Abstract
Rational
More research has recently been focused on multigenerational toxicogenomics impacts. Such studies rely on behavioral as well as genetic and epigenetic analyses using various biotechniques. Of these technologies, qRT-PCR is considered as a mature discovery and validation tool. Nevertheless, the interpretation of the resulting gene expression necessitates the establishment of reliable internal controls for normalization. No study has been performed to identify reliable reference genes in multigenerational settings.
Objectives
The primary aim was to evaluate the stability of 16 reference gene candidates in C. elegans exposed to nicotine and their two subsequent generations for determining the most reliable reference genes for multigenerational study.
Methods
We exposed C. elegans to nicotine in the F0 generation, and investigated the relative stabilities of 16 housekeeping genes in L4 larvae across three generations (F0, F1, and F2) using five statistical approaches (geNorm, ΔCt method, NormFinder, BestKeeper, and ReFinder).
Results
GeNorm shows that CDC-42 and Y45F10D.4 were the most stable reference genes. Based on NormFinder, TBA-1, EIF3.C, ARP-6, CDC-42, and MDH-2 may serve the top reliable reference genes. Comparative ΔCt method ranked TBA-1, CDC-42, EIF3.C, ARP-6, and Y45F10D.4 as the most stable reference genes. BestKeeper shows that Y45F10D.4, F35G12.2, TBA-1, CDC-42, and CSQ-1were better reference genes. Overall, TBA-1, CDC-42, EIF3.C, ARP-6, and Y45F10D.4 were the most reliable reference genes for mutigenerational nicotine-exposed study.
Conclusions
Of the 16 tested gene candidates, TBA-1 and CDC-42 were the two most stable reference genes for performing reliable gene expression normalization in the multigenerational impact of nicotine exposure.
Keywords: qRT-PCR, reference genes, multi-generational, C. elegans, nicotine, drugs of abuse
Introduction
Transcriptome studies have revolutionized molecular biology. Despite the increasing popularity of some advanced “discovery” technologies such as next generation sequencing (NGS) (e.g. RNA-seq), those high-throughput, sensitive technologies are still in a juvenile stage. Major drawbacks are attributed to the absence of standardized data analyses approaches and inability to distinguish between signal and noise (Pertea, 2012). Inconsistencies in the data are further corrected and validated via more established technologies such as qRT-PCR that has been serving as a valuable mature tool for the validation of various transcriptome-related micro-arrays and NGS (Git et al., 2010).
qRT-PCR is a mature biotechnique with both advantages and limitations. Efforts to correct for biases and variations caused by experimental errors and data handling have long been investigated and reported (Lefever et al., 2009). In qRT-PCR, such can be accounted for by many factors, including the total RNA quantity and integrity, enzymatic efficiencies, total transcriptional status of cells or organisms as a whole, enzymatic inefficiencies as well as pipetting errors (Ginzinger, 2002). To correct some of these false positive results, genes of interest are normalized to reference genes that have almost constant expression levels in the tested environmental condition. The choice of a reference gene is not so trivial. It has been concluded that there is no “universally suitable reference gene”. With this in mind, control genes should be selected based on the nature of the investigations and are expected to be resistant to the induced perturbation and modification (Hruz et al., 2011).
A lot of studies have been done to investigate the mechanism of action of nicotine in different organisms (e.g. cell culture, rats, mice, drosophila, zebrafish, C. elegans) (Matta et al., 2007). Of the 4000 chemicals in tobacco smoking, nicotine has received a lot of attention and research due to its addictive and toxic properties (CDC, 1988, 2010). Unfortunately, addiction is a universally notorious disease that affects millions worldwide. Despite concentrated efforts to limit nicotine exposure, the rate of tobacco smoking remains high in many developing countries and particularly among youth and children (WHO, 2012). The obscurity of the molecular mechanisms of maladaptive neuroplasticity like addiction, especially on children, necessitates further in depth research to understand the extent of physiological disruptions. Our ongoing study implies the extension of addictive behavioral and molecular biomarkers across generations. Such an association is expected to trigger further replications and more in depth experiments involving protein coding as well as non-coding genes. For reasons described below, we employed C. elegans as our model organism to investigate the systemic mechanism of action of nicotine.
C. elegans is one of the major model organisms (Brenner, 1974) which can be easily and economically maintained. Research on C. elegans is free of ethical concern and has contributed to advances in the biomedical fields. Up to 80% of its genome is homologous to that of humans (Beitel et al., 1990) and is characterized by fewer genetic redundancies in coding and non-coding sequences (Kazazian, 2004; Kirienko et al., 2010). So far, extensive toxicogenomics research has been conducted on C. elegans in specific developmental stages and in response to different treatments (Karp et al., 2011; Lant and Storey, 2010; Pincus et al., 2011; Viñuela et al., 2010). However, correct interpretations and extrapolations on the genetic level necessitate reliable and sensitive control reference genes. With transgenerational nicotine addiction being the main focus of our research, the goal of this study was to identify reliable reference gene candidates for gene expression analysis at a multigenerational aspect.
In this study, we compiled a list of reference gene candidates from previous publications that included both protein coding and RNA genes. We were interested in investigating the relative stabilities of the selected genes in response to nicotine exposure across three generations. In our experiment, wild type L1 worms (N2) were distributed into three treatment groups: 0μM (control), 20μM and 20mM nicotine NGM plates. Worms were exposed to nicotine until early L4 stage (~30 hours). Exposure was restricted to the F0 generation, but we continued sampling L4 worms in both F1 and F2 generations. Among the sixteen selected genes, we aimed to determine the most reliable gene candidate(s) that can be used in nicotine related transgenerational molecular studies. To accomplish our objective, we used four of the most popular reference gene analysis software: geNorm, NormFinder, comparative ΔCt method, and BestKeeper. Taking all into consideration, the most stable gene(s) candidate was (were) determined by an overall comprehensive ranking approach (Xie et al., 2012).
As a summary, recent evidences show that environmental exposure can cause multigenerational impacts on animal growth and development and even some diseases (Contreras et al., 2012; Tominaga et al., 2003; Yu et al., 2012). On the other hand, several other reports have demonstrated that chemicals may induce transgenerational alterations in gene expressions (Ashe et al., 2012; Braunschweig et al., 2012; Manikkam et al., 2012). However, no study has been performed to examine the effect of any chemical on housekeeping genes and thus no reliable reference genes exist for mutigenerational investigations. In this study, we employed C. elegans as an animal model system to evaluate and identify the most reliable reference genes for future mutigenerational toxicogenomics approaches and gene expression analyses related to nicotine addiction.
Material and methods
Chemicals and C. elegans strains
Nicotine was purchased from Acros Organics (New Jersey, USA). 1 M and 0.001 M stocks were prepared by diluting nicotine in phosphate buffer. From the two stock solutions, nicotine was then added into the NGM medium, after the addition of cholesterol, CaCl2, MgSO4, and KH2PO4, to give final concentrations of 20 μM and 20 mM, respectively.
C. elegans hermaphrodite N2 Bristol wild type was used. Worms were constantly transferred via chunking method to a new NGM plate freshly seeded with OP50.
