Abstract
8-Oxo-7,8-dihydroguanine (8-oxoG) is one of the most abundant DNA base lesions induced by reactive oxygen species (ROS). Accumulation of 8-oxoG in the mammalian genome is considered a marker of oxidative stress, to be causally linked to inflammation, and is thought to contribute to aging processes and various aging-related diseases. Unexpectedly, mice that lack 8-oxoguanine DNA glycosylase-1 (OGG1) activity and accumulate 8-oxoG in their genome have a normal phenotype and longevity; in fact, they show increased resistance to both inflammation and oxidative stress. OGG1 excises and generates free 8-oxoG base during DNA base-excision repair (BER) processes. In the present study, we report that in the presence of the 8-oxoG base, OGG1 physically interacts with guanine nucleotide-free and GDP-bound Rac1 protein. This interaction results in rapid GDP→GTP, but not GTP→GDP, exchange in vitro. Importantly, a rise in the intracellular 8-oxoG base levels increases the proportion of GTP-bound Rac1. In turn Rac1-GTP mediates an increase in ROS levels via nuclear membrane-associated NADPH oxidase type 4. These results show a novel mechanism by which OGG1 in complex with 8-oxoG is linked to redox signaling and cellular responses.
Keywords: 8-oxoguanine DNA glycosylase-1, 8-oxoguanine, Rac1 GTPase
Introduction
Environmental pollutants, accidental chemical exposures, and ionizing and ultraviolet radiation increase the cellular levels of reactive oxygen species (ROS), directly or via activation of oxido-reductases and/or induction of mitochondrial dysfunction. ROS from various sources and mitochondria (with or without mitochondrial dysfunction) are modulators of cellular signaling pathways [1]. On the other hand, modify proteins, lipids, and DNA [reviewed in [2]. Among DNA and RNA bases, guanine is the most susceptible to oxidation [3]. One of the most frequent oxidation products of guanine in DNA is 8-oxo-7,8-dihydroguanine (8-oxoG) [4]. It is recognized and excised as a base by 8-oxoguanine DNA glycosylase-1 (OGG1) during the DNA base excision repair (BER) pathway, both in the nucleus and in mitochondria [5, 6]. OGG1-initiated BER encompasses four key steps, including damaged base recognition and excision, 2′-deoxyribose-3′-phosphate end processing by AP endonuclease 1 (APE1), filling in the nucleotide gap by DNA polymerase β, and nick-sealing by DNA ligase [5]. OGG1’s repair activity is modulated by post-translational modifications, including phosphorylation [7], acetylation [8], and by interactions with canonical repair and non-repair proteins [9]. Studies have also unveiled a redox-dependent mechanism for the regulation of OGG1 activity [10].
Accumulation of 8-oxoG in DNA has conventionally been associated with inflammatory processes, various diseases, accelerated telomere shortening, and aging processes [11–13]. In addition, unrepaired 8-oxoG is potentially one of the most mutagenic lesions among oxidatively modified DNA bases, because its pairing with A will cause a GC=>TA mutation [14]. Unexpectedly, Ogg1 knock out (Ogg1−/−) mice have an unaltered lifespan, show only moderate increases in tumor formation, and exhibit slight patho-physiological changes despite the supraphysiological levels of 8-oxoG in their DNA [15, 16]. Furthermore, Ogg1−/− mice showed an increased resistance to inflammation [17, 18] and lack of Ogg1 activity protected mice from the trinucleotide repeat expansions underlying Huntington’s disease [19]. It also appears that a lack of Ogg1′ activity is accompanied by dysfunction of signaling pathway(s) linking oxidant stress to cellular responses. These observations raise the possibility that the 8-oxoG base released from the genome by OGG1 could have physiological/patho-physiological relevance. Previous studies have implied roles for OGG1 in multiple cellular processes in addition to that of being a canonical DNA BER protein [6, 20]. For example, it has been shown that OGG1 co-localizes with centrioles (microtubule organizing center), microtubule networks, and mitotic chromosomes, [7, 21, 22]. Moreover, we have previously shown that OGG1 binds its excision product, the 8-oxoG base, but not 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG) or 2,6- diamino-4-hydroxy-5-formamidopyrimidine (FapyG) or other intact or oxidized nucleotides and nucleosides, at a site independent from its catalytic active site. In complex with the 8-oxoG base, OGG1 interacts with canonical Ras family members and induces guanine nucleotide exchange [23]. Activated Ras then initiates signal transduction via Raf1-MEK1,2/ERK1,2, leading to the transcriptional activation of genes as shown by microarray analysis (NCBI, GEO # GSE26813) [23]. These data and those showing the implication of OGG1 in chromatin remodeling and transcriptional initiation [24, 25] imply a role of OGG1 in 8-oxoG-dependent regulation of cellular signaling. Elegant studies have demonstrated that a DNA repair product of nucleotide excision repair (NER) plays a role in the cellular response to DNA damage [26] and a failure in degradation of this NER product by endo/exonuclease(s) has been associated with various immune disorders [27].
In a previous study using biochemical and cell biological assays, we showed that in the presence of molecular oxygen the 8-oxoG base is transformed to a catalase- and glutathione peroxidase-sensitive hydroperoxide. It rapidly oxidizes Amplex red to resorufin, H2DCF to DCF, Fe2+ to Fe3+, and GSH to GSSG and when added to cells caused an oxidative burst [28]. Interestingly, the addition of 8-oxoG base to cells also increases cellular ROS levels; however, the kinetics are different from those induced by the hydroperoxide form of 8-oxoG. The present study hypothesizes that the increase in cellular ROS levels by 8-oxoG base is due to the activation of cellular oxido-reductases, which may require small GTPase(s). Here we document that in the presence of 8-oxoG base the OGG1 protein physically interacted with GDP-bound Rac1. This interaction catalyzed a guanine nucleotide exchange and increased the GTP-bound form of Rac1 in vitro, in cultured cells and the tissue environment (animal model). In turn, Rac1-GTP resulted in a transient increase in ROS levels via nuclear membrane-associated NADPH oxidase type 4. Our novel observations suggest that the OGG1-mediated repair of 8-oxoG not only prevents mutagenesis, but also induces cellular responses via a Rac1-dependent mechanism.
Material and Methods
Cells cultures
A549 type II alveolar epithelial cells (ATCC # CCL-185) were cultured in Ham’s F12 (GIBCO-BRL). Human embryonic fibroblasts (MRC-5) were grown in EMEM (GIBCO-BRL). All media were supplemented with 10% FBS (Atlanta Biologicals, Lawrenceville, GA), penicillin (100 units/ml; GIBCO-BRL), and streptomycin (100 μg/ml; GIBCO-BRL).
Animals and challenge
Six- to 8-week-old female BALB/c mice (purchased from Harlan Sprague-Dawley; San Diego, CA) were used for these studies. While mice were under mild anesthesia, their lungs were challenged intranasally with 8-oxoG [29]. The animals were sacrificed and their lungs were homogenized in a buffer (25 mM Tris-HCl/pH 7.5/, 150 mM NaCl, 1% NP-40, 1 mM DTT, 5% glycerol, 20 mM NaF, 1 mM sodium orthovanadate, 1 μg/ml leupeptin and 1 μg/ml aprotinin) and GTP bound levels of Rac determined as described in “Assessment of GTP-bound Rac1 levels.” All experiments were performed according to the NIH Guidelines for the Care and Use of Experimental Animals. The protocol used was approved by the University of Texas Medical Branch Animal Care and Use Committee (#0807044A).
Reagents
We obtained reagents from the following sources: 8-oxoG (Cayman Chemical, Ann Arbor, MI); diphenyleneiodonium chloride (DPI), 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG) and guanine (Sigma-Aldrich, St. Louis, MO); 8-oxo-7,8-dihydroadenine (8-OH-Ade; Biolog Life Science Institute, Bremen, Germany); 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG) from Dr. Miral Dizdaroglu (Chemical Science and Technology Laboratory, National Institute of Standards and Technology, Gaithersburg, MD); N-acetyl-L-cysteine, Rac1 antibody (Ab; Thermo Fisher Scientific, Rockford, IL); Rac2 and Rac3-, NADPH oxidase subunit Abs (Epitomics, Burlingame, CA); recombinant human Rac1 protein (Cytoskeleton, Denver, CO); OGG1 Ab (Abcam, Cambridge, MA); recombinant human OGG1 (Novus Biological, Littleton, CO) HRP-conjugated anti-rabbit Ab (Southern Biotech, Birmingham, AL); 3,3′,5,5′-Tetramethylbenzidine (TMB) substrate (eBioscience, San Diego, CA); H2SO4 (Fisher Scientific, Fair Lawn, NJ); (Mant)-GTP (2′-(or-3′)-O-(N-methylanthraniloyl)guanosine 5′-triphosphate, trisodium salt, MantGTP) and (Mant)-GDP (2′-(or-3′)-O-(N-methylanthraniloyl)guanosine 5′-diphosphate, disodium salt, and MantGDP) (Invitrogen, Carlsbad, CA). GDP and GTPγS were purchased from Cytoskeleton.
Assessment of cellular ROS levels
To asses cellular ROS generation the hydrogen peroxide sensor pHyPer (Evrogen, Axxora Inc., Farmingdale, NY) was used [30, 31]. Vectors include pHyPer-Cyto (without targeting signal); pHyPer-dMito (mitochondria targeting signal fused to HyPer N-terminus) or pHyPer-Nuc (nuclear localization signal fused to the C-terminus of HyPer). Vectors were introduced into cells by using Lipofectamine™ 2000 as described by the manufacturer’s instructions (Invitrogen, Life Technologies Corporation). Seventy two hours after transfection cells were incubated in medium containing 1% FBS for 4h and challenged with 8-oxoG (10 μM) or H2O2 (10 μM). At times 0, 12, 20 and 60 min post-exposure cells were fixed in formalin (3.7%), dried and mounted on microscope slides. Images were taken by NIKON Eclipse Ti System.
