Abstract
Purpose
Killer-cell immunoglobulin-like receptors (KIRs) that regulate natural-killer cells are highly polymorphic. Some KIR2DL1 alleles encode receptors that have stronger signaling function than others. We tested the hypothesis that the clinical outcomes of allogeneic hematopoietic stem-cell transplantation (HSCT) could be affected by donor KIR2DL1 polymorphism.
Patients and Methods
All 313 pediatric patients received allogeneic HSCT at a single institution. Donor KIR2DL1 functional allele typing was retrospectively performed using single nucleotide polymorphism assay.
Results
Patients who received a donor graft containing the functionally stronger KIR2DL1 allele with arginine at amino acid position 245 (KIR2DL1-R245) had better survival (P = .0004) and lower cumulative incidence of disease progression (P = .001) than those patients who received a donor graft that contained only the functionally weaker KIR2DL1 allele with cysteine at the same position (KIR2DL1-C245). The effect of KIR2DL1 allelic polymorphism was similar in patients with acute myeloid leukemia or acute lymphoblastic leukemia among all allele groups (P ≥ .71). Patients who received a KIR2DL1-R245–positive graft with HLA-C receptor-ligand mismatch had the best survival (P = .00003) and lowest risk of leukemia progression (P = .0005) compared with those who received a KIR2DL1-C245 homozygous graft.
Conclusion
Donor KIR2DL1 allelic polymorphism affects recipient outcomes after allogeneic HSCT. These findings have substantial implications for prognostication and donor selection.
INTRODUCTION
Allogeneic hematopoietic stem-cell transplantation (HSCT) is a curative treatment for many malignancies or genetic diseases. Donor selection is a critical element for successful transplantation. In addition to the consideration of donor's health status and HLA matching, routine typing of killer cell immunoglobulin like receptors (KIRs) has recently been proposed.1–3
KIRs are present on the surface of natural killer (NK) cells and a subset of cytotoxic T cells.4 The receptors recognize class I HLA molecules on targets cells to control the effector cell activity. Analogous to T cells, NK cells may facilitate engraftment, combat infection, and control cancer recurrence in HSCT; however, NK cells may reduce rather than increase the risk of graft-versus-host disease (GvHD).5,6 Recent clinical data have confirmed that both inhibitory and activating KIRs are important determinants in the outcome of HSCT in both malignant and nonmalignant disorders.7–9 The KIR genes are encoded on chromosome 19q13 and are highly polymorphic.10–12 Fifteen members (plus two pseudogenes) of the KIR family have been identified in humans thus far. There is substantial variability among people in KIR haplotype content, gene expression, and allele function.
KIR2DL1 is one of the KIR family members known to be important in HSCT and is present in approximately 90% to 95% of healthy individuals.13–15 The KIR2DL1 receptor inhibits NK cell functions as well as educates NK cells for “missing self” recognition of its ligand group 2 HLA-CLys80 (C2).16,17 Based on this biologic principle, some HSCT studies have shown that the presence of donor KIR2DL1 and the absence of recipient C2 ligand (ie, KIR2DL1 receptor-ligand mismatch) were associated with favorable outcomes.2,18–23 Similar findings were observed for the corresponding receptors KIR2DL2 and KIR2DL3 that recognize group 1 HLA-CAsn80 (C1). However, there were conflicting reports from different transplantation centers, in part because prior NK alloreactivity models did not take into account the functional heterogeneity in KIR alleles. Until recently, all KIR alleles of the same family have been considered functionally identical in HSCT analyses.9 Although significant variability in terms of function as well as surface expression among different alleles of the same KIR gene has been reported,24–27 the effect of KIR allelic polymorphism in HSCT remains unknown. We recently reported28 that KIR2DL1 alleles with arginine in position 245 of the transmembrane domain (KIR2DL1-R245) are stronger than those with cysteine in the same position (KIR2DL1-C245). Specifically, KIR2DL1-R245 can recruit more Src-homology-2 domain-containing protein tyrosine phosphatase 2 and beta-arrestin 2, shows higher inhibition of lipid raft polarization at the immune synapse, and has less downregulation of cell-surface expression on interaction with its ligand. Based on this knowledge of the molecular determinants, we developed a high-throughput method to type different KIR2DL1 functional allelic groups.29 In this study, we investigated the influence of donor KIR2DL1-R245 and KIR2DL1-C245 on the outcome of allogeneic HSCT. Furthermore, we examined the effect of receptor-ligand mismatch in the context of KIR2DL1 allelic polymorphism.