Egg synchronization was done via bleaching (Sulston and Hodgkin, 1988). Briefly, M9 buffer was used to wash adult gravid worms off the plate into 15 ml falcon tubes. Then the falcon tube was centrifuged at 2000 rpm for 2 minutes, respectively. After discarding the supernatant, the wash was repeated. Then, 5 ml of synchronization solution (70% dH2O, 10% NaOH, and 20% bleach) was added. The tubes were vigorously shaken (or vortexed) for a maximum of 5 minutes until the adult worms burst leaving the eggs dispersed in solution. The tubes were then spun at 2000 for 2 minutes. The supernatant was removed and three to four 5-ml M9 washes followed leaving the last wash without centrifugation. The tubes with the suspended eggs were placed on a shaker in the 20°C incubator for 14–18 hours. After hatching, the L1 larvae were pooled and randomly transferred to the different treatment groups.
The three treatment groups included the control group, the 20μM and 20mM nicotine treatment groups. L1 larvae of the F0 generation were incubated at 20°C on seeded control and treatment plates for about 31 hours until the end of L3-the beginning of L4 stage. From each plate, worms were unequally harvested off the plates into two eppendorf tubes. The one with the larger pellet was intermittently centrifuged two times at 2000 rpm then 3000 rpm to separate the worms from bacteria and debris. Consequentially, the pellet was flash frozen in liquid nitrogen, and then stored at −80°C until molecular analysis. As for the eppendorf with the smaller pellet, the L4 worms were then transferred into OP50-seeded NGM plates, left to dry, then sealed and placed back in the 20°C incubator to grow until egg-laying peaked (around second day of adulthood). Adults were then collected for synchronization to gather the eggs for the subsequent generations. The whole procedure was repeated until the L4 stage of the F2 generation was reached.
RNA extraction and qRT-PCR
Total RNA extraction was performed according to the protocol using mirVana™ miRNA Isolation Kit (Ambion, Austin, TX). Briefly, the sample was denatured using a lysis buffer. RNA was then separated from DNA and other proteins via acid-phenol extraction. Then, ethanol was added to the sample followed by passing through a glass-filter. Several washes preceded the elution of the RNA with low ionic strength solution.
RNA quantification and evaluation were done using the NanoDrop ND-1000 Micro-Volume UV/Vis Spectrophotometer (NanoDrop Technologies, Wilmington, DE) and were based on the concentration (ng/μL) and absorbance ratios of 260/280 and 260/230.
Reverse transcription was performed using TaqMan microRNA Reverse Transcription kit from Applied Biosystems (Foster City, CA) to reverse-transcribe RNA to cDNA for both protein coding genes and small RNAs. The poly-T was used for protein-coding genes and specific primers were used for small RNAs. For each reaction, the final reaction volume was 15 μL and included 1000ng of total RNAs, 0.19μL RNase inhibitor (20U/μL), 0.15mu;L of 100mM dNTPs, 1.5mu;L of reverse transcription buffer (10X), 2mu;L of primer mix, and 1mu;L of reverse transcriptase (50U/mu;L). The samples were then run via thermal cycler using the program: 16°C for 30 min followed by 42°C for 30 min, 85°C for 5 min and were finally held at 4°C. The samples were diluted in 80mu;L DNase/RNase-free water for subsequent qRT-PCR.
The expression levels of selected genes were analyzed after performing qRT-PCR on 96-well-plate using the 7300 Real-Time PCR System (Applied Biosystem) using the SYBR Green PCR master mix from SuperArray Bioscience Corp. (Frederick, MD). Specific reverse and forward primers were used for each tested gene (Table 1). Briefly, each well carried a 20mu;L reaction resulting from the combination of 7mu;L DNase/RNase free water, 10mu;L SYBR Green master mix, 1mu;L cDNA, 2mu;L primers. A minimum of three biological replicates with two technical replicates were run. The qRT-PCR program was started at 95°C for 10 min for enzyme activation followed by denaturation for 15 sec at 95°C and an annealing/extension step for 60 sec at 60°C. The latter 2 steps were repeated for 40 cycles.
Table 1.
Properties of the sixteen reference gene candidates.
| Gene Symbol | Locus tag | Gene description | Forward primer | Reverse primer |
|---|---|---|---|---|
| CDC-42 | R07G3.1 | Cell Division Cycle related | AGCCATTCTGGCCGCTCTCG | GCAACCGCTTCTCGTTTGGC |
|
|
||||
| PMP-3 | C54G10.3 | Peroxisomal Membrane Protein related | TGGCCGGATGATGGTGTCGC | ACGAACAATGCCAAAGGCCAGC |
|
|
||||
| EIF-3.C | T23D8.4 | Eukaryotic Initiation Factor | ACACTTGACGAGCCCACCGAC | TGCCGCTCGTTCCTTCCTGG |
|
|
||||
| ARP-6 | C08B11.6 | Spliceosome-Associated Protein family member (sap-49) | TGGCGGATCGTCGTGCTTCC | ACGAGTCTCCTCGTTCGTCCCA |
|
|
||||
| ACT-2 | T04C12.5 | ACTin | GCGCAAGTACTCCGTCTGGATCG | GGGTGTGAAAATCCGTAAGGCAGA |
|
|
||||
| CSQ-1 | F40E10.3 | Calsequestrin | GCCTTGCGCTAGTGGTTGTGC | GCTCTGAGTCGTCCTCTTCCACG |
|
|
||||
| Y45F10D.4 | Y45F10D.4 | Putative iron-sulfur cluster assembly enzyme | CGAGAACCCGCGAAATGTCGGA | CGGTTGCCAGGGAAGATGAGGC |
|
|
||||
| TBA-1 | F26E4.8 | TuBulin, Alpha family member | TCAACACTGCCATCGCCGCC | TCCAAGCGAGACCAGGCTTCAG |
|
|
||||
| MDH-2 | F20H11.3 | Malate DeHydrogenase | TGGAGCTGCCGGAGGAATTGG | TCAGCGTTCTCAACGGCGGC |
|
|
||||
| AMA-1 | F36A4.7 | AMAnitin resistant family member | CGGATGGAGGAGCATCGCCG | CAGCGGCTGGGGAAGTTGGC |
|
|
||||
| F35G12.2 | F35G12.2 | ortholog of mitochondrial NAD+-isocitrate dehydrogenase. | ACTGCGTTCATCCGTGCCGC | TGCGGTCCTCGAGCTCCTTC |
|
|
||||
| RBD-1 | T23F6.4 | RBD(RNA binding domain)protein | GGTCAGATTTCCGATGCGTCGCT | ACTTGCTCCAGGCTCTCGGC |
|
|
||||
| U6 | CELE_F35C11.9 | snRNA involved in mRNA splicing | CAGAGAAGATTAGCATGGCCC | TTGGAACGCTTCACGAATTTGC |
|
|
||||
| 18s rRNA | CELE_F31C3.7 | rRNA subunit | TTCTTCCATGTCCGGGATAG | CCCCACTCTTCTCGAATCAG |
|
|
||||
| Ce234.1 | DQ789547 | C/D box snoRNA | GGTTACGGTAGCCGAGTCAG | GCCATAACTGTTCACCGTCG |
|
|
||||
| U18 | Z75111 | snoRNA | TGATGATCACAAATCCGTGTTTC | GCTCAGCCGGTTTTCTATCG |
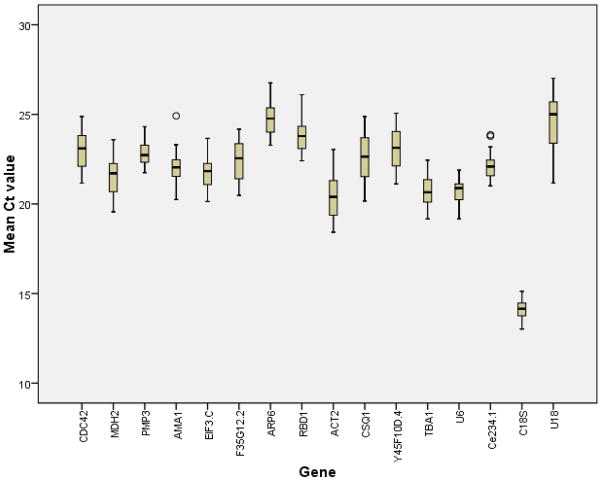
Primer specificity and efficiency have been previously calculated. Moreover, descriptive statistics (i.e. mean, SD) were calculated via SPSS for the raw Ct values of each gene candidate. Boxplot graphs were done via SPSS20 (Figure 1; Table 2).