Changes in cellular ROS levels were also determined by using the fluorogenic probe carboxymethyl 2′-7′-dihydro-dichlorofluorescein-DA (H2DCF; Molecular Probes, Eugene, OR) [32]. Briefly, cells were grown to 70% confluence and starved in medium containing 1% FBS for 4h, and 50 μM H2DCF-DA was added at 37°C for 30 minutes. Cells were then washed twice with PBS and exposed to nucleotides, nucleosides, or solvent. Changes in DCF fluorescence were recorded in an FLx800 (Bio-Tek Instruments Inc., Winooski, VT) microplate reader at 485 nm excitation and 528 nm emission. In selected experiments, changes in cellular ROS levels were determined by flow cytometry (BD FACSCanto flow cytometer; BD Biosciences, Franklin Lakes, NJ).
Protein-protein binding assays
To determine interactions between OGG1 and Rac1, we used enzyme-linked immunosorbent assays (ELISA). Briefly, Rac1 antibody-coated wells were washed with PBS-T (2.68 mM KCl, 1.47 mM KH2PO4, 136.8 mM NaCl, 9.58 mM NaH2PO4, 0.05% Tween-20), and then guanine nucleotide free (empty) Rac1 (5.3 pmol), GDP-, or GTP-loaded Rac1 protein (5.3 pmol) was added to parallel wells in PBS-T alone or together with OGG1 (5.3 pmol) and 8-oxoG (5.3 pmol) for 1 h at room temperature. Unbound proteins were removed by washing before incubation with anti-OGG1 Ab (1 h). HRP-conjugated secondary Ab was added for 45 min, and color was developed by using 3,3′,5,5′-Tetramethylbenzidine (TMB) substrate. Absorbance was determined on a SpectraMax 190 Microplate Reader (Molecular Devices, Sunnyvale, CA). To confirm protein-protein interactions by His-affinity pulldown assays, nickel-nitrilotriacetic acid- (Ni-NTA)-agarose beads (Qiagen Inc., Valencia, Ca) were mixed with His-Rac1 protein (6 pmol) (Cytoskeleton) in 300 μl interaction buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, 0.05% Tween 20, pH 7.5) [33]. After a 30 min incubation at 4°C, His-Rac1-bound beads were washed 3 times, and equimolar, non-tagged OGG1 alone or OGG1 (6 pmol) plus 8-oxoG (6 pmol) was added to the interaction buffer. Samples were incubated for 30 min at 4°C, then washed twice with interaction buffer, and proteins eluted with Laemmli buffer (0.125 M Tris-HCl, 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, pH 6.8) at 100°C for 5 min. The eluants were analyzed on Western immunoblots as described below.
Assessment of GTP-bound Rac1 levels
Changes in the levels of GTP-bound Rac1 were analyzed by using the Active Rac1 pull-down and detection kit (Pierce, Thermo Scientific Inc) per the manufacturer’s instructions with slight modifications. Briefly, cells were washed once with ice-cold TBS and lysed with 1x Lysis/Binding/Washing buffer (25 mM Tris-HCl/pH 7.5/, 150 mM NaCl, 1% NP-40, 1 mM DTT, 5% glycerol, 20 mM NaF, 1 mM sodium orthovanadate, 1 μg/ml leupeptin and 1 μg/ml aprotinin). Cell/tissue extracts were cleared by centrifugation, and GTP-bound Rac1 was captured by the Rac-binding domain of p21/Cdc42/Rac1-activated kinase 1 bound to GST beads [34]. GST beads were washed with lysis buffer and bound proteins were fractionated on a 4 to 20% PAGE. Changes in Rac1 levels were determined by Western immunoblot analysis.
Guanine nucleotide exchange assays
The GDP-GTP and GTP-GDP exchange on Rac1 was determined by real-time fluorescence spectroscopic analysis [35]. Rac1 (6 pmol) was loaded with the nucleotide analog (2′-(or-3′)-O-(N-methylanthraniloyl)guanosine 5′-triphosphate (Mant)-GTP (MantGTP) or GDP (MantGDP) in exchange buffer containing 20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM dithiothreitol, 50 μg of bovine serum albumin for 30 min. In the case of GDP-GTP exchange, Rac-1-MantGDP and OGG1 protein (6 pmol) + 8-oxoG base (6 pmol) were mixed with untagged GTP. A similar strategy was used to monitor GTP-GDP exchange. Kinetic changes in the fluorescence of Rac1-MantGDP or Rac1-MantGTP were determined by using a POLARstar Omega reader (BMG: Bio Medical Gurrat; LABTECH). Curves were fitted by using MS Excel. The half-life of Rac1-MantGDP was determined using POLARstar Omega software.
Western blot analysis
Cells were harvested and incubated for 15 min with lysis buffer (25 mM Tris-HCl, pH 7.5; 150 mM NaCl; 1% NP-40; 1 mM DTT; 5% glycerol; 20 mM NaF; 1 mM sodium orthovanadate; 1 μg/ml leupeptin and 1 μg/ml aprotinin). The lysates were clarified by centrifugation, and the supernatants were collected. Protein samples were mixed with sample loading buffer, heated for 5 min at 95°C, and separated by 4–20% SDS-PAGE. Proteins were transferred to Hybond-ECL nitrocellulose (Amersham Biosciences UK Ltd, Uppsala, Sweden) membrane by electroblotting. The membranes were then blocked with 3% BSA in TBS containing 0.1% Tween (TBS-T) for 3 h and incubated overnight at 4°C with the primary anti-Rac1 Ab diluted in 3% BSA in TBS-T. The blots were then washed 4x with TBS-T and incubated for 1 h with HRP-conjugated secondary HPR-conjugated Ab (anti-mouse IgG, GE Healthcare UK Ltd, Pittsburgh, PA) in 5% non-fat dry milk in TBS-T. After washing, immunoreactive bands on membranes were visualized by chemiluminescence using an ECL substrate (Amersham Biosciences).
siRNA ablation of gene expression
siRNAs were introduced into A549 and MRC-5 cells by using INTERFERin™ (Polyplus-transfection Inc., New York, NY) transfection reagent per the manufacturer’s instructions. Briefly, siRNAs (20 nM final concentrations, as determined in preliminary studies) were mixed with INTERFERin™ transfection reagent and added to cells. After 3 h incubation in serum-free medium, growth medium was added for 72 h. Control and Rac1 siRNAs (siGENOME SMARTpool) were obtained from Dharmacon (Thermo Fisher Scientific, Inc.); control and p22phox siRNA were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). siRNA to OGG1 (siGENOME SMARTpool) was obtained from Dharmacon. OGG1 was depleted via a simultaneous siRNA transfection and plating method [23]. Depletion of the target genes’ mRNA levels was determined by qRT-PCR and Western blot analysis.
Quantitative real-time PCR
qRT-PCR was done by the SYBRGreen method using an ABI 7000 System equipment and software (Applied Biosystems, Foster City, CA) per the manufacturer’s recommended protocol.. The thermal profile was: 50°C for 2 min, 95°C for 10 min, and 45 cycles of 95°C for 15 sec, followed by 60°C for 1 min. A dissociation stage was added at the end of the run to verify the primers’ specificity (95°C for 15 sec, 60°C for 20 sec and 90°C for 15 sec). Expression levels (fold change) were determined by the delta-delta Ct method (33Ct) [36, 37]. Primers: p22phox, F: 5′-AACGAGCAGGCGCTGGCGTCCG-3′; R: 5′-GCTTGGGCTCGATG GGCGTCCACT-3′; Rac1, F: 5′-CTGATGCAGGCCATCAAGT-3′ R: 5′-CAGGAAATGCATTGGTTG TG-3′; OGG1, F: 5′-GCATCGTACTCTAGCCTCCA-3′ R: 5′-GCTCTTGTCTCCTCGGTACA-3′; NOX4, F: 5′-CTGGAGGAGCTGGCTCGCCAACGAAG-3′; R: 5′-GTGATCATGAGGAATAGCACCACCACCATGCAG-3′; GAPDH, F: 5′-GAAGGTGAAGGTCGGAGT-3′; R: 5′-GAAGATGGTGATGGGATTTC-3′
Microscopic imaging
Cells on microscope coverslips were mock-treated or pulsed for 30 min with 10 μM 8-oxoG base, fixed in 4% paraformaldehyde at 4°C and then permeabilized with Triton X100 for 30 min at 37°C. The cells were then incubated for overnight at 4°C with primary antibody to OGG1 (1:200), Rac1 (1:400), and NADPH oxidase type 4 antibody (1:300). After washing (PBS-Tween 20: PBS-T) cells were incubated for 1 h at room temperature with Alexa 488-, Alexa-594 and/or Texas Red-conjugated secondary antibodies. Nuclei of cells were stained for 15 min with DAPI (4′6-diamidino-2-phenylindole dihydrochloride; 10 ng/mL). Cells were then mounted in anti-fade medium (Dako Inc. Carpinteria, CA) on a microscope slide. Microscopy was performed on a NIKON Eclipse Ti System. Magnification: x125. Co-localization was visualized by superimposition of green and red images by using Nikon NIS Elements Version 3.5 (NIKON Instruments, Tokyo, Japan).