PATIENTS AND METHODS
Patients and Donors
KIR2DL1 functional allelotyping was performed using DNA samples from 313 donors for patients who underwent allogeneic HSCT at St Jude Children's Research Hospital over the 10-year period from January 1, 2000, to January 1, 2010. Of the 313 patients, their median age at the time of transplantation was 9.9 years (range, 1 month to 18.6 years). The primary diagnoses included 231 hematologic malignancies (lymphoid, 116 patients; myeloid, 115 patients; total, 74%), 25 solid tumors (embryonal tumors, 19 patients; bone sarcomas, six patients; total, 8%), and 57 nonmalignant diseases (marrow failure, 10 patients; genetic diseases, 47 patients; total, 18%). No patients were excluded based on primary disease type to allow examination of generalizability of our findings. Of the 256 patients with cancer, 143 patients (56%) were in remission and 113 (44%) had persistent disease at the time of transplantation. About half (175 patients; 56%) received a conditioning regimen including total-body irradiation. The majority (76.7%) was myeloablative, defined as total-body irradiation ≥ 5 Gy in single dose or ≥ 8 Gy fractionated; busulfan more than 9 mg/kg; or melphelan more than 150 mg/m2. The grafts were obtained from matched-sibling donors (27%), matched-unrelated donors (31%), or haploidentical donors (41%). General supportive care for transplantation recipients at our institution has been described recently.30 This study was approved by the institutional review board.
Laboratory Assay for KIR and HLA Genotyping
The single-nucleotide polymorphism (SNP) assay for KIR2DL1 functional allele typing was performed on an HT7900 Sequence Detection System (Applied Biosystems, South San Francisco, CA) as described previously.29 The probes were designed based on a single-nucleotide difference at amino acid position 245 in the mature protein. The sequence of the probes was 6FAM-CATCGCTGGTGCTC-MGBNFQ and VIC-CATTGCTGGTGCTCC-MGBNFQ. A universal primer set was designed that could specifically amplify all the alleles of KIR2DL1: forward primer 5′-CTCTTCATCCTCCTCTTCTTTC-3′ and reverse primer 5′-GAAAGTCCTGCCTCTGTGGC-3′. Each reaction mix (total volume, 25 μL) contained 250 nmol/L probes, 100 ng donor's genomic DNA, 5 pM forward and reverse primer, and 1× TaqMan genotyping master mix. The allelic discrimination assay software SDS2.3 (Applied Biosystems) was used to set up the background in the preread program. Then, absolute quantification program SDS2.3 was run to amplify the samples. The polymerase chain reaction steps were stage 1: 2 minutes at 50°C; stage 2: 10 minutes at 95°C; stage 3: 95°C for 15 seconds and 60°C for 1 minute. Stage 3 was repeated for 45 cycles. Finally, the allelic discrimination assay program was used for postreading. HLA typing for HLA-A, -B, -C, and -DRB1 was performed by DNA methods.30,31 KIR genotyping for haplotype A and B assignment was performed using polymerase chain reactions with specific primers as described previously.32
End Points and Statistical Analysis
The χ2 or Fisher's exact test was used to compare baseline variables among various KIR2DL1 allelic groups. Overall survival, progression-free survival (PFS) and grade II to IV GvHD probabilities were estimated using the Kaplan-Meier method. Overall survival was defined as the time from transplantation until death from any cause, censoring those alive at last follow-up. PFS was defined as the time from transplantation until the first event (relapse, progression, or death from any cause), censoring patients alive and progression free at last follow-up. PFS, rather than disease-free survival, was estimated, because 44% of the patients had active disease at the time of HSCT. The cumulative incidences of events were estimated using the method of Kelbfleisch and Prentice. In the estimation of cumulative incidence of disease progression, deaths unrelated to disease were considered competing events. Univariate analyses of survival probabilities were performed using log-rank tests. Cumulative incidences were compared using Gray's tests. The parameters associated with outcomes in univariate analyses at a nominal level of 0.15 were included in multivariate analyses using Cox proportional hazards regression models. The assumption of proportional hazard was confirmed in all analyses.
