Antimicrob Agents Chemother. 2007;51(8):2733–2740. doi: 10.1128/AAC.01249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Darouiche RO, Mansouri MD, Gawande PV, Madhyastha S. Antimicrobial and antibiofilm efficacy of triclosan and DispersinB combination. J Antimicrob Chemother. 2009;64(1):88–93. doi: 10.1093/jac/dkp158. [DOI] [PubMed] [Google Scholar]
- 71.Alkawash MA, Soothill JS, Schiller NL. Alginate lyase enhances antibiotic killing of mucoid Pseudomonas aeruginosa in biofilms. APMIS. 2006;114(2):131–138. doi: 10.1111/j.1600-0463.2006.apm_356.x. [DOI] [PubMed] [Google Scholar]
- 72.Alipour M, Suntres ZE, Omri A. Importance of DNase and alginate lyase for enhancing free and liposome encapsulated aminoglycoside activity against Pseudomonas aeruginosa. J Antimicrob Chemother. 2009;64(2):317–325. doi: 10.1093/jac/dkp165. [DOI] [PubMed] [Google Scholar]
- 73.Bayer a S, Park S, Ramos MC, Nast CC, Eftekhar F, Schiller NL. Effects of alginase on the natural history and antibiotic therapy of experimental endocarditis caused by mucoid Pseudomonas aeruginosa. Infect Immun. 1992;60(10):3979–3985. doi: 10.1128/iai.60.10.3979-3985.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295(5559):1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 75.Tetz VV, Tetz GV. Effect of extracellular DNA destruction by DNase I on characteristics of forming biofilms. DNA Cell Biol. 2010;29(8):399–405. doi: 10.1089/dna.2009.1011. [DOI] [PubMed] [Google Scholar]
- 76.Kiedrowski MR, Kavanaugh JS, Malone CL, et al. Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS One. 2011;6(11):e26714. doi: 10.1371/journal.pone.0026714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mann EE, Rice KC, Boles BR, et al. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One. 2009;4(6):e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **78.Kaplan JB, Lovetri K, Cardona ST, et al. Recombinant human DNase I decreases biofilm and increases antimicrobial susceptibility in staphylococci. J Antibiot (Tokyo) 2012;65(2):73–77. doi: 10.1038/ja.2011.113. This study showed that recombinnt human DNase I was capable of both inhibiting and dispersing S. aureus biofilms. Additionally, treatment with rhDNase greatly increased S. aureus biocide susceptibility.
- 79.Hall-Stoodley L, Nistico L, Sambanthamoorthy K, et al. Characterization of biofilm matrix, degradation by DNase treatment and evidence of capsule downregulation in Streptococcus pneumoniae clinical isolates. BMC Microbiol. 2008;8:173. doi: 10.1186/1471-2180-8-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Parsiegla G, Noguere C, Santell L, Lazarus RA, Bourne Y. The structure of human DNase I bound to magnesium and phosphate ions points to a catalytic mechanism common to members of the DNase I-like superfamily. Biochemistry. 2012;51(51):10250–10258. doi: 10.1021/bi300873f. [DOI] [PubMed] [Google Scholar]
- 81.Hoiby N, Ciofu O, Bjarnsholt T. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol. 2010;5(11):1663–1674. doi: 10.2217/fmb.10.125. [DOI] [PubMed] [Google Scholar]
- 82.Frederiksen B, Pressler T, Hansen A, Koch C, Hoiby N. Effect of aerosolized rhDNase (Pulmozyme) on pulmonary colonization in patients with cystic fibrosis. Acta Paediatr. 2006;95(9):1070–1074. doi: 10.1080/08035250600752466. [DOI] [PubMed] [Google Scholar]
- 83.Boles BR, Horswill a R. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008;4(4):e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Iwase T, Uehara Y, Shinji H, et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465(7296):346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 85.Park JH, Lee JH, Cho MH, Herzberg M, Lee J. Acceleration of protease effect on Staphylococcus aureus biofilm dispersal. FEMS Microbiol Lett. 2012;335(1):31–38. doi: 10.1111/j.1574-6968.2012.02635.x. [DOI] [PubMed] [Google Scholar]
- 86.Selan L, Berlutti F, Passariello C, Comodi Ballanti M R, Thaller MC. Proteolytic enzymes: a new treatment strategy for prosthetic infections? Antimicrob Agents Chemother. 1993;37(12):2618–2621. doi: 10.1128/aac.37.12.2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Longhi C, Scoarughi GL, Poggiali F, et al. Protease treatment affects both invasion ability and biofilm formation in Listeria monocytogenes. Microb Pathog. 2008;45(1):45–52. doi: 10.1016/j.micpath.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 88.Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol. 2004;186(14):4466–4475. doi: 10.1128/JB.186.14.4466-4475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **89.Digiandomenico A, Warrener P, Hamilton M, et al. Identification of broadly protective human antibodies to Pseudomonas aeruginosa exopolysaccharide Psl by phenotypic screening. J Exp Med. 2012;209(7):1273–1287. doi: 10.1084/jem.20120033. The authors used phage libraries to identify three monoclonal antibodies capable of binding distinct epitopes on Psl, an exopolysaccharide important for P. aeruginosa biofilm formation.
- 90.Maira-Litran T, Kropec A, Goldmann DA, Pier GB. Comparative opsonic and protective activities of Staphylococcus aureus conjugate vaccines containing native or deacetylated Staphylococcal Poly-N-acetyl-beta-(1-6)-glucosamine. Infect Immun. 2005;73(10):6752–6762. doi: 10.1128/IAI.73.10.6752-6762.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]