The overlap coefficient of proteins was calculated according to Manders [38]. R = ΣS1 × S2/√Σ(S1)2 × Σ(S21)2 where S1 represents the signal intensity of pixels in channel 1 and S2 represents signal intensity of pixels in channel 2. This coefficient is not sensitive to the limitations of typical fluorescence imaging, such as efficiency of hybridization, sample photobleaching, and camera quantum efficiency [39]. The overlap coefficients k1 and k2 split the value of co-localization into a pair of separate parameters: k1 = ΣS1 × S2/Σ(S1)2; k2 = ΣS1 × S2/Σ(S2)2, where S1 represents the signal intensity of pixels in channel 1 and S2 represents signal intensity of pixels in channel 2 [38].
Preparation of 8-oxoG Solution
8-OxoG is provided as a hydroacetate salt, and was dissolved in 10 mM NaOH (4 mM final concentration). An 8-OxoG stock solution was prepared freshly, diluted in PBS (w/o Ca2+/Mg2+, pH: 7.4), and used within 1 h for experiments. All other intact and oxidized nucleotide bases and nucleosides were dissolved in the same manner.
Assessment of 8-oxoG’s Cellular Uptake
MRC-5 cells at 80% confluence were washed, and PBS containing 10 μM of 8-oxoG (final concentration) was added. Aliquots were removed at 0, 1, 30 and 60 min, lyophilized, and re-dissolved in 200 μl of 10 mM NaOH. An aliquot of 2.8 nmol of 8-oxoG-13C3, 15N was added to 20 μl of this solution as an internal standard. The sample was lyophilized overnight and then derivatized and analyzed by gas chromatography/mass spectrometry (GC/MS) to determine the level of 8-oxoG [40].
Statistical Analysis
The data are expressed as the mean ± SD. Results were analyzed for significant differences using ANOVA procedures and Student’s t-tests (Sigma Plot 11.0). Differences were considered significant at p<0.05 (*p<0.05, **p<0.01, ***p<0.001, ****p< or =0.0001).
Results
Rac1 GDP-GTP exchange by OGG1 in the presence of 8-oxoG
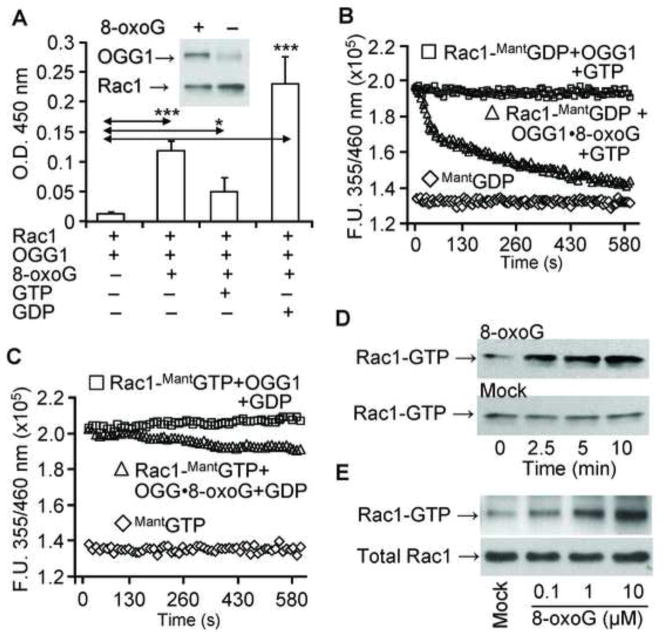
First we examined whether OGG1 in the presence of 8-oxoG [OGG1(8-oxoG)] interacts with Rac1 in vitro. The result summarized in Fig. 1A shows that OGG1 protein alone interacted poorly with guanine nucleotide-free (empty) Rac1 protein. However, in the presence of 8-oxoG, physical interactions between OGG1 and empty Rac1 were significantly increased, as shown by ELISA and Ni-NTA pull-down assays (Fig. 1A, and Fig. 1A inset). OGG1(8-oxoG) bound most extensively to GDP-loaded Rac1 (Fig. 1A), while binding to Rac1-GTP was significantly lower than to both empty and GDP-Rac1 (Fig. 1A). Together these data reveal an unexpected physical interaction of the OGG1(8-oxoG) complex with Rac1, and also suggest that the conformation of GDP-bound Rac1 allows the most stable interaction with OGG1(8-oxoG).
Figure 1.
Interaction of Rac1 with OGG1 protein in the presence of 8-oxoG base and guanine nucleotide exchange. (A) Physical interaction of OGG1 protein with Rac1. Rac1 protein was added to Rac1 Ab-coated wells and incubated with OGG1±8-oxoG in the presence or absence of GDP or GTP. OGG1 binding was detected by HRP conjugated Ab at 450 nm (Materials and Methods). Results are means ±SEM (n = 3–7). * = p<0.05 *** = p<0.001. Inset: Interaction of OGG1 with Rac1 shown by pull-down assay. Rac1 was bound to Ni-agarose beads, and OGG1 ± 8-oxoG was added. After washing, the amount of OGG1 bound to Rac1 was analyzed by Western blot (Materials and Methods). (B) Exchange of Rac1-bound GDP for GTP by OGG1 in the presence of the 8-oxoG base. Rac1 protein was loaded with MantGDP, and nucleotide exchange was initiated by adding OGG1 + 8-oxoG and GTP (6 pmol) (△). In controls, GTP alone was added to Rac1-MantGDP (□). Fluorescence of MantGDP alone (◇). (C) OGG1 does not catalyze Rac1-GTP- GDP exchange. Rac1 protein was loaded with MantGTP and OGG1 + 8-oxoG and GDP (▲) or GDP alone (■) was added. Fluorescence of MantGTP alone (◇). In B and C, changes in fluorescence of Rac1-MantGDP and Rac1-MantGTP were determined by real-time measurements by using a POLARstar Omega (BMG Germany). Curves were fitted by using MS Excel (Materials and Methods) n = 3–5. (D, E) Time- and dose-dependent increase in Rac1-GTP levels in cell extracts upon addition of 8-oxoG base. D, Cell extracts were prepared, 1 μM 8-oxoG base was added (upper panel) or not (lower panel), and aliquots (250 μg) were taken at 0, 2.5, 5 and 10 min. E, increasing (0, 0.1, 1 and 10 μM) 8-oxoG was added to cell extracts. In D and E, Rac1-GTP levels were determined by using active Rac1 GST pull-down assays (Material and Methods). The total Rac1 level is shown in 20-μg cell extracts.
To examine the possibility that OGG1(8-oxoG) may serve as a guanine nucleotide exchange factor (GEF) and increase Rac1-GTP levels, we performed guanine nucleotide exchange assays utilizing fluorescently labeled guanine nucleotides (MantGDP and MantGTP) [35]. When Rac1 protein was loaded with MantGDP (1:1 molar ratio), the fluorescence intensity of MantGDP was increased from 1.36 ×105 fluorescence unit (FU) to 1.9 ×105 FU, as determined by spectroscopic analysis (Fig. 1B). Upon addition of OGG1 and 8-oxoG along with unlabeled GTP to Rac1-MantGDP, the fluorescence rapidly decreased, indicating that Rac1-bound MantGDP was replaced by non-fluorescent GTP (Fig. 1B). The release of MantGDP and replacement with GTP was rapid, as during a 45-sec time period more that 50% of MantGDP was exchanged for GTP (Fig. 1B). In controls, OGG1 plus GTP did not change Rac1-MantGDP fluorescence (Fig. 1B), and OGG1 alone also had no effect (data not shown). Next, we found that the fluorescence intensity of MantGTP was increased (from 1.38 ×105 FU to 2.02 ×105 FU) (Fig. 1C) when bound to Rac1 in a manner similar to that observed for Rac1-MantGDP (Fig. 1B). Addition of OGG1 plus 8-oxoG along with non-labeled GDP resulted in a slow guanine nucleotide exchange (Fig. 1C). OGG1 caused no change in Rac1-MantGTP fluorescence in the presence of GDP; OGG1 alone also had no effect (data not shown). These data are in line with the poor interaction of OGG1 with Rac1-GTP (Fig. 1A). These results thus show that OGG1 in complex with 8-oxoG catalyzes the release of GDP efficiently, and may function as a GEF in the intracellular environment.
To examine this possibility, we further investigated whether OGG1 protein in the presence of 8-oxoG base increases Rac1-GTP levels in cell extracts. Our results showed that addition of 8-oxoG base to the extract from A549 cells increased Rac1-GTP levels in a time-dependent manner (Fig. 1D, upper panel). There was no change in the level of Rac1-GTP with cell lysates without OGG1 (Fig. 1D, lower panel). Our results also showed that the lowest dose of the 8-oxoG base that increased levels of Rac1-GTP was 0.1 μM (Fig. 1E). There was no concentration-dependent increase in Rac1-GTP levels above 10 μM of 8-oxoG (data not shown), possibly due to 8-oxoG’s low solubility at physiological pH [3]. Together these data suggest that OGG1 in the presence 8-oxoG base catalyzes the replacement of Rac1-bound GDP by GTP in cellulo.