Figure 1.
The average Ct values calculated from raw qRT-PCR output for the 16 reference gene candidates in L4 C. elegans (N2). 50% of the values are included in the box. The median is represented by the line in the box. The interquartile range is bordered by the upper and lower edges, which indicate the 75th and 25th percentiles, respectively. The whiskers are inclusive of the maximal and minimal values, but exclusive of the outliers, represented as circles.
Table 2.
Overall descriptive statistics of the raw Ct values for each candidate gene among all nicotine treatment groups in L4 C. elegans.
| N | Minimum | Maximum | Mean | SD | Median | |
|---|---|---|---|---|---|---|
| CDC42 | 56 | 21.16 | 24.88 | 23.02 | 0.97 | 23.10 |
|
|
||||||
| MDH2 | 56 | 19.56 | 23.58 | 21.52 | 0.99 | 21.71 |
|
|
||||||
| PMP3 | 56 | 21.74 | 24.31 | 22.83 | 0.63 | 22.72 |
|
|
||||||
| AMA1 | 56 | 20.25 | 24.91 | 22.04 | 0.76 | 22.05 |
|
|
||||||
| EIF3.C | 56 | 20.14 | 23.66 | 21.65 | 0.83 | 21.83 |
|
|
||||||
| F35G12.2 | 56 | 20.48 | 24.17 | 22.40 | 1.11 | 22.55 |
|
|
||||||
| ARP6 | 56 | 23.27 | 26.76 | 24.77 | 0.95 | 24.77 |
|
|
||||||
| RBD1 | 56 | 22.41 | 26.10 | 23.76 | 0.81 | 23.79 |
|
|
||||||
| ACT2 | 56 | 18.43 | 23.05 | 20.47 | 1.29 | 20.39 |
|
|
||||||
| U6 | 56 | 19.17 | 21.89 | 20.74 | 0.61 | 20.88 |
|
|
||||||
| CSQ1 | 56 | 20.17 | 24.88 | 22.59 | 1.28 | 22.64 |
|
|
||||||
| Ce234.1 | 56 | 21.01 | 23.86 | 22.09 | 0.64 | 22.09 |
|
|
||||||
| Y45F10D.4 | 56 | 21.12 | 25.07 | 23.05 | 1.08 | 23.13 |
|
|
||||||
| 18s rRNA | 56 | 13.01 | 15.13 | 14.14 | 0.49 | 14.16 |
|
|
||||||
| TBA1 | 56 | 19.17 | 22.44 | 20.70 | 0.84 | 20.65 |
|
|
||||||
| U18 | 56 | 21.17 | 27.02 | 24.56 | 1.48 | 25.00 |
Determination of gene stability
Five different statistical approaches (geNorm, ΔCt method, NormFinder, BestKeeper, and ReFinder) were employed to determine the stability of each tested reference gene candidate.
The geNorm (Vandesompele et al., 2002) applet allows the determination of the most stable reference gene(s) based on pairwise comparisons between each gene with all other candidates. The variation in the expression level of each gene was calculated as the geometric mean of the standard deviation (SD) relative to all other genes. Such a stability index is described as the ‘M-value’. Ranking is achieved after sequential elimination of most variable gene, followed by recalculation of the ‘M-value’. Finally, genes with the lowest ‘M-value’ will be ranked with highest stability in comparison with the other tested genes. Conceptually, geNorm assumes that an ideal-gene pair will have the least variation in expression in all samples regardless of experimental conditions. GeNorm goes beyond that to estimate the minimal n (e.g. number of genes) needed to perform reliable normalization. This is based on pairwise variation [Vn/Vn+1] calculated for each gene pair normalization factors [NFn, NFn+1]. Through this approach, the need for the inclusion of an additional reference gene would be reflected by a high variation (i.e. >0.15 established cutoff value), and vice versa.
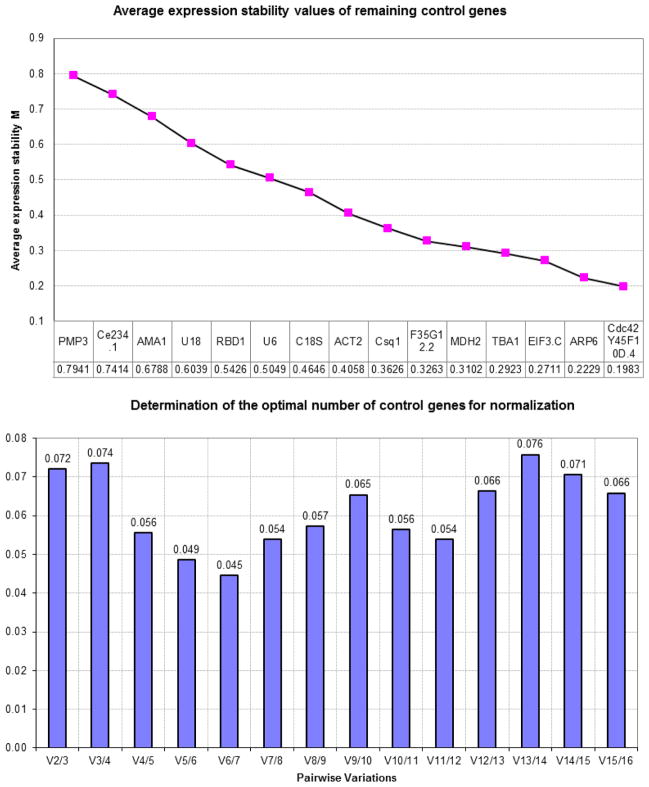
To prepare the input for geNorm, relative quantification from raw Ct values among all samples was done for each gene. Briefly, the smallest Ct value was determined for each gene among all samples. Then, this value was subtracted from all the other Ct values related to this gene. Therefore, the minimal value would be zero. Then, each value is transformed using the formula: 2^(Ctoriginical-Ctmin). The resulting converted data were used as input for geNorm with the names of the genes and samples in the first row and column, respectively. Together, they were saved in the provided input directory. After loading the input file into geNorm, the analysis was run and two charts are automatically generated as shown in Figure 2.