We used the Memphis receptor-ligand mismatch model to examine the statistical interactions between KIR2DL1 allele polymorphism and its HLA-C ligand effects. HLA-C ligands are dimorphic, as the C2 group is the ligand for KIR2DL1 and the C1 group is the ligand for KIR2DL2/3. If a patient had HLA C1/C1 and the donor was KIR2DL1-positive, or if a patient had HLA C2/C2 and the donor was KIR2DL2/3-positive, then they were classified as receptor-ligand mismatched. In contrast, the donors for patients who were C1/C2 were defined as HLA-C receptor-ligand matched. All the reported P values are two-sided and are considered significant if less than .05. Statistical analyses were performed with SAS software version 9.2 and R version 2.14.0.
RESULTS
Patient and Donor Characteristics
The patient and donor characteristics, stratified by donor KIR2DL1 functional allelic groups, are summarized in Table 1. Among the 313 donors, 215 were homozygous for KIR2DL1-R245 (RR; RR, 68.7%), 22 were homozygous for KIR2DL1-C245 (CC; CC, 7%), and 76 were heterozygous for KIR2DL1-R245/C245 (RC; RC, 24.3%). As expected, KIR2DL1-R245 was associated with centromeric A (Cen A) –containing haplotypes and KIR2DL1-C245 with Cen-B haplotypes (P < .00001); otherwise, there was no statistically significant difference among the three allelic groups in any demographic or transplantation variables (all P > .23).
Table 1.
Patient and Donor Characteristics Stratified by Donor KIR2DL1 Functional Allelic Groups
| Characteristic | Total |
Donor KIR2DL1 Alleles |
P | ||||||
|---|---|---|---|---|---|---|---|---|---|
| RR |
RC |
CC |
|||||||
| No. of Patients | % | No. of Patients | % | No. of Patients | % | No. of Patients | % | ||
| No. of patients | 313 | 215 | 68.7 | 76 | 24.3 | 22 | 7.0 | ||
| Donor KIR haplotype | < .001 | ||||||||
| Cen A/A | 104 | 33.4 | 89 | 41.8 | 15 | 19.7 | 0 | 0 | |
| Cen A/B | 117 | 37.6 | 80 | 37.6 | 30 | 39.5 | 7 | 31.8 | |
| Cen B/B | 90 | 28.9 | 44 | 20.6 | 31 | 40.8 | 15 | 68.2 | |
| Age, years | .68 | ||||||||
| < 10 | 158 | 50.5 | 112 | 52.1 | 35 | 46.1 | 11 | 50 | |
| > 10 | 155 | 49.5 | 103 | 47.9 | 41 | 53.9 | 11 | 50 | |
| Sex | .45 | ||||||||
| Female | 113 | 36.1 | 73 | 34 | 32 | 42.1 | 8 | 36.4 | |
| Male | 200 | 63.9 | 142 | 66 | 44 | 57.9 | 14 | 63.6 | |
| Race | .29 | ||||||||
| White | 198 | 63.3 | 140 | 65.1 | 41 | 53.9 | 17 | 77.3 | |