Increased Rac1-GTP levels in 8-oxoG-exposed cultured cells and an animal model
Next, we explored whether exposing cells to the 8-oxoG base changes cellular Rac1-GTP levels. MRC-5 and A549 cells were maintained in low serum (1%)-containing medium for 24 h and exposed to 8-oxoG base (10 μM) for increasing time intervals. The results, summarized in Fig. 2A, showed a rapid increase in Rac1-GTP levels in MRC-5 cells between 1 and 5 min of treatment, which peaked at 5 min and then decreased nearly to basal levels by 30 min. Similar results were obtained by using A549 cells (Fig. 2B). Fig. 2C shows the percent changes in Rac-1 GTP levels after 0, 1, 3, 5, 15 and 30 min of 8-oxo-G exposure. At time 0, the percentage of GTP-bound Rac1 was 0.78± 0.2 % and 0.74± 0.1% in MRC-5 and A459 cells, respectively, whereas after a 5-min exposure to 8-oxoG, the percentage of GTP- bound Rac1 levels increased to 6.5± 2% (MRC-5) and 7.05± 1,7% (A459) cells. To further confirm that the 8-oxoG-induced increase in Rac1-GTP levels in cellulo required OGG1, we decreased its levels with siRNA [23]. In OGG1-depleted cells (MRC-5, A549), there was no significant increase in Rac1-GTP levels after 8-oxoG addition (Fig. 2D, upper and lower panels) when compared to Rac1-GTP levels in the OGG1-expressing cells. The extent of OGG1 depletion was confirmed by Western blot analysis (Fig. 2E). Importantly, the guanine base, FapyG (another BER product of OGG1) and 8-OH-Ade or uric acid did not increase Rac1-GTP levels in MRC5 and A549 cells (data not shown). On the other hand, 8-oxodG slightly inhibited Rac1 activation (data not shown), as shown in previous studies [41]. MRC5 and A549 cells abundantly express Rac1, whereas the expression of Rac3 (closely related to Rac1 in functions and nucleotide binding; rev in [42]) was approximately 22- and 43-times lower than that of Rac1 in MRC-5 and A549 cells, respectively (Fig. 2F). Rac2, primarily expressed in myeloid cells [42], was not detectable in these cells at the protein level (Fig. 2F). Therefore, we were unable to show changes in Rac2-GTP or Rac3-GTP levels in these cells (data not shown). In control experiments, we demonstrated a rapid decrease in 8-oxoG base levels in cell supernatant, suggesting that 8-oxoG is taken up by cells (Fig. 2G). Together, these data demonstrate that the 8-oxoG base is unique in increasing GTP-bound Rac-1 levels, and suggest that OGG1(8-oxoG) interacts with Rac1-GDP and catalyzes guanine nucleotide exchange in cellulo.
Figure 2.
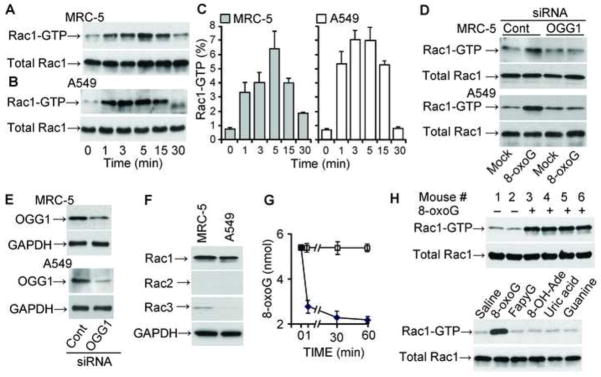
Increase Rac1-GTP levels in OGG1-expressing cells upon 8-oxoG exposures. (A, B) Changes in Rac1-GTP levels in MRC-5 (A) and A549 (B) cells. The 8-oxoG base (10 μM) was added to cells and extracts were made at indicated times points. Rac1-GTP levels were determined by GST pull-down assays (Materials and Methods). Upper panels: Rac1-GTP levels were determined in 100 μg cell extract. Lower panel: total Rac1 protein levels in 10 μg cell lysate. n = 3–8. (C) Graphical depiction of the percentage of Rac-1GTP levels. Rac1-GTP band intensities were determined by densitometry (Image J 1.44), and the percentage changes were calculated by using MS Excel. (D) Depletion of OGG1 expression decreased 8-oxoG exposure-induced Rac1-GTP levels. MRC-5 and A549 cells were transfected with OGG1 siRNA or control siRNA (Materials and Methods) and then exposed to 8-oxoG (10 μM) for 10 min. Rac1-GTP levels were determined as in the legends to panels A and B. (E) siRNA-mediated decrease in OGG1 levels. GAPDH is shown for equal protein loading. (F) Expression of Rac family proteins in MRC-5 and A549 cells. Cells were lysed and protein levels were determined in 10 μg (Rac1); and 25 μg (Rac2 and Rac3) per lane extract by immunoblotting. Results of a representative set of experiments are shown. GAPDH is shown for equal protein loading. (G) Cellular uptake of 8-oxoG base. The medium of MRC5 monolayers was replaced with PBS (pH 7.4), and 10 μM (final concentration) of 8-oxoG was added. Aliquots of the medium were taken at 0, 1, 30 and 60 min. In controls (□), 8-oxoG was added to PBS in the absence of cells. The 8-oxoG content in the aliquots was determined by GC/MS [40]. (H) Activation of Rac1 by 8-oxoG challenge in lungs. The lungs of mice were challenged intranasally with saline ± 1 μM 8-oxoG. In controls, mice were challenged with 1 μM of FapyG, 8-oxo-Ade, uric acid or guanine. At 15 min thereafter Rac1-GTP levels were determined by GST pull-down assays in 125 μg lung extracts.
Next, we examined whether 8-oxoG base increases Rac1-GTP levels in a tissue environment. The 8-oxoG base was instilled into the lungs of mice, and Rac1-GTP levels were determined as described in Materials and Methods. Compared to saline-challenged lungs (Fig. 2H, mice # 1 and 2) there was a robust increase in Rac1-GTP levels at 15 min post-challenge (Fig. 2H, mice # 3, 4, 5, and 6), consistent with our in vitro and in cellulo studies. Specificity of Rac1 activation by 8-oxoG exposure is supported by data showing that FapyG, 8-OH-Ade, uric acid and guanine base did not alter Rac1-GTP levels in lungs (Fig. 2H, lower panels). 8-OxodG slightly lowered Rac1-GTP levels (data not shown) as documented previously [41]. As shown by immunoblot analysis, Rac1 is abundantly expressed in lung tissues (Fig. 2H, total Rac1), while Rac2 and Rac3 expression was nearly undetectable (data not shown). Although the significance of Rac1 activation in lungs by the 8-oxoG base is yet to be defined, these results are in line with those generated with normal diploid lung fibroblasts (MRC5) and A549 cells (type II alveolar lung epithelial cells).
Transient increase in ROS levels in OGG1-expressing, 8-oxoG-exposed cells
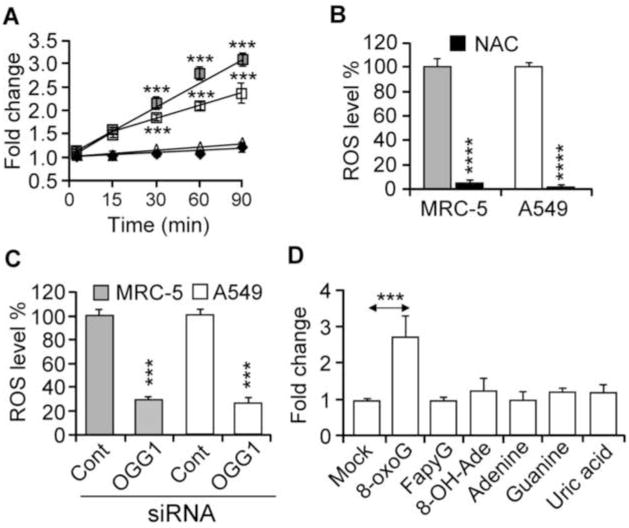
Rac1 is a multifunctional GTPase, e.g., it is clearly involved in the modulation of cellular redox state via regulating NADPH oxidase(s) activities [42]. Here we show that the addition of 8-oxoG to the cells increased the intracellular ROS levels (Fig. 3A–D). Pre-treating the cells with N-acetyl-L-cysteine (NAC; 10 mM, for 3 h), a precursor of glutathione biosynthesis and a scavenger of oxygen radicals [43], prevented the 8-oxoG-mediated increase in intracellular ROS levels (Fig. 3B). 8-OxoG induced a significantly lower levels of ROS in OGG1-depleted cells (Fig. 3C), consistent with decreased Rac1 activation (Fig. 2D). The 8-oxoG base is unique in increasing ROS levels, because neither the guanine base, FapyG, 8-OH-Ade, adenine or uric acid (deaminated form of 8-oxoG) has this activity (Fig. 3D).
Figure 3.
Changes in ROS levels in OGG1-expressing cells after exposure to 8-oxoG base. (A) Kinetic changes in ROS levels in MRC-5 and A459 cells after 8-oxoG treatment. □, A549; ■, MRC-5; △, mock-treated A549; ◇, mock-treated MRC-5. (B) 8-OxoG base exposure failed to increase ROS levels in N-acetyl-L-cysteine (NAC)-pretreated cells. (C) Depletion of OGG1 prevented the 8-oxoG-mediated increase in ROS levels. Cells were transfected with OGG1 siRNA (Materials and Methods) and exposed to 8-oxoG. ROS levels were determined at 30 min. (D) Exposure of cells to FapyG, 8-oxo-Ade, adenine, guanine or uric acid failed to change intracellular ROS levels. In panels A, B, C, and D, cells at 80% confluence were kept in 0.1% serum-containing media for 4 h, H2DCF-DA-loaded, then exposed for 30 min before DCF fluorescence was assessed (Materials and Methods). 8-oxoG, 8-oxo-7,8-dihydroguanine; FapyG, 2,6-diamino-4-hydroxy-5-formamidopyrimidine; 8-oxoAde, 8-oxo-7,8-dihydroadenine. Results are means ± SEM (n=7–15) ***= p<0.001, **** = p< 0.0001.