Figure 2.
Top: geNorm ranking of the most stable gene candidates among all treatment groups and generations. Bottom: GeNorm-based pair-wise variation value (V value) among the candidate genes. The cut-off value being 0.15. All values were below cutoff. Hence, the combination of two reference genes is enough to be used for normalization of qRT-PCR expression levels.
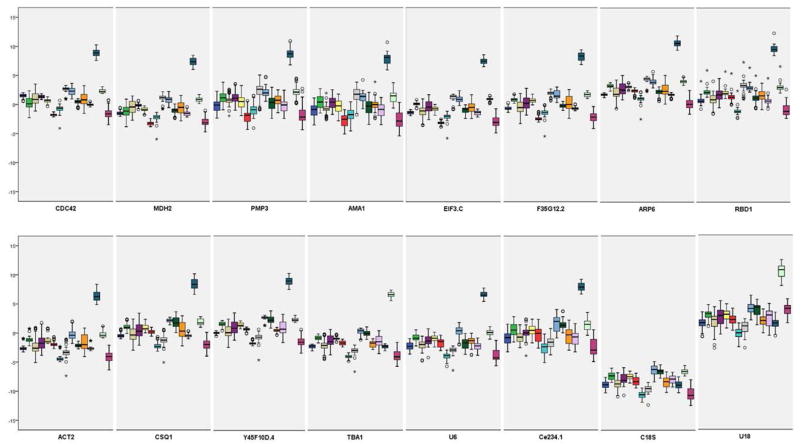
The comparative delta Ct method (Silver et al., 2006) is a relatively similar approach that depends on pairwise comparisons between genes. This method can be easily done on an excel spreadsheet without the help of a designed program. In addition, its development facilitated gene expression normalization for experiments with non-ideal sample sizes and purity. Simply, a set of comparisons is performed where each gene is compared against all other gene candidates. The ΔCt is calculated for every gene pair across all the samples in the treatment groups. For every gene pair, the mean ΔCt and SD are calculated. A high SD reflects that one or both genes are not stable. Then, an overall average SD is calculated for every gene being compared against all others (i.e. gene pair set). Including more genes into the comparison will allow for the selection of the one with the least variability. Thus, the gene with the least SD will be the top-ranked candidate for normalization. Calculations for the comparative ΔCt method were done using excel spreadsheet as described above. Boxplots were generated via SPSS20. For each gene set, different colors represent different ‘gene pairs’ as shown in Figure 3 and Table 4.
Figure 3.
A box-plot graph representing the values of gene expression of the 16 reference gene candidates. Expression levels were calculated from each “pair of genes” in each group. 50% of the values are included in the box. The median is represented by the line in the box. The interquartile range is bordered by the upper and lower edges, which indicate the 75th and 25th percentiles, respectively. The whiskers are inclusive of the maximal and minimal values, but exclusive of the outliers, represented as circles and asterisks. Different “gene pairs” are shown as different colors. The y-axis represents the ΔCt values between each gene pair/group, while the x-axis shows the 16 reference candidates.
Table 4.
A summary of the pair-wise mean and SD calculations for each of the reference gene candidates. The last column on the left is the average SD for each candidate. The latter was used in the Delta-Ct-based method to identify the most stable genes.
| Gene | Pair 1 | Pair 2 | Pair 3 | Pair 4 | Pair 5 | Pair 6 | Pair 7 | Pair 8 | Pair 9 | Pair 10 | Pair 11 | Pair 12 | Pair 13 | Pair 14 | Pair 15 | Avg. SD | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TBA1 | Mean | −2.31 | −0.81 | −2.12 | −1.33 | −0.94 | −1.70 | −4.07 | −3.05 | 0.23 | −0.04 | −1.89 | −1.39 | −2.34 | 6.56 | −3.85 | |
|
|
|||||||||||||||||
| SD | 0.32 | 0.37 | 1.10 | 1.05 | 0.27 | 0.38 | 0.36 | 0.67 | 0.56 | 0.53 | 0.54 | 1.03 | 0.34 | 0.48 | 0.93 | 0.59 | |
|
|
|||||||||||||||||
| CDC42 | Mean | 1.50 | 0.19 | 0.98 | 1.37 | 0.62 | −1.76 | −0.74 | 2.55 | 2.28 | 0.42 | 0.93 | −0.03 | 8.88 | 2.31 | −1.54 | |
|
|
|||||||||||||||||
| SD | 0.34 | 1.17 | 1.09 | 0.29 | 0.35 | 0.20 | 0.65 | 0.58 | 0.68 | 0.42 | 1.22 | 0.20 | 0.66 | 0.32 | 0.91 | 0.61 | |
|
|
|||||||||||||||||
| EIF3.C | Mean | −1.37 | 0.13 | −1.18 | −0.39 | −0.75 | −3.13 | −2.11 | 1.17 | 0.91 | −0.95 | −0.44 | −1.40 | 7.51 | 0.94 | −2.91 | |
|
|
|||||||||||||||||
| SD | 0.29 | 0.29 | 1.07 | 1.01 | 0.46 | 0.29 | 0.65 | 0.71 | 0.56 | 0.60 | 1.04 | 0.37 | 0.54 | 0.27 | 0.95 | 0.61 | |
|
|
|||||||||||||||||
| ARP6 | Mean | 1.76 | 3.26 | 1.95 | 2.74 | 3.13 | 2.37 | 1.02 | 4.30 | 4.03 | 2.18 | 2.68 | 1.73 | 10.63 | 4.07 | 0.22 | |
|
|
|||||||||||||||||
| SD | 0.20 | 0.35 | 1.13 | 1.02 | 0.29 | 0.40 | 0.62 | 0.69 | 0.62 | 0.55 | 1.20 | 0.27 | 0.65 | 0.36 | 0.87 | 0.61 | |
|
|
|||||||||||||||||
| Y45F10D.4 | Mean | 0.03 | 1.53 | 0.22 | 1.01 | 1.40 | 0.65 | −1.73 | −0.71 | 2.57 | 2.31 | 0.45 | 0.96 | 8.91 | 2.34 | −1.51 | |
|
|
|||||||||||||||||
| SD | 0.20 | 0.38 | 1.29 | 1.20 | 0.37 | 0.25 | 0.27 | 0.76 | 0.46 | 0.73 | 0.32 | 1.30 | 0.74 | 0.34 | 0.86 | 0.63 | |
|
|
|||||||||||||||||
| MDH2 | Mean | −1.50 | −1.31 | −0.52 | −0.13 | −0.88 | −3.26 | −2.24 | 1.04 | 0.78 | −1.08 | −0.57 | −1.53 | 7.38 | 0.81 | −3.04 | |
|
|
|||||||||||||||||
| SD | 0.34 | 1.18 | 1.09 | 0.29 | 0.37 | 0.35 | 0.69 | 0.69 | 0.63 | 0.55 | 1.14 | 0.38 | 0.66 | 0.37 | 0.81 | 0.64 | |
|
|