| Black | 57 | 18.2 | 38 | 17.7 | 17 | 22.4 | 2 | 9.1 | |
| Other | 58 | 18.5 | 37 | 17.2 | 18 | 23.7 | 3 | 13.6 | |
| Primary disease | .44 | ||||||||
| Hematologic malignancies | 231 | 73.8 | 157 | 73 | 54 | 71 | 20 | 90.9 | |
| Solid tumors | 25 | 8 | 18 | 8.4 | 6 | 7.9 | 1 | 4.5 | |
| Nonmalignant diseases | 57 | 18.2 | 40 | 18.6 | 16 | 21.1 | 1 | 4.5 | |
| Acute leukemia | .89 | ||||||||
| ALL | 98 | 54.4 | 67 | 54.5 | 21 | 56.8 | 10 | 50 | |
| AML | 82 | 45.6 | 56 | 45.5 | 16 | 43.2 | 10 | 50 | |
| Malignant disease status | .73 | ||||||||
| CR1 | 77 | 30.1 | 54 | 30.9 | 16 | 26.7 | 7 | 33.3 | |
| CR2 | 52 | 20.3 | 33 | 18.9 | 16 | 26.7 | 3 | 14.3 | |
| CR3/CR4 | 14 | 5.5 | 9 | 5.1 | 5 | 8.3 | 0 | 0 | |
| Refractory/progressive | 113 | 44.1 | 79 | 45.1 | 23 | 38.3 | 11 | 52.4 | |
| Conditioning type | .40 | ||||||||
| TBI containing | 175 | 55.9 | 116 | 54 | 44 | 57.9 | 15 | 68.2 | |
| Non-TBI | 138 | 44.1 | 99 | 46 | 32 | 42.1 | 7 | 31.8 | |
| Conditioning intensity | .93 | ||||||||
| Myeloablative | 240 | 76.7 | 164 | 76.3 | 58 | 76.3 | 18 | 81.8 | |
| Nonmyeloablative | 73 | 23.3 | 51 | 23.7 | 18 | 23.7 | 4 | 18.2 | |
| Donor type | .56 | ||||||||
| Sibling | 86 | 27.5 | 57 | 26.5 | 22 | 29 | 7 | 31.8 | |
| Unrelated | 98 | 31.3 | 73 | 34 | 21 | 27.6 | 4 | 18.2 | |
| Haploidentical | 129 | 41.2 | 85 | 39.5 | 33 | 43.4 | 11 | 50 | |
| T cell depleted | .9 | ||||||||
| Yes | 154 | 49.2 | 105 | 48.8 | 37 | 48.7 | 12 | 54.5 | |
| No | 159 | 50.8 | 110 | 51.2 | 39 | 51.3 | 10 | 45.5 | |
| Donor CMV status | .23 | ||||||||
| Positive | 165 | 52.7 | 108 | 50.2 | 44 | 57.9 | 13 | 59.1 | |
| Negative | 137 | 43.8 | 96 | 44.7 | 32 | 42.1 | 9 | 40.9 | |
| Unknown | 11 | 3.5 | 11 | 5.1 | 0 | 0 | 0 | 0 | |
Abbreviations: ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; CC, KIR2DL1-C245 homozygous; Cen, centromeric; CMV, cytomegalovirus; CR, complete remission; KIR, killer-cell immunoglobulin-like receptors; RC, KIR2DL1-R245/C245 heterozygous; RR, KIR2DL1-R245 homozygous; TBI, total-body irradiation.
Overall Survival
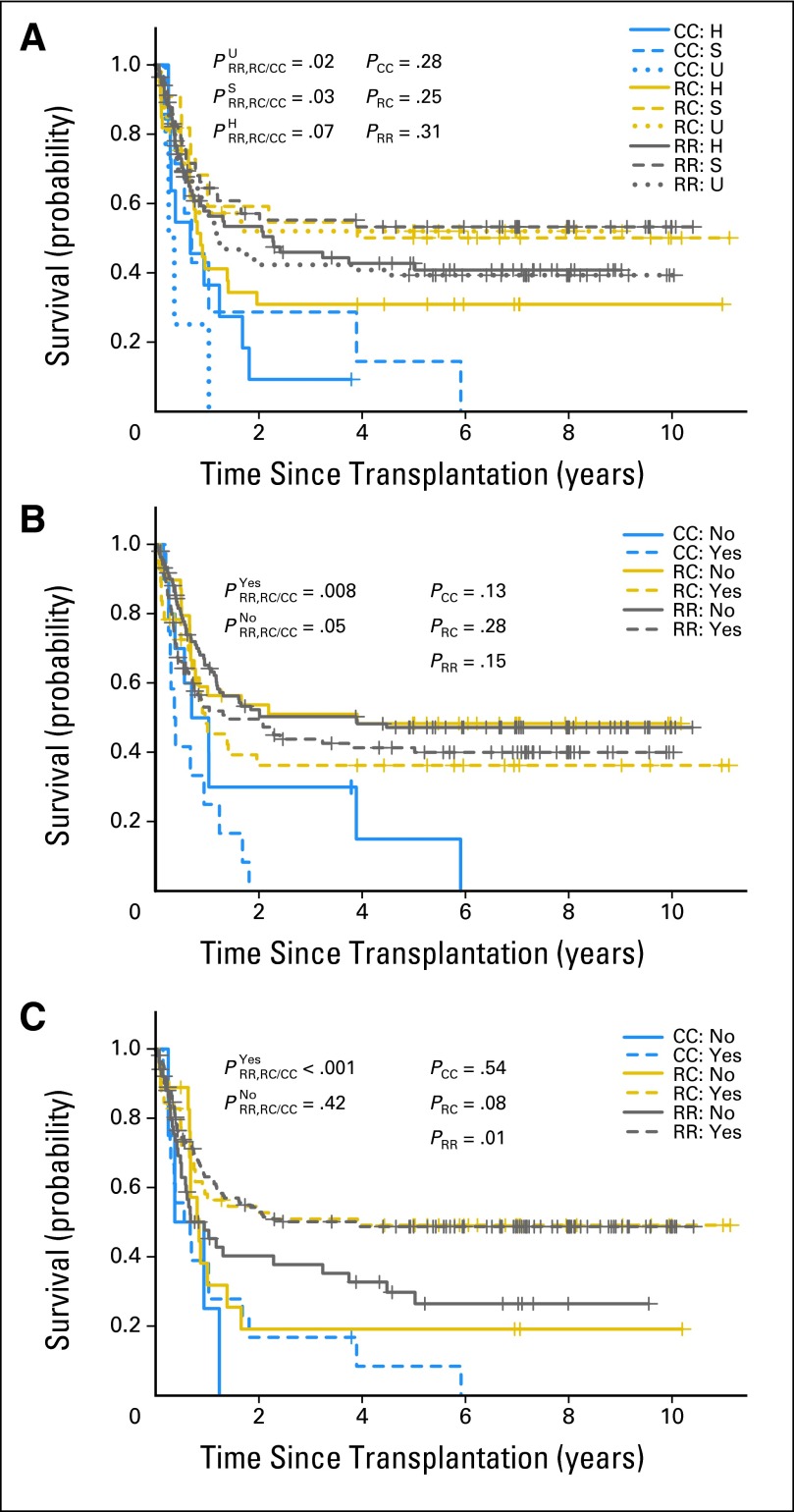