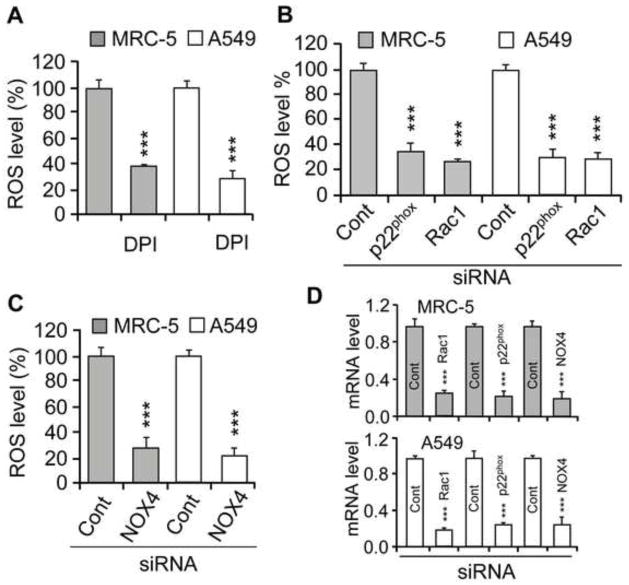
Treating the cells with DPI (10 μM, determined in preliminary studies), an NADPH oxidase (NOX) inhibitor, before 8-oxoG addition decreased cellular ROS levels by 62±3% and 70±7% in MRC-5 and A549 cells, respectively, compared to cells exposed to 8-oxoG alone (Fig. 4A). p22phox is an essential regulatory subunit of NOX1-4 [42]. siRNA-mediated depletion of p22phox lowered ROS levels by 64±8% (MRC-5 cells) and 69±6% (A549 cells) in 8-oxoG-exposed cells, which further confirmed the involvement of NOXs in ROS generation (Fig. 4B). siRNA to NOX 1, 2 and 3 only somewhat (data not shown), but NOX4 siRNA significantly decreased (67±9.5% for MRC5; 76±4% for A549) ROS levels (Fig. 4C). Depletion of Rac1 decreased ROS levels by 72±3% in MRC-5 and 68±7% in A549 cells after 8-oxoG exposure (Fig. 4B). The siRNA-mediated decrease in Rac1, p22phox and NOX4 expression is shown in Fig. 4D.
Figure 4.
An increase in cellular ROS levels in 8-oxoG-exposed cells. (A) DPI pre-treatment of cells lowered ROS levels in MRC-5 and A549 cells after exposure to 8-oxoG. Cells were pretreated with DPI (15 μM) for 1 h and then H2DCF-DA-loaded and exposed to 8-oxo-G. Changes in ROS levels were determined after 30 min (Material and Methods). (B, C) Depletion of p22phox, NOX4 or Rac1 decreased ROS levels. Parallel cultures of cells (MRC-5, A549) were transfected with siRNA to p22phox, NOX4 or Rac1, and 48 h later cells were exposed to 8-oxoG base (10 μM). Changes in ROS levels were determined as in the legend to panel A. (D) mRNA levels of p22phox, NOX4 and Rac1 48 h after siRNA transfection of cells. RNA isolation and qRT-PCR were carried out as in Material and Methods. Abbreviations: DPI, diphenyleneiodonium chloride; Cont, control; p22phox, NOX4, NADPH oxidase type 4; siRNA, small interfering RNA. Results are means ±SEM (n=3–6) ***p<0.001.
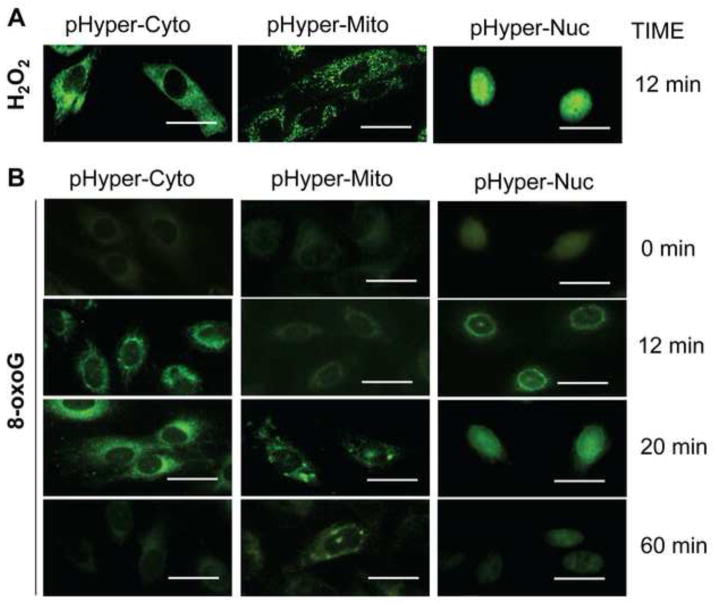
NOX type 4 (NOX4) have been reported to be localized to cytoplasmic compartments and nuclear membranes, and are involved in multiple cellular processes, including localized redox modulation, and cellular signaling [42, 44]. To determine the cellular localization of NOX4 that generate ROS in response to 8-oxoG exposure, cells were transfected with the biosensor pHyPers [30, 31], expressed in cytoplasm (pHyPer-Cyto) or targeted to nucleus (pHyPer-Nuc), and mitochondria (pHyPer-Mito). Seventy two h later cells were challenged with H2O2 (as control; 10 μM) or 8-oxoG (10 μM). Intracellular site of fluorescence were recorded by microscopic imaging. As shown in Fig. 5A, H2O2-induced pHyPer signal was localized to cytoplasm, mitochondria and nuclei of cells. When cells were exposed to 8-oxoG pHyPer-Nuc fluorescence appeared first at the perinuclear region (10 to 12 min post-exposure) and then a nuclear fluorescence was observed (~20 min). pHyPer-Nuc signal decreased to the basal level by 60 min. pHyPer-Cyto-mediated fluorescence was initiated around the nuclei of cells (10 to 12 min after post-exposure) then spread to the cytoplasm (20 min; Fig. 5). The pHyPer-Mito fluorescence appeared at 20 min after 8-oxoG addition. Interestingly, only few mitochondria showed fluorescence suggesting that pHyPer-Mit oxidation is a secondary event and possibly due to ROS generated by NOX4. There were no increased fluorescence of biosensors observed at 60 min post exposure with 8-oxoG. Together these data suggest that NOX4 generating ROS is localized to the nuclear membrane and 8-oxoG exposure-induced ROS generation is transient.
Figure 5.
Transient and localized changes in cellular ROS levels in response to 8-oxoG exposure. Cells were transfected with pHyPer-Cyto, pHyPer-dMito or pHyPer-Nuc and 72h later challenged with 8-oxoG (10 μM) or H2O2 (10 μM). At times indicated, cells were washed in PBS, fixed with formalin (3.7%), dryed and mounted on microscope slides. Images were taken by a NIKON Eclipse Ti System (magnification x125). (A), H2O2 exposure-induced changes in fluorescence in sub-cellular compartments of cells. (B), Localized ROS generation by 8-oxoG exposure of cells. Scale bars: 20 μm.
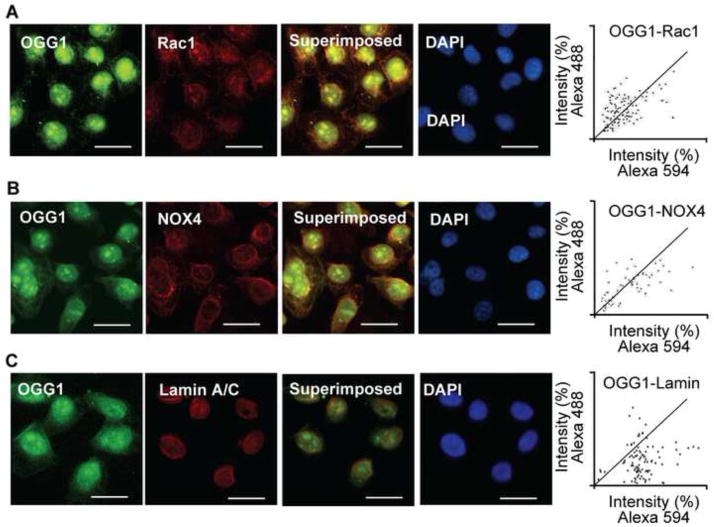
To obtain an insight on close proximity of OGG1, Rac1 and NOX4 at perinuclear regions, we immuno-stained A549 cells by using specific antibodies to these proteins. Fig. 6A, B and C show that OGG1 was primarily localized to the cell nuclei, but a fraction of it was associated with the nuclear membrane. Microscopic imaging showed that like NOX4, Rac1 is associated with the nuclear membrane. Lamin A/C antibody stained the nuclear membrane exclusively (Fig. 6C). We utilized digital imaging tools to examine the co-localization of OGG1 with Rac1 and NOX4 in images obtained by fluorescence microscopy. The fluorescence intensities emitted by OGG1, Rac1 and NOX4 were different, so we employed and calculated overlap coefficients according to Manders, a method which allows a reliable estimation of close proximity localization of proteins [38, 39]. Manders’s formula (Materials and Methods) showed a close proximity localization of OGG1 and Rac1 (Mander’s overlap=0.935431; overlap coefficients k1=0.886159 and overlap coefficients k2=0.987443) and NOX4 (Mander’s overlap=0.909245, overlap coefficients k1=0.872817, and overlap coefficients k2=0.980908). In controls, OGG1 was localized to the nuclear membrane, like lamin A/C; however, according to the Manders’ equation (overlap=0.898948, coefficients k1=1.341340, coefficients k2=0.602463) showed that OGG1 and lamin A/C are not in close proximity. The close proximity of OGG1 with Rac-1 (Fig. 6A) and NOX4 (Fig. 6B) is consistent with an increase in Rac1-GTP levels and the observed nuclear membrane-associated ROS generation (Fig. 5).