|||||||||||||||||
| F35G12.2 | Mean | −0.62 | 0.88 | −0.43 | 0.36 | 0.75 | −2.37 | −1.36 | 1.93 | 1.66 | −0.19 | 0.31 | −0.65 | 8.26 | 1.70 | −2.16 | |
|
|
|||||||||||||||||
| SD | 0.35 | 0.37 | 1.33 | 1.22 | 0.46 | 0.40 | 0.77 | 0.42 | 0.72 | 0.34 | 1.29 | 0.25 | 0.75 | 0.38 | 0.80 | 0.66 | |
|
|
|||||||||||||||||
| 18s rRNA | Mean | −8.88 | −7.38 | −8.68 | −7.89 | −7.51 | −8.26 | −10.63 | −9.61 | −6.33 | −6.60 | −8.45 | −7.95 | −8.91 | −6.56 | −10.42 | |
|
|
|||||||||||||||||
| SD | 0.66 | 0.66 | 0.89 | 0.88 | 0.54 | 0.75 | 0.65 | 0.64 | 0.90 | 0.46 | 0.92 | 0.71 | 0.74 | 0.48 | 1.13 | 0.73 | |
|
|
|||||||||||||||||
| U6 | Mean | −2.28 | −0.78 | −2.09 | −1.30 | −0.91 | −1.66 | −4.03 | −3.02 | 0.27 | −1.86 | −1.35 | −2.31 | 6.60 | 0.04 | −3.82 | |
|
|
|||||||||||||||||
| SD | 0.68 | 0.63 | 0.83 | 0.82 | 0.56 | 0.72 | 0.62 | 0.63 | 0.96 | 0.97 | 0.76 | 0.73 | 0.46 | 0.53 | 1.11 | 0.73 | |
|
|
|||||||||||||||||
| CSQ1 | Mean | −0.42 | 1.08 | −0.23 | 0.56 | 0.95 | 0.19 | −2.18 | −1.16 | 2.12 | 1.86 | 0.50 | −0.45 | 8.45 | 1.89 | −1.96 | |
|
|
|||||||||||||||||
| SD | 0.42 | 0.55 | 1.52 | 1.42 | 0.60 | 0.34 | 0.55 | 0.94 | 0.33 | 0.97 | 1.48 | 0.32 | 0.92 | 0.54 | 0.91 | 0.78 | |
|
|
|||||||||||||||||
| RBD1 | Mean | 0.74 | 2.24 | 0.93 | 1.72 | 2.11 | 1.36 | −1.02 | 3.28 | 3.02 | 1.16 | 1.67 | 0.71 | 9.61 | 3.05 | −0.80 | |
|
|
|||||||||||||||||
| SD | 0.65 | 0.69 | 1.01 | 0.89 | 0.65 | 0.77 | 0.62 | 1.03 | 0.63 | 0.94 | 1.00 | 0.76 | 0.64 | 0.67 | 1.05 | 0.80 | |
|
|
|||||||||||||||||
| ACT2 | Mean | −2.55 | −1.04 | −2.35 | −1.56 | −1.17 | −1.93 | −4.30 | −3.28 | −0.27 | −2.12 | −1.62 | −2.57 | 6.33 | −0.23 | −4.09 | |
|
|
|||||||||||||||||
| SD | 0.58 | 0.69 | 1.55 | 1.48 | 0.71 | 0.42 | 0.69 | 1.03 | 0.96 | 0.33 | 1.43 | 0.46 | 0.90 | 0.56 | 0.98 | 0.85 | |
|
|
|||||||||||||||||
| U18 | Mean | 1.54 | 3.04 | 1.73 | 2.52 | 2.91 | 2.16 | −0.22 | 0.80 | 4.09 | 3.82 | 1.96 | 2.47 | 1.51 | 10.42 | 3.85 | |
|
|
|||||||||||||||||
| SD | 0.91 | 0.81 | 1.65 | 1.51 | 0.95 | 0.80 | 0.87 | 1.05 | 0.98 | 1.11 | 0.91 | 1.50 | 0.86 | 1.13 | 0.93 | 1.06 | |
|
|
|||||||||||||||||
| AMA1 | Mean | −0.98 | 0.52 | −0.79 | 0.39 | −0.36 | −2.74 | −1.72 | 1.56 | 1.30 | −0.56 | −0.06 | −1.01 | 7.89 | 1.33 | −2.52 | |
|
|
|||||||||||||||||
| SD | 1.09 | 1.09 | 0.82 | 1.01 | 1.22 | 1.02 | 0.89 | 1.48 | 0.82 | 1.42 | 0.98 | 1.20 | 0.88 | 1.05 | 1.51 | 1.10 | |
|
|
|||||||||||||||||
| Ce234.1 | Mean | −0.93 | 0.57 | −0.73 | 0.06 | 0.44 | −0.31 | −2.68 | −1.67 | 1.62 | 1.35 | −0.50 | −0.96 | 7.95 | 1.39 | −2.47 | |
|
|
|||||||||||||||||
| SD | 1.22 | 1.14 | 0.89 | 0.98 | 1.04 | 1.29 | 1.20 | 1.00 | 1.43 | 0.76 | 1.48 | 1.30 | 0.71 | 1.03 | 1.50 | 1.13 | |
|
|
|||||||||||||||||
| PMP3 | Mean | −0.19 | 1.31 | 0.79 | 1.18 | 0.43 | −1.95 | −0.93 | 2.35 | 2.09 | 0.23 | 0.73 | −0.22 | 8.68 | 2.12 | −1.73 | |
|
|
|||||||||||||||||
| SD | 1.17 | 1.18 | 0.82 | 1.07 | 1.33 | 1.13 | 1.01 | 1.55 | 0.83 | 1.52 | 0.89 | 1.29 | 0.89 | 1.10 | 1.65 | 1.16 | |
Whereas pairwise comparison approaches focus on intra-group variation with less, if any, consideration on the inter-group variation, NormFinder (Andersen et al., 2004) ranks gene stability based on minimal variation of samples not only among all treatment groups, but also within each group. NormFinder prevents the exclusion of stable genes with different expression levels that would otherwise be ranked as one of the least stable through pairwise comparison. In addition false positive results caused by co-regulated genes with similar expression patterns would be avoided. Through NormFinder, a top-ranked gene would introduce the least systemic error when used for normalization.
Another excel-based applet is BestKeeper (Pfaffl et al., 2004) that allows the analysis of 10 reference gene candidates as well as target genes for many samples. For that, we excluded the 6 least stable genes (AMA-1, RBD-1, PMP-3, ACT-2, Ce234.1, and U18) based on geNorm, NormFinder, and delta Ct method. Its ranking is a result of a stepwise process that starts with the exclusion of genes with expressions having an SD>1. To analyze the relationship of candidate genes with one another, a series of pairwise comparisons between each pair is calculated as Pearson’s correlation coefficient [r]. Then, based on the most highly correlated genes, the geometric mean of the Ct values is used to calculate an index. After a pairwise-correlation analysis of each candidate gene with BestKeeper index, genes with the highest statistically significant correlation coefficient represent the most stable genes.