Compared with the patients who received a CC donor graft, the risk of death after HSCT was significantly lower in those who received an RR graft (hazard ratio [HR], 0.4; 95% CI, 0.25 to 0.64; P = .0001) or an RC graft (HR, 0.42; 95% CI, 0.25 to 0.71; P = .0013; Table 2). Thus, the Kaplan-Meier estimates of survival probability in the patients who received an RR or RC graft were significantly higher than the estimates for those with a CC graft (PRR,RC/CC = .0004; Fig 1A). There was no statistical difference in survival in the patients who received an RR graft and those who received an RC graft (PRR/RC =.81; Fig 1A). Among the 256 patients with malignant disorders, the patients who received a CC graft had the worst survival rates (PRR,RC/CC = .001; Fig 1B). When the analysis was further limited to the 231 patients with hematologic malignancies, the survival probability was still significantly higher in patients who received an RR or RC graft than in those who received a CC graft (PRR,RC/CC =.0007; Fig 1C). When compared with the patients with acute myeloid leukemia (AML), patients with acute lymphoblastic leukemia (ALL) had similar survival probabilities among all three donor KIR2DL1 allelic groups (all P ≥ .71; Fig 1D). The effects of donor KIR2DL1 alleles were comparable in the following subsets: sibling, unrelated, and haploidentical HSCT (Appendix Fig A1A; online-only); T-cell depleted and replete grafts (Appendix Fig A1B); and myeloablative HSCT (Appendix Fig A1C). The difference in nonmyeoablative HSCT was not significant statistically (PRR,RC/CC = .42).
Table 2.
Univariate Analysis of Mortality, PFS, and Acute GvHD
| Variable | Mortality |
PFS |
Grade II-IV Acute GvHD |
||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | |
| Donor KIR2DL1 | .0004 | .0013 | .62 | ||||||
| CC | 1 | 1 | 1 | ||||||
| RC | 0.42 | 0.25 to 0.71 | .0013 | 0.48 | 0.28 to 0.82 | .0075 | 1.12 | 0.46 to 2.75 | .8 |
| RR | 0.4 | 0.25 to 0.64 | .0001 | 0.42 | 0.26 to 0.68 | .0003 | 0.89 | 0.38 to 2.06 | .78 |
| Donor KIR haplotype | .92 | .34 | .16 | ||||||
| Cen A/A | 1 | 1 | 1 | ||||||
| Cen A/B | 1.03 | 0.72 to 1.47 | .87 | 1.11 | 0.76 to 1.61 | .59 | 1.28 | 0.75 to 2.2 | .36 |
| Cen B/B | 1.08 | 0.74 to 1.56 | .69 | 1.33 | 0.9 to 1.95 | .15 | 1.68 | 0.98 to 2.88 | .057 |
| Age, years | |||||||||
| < 10 | 1 | 1 | 1 | ||||||
| > 10 | 1.33 | 0.99 to 1.8 | .06 | 1.17 | 0.85 to 1.59 | .33 | 1.5 | 0.97 to 2.31 | .067 |
| Sex | |||||||||
| Female | 1 | 1 | 1 | ||||||
| Male | 0.95 | 0.7 to 1.29 | .74 | 1.04 | 0.76 to 1.43 | .79 | 1 | 0.64 to 1.56 | .99 |
| Race | .054 | .11 | .048 | ||||||
| Black | 1 | 1 | 1 | ||||||