Figure 6.
Co-localization of Rac1 and OGG1 with NADPH oxidase 4 in nuclear membrane. A549 cells were cultured on microscope coverslips, then incubated with Abs to OGG1, Rac1 (panel A) and/or NOX4 (panel B). Panel C shows OGG1’s localization to the nuclear membrane based on Lamin A/C. Binding of primary Abs was visualized by using Alexa 488- and Alexa 594-conjugated secondary Abs. Staining of cells was carried out as described in Materials and Methods.. Images were captured with a NIKON Eclipse Ti System (magnification: 125). Co-localization was visualized by superimposition of green and red images using Nikon NIS Elements Version 3.5 (NIKON Instruments, Tokyo, Japan). Scale bars: 20 μm. Mander’s overlap coefficient is 0.935431 for OGG1 and Rac1 (upper right panel); 0.909245 for OGG1 and NOX4 (middle right panel). DAPI = 4′6-diamidino-2-phenylindole dihydrochloride.
Discussion
Among multiple types of DNA base damage generated by ROS, 8-oxoG has received the most attention, as its accumulation in the genome has been associated with increased mutations, various diseases, and aging processes. 8-OxoG is excised from DNA via the OGG1-initiated BER pathway, producing an 8-oxoG base which, when measured in serum and urine is one of the best markers of oxidative exposures. Here we show that in the presence of the 8-oxoG base, OGG1 interacts with the GDP-bound Rac1 protein and promotes the exchange of GDP for GTP in vitro. Importantly, in OGG1-expressing cells the 8-oxoG base increased Rac1-GTP levels. Activated Rac1 then facilitated a spatially controlled increase in cellular ROS levels via NADPH oxidase type 4, which is associated with the nuclear membrane. These results suggest that OGG1, in association with the 8-oxoG base, has a role in cellular physiological and/or patho-physiological signaling via the activation of Rac1.
Our recent study showed that OGG1 binds to the free 8-oxoG base with high affinity [binding constant (Kd) of 0.56±0.19 nM] in the absence of duplex DNA [23], which was unexpected for binding to a product of an enzyme. It has also been shown that interaction of OGG1 with 8-oxoG allowed its binding with canonical Ras family proteins and, importantly, induced guanine nucleotide exchange [23]. In the present study, OGG1 protein alone showed poor binding to guanine nucleotide-free (empty) Rac1 protein, but the 8-oxoG base increased this interaction. On the other hand, OGG1 showed poor interactions with Rac1-GTP in the presence of 8-oxoG – however, less than with the empty protein. These observations resemble those showing binding of OGG1(8-oxoG) with canonical Ras family proteins [23] and those reporting interaction between Rac1 and GEFs such as T lymphoma invasion and metastasis (Tiam) protein [45], Son of sevenless 1 (SOS1) [46], or Vav2 (a homolog of the vav proto-ocogene) [47]. The interaction of OGG1 with GDP-loaded Rac1 suggested that 8-oxoG binding results in a conformational change which allows OGG1 to bind to Rac1. In support of this hypothesis, it has been shown that GEFs require either post-translational modification(s) or binding to regulatory molecules for interaction and catalysis of guanine nucleotide exchange on Rac1 GTPases [48, 49]. Tiam1’s interaction with Rac1 and its GEF activity are increased by association with phosphoinositides in its N-terminal pleckstrin homology domain [50]. The GEF activity of Ras guanine nucleotide release factor 2 requires Ca2+/calmodulin binding to its IQ motif structure [48, 49]. The activity of Ras guanine nucleotide release protein is regulated by Ca2+- and diacylglycerol [51]. Together these data suggest that the OGG1 protein is functionally similar to other Rac1 GEFs, as it requires binding of a co-factor (8-oxoG) to gain the proper conformation necessary for its binding to Rac1-GDP.
During guanine nucleotide exchange, GEFs first interact with GDP-bound GTPase and dissociate GDP at an increased rate, and then the bound GTP promotes the release of GEF by the GTPase [52, 53]. In the presence of 8-oxoG, OGG1 was effective only in promoting GDP release in parallel with GTP incorporation onto Rac1 in vitro. OGG1 did not promote the release of GTP, consistent with its poor interaction with Rac1-GTP indicating that OGG1 functions as a Rac1 GEF similar to Tiam1 [45], SOS1 [46] or Vav2 [47].
An increase in the cellular level of 8-oxoG base rapidly enhanced the GTP-bound level of Rac1. The results obtained by using cultured cells and those generated in the lungs are consistent. Changes in Rac1-GTP levels were time- and 8-oxoG dose-dependent. Importantly, an increase in Rac1-GTP levels did not take place in cells lacking OGG1, suggesting that OGG1 is the primary protein that binds the 8-oxoG base, and is required for Rac1 GDP-GTP exchange. The increase in Rac1-GTP levels upon 8-oxoG addition to the lungs was specific, as it did not occur after challenge with 8-OH-Ade, uric acid, guanine, 8-oxodG or FapyG (the latter two are equally good substrates for OGG1 in DNA [6]).
A549 and MRC-5 cells as well as lung tissues abundantly express Rac1, while Rac2 was nearly undetectable. Expression of Rac3 protein (closely related to Rac1 with respect to function and nucleotide binding) was low in these cells, as well as in mouse lungs. Accordingly, we were unable to show Rac2 or Rac3 activation in either cultured cells or mouse lungs. These data are consistent with those showing that Rac1 is ubiquitously expressed in non-phagocytic cells, including lung epithelial cells and fibroblasts. Rac2 is expressed primarily in myeloid cells (e.g., human neutrophils [54]), and Rac3 is predominantly found in the central nervous system [42].
The physiological functions of the Rac1 family GTPases are diverse, including modulation of the cellular redox state, regulation of cell movements, cellular signaling, gene expression, and cell differentiation (rev in [42]). Rac1 is one of the three Rac family molecules that control NADPH oxidase activity (NOX1, NOX2 and NOX4 as well as NOX3) both in phagocytes and non-phagocytic cells [42, 55]. To gain insight into the possible biological consequences of OGG1(8-oxoG)-mediated Rac1 activation, we examined changes in the cellular redox state. After 8-oxoG addition to cells, there was a >2-fold increase in cellular ROS levels. This effect of 8-oxoG was specific, as FapyG, guanine, 8-oxo-Ade, adenine and uric acid showed no such properties. In contrast to 8-hydroperoxyguanine, the 8-oxoG base has no direct redox properties [28], and thus our present data may mean that the 8-oxoG base increases cellular ROS levels in OGG1-expressing cells via Rac1 activation. This observation was supported by data showing that siRNA-mediated depletion of Rac1 itself, NOX4 or the p22phox regulatory subunit of NOXs (rev in [42, 55]) significantly decreased ROS levels upon 8-oxoG addition. These observations are consistent with those showing that both A549 and MRC5 cells express NOX4 [42, 55]. We found that 8-oxodG decreased Rac1-GTP levels (data not shown) – an observation in good agreement with previous studies showing a decrease in NOX activity by 8-oxodG [41].
Redox sensors (pHyPer-Nuc and pHyPer-Cyto) showed changes in ROS levels first at the perinuclear region of cells, which seems suggestive of localized ROS generation. These results are supported by our data showing that Rac1, NOX4, as well as OGG1 are in close proximity in the nuclear membrane of A549 cells. Similarly Rac1, NOX4 and OGG1 proteins were localized to the nuclear membrane of MRC5 cells (data not shown). Co-localization in the nuclear membrane of OGG1 with Rac1, and NOX4 (based on Manders overlap coefficients [38]) are novel observations, while another report has already documented the nuclear membrane localization of NOX4 (and NOX1, 2) and their activators of Rac1, as well as p22phox (and p47phox), and their contribution to localized ROS generation [56]. Although the biological significance of our findings will be the focus of future investigations, at first glance it seems feasible that ROS could alter OGG1’s repair activity as well as subcellular localization in response to the cell’s need to repair oxidatively damaged DNA [21, 57]. It has been shown that OGG1 is redistributed in response to locally generated oxidative stress within the nucleus, as shown by the accumulation of OGG1-GFP specifically at sites of laser-targeted ROS/DNA damage [58]. On the other hand, ROS generated locally could produce oxidative modifications at cysteine residues, especially those at 253 and 255, in close proximity to OGG1’s active site [59], which may thereby result in decreased OGG1’s activity. This hypothesis is consistent with functional studies observing decreases in OGG1’s activity by oxidative modifications at cysteine residues [10]. It has also been shown that NOX4-derived ROS cause DNA oxidation resulting in 8-oxoG and alterations in gene expression, especially those associated with inflammation [44, 60].
OGG1 is considered to be a canonical DNA BER protein [5]; however, our unexpected observations show that OGG1(8-oxoG) functions as a Rac1 GEF. Although we only show that activated Rac1 increases ROS levels via NOX4, it could also be involved in other cellular processes such as regulation of the cytoskeletal network. Indeed, OGG1 was also observed to be associated with the microtubule organizing center and microtubule network [7, 21]. Thus, by activating Rac1 OGG1 may participate in the organization of microtubules and the mitotic spindle assembly and centrosomes, and thereby could play a critical role in cell cycle progression, including mitosis as previously proposed [21]. These data point to controlled DNA damage and repair in physiologic cellular signaling as shown by others [24, 25, 61] and the OGG1(8-oxoG) may be among these.