Results
Descriptive statistics for the expression levels of reference gene candidates
Figure 1 demonstrates the expression levels of each reference gene candidate. The expression levels were calculated from the original Ct values for all samples belonging to three nicotine treatment groups (control, 20μM, and 20mM) across all three generations (F0, F1 and F2). 18s rRNA, ACT-2, and TBA-1 had the least median Ct values with Ctmedian of 14.16, 20.39, and 20.65, respectively. Whereas, RBD-1, ARP-6, and U18 had highest Ct values with Ctmedian values of 23.79, 24.77, and 25.00, respectively. However, looking at the variations in the Ct values among treatment groups and generations, it appears that the least variable genes were U18, U6 and PMP-3 with standard deviation (SD) values of 0.49, 0.61, and 0.63, respectively. Conversely, the three most variable genes were CSQ-1, ACT-2, and U18 with SD values of 1.28, 1.29, and 1.48, respectively. Of the 16 tested genes, U18 would not be a reliable reference gene as it had the lowest and the most variable expression levels among all the samples. Additionally, ACT-2 would not be a reliable reference gene because its expression level varied greatly among different treatments and across different generations.
Generally speaking, a good reference gene should have an expression level that is in the similar range relative to the targeted genes (Cappelli et al., 2008). Although 18S rRNA had a relatively stable expression level, it might not be considered as a suitable reference gene because its expression is too high. Thus, simple statistical criteria based solely on numerical values may mask genomic context. More measures should be taken into account when selecting the top reference gene(s) from the candidate list for particular experimental settings. With this in mind, we took advantage of five previously established statistical approaches (geNorm, NormFinder, BestKeeper, comparative ΔCt method, and comprehensive ranking) to evaluate each individual reference gene candidate. This facilitated the final determination of more reliable reference genes for qRT-PCR normalization in C. elegans across three generations after parental nicotine exposure.
Reference gene ranking based on geNorm
GeNorm ranks the reference genes based on the stability value (M value). The lower the M-value, the more stable the gene. Figure 2 clearly shows that CDC-42 and Y45F10D.4 were the most stable genes among the reference gene candidates with the least M-value of 0.198. ARP-6 (0.223), EIF3.C (0.271), and TBA-1 (0.292) had close M-values. The least stable genes were RBD-1 (0.542), U18 (0.603), AMA-1(0.679), Ce234.1 (0.741) and PMP-3 (0.794). The rank of Y45F10D.4 was consistent with previous studies using IIS-mutants, dauers and L3 worms (Hoogewijs et al., 2008) as well as L4 worms treated with copper oxide (Zhang et al., 2012). However, a drastic change in PMP-3 stability index was evident as it was ranked as the least stable gene in our experimental settings. The rank of CDC-42 was consistent with one study (Hoogewijs et al., 2008), but not the other (Zhang et al., 2012).
In order to examine the minimal number of genes required for reliable normalization, the V-value for all the gene pairs was calculated and all were less than the default cutoff value (0.15) (Figure 2). This suggests that the introduction of a new gene was not associated with high variation in the relative expression levels. Thus, taking both indices (M and V-values) together, it can be inferred that CDC-42 and Y45F10D.4 are enough for a reliable normalization (Figure 2).
Reference gene ranking based on NormFinder
Based on NormFinder, TBA-1(0.18), EIF3.C (0.22), ARP-6 (0.27), CDC-42 (0.29), and MDH-2 (0.31) show the lowest stability values (Table 5) and may serve as the top five reliable reference genes. This rank was similar to that of geNorm, although the exact order was not identical. The inclusion of TBA-1, EIF3.C, ARP-6, and CDC-42 among the top-ranked genes was common to both analyses. Previous reports using the same methods placed TBA-1 and EIF3.C among the top five stable genes (Zhang et al., 2012). As for the least stable genes, our results show that ACT-2 (0.71), U18 (0.93), AMA-1 (0.95), Ce234.1 (1.00), and PMP-3(1.04) were ranked last. Interestingly, the lowest four genes were ordered exactly like geNorm as mentioned above. AMA-1 was also found among the least stable with other experimental conditions, but this was not the case for PMP-3 (Zhang et al., 2012).
Table 5.
A summary for the different rankings of the 16 candidate genes derived from 5 methods in response to nicotine exposure in L4 C. elegans.
| Comprehensive ranking | Delta Ct method | BestKeeper | NormFinder | GeNorm | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Stability value | Genes | Mean SD | Gene | SD [Ct] | Gene | coeff. of corr. [r] | Gene | Stability value | Gene | M-value |
| TBA1 | 2.51 | TBA1 | 0.59 | 18s rRNA | 0.40 | Y45F10D.4 | 0.99 | TBA1 | 0.18 | CDC42 | Y45F10D.4 | 0.20 |
| CDC42 | 2.99 | CDC42 | 0.61 | U6 | 0.49 | F35G12.2 | 0.99 | EIF3.C | 0.22 | 3 | 3 |
| EIF3.C | 3.60 | EIF3.C | 0.61 | EIF3.C | 0.69 | TBA1 | 0.98 | ARP6 | 0.27 | ARP6 | 0.22 |
| ARP6 | 4.24 | ARP6 | 0.61 | TBA1 | 0.70 | CDC42 | 0.98 | CDC42 | 0.29 | EIF3.C | 0.27 |
| Y45F10D.4 | 4.36 | Y45F10D.4 | 0.63 | ARP6 | 0.79 | CSQ1 | 0.97 | MDH2 | 0.31 | TBA1 | 0.29 |
| 18s rRNA | 5.03 | MDH2 | 0.64 | CDC42 | 0.83 | ARP6 | 0.97 | Y45F10D.4 | 0.38 | MDH2 | 0.31 |
| U6 | 6.50 | F35G12.2 | 0.66 | MDH2 | 0.84 | EIF3.C | 0.97 | F35G12.2 | 0.41 | F35G12.2 | 0.33 |
| MDH2 | 6.67 | 18s rRNA | 0.73 | Y45F10D.4 | 0.93 | MDH2 | 0.97 | 18s rRNA | 0.42 | CSQ1 | 0.36 |
| F35G12.2 | 8.17 | U6 | 0.73 | F35G12.2 | 0.97 | 18s rRNA | 0.86 | U6 | 0.42 | ACT2 | 0.41 |
| RBD1 | 9.43 | CSQ1 | 0.78 | CSQ1 | 1.11 | U6 | 0.80 | RBD1 | 0.53 | 18s rRNA | 0.47 |
| Ce234.1 | 10.03 | RBD1 | 0.80 | CSQ1 | 0.64 | U6 | 0.51 | ||||
| CSQ1 | 10.72 | ACT2 | 0.85 | ACT2 | 0.71 | RBD1 | 0.54 | ||||
| AMA1 | 10.82 | U18 | 1.06 | U18 | 0.93 | U18 | 0.60 | ||||
| PMP3 | 11.31 | AMA1 | 1.10 | AMA1 | 0.95 | AMA1 | 0.68 | ||||
| ACT2 | 11.61 | Ce234.1 | 1.13 | Ce234.1 | 1.00 | Ce234.1 | 0.74 | ||||
| U18 | 13.69 | PMP3 | 1.16 | PMP3 | 1.04 | PMP3 | 0.79 | ||||
Reference gene ranking based on comparative ΔCt method
Comparative ΔCt method ranked TBA-1(0.595), CDC-42 (0.606), EIF3.C (0.607), ARP-6 (0.614), and Y45F10D.4 (0.631) as the most stable reference genes among the 16 candidate genes (Table 5). Although the order was slightly different, it was similar to the top five genes ranked in geNorm and top four genes ranked in NormFinder. On the other hand, the least stable genes were ACT-2 (0.852), U18 (1.064), AMA-1(1.098), Ce-234.1(1.131), PMP-3(1.162) (Figure 3; Table 4). This results were consistent with results from NormFinder and geNorm. Despite the fact that this method depends on a simpler statistical methodology, it agreed with other sophisticated approaches. Comparing our results with studies that used the ΔCt method, TBA-1, EIF-3 and Y45F10D.4 were also among the more stable genes (Zhang et al., 2012). Also, AMA-1 was of the least reliable genes for normalization, while ARP-6, and CDC-42 were among the least stable in their study (Zhang et al., 2012).