| Other | 0.66 | 0.38 to 1.14 | .14 | 0.61 | 0.34 to 1.08 | .092 | 1.18 | 0.51 to 2.74 | .69 |
| White | 1.14 | 0.77 to 1.69 | .51 | 1.01 | 0.67 to 1.52 | .98 | 1.99 | 1.02 to 3.87 | .044 |
| Primary diseases | .0001 | .0064 | |||||||
| Nonmalignant | 1 | 1 | |||||||
| Hematologic malignancy | 2.32 | 1.4 to 3.84 | .0011 | 1 | 1.4 | 0.75 to 2.61 | .29 | ||
| Solid tumors | 4.26 | 2.26 to 8.01 | .0001 | 1.67 | 1.07 to 2.6 | .024 | 3.27 | 1.46 to 7.31 | .0039 |
| Hematologic malignancies | |||||||||
| ALL | 1 | 1 | 1 | ||||||
| AML | 1.04 | 0.71 to 1.52 | .85 | 1.07 | 0.73 to 1.56 | .72 | 1.17 | 0.66 to 2.06 | .59 |
| Malignant diseases status | .7 | .079 | .39 | ||||||
| CR1 | 1 | 1 | 1 | ||||||
| CR2 | 1.31 | 0.82 to 2.11 | .26 | 1.38 | 0.87 to 2.21 | .17 | 0.64 | 0.3 to 1.41 | .27 |
| CR3/CR4 | 1.09 | 0.51 to 2.35 | .82 | 1.32 | 0.64 to 2.74 | .45 | 0.55 | 0.13 to 2.34 | .42 |
| Refractory/progressive | 1.2 | 0.82 to 1.76 | .35 | 1.67 | 1.13 to 2.46 | .01 | 1.1 | 0.66 to 1.83 | .7 |
| Conditioning type | |||||||||
| Non-TBI | 1 | 1 | 1 | ||||||
| TBI based | 1.06 | 0.78 to 1.43 | .7 | 0.78 | 0.57 to 1.07 | .13 | 0.89 | 0.58 to 1.37 | .59 |
| Conditioning intensity | |||||||||
| Nonmyeloablative | 1 | 1 | 1 | ||||||
| Myeloablative | 0.61 | 0.44 to 0.85 | .0034 | 0.57 | 0.41 to 0.8 | .001 | 0.52 | 0.33 to 0.82 | .0052 |
| Donor type | .19 | .26 | .61 | ||||||
| Haploidentical | 1 | 1 | 1 | ||||||
| Sibling | 0.71 | 0.49 to 1.03 | .069 | 0.75 | 0.51 to 1.1 | .14 | 0.9 | 0.53 to 1.55 | .71 |
| Unrelated | 0.9 | 0.64 to 1.27 | .56 | 0.79 | 0.55 to 1.14 | .21 | 1.19 | 0.72 to 1.95 | .5 |
| T cell depleted | |||||||||
| No | 1 | 1 | 1 | ||||||
| Yes | 1.38 | 1.02 to 1.86 | .034 | 1.29 | 0.95 to 1.76 | .1 | 1.18 | 0.77 to 1.81 | .45 |
| Donor CMV status | .19 | .57 | .23 | ||||||
| Negative | 1 | 1 | 1 | ||||||
| Positive | 1.26 | 0.93 to 1.71 | .13 | 1.17 | 0.85 to 1.6 | .33 | 1.45 | 0.92 to 2.28 | .11 |
| Unknown | 0.72 | 0.29 to 1.78 | .48 | 0.88 | 0.32 to 2.42 | .81 | 1.71 | 0.6 to 4.85 | .31 |
Abbreviations: ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; CC, KIR2DL1-C245 homozygous; Cen, centromeric; CMV, cytomegalovirus; GvHD, graft-versus-host disease; HR, hazard ratio; KIR, killer-cell immunoglobulin-like receptors; PFS, progression-free survival; RC, KIR2DL1-R245/C245 heterozygous; RR, KIR2DL1-R245 homozygous; TBI, total-body irradiation.
Fig 1.