In conclusion, while the biological significance of our unexpected findings has yet to be fully elucidated, it appears that activation of the small GTPase Rac1, leading to localized ROS generation could be part of a physiological DNA damage/repair response initiated by OGG1. As the OGG1(8-oxoG) not only activates Rac1 but also the canonical Ras family GTPases [23], it could be that Rac1/NOX4/ROS and Ras-mediated signaling are involved in maintaining cellular homeostasis. We also speculate that a failure in the control of OGG1 activity may lead to excessive release of 8-oxoG from DNA, resulting in unscheduled activation of Rac and Ras family GTPases that could result in pathophysiological cellular responses, contributing to diseases, cellular senescence and aging processes.
Research Highlights.
8-oxoguanine DNA glycosylase-1 (OGG1) excises it from DNA and generates free 8-oxoG base.
In the presence 8-oxoG base, OGG1 interacts with GDP-bound Rac1 and induces GDP→GTP exchange.
Rac1-GTP mediates a localized increase in ROS levels via NADPH oxidase type 4.
Acknowledgments
This work was supported by grants NIEHS RO1 ES018948 (IB), NIA/AG 021830 (IB) NIAID/AI062885 (IB), NINDS R01NS073976 (THK, IB) and the NHLBI Proteomic Center, N01HV00245 (Dr. A. Kurosky). L. Aguilera-Aguirre is an Environmental Toxicology Research Training Fellow (NIEHS T32 ES007254). The work was also supported by the TAMOP 4.2.1/B-09/1/KONV-2010-0007 project (AB), which is co-financed by the European Union and the European Social Fund. AB was also supported by the Janos Bolyai Fellowship from the Hungarian Academy of Sciences. We thank Drs. Miral Dizdaroglu and Pawel Jaruga (Chemical Science and Technology Laboratory, National Institute of Standards and Technology, Gaithersburg, MD USA) for GC/MS measurements of the 8-oxoG and for the gift of FapyG. We thank Drs. Sankar Mitra, and Miral Dizdaroglu for their scientific input, and Dr. David Konkel (Department of Biochemistry and Molecular Biology) and Mardelle Susman (Department of Microbiology and Immunology) for critically editing the manuscript. We also thank the anonymous reviewers for their suggestions, thoughts and concept, which improved the quality of our paper.
Abbreviations
- 8-oxoG
8-Oxo-7,8-dihydroguanine
- 8-oxodG
8-oxo-7,8-dihydro-2′-deoxyguanosine
- 8-OH-Ade
8-oxo-7,8-dihydroadenine
- APE1
apurinic/apyrimidinic endonuclease 1
- DNA BER
base excision repair
- FapyG
2,6- diamino-4-hydroxy-5-formamidopyrimidine
- FU
fluorescence unit
- GEF
guanine nucleotide exchange factor
- GC/MS
gas chromatography/mass spectrometry
- GST
Glutathione-S-Transferase
- H2DCF-DA
2′-7′-dihydro-dichlorofluorescein diacetate
- MantGTP
GTP (2′-(or-3′)-O-(N-methylanthraniloyl)guanosine 5′-triphosphate
- MantGDP
GDP (2′-(or-3′)-O-(N-methylanthraniloyl)guanosine 5′-diphosphate
- NAC
N-acetyl-L-cysteine
- NOX4
NADPH oxidase type 4
- OGG1
8-oxoguanine DNA glycosylase-1
- p22phox
regulatory subunit of NOXs
- Rac1-GTPase
Ras-related C3 botulinum toxin substrate 1
- Rac1-GTP
GTP bound Rac1-GTPase
- Rac1-GDP
GDP-bound Rac1-GTPase
Footnotes
Conflict of interest statement
There are no conflicts of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.de Moura MB, dos Santos LS, Van Houten B. Mitochondrial dysfunction in neurodegenerative diseases and cancer. Environ Mol Mutagen. 2010;51:391–405. doi: 10.1002/em.20575. [DOI] [PubMed] [Google Scholar]
- 2.D’Autreaux B, Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 3.Steenken S. Purine bases, nucleosides, and nucleotides: aqueous solution redox chemistry and transformation reactions of their radical cations and e3 and OH adducts. Chem Rev. 1989;89:503–520. [Google Scholar]
- 4.Radak Z, Boldogh I. 8-Oxo-7,8-dihydroguanine: links to gene expression, aging, and defense against oxidative stress. Free Radic Biol Med. 2010;49:587–596. doi: 10.1016/j.freeradbiomed.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitra S, Izumi T, Boldogh I, Bhakat KK, Hill JW, Hazra TK. Choreography of oxidative damage repair in mammalian genomes. Free Radic Biol Med. 2002;33:15–28. doi: 10.1016/s0891-5849(02)00819-5. [DOI] [PubMed] [Google Scholar]
- 6.Dizdaroglu M, Kirkali G, Jaruga P. Formamidopyrimidines in DNA: mechanisms of formation, repair, and biological effects. Free Radic Biol Med. 2008;45:1610–1621. doi: 10.1016/j.freeradbiomed.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Dantzer F, Luna L, Bjoras M, Seeberg E. Human OGG1 undergoes serine phosphorylation and associates with the nuclear matrix and mitotic chromatin in vivo. Nucleic Acids Res. 2002;30:2349–2357. doi: 10.1093/nar/30.11.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhakat KK, Mokkapati SK, Boldogh I, Hazra TK, Mitra S. Acetylation of human 8-oxoguanine-DNA glycosylase by p300 and its role in 8-oxoguanine repair in vivo. Mol Cell Biol. 2006;26:1654–1665. doi: 10.1128/MCB.26.5.1654-1665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hegde MLHP, Rao KS, Mitra S. Oxidative genome damage and its repair in neurodegenerative diseases: function of transition metals as a double-edged sword. J Alzheimers Dis. 2011;24:183–198. doi: 10.3233/JAD-2011-110281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bravard A, Vacher M, Gouget B, Coutant A, de Boisferon FH, Marsin S, Chevillard S, Radicella JP. Redox regulation of human OGG1 activity in response to cellular oxidative stress. Mol Cell Biol. 2006;26:7430–7436. doi: 10.1128/MCB.00624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Markesbery WR, Lovell MA. DNA oxidation in Alzheimer’s disease. Antioxid Redox Signal. 2006;8:2039–2045. doi: 10.1089/ars.2006.8.2039. [DOI] [PubMed] [Google Scholar]
- 13.Radak Z, Bori Z, Koltai E, Fatouros IG, Jamurtas AZ, Douroudos II, Terzis G, Nikolaidis MG, Chatzinikolaou A, Sovatzidis A, Kumagai S, Naito H, Boldogh I. Age-dependent changes in 8-oxoguanine-DNA glycosylase activity are modulated by adaptive responses to physical exercise in human skeletal muscle. Free Radic Biol Med. 2011;51:417–423. doi: 10.1016/j.freeradbiomed.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishimura S. Involvement of mammalian OGG1(MMH) in excision of the 8-hydroxyguanine residue in DNA. Free Radic Biol Med. 2002;32:813–821. doi: 10.1016/s0891-5849(02)00778-5. [DOI] [PubMed] [Google Scholar]
- 15.Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci U S A. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minowa O, Arai T, Hirano M, Monden Y, Nakai S, Fukuda M, Itoh M, Takano H, Hippou Y, Aburatani H, Masumura K, Nohmi T, Nishimura S, Noda T. Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc Natl Acad Sci U S A. 2000;97:4156–4161. doi: 10.1073/pnas.050404497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mabley JG, Pacher P, Deb A, Wallace R, Elder RH, Szabo C. Potential role for 8-oxoguanine DNA glycosylase in regulating inflammation. Faseb J. 2005;19:290–292. doi: 10.1096/fj.04-2278fje. [DOI] [PubMed] [Google Scholar]
- 18.Li G, Yuan K, Yan C, Fox J, 3rd, Gaid M, Breitwieser W, Bansal AK, Zeng H, Gao H, Wu M. 8-Oxoguanine-DNA glycosylase 1 deficiency modifies allergic airway inflammation by regulating STAT6 and IL-4 in cells and in mice. Free Radic Biol Med. 2012;52:392–401. doi: 10.1016/j.freeradbiomed.2011.10.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitra S. DNA glycosylases: specificity and mechanisms. Prog Nucleic Acid Res Mol Biol. 2001;68:189–192. doi: 10.1016/s0079-6603(01)68099-1. [DOI] [PubMed] [Google Scholar]
- 21.Conlon KA, Zharkov DO, Berrios M. Cell cycle regulation of the murine 8-oxoguanine DNA glycosylase (mOGG1): mOGG1 associates with microtubules during interphase and mitosis. DNA Repair (Amst) 2004;3:1601–1615. doi: 10.1016/j.dnarep.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 22.Szczesny B, Hazra TK, Papaconstantinou J, Mitra S, Boldogh I. Age-dependent deficiency in import of mitochondrial DNA glycosylases required for repair of oxidatively damaged bases. Proc Natl Acad Sci U S A. 2003;100:10670–10675. doi: 10.1073/pnas.1932854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boldogh I, Hajas G, Aguilera-Aguirre L, Hegde ML, Radak Z, Bacsi A, Sur S, Hazra TK, Mitra S. Activation of ras signaling pathway by 8-oxoguanine DNA glycosylase bound to its excision product, 8-oxoguanine. J Biol Chem. 2012;287:20769–20773. doi: 10.1074/jbc.C112.364620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 25.Amente S, Bertoni A, Morano A, Lania L, Avvedimento EV, Majello B. LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription. Oncogene. 2011;29:3691–3702. doi: 10.1038/onc.2010.120. [DOI] [PubMed] [Google Scholar]