Reference gene ranking based on BestKeeper
BestKeeper calculations depend on two criteria to deduce suitable reference genes. The initial analysis was based on the SD values and ranked 18s rRNA (0.40), U6 (0.49), EIF3.C (0.69), TBA-1(0.70), ARP-6 (0.79) with the least variable expression levels (Table 3). The results obtained from BestKeeper did not completely agree with those obtained from geNorm, NormFinder, and ΔCt method. Despite its relatively stable expression, 18s rRNA had a much higher expression level compared to other genes and it was therefore not a good candidate. However, when considering the index based on pairwise correlation calculations (i.e. r-coefficients), Y45F10D.4 (0.989), F35G12.2 (0.986), TBA-1(0.980), CDC-42 (0.978), and CSQ-1(0.971) were ranked as the best (Table 3). Taking both criteria into consideration, Y45F10D.4 and F35G12.2 had the highest (r-value); however, together with CSQ-1, they had the most variable expression levels among the treatment groups and generations (SDY45F10D.4=0.92, SDF35G12.2=0.97, SDCSQ-1=1.11). As a conclusion, the expression levels of TBA-1(SD=0.70) and CDC-42 (SD=0.83) were relatively stable and highly correlated with the BestKeeper index at P=0.001. This result was consistent with results from geNorm and NormFinder. Additionally, TBA-1 was also among the five most stable genes ranked by BestKeeper in a previous study on L4 worms exposed to nanoparticle treatment (Zhang et al., 2012).
Table 3.
Ranking of most stable reference genes based on BestKeeper.
| Gene | n | GM [CP] | AR [CP] | min [CP] | max [CP] | SD [±CP] | CV [%CP] | [r] | P value | Ranking based on | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SD | [r] | ||||||||||
| ARP6 | 56 | 24.757 | 24.774 | 23.274 | 26.762 | 0.793 | 3.199 | 0.968 | 0.001 | 18s rRNA | Y45F10D.4 |
| 18s rRNA | 56 | 14.133 | 14.141 | 13.015 | 15.129 | 0.397 | 2.810 | 0.857 | 0.001 | U6 | F35G12.2 |
| CDC42 | 56 | 22.999 | 23.019 | 21.163 | 24.883 | 0.833 | 3.619 | 0.978 | 0.001 | EIF3.C | TBA1 |
| CSQ1 | 56 | 22.560 | 22.595 | 20.165 | 24.880 | 1.109 | 4.907 | 0.971 | 0.001 | TBA1 | CDC42 |
| EIF3.C | 56 | 21.632 | 21.648 | 20.135 | 23.661 | 0.687 | 3.172 | 0.968 | 0.001 | ARP6 | CSQ1 |
| F35G12.2 | 56 | 22.373 | 22.400 | 20.478 | 24.173 | 0.971 | 4.336 | 0.986 | 0.001 | CDC42 | ARP6 |
| MDH2 | 56 | 21.496 | 21.518 | 19.560 | 23.585 | 0.838 | 3.893 | 0.966 | 0.001 | MDH2 | EIF3.C |
| TBA1 | 56 | 20.688 | 20.705 | 19.172 | 22.444 | 0.705 | 3.404 | 0.980 | 0.001 | Y45F10D.4 | MDH2 |
| U6 | 56 | 20.731 | 20.740 | 19.168 | 21.887 | 0.490 | 2.362 | 0.799 | 0.001 | F35G12.2 | 18s rRNA |
| Y45F10D.4 | 56 | 23.022 | 23.046 | 21.124 | 25.067 | 0.929 | 4.031 | 0.989 | 0.001 | CSQ1 | U6 |
Comprehensive ranking
Taking advantage of the different angles covered by the four different statistical methods, we used RefFinder software (Xie et al., 2012) that accommodates all the logarithms to finally provide an overall comprehensive ranking for the stability of the sixteen gene targets. As shown in Table 5, TBA-1 (2.51), CDC-42 (2.99), EIF3.C (3.60), ARP-6 (4.24), and Y45F10D.4 (4.36) were the most stable housekeeping genes for reference genes in mutigenerational nicotine-exposed study. TBA-1 and Y45F10D.4 were also among the top five enlisted genes (Zhang et al., 2012). On the other hand, the least stable genes were CSQ-1(10.72), AMA-1(10.82), PMP-3(11.31), ACT-2 (11.61), and U18 (13.69). The stability index for CSQ-1 and AMA-1 was consistent with previous results in response to nanoparticle treatment (Zhang et al., 2012). The radical shift in PMP-3 remained evident in the comprehensive ranking as it was of the least stable genes in our experimental settings.
Discussion
Previous studies involved in choosing reliable reference genes for qRT-PCR normalization have already been conducted in C. elegans (Hoogewijs et al., 2008; Zhang et al., 2012). However, none has evaluated reference genes in multigenerational investigations as a function of environmental condition. Choosing a proper reference gene remains one of the golden rules to increase the sensitivity and credibility of data interpretation. Generally, there are two types of approaches to tackle the issue: the top-bottom model is not restricted to a set of genes and starts with a high-throughput investigation from genome-wide background (e.g. microarray). On the other hand, a bottom–top model starts with a handful of genes with conserved basic roles and hypothesized to be of relatively constant expression levels (Hruz et al., 2011). We were interested in identifying suitable reference genes in C. elegans in response to nicotine. Nicotine is one of the major drugs of abuse with high rates of primary and secondary exposures. Here, we evaluated the expression levels of sixteen housekeeping genes, including four small RNA genes, across multiple generations in response to parental nicotine exposure.
We treated C. elegans hermaphrodites (N2) with two nicotine concentrations from L1 to the beginning of L4 stage. We collected worms at L4 stage from F0, F1, and F2 generations. All the samples from the three treatment groups (control, and nicotine-treated) were used to investigate the expression levels of sixteen selected genes. Based on our results, particularly from the comprehensive ranking, it appears that TBA-1, CDC-42, EIF3.C, ARP-6 and Y45F10D.4 were the most reliable reference genes among the sixteen gene candidates. Based on outputs from the different methodologies, all except for BestKeeper considered TBA-1, CDC-42, EIF3.C, ARP-6 as the most reliable reference genes. When considering results from all methods, including BestKeeper, TBA-1 and CDC-42 would be the most reliable reference genes to study the transgenerational effect of C. elegans exposed to nicotine. Based on results from geNorm, the combination of two reference genes from our list is sufficient for reliable normalization. Thus, we recommend the combination of TBA-1 with any other gene of the top five genes mentioned above. PMP-3, AMA-1, and U18 were the least stable and would not be recommended to be used for normalization.