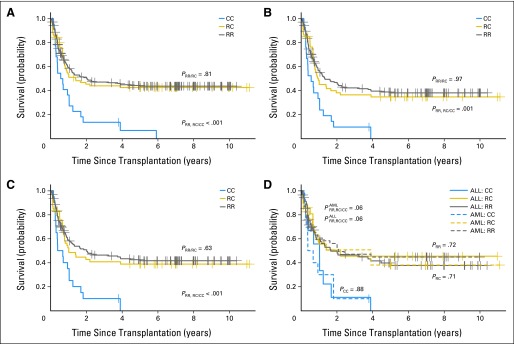
Survival of patients stratified by donor KIR2DL1 functional allelic groups. Shown are the overall survival of (A) the entire cohort of 313 transplantation recipients, (B) the 256 patients with malignant disease, (C) the 231 patients with a hematologic malignancy, and (D) the patients with acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML) displayed separately after receiving a KIR2DL1-R245 homozygous (RR), KIR2DL1-R245/C245 heterozygous (RC), or KIR2DL1-C245 homozygous (CC) graft. PRR/RC, P value between RR and RC group; PRR,RC/CC, P value comparing RR and RC groups with CC group; ALL:RR, patients with ALL who received an RR donor graft; ALL:RC, patients with ALL who received an RC donor graft; ALL:CC, patients with ALL who received a CC donor graft; AML:RR, patients with AML who received an RR donor graft; AML:RC, patients with AML who received an RC donor graft; AML:CC, patients with AML who received a CC donor graft; P×RR,RC/CC, P value when comparing RR and RC groups with CC group for patients with the diagnosis of AML or ALL as indicated; PRR, PRC, and PCC, P values when comparing ALL and AML patients who received a similar RR, RC, or CC donor graft.
Other than donor KIR2DL1 polymorphism, we found that primary disease (P = .0001), T-cell depletion (P = .034), and conditioning intensity (P = .0034) were also statistically significant in association with patient survival in univariate analysis (Table 2). After adjusting for all these non-KIR2DL1 factors, we found that survival was still significantly higher in patients receiving an RR (HR, 0.4; 95% CI, 0.25 to 0.64; P = .0001) or RC graft (HR, 0.44; 95% CI, 0.26 to 0.74; P = .0024) than in those who received a CC graft (Table 3).
Table 3.
Multivariate Analysis of Mortality, PFS, and Acute GvHD
| Variable | Mortality |
PFS |
Grade II-IV Acute GvHD |
||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | |
| Donor KIR2DL1 | |||||||||
| CC | 1 | 1 | 1 | ||||||
| RC | 0.44 | 0.26 to 0.74 | .0024 | 0.45 | 0.26 to 0.78 | .0049 | 0.59 | 0.23 to 1.49 | .26 |
| RR | 0.4 | 0.25 to 0.64 | .0001 | 0.38 | 0.23 to 0.63 | .0001 | 0.46 | 0.19 to 1.09 | .08 |
| Age, years | |||||||||
| < 10 | 1 | 1 | |||||||
| > 10 | 1.2 | 0.88 to 1.63 | .24 | 1.54 | 0.98 to 2.42 | .06 | |||
| Race | |||||||||
| Black | 1 | 1 | 1 | ||||||
| Other | 0.66 | 0.38 to 1.15 | .14 | 0.63 | 0.35 to 1.15 | .13 | 0.92 | 0.39 to 2.2 | .85 |
| White | 1.05 | 0.7 to 1.57 | .81 | 1.07 | 0.69 to 1.66 | .75 | 1.48 | 0.75 to 2.92 | .26 |
| Primary diseases | |||||||||
| Nonmalignant | 1 | 1 | |||||||
| Hematologic malignancy | 1.94 | 1.15 to 3.27 | .013 | 1 | 1.54 | 0.8 to 2.96 | .2 | ||
| Solid tumors | 3.55 | 1.83 to 6.89 | .0001 | 1.49 | 0.87 to 2.54 | .15 | 2.98 | 1.25 to 7.13 | .01 |
| Malignant diseases status | |||||||||
| CR1 | 1 | ||||||||
| CR2 | 1.29 | 0.8 to 2.07 | .3 | ||||||
| CR3/CR4 | 1.31 | 0.62 to 2.76 | .48 | ||||||
| Refractory/progressive | 1.34 | 0.88 to 2.02 | .17 | ||||||
| Conditioning intensity | |||||||||
| Nonmyeloablative | 1 | 1 | 1 | ||||||
| Myeloablative | 0.7 | 0.49 to 1.0 | .049 | 0.67 | 0.46 to 0.99 | .046 | 0.44 | 0.27 to 0.72 | .001 |
| T cell depleted | |||||||||
| No | 1 | 1 | |||||||
| Yes | 1.45 | 1.05 to 1.99 | .023 | 1.3 | 0.92 to 1.85 | .14 | |||
Abbreviations: CC, KIR2DL1-C245 homozygous; GvHD, graft-versus-host disease; HR, hazard ratio; PFS, progression-free survival; RC, KIR2DL1-R245/C245 heterozygous; RR, KIR2DL1-R245 homozygous.