- 26.Kemp MG, Reardon JT, Lindsey-Boltz LA, Sancar A. Mechanism of release and fate of excised oligonucleotides during nucleotide excision repair. J Biol Chem. 2012;287 doi: 10.1074/jbc.M112.374447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Napirei M, Karsunky H, Zevnik B, Stephan H, Mannherz HG, Moroy T. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat Genet. 2000;25:177–181. doi: 10.1038/76032. [DOI] [PubMed] [Google Scholar]
- 28.Hajas G, Bacsi A, Aguilerra-Aguirre L, German P, Radak Z, Sur S, Hazra TK, Boldogh I. Biochemical identification of a hydroperoxide derivative of the free 8-oxo-7,8-dihydroguanine base. Free Radic Biol Med. 2012;52:749–756. doi: 10.1016/j.freeradbiomed.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boldogh I, Bacsi A, Choudhury BK, Dharajiya N, Alam R, Hazra TK, Mitra S, Goldblum RM, Sur S. ROS generated by pollen NADPH oxidase provide a signal that augments antigen-induced allergic airway inflammation. J Clin Invest. 2005;115:2169–2179. doi: 10.1172/JCI24422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, Lukyanov S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 31.Malinouski M, Zhou Y, Belousov VV, Hatfield DL, Gladyshev VN. Hydrogen peroxide probes directed to different cellular compartments. PLoS One. 2011;6:e14564. doi: 10.1371/journal.pone.0014564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bacsi A, Chodaczek G, Hazra TK, Konkel D, Boldogh I. Increased ROS generation in subsets of OGG1 knockout fibroblast cells. Mech Ageing Dev. 2007;128:637–649. doi: 10.1016/j.mad.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin J, Xie Y, Wang B, Hoshino M, Wolff DW, Zhao J, Scofield MA, Dowd FJ, Lin MF, Tu Y. Upregulation of PIP3-dependent Rac exchanger 1 (P-Rex1) promotes prostate cancer metastasis. Oncogene. 2009;28:1853–1863. doi: 10.1038/onc.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benard V, Bokoch GM. Assay of Cdc42, Rac, and Rho GTPase activation by affinity methods. Methods Enzymol. 2002;345:349–359. doi: 10.1016/s0076-6879(02)45028-8. [DOI] [PubMed] [Google Scholar]
- 35.Zhang B, Zhang Y, Wang Z, Zheng Y. The role of Mg2+ cofactor in the guanine nucleotide exchange and GTP hydrolysis reactions of Rho family GTP-binding proteins. J Biol Chem. 2000;275:25299–25307. doi: 10.1074/jbc.M001027200. [DOI] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Aguilera-Aguirre L, Bacsi A, Saavedra-Molina A, Kurosky A, Sur S, Boldogh I. Mitochondrial dysfunction increases allergic airway inflammation. Journal of Immunology. 2009;183:5379–5387. doi: 10.4049/jimmunol.0900228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manders EM, Verbeek FJ, Aten JA. Measurement of co-localization of objects in dual-colour confocal images. Journal of Microscopy. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 39.Zinchuk V, Zinchuk O, Okada T. Quantitative colocalization analysis of multicolor confocal immunofluorescence microscopy images: pushing pixels to explore biological phenomena. Acta Histochem Cytochem. 2007;40:101–111. doi: 10.1267/ahc.07002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dizdaroglu M, Jaruga P, Rodriguez H. Measurement of 8-hydroxy-2′-deoxyguanosine in DNA by high-performance liquid chromatography-mass spectrometry: comparison with measurement by gas chromatography-mass spectrometry. Nucleic Acids Res. 2001;29:E12. doi: 10.1093/nar/29.3.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim HJ, Yoon SH, Ryu HO, Yoon BH, Choi S, Ye SK, Chung MH. 8-oxo-7,8-dihydroguanosine triphosphate(8-oxoGTP) down-regulates respiratory burst of neutrophils by antagonizing GTP toward Rac, a small GTP binding protein. Free Radic Res. 2007;41:655–662. doi: 10.1080/10715760701250270. [DOI] [PubMed] [Google Scholar]
- 42.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 43.Das GC, Bacsi A, Shrivastav M, Hazra TK, Boldogh I. Enhanced gamma-glutamylcysteine synthetase activity decreases drug-induced oxidative stress levels and cytotoxicity. Mol Carcinog. 2006;45:635–647. doi: 10.1002/mc.20184. [DOI] [PubMed] [Google Scholar]
- 44.Gordillo G, Fang H, Park H, Roy S. Nox-4-dependent nuclear H2O2 drives DNA oxidation resulting in 8-OHdG as urinary biomarker and hemangioendothelioma formation. Antioxid Redox Signal. 2010;12:933–943. doi: 10.1089/ars.2009.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haeusler LC, Blumenstein L, Stege P, Dvorsky R, Ahmadian MR. Comparative functional analysis of the Rac GTPases. FEBS Lett. 2003;555:556–560. doi: 10.1016/s0014-5793(03)01351-6. [DOI] [PubMed] [Google Scholar]
- 46.Khanday FA, Santhanam L, Kasuno K, Yamamori T, Naqvi A, Dericco J, Bugayenko A, Mattagajasingh I, Disanza A, Scita G, Irani K. Sos-mediated activation of rac1 by p66shc. J Cell Biol. 2006;172:817–822. doi: 10.1083/jcb.200506001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sauzeau V, Sevilla MA, Montero MJ, Bustelo XR. The Rho/Rac exchange factor Vav2 controls nitric oxide-dependent responses in mouse vascular smooth muscle cells. J Clin Invest. 2010;120:315–330. doi: 10.1172/JCI38356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan WT, Koch CA, de Hoog CL, Fam NP, Moran MF. The exchange factor Ras-GRF2 activates Ras-dependent and Rac-dependent mitogen-activated protein kinase pathways. Curr Biol. 1998;8:935–938. doi: 10.1016/s0960-9822(07)00376-4. [DOI] [PubMed] [Google Scholar]
- 49.Innocenti M, Zippel R, Brambilla R, Sturani E. CDC25(Mm)/Ras-GRF1 regulates both Ras and Rac signaling pathways. FEBS Lett. 1999;460:357–362. doi: 10.1016/s0014-5793(99)01374-5. [DOI] [PubMed] [Google Scholar]
- 50.Fleming IN, Gray A, Downes CP. Regulation of the Rac1-specific exchange factor Tiam1 involves both phosphoinositide 3-kinase-dependent and -independent components. Biochem J. 2000;351:173–182. doi: 10.1042/0264-6021:3510173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buday L, Downward J. Many faces of Ras activation. Biochim Biophys Acta. 2008;1786:178–187. doi: 10.1016/j.bbcan.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 52.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 53.Boriack-Sjodin PA, Margarit SM, Bar-Sagi D, Kuriyan J. The structural basis of the activation of Ras by Sos. Nature. 1998;394:337–343. doi: 10.1038/28548. [DOI] [PubMed] [Google Scholar]
- 54.Hordijk PL. Regulation of NADPH oxidases: the role of Rac proteins. Circ Res. 2006;98:453–462. doi: 10.1161/01.RES.0000204727.46710.5e. [DOI] [PubMed] [Google Scholar]
- 55.Lambeth JD, Kawahara T, Diebold B. Regulation of Nox and Duox enzymatic activity and expression. Free Radic Biol Med. 2007;43:319–331. doi: 10.1016/j.freeradbiomed.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spencer NY, Yan Z, Boudreau RL, Zhang Y, Luo M, Li Q, Tian X, Shah AM, Davisson RL, Davidson B, Banfi B, Engelhardt JF. Control of hepatic nuclear superoxide production by glucose 6-phosphate dehydrogenase and NADPH oxidase-4. J Biol Chem. 2011;286:8977–8987. doi: 10.1074/jbc.M110.193821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szczesny B, Bhakat KK, Mitra S, Boldogh I. Age-dependent modulation of DNA repair enzymes by covalent modification and subcellular distribution. Mech Ageing Dev. 2004;125:755–765. doi: 10.1016/j.mad.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 58.Zielinska A, Davies OT, Meldrum RA, Hodges NJ. Direct visualization of repair of oxidative damage by OGG1 in the nuclei of live cells. J Biochem Mol Toxicol. 2011;25:1–7. doi: 10.1002/jbt.20346. [DOI] [PubMed] [Google Scholar]
- 59.Qi Y, Spong MC, Nam K, Banerjee A, Jiralerspong S, Karplus M, Verdine GL. Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme. Nature. 2009;462:762–766. doi: 10.1038/nature08561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weyemi U, Dupuy C. The emerging role of ROS-generating NADPH oxidase NOX4 in DNA-damage responses. Mutat Res. 2012;751:77–81. doi: 10.1016/j.mrrev.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 61.Al-Mehdi AB, Pastukh VM, Swiger BM, Reed DJ, Patel MR, Bardwell GC, Pastukh VV, Alexeyev MF, Gillespie MN. Perinuclear mitochondrial clustering creates an oxidant-rich nuclear domain required for hypoxia-induced transcription. Sci Signal. 2012;5:ra47. doi: 10.1126/scisignal.2002712. [DOI] [PMC free article] [PubMed] [Google Scholar]