Our results partially agree with previous studies (Hoogewijs et al., 2008; Zhang et al., 2012) where TBA-1, CDC-42 and Y45F10D.4 were the most reliable reference genes. However, other genes, such as PMP-3, were the most reliable reference gene in other reports (Hoogewijs et al., 2008; Zhang et al., 2012), but were among the least stable genes in our study. This suggests that housekeeping genes are differentially affected in a context-dependent manner and that assessing potential reference genes should precede expression profile analysis.
Although reference genes related studies are not novel, the replication of such a concept using different treatment conditions and developmental conditions is important for future meta-analyses. This allows to test whether an ideal universal reference gene exists or to further confirm the concept of condition-specific reference gene selection.
Acknowledgments
The project described was partially supported by Grant Number R03DA032515 from the National Institute on Drug Abuse. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute on Drug Abuse or the National Institutes of Health.
Footnotes
Conflict of interest: All authors declare no conflict of interests and had no disclosures.
References
- Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, Pintacuda G, Sakaguchi A, Sarkies P, Ahmed S, Miska EA. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitel GJ, Clark SG, Horvitz HR. Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature. 1990;348:503–509. doi: 10.1038/348503a0. [DOI] [PubMed] [Google Scholar]
- Braunschweig M, Jagannathan V, Gutzwiller A, Bee G. Investigations on Transgenerational Epigenetic Response Down the Male Line in F2 Pigs. PloS one. 2012;7:e30583. doi: 10.1371/journal.pone.0030583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelli K, Felicetti M, Capomaccio S, Spinsanti G, Silvestrelli M, Supplizi AV. Exercise induced stress in horses: selection of the most stable reference genes for quantitative RT-PCR normalization. BMC molecular biology. 2008;9:49. doi: 10.1186/1471-2199-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. The Health Consequences of Smoking - Nicotine Addiction: A Report of the Surgeon General, Center for Health Promotion and Education. Office on Smoking and Health; 1988. (DHHS Publication No. (CDC) 88-8406). Retrieved from http://profiles.nlm.nih.gov/NN/B/B/Z/D/ [Google Scholar]
- CDC. The Biology and Behavioral Basis for Smoking-Attributable Disease: A Report of the Surgeon General. Atlanta: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health; 2010. Retrieved from http://www.surgeongeneral.gov/library/reports/tobaccosmoke/full_report.pdf. [Google Scholar]
- Contreras EQ, Cho M, Zhu H, Puppala HL, Escalera G, Zhong W, Colvin VL. Toxicity of quantum dots and cadmium salt to Caenorhabditis elegans after multigenerational exposure. Environmental Science & Technology. 2012 doi: 10.1021/es3036785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginzinger DG. Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Experimental hematology. 2002;30:503–512. doi: 10.1016/s0301-472x(02)00806-8. [DOI] [PubMed] [Google Scholar]
- Git A, Dvinge H, Salmon-Divon M, Osborne M, Kutter C, Hadfield J, Bertone P, Caldas C. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA. 2010;16:991–1006. doi: 10.1261/rna.1947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC molecular biology. 2008;9:9. doi: 10.1186/1471-2199-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Wyss M, Docquier M, Pfaffl MW, Masanetz S, Borghi L, Verbrugghe P, Kalaydjieva L, Bleuler S, Laule O, Descombes P, Gruissem W, Zimmermann P. RefGenes: identification of reliable and condition specific reference genes for RT-qPCR data normalization. BMC Genomics. 2011;12:156. doi: 10.1186/1471-2164-12-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp X, Hammell M, Ow MC, Ambros V. Effect of life history on microRNA expression during C. elegans development. RNA. 2011;17:639–651. doi: 10.1261/rna.2310111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kirienko NV, Mani K, Fay DS. Cancer models in Caenorhabditis elegans. Dev Dyn. 2010;239:1413–1448. doi: 10.1002/dvdy.22247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lant B, Storey KB. An overview of stress response and hypometabolic strategies in Caenorhabditis elegans: conserved and contrasting signals with the mammalian system. International journal of biological sciences. 2010;6:9–50. doi: 10.7150/ijbs.6.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefever S, Hellemans J, Pattyn F, Przybylski DR, Taylor C, Geurts R, Untergasser A, Vandesompele J. RDML: structured language and reporting guidelines for real-time quantitative PCR data. Nucleic Acids Res. 2009;37:2065–2069. doi: 10.1093/nar/gkp056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manikkam M, Guerrero-Bosagna C, Tracey R, Haque MM, Skinner MK. Transgenerational Actions of Environmental Compounds on Reproductive Disease and Identification of Epigenetic Biomarkers of Ancestral Exposures. PloS one. 2012;7:e31901. doi: 10.1371/journal.pone.0031901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matta SG, Balfour DJ, Benowitz NL, Boyd RT, Buccafusco JJ, Caggiula AR, Craig CR, Collins AC, Damaj MI, Donny EC, Gardiner PS, Grady SR, Heberlein U, Leonard SS, Levin ED, Lukas RJ, Markou A, Marks MJ, McCallum SE, Parameswaran N, Perkins KA, Picciotto MR, Quik M, Rose JE, Rothenfluh A, Schafer WR, Stolerman IP, Tyndale RF, Wehner JM, Zirger JM. Guidelines on nicotine dose selection for in vivo research. Psychopharmacology. 2007;190:269–319. doi: 10.1007/s00213-006-0441-0. [DOI] [PubMed] [Google Scholar]
- Pertea M. The Human Transcriptome: An Unfinished Story. Genes. 2012;3:344–360. doi: 10.3390/genes3030344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnology letters. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Pincus Z, Smith-Vikos T, Slack FJ. MicroRNA predictors of longevity in Caenorhabditis elegans. PLoS Genet. 2011;7:e1002306. doi: 10.1371/journal.pgen.1002306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC molecular biology. 2006;7:33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J, Hodgkin J. In: The Nematode Caenorhabditis elegans. Wood WB, editor. New York: Cold Spring Harbor Laboratory Press; 1988. p. 587. [Google Scholar]
- Tominaga N, Kohra S, Iguchi T, Arizonoc K. A Multi-Generation Sublethal Assay of Phenols Using the Nematode Caenorhabditis elegans. J Health Sci. 2003;49:459–463. [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viñuela A, Snoek LB, Riksen JAG, Kammenga JE. Genome-Wide Gene Expression Analysis in Response to Organophosphorus Pesticide Chlorpyrifos and Diazinon in C. elegans. PloS one. 2010;5:e12145. doi: 10.1371/journal.pone.0012145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Global Health Observatory. World Health Organization; 2012. World Health Statistics. [Google Scholar]
- Xie F, Xiao P, Chen D, Xu L, Zhang B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Molecular Biology. 2012;80:75–84. doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- Yu Z, Chen X, Zhang J, Wang R, Yin D. Transgenerational effects of heavy metals on L3 larva of Caenorhabditis elegans with greater behavior and growth inhibitions in the progeny. Ecotoxicology and environmental safety. 2012 doi: 10.1016/j.ecoenv.2012.11.012. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen D, Smith MA, Zhang B, Pan X. Selection of reliable reference genes in Caenorhabditis elegans for analysis of nanotoxicity. PloS one. 2012;7:e31849. doi: 10.1371/journal.pone.0031849. [DOI] [PMC free article] [PubMed] [Google Scholar]