Progression-Free Survival
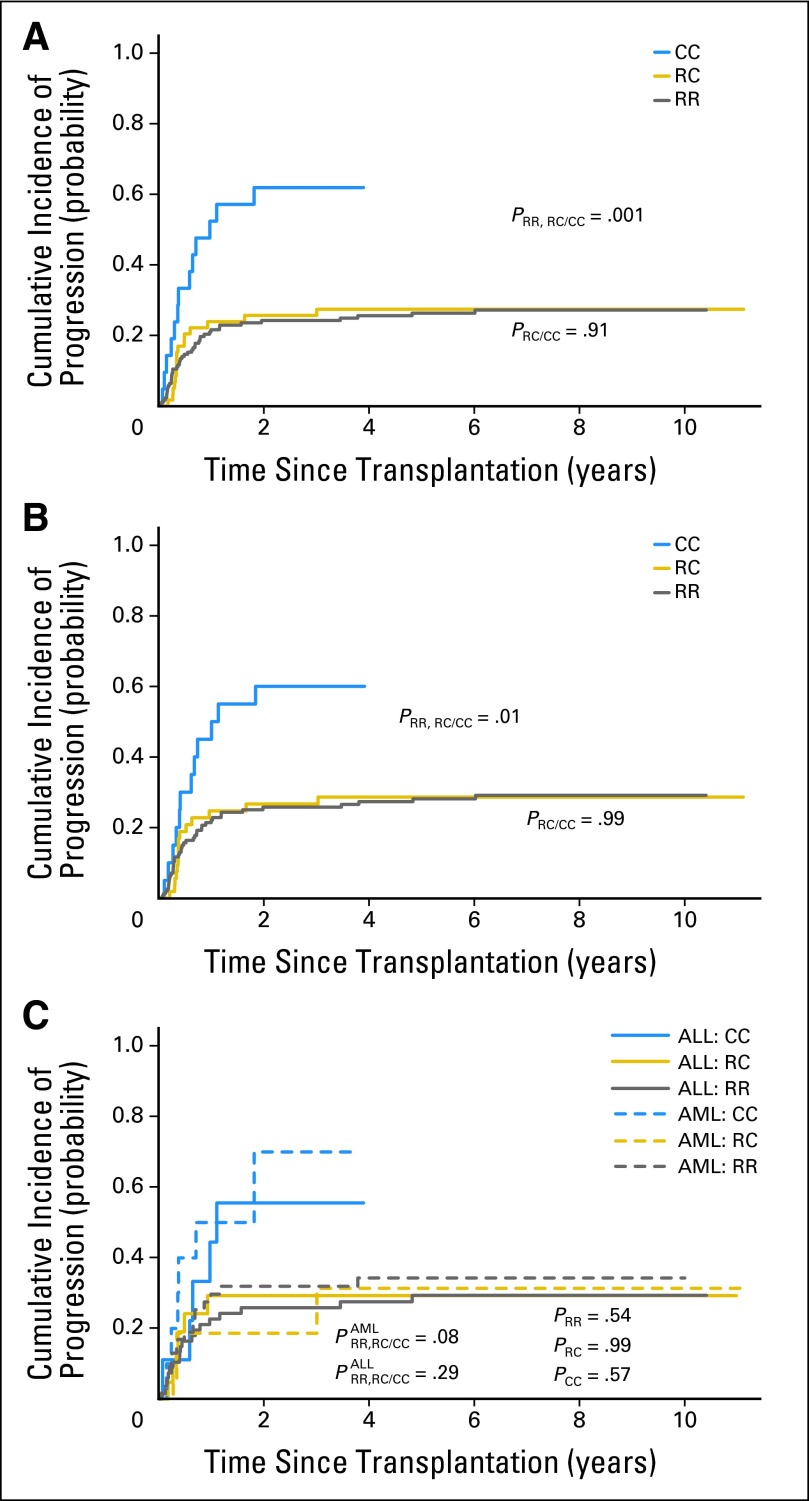
In univariate analysis, the PFS in the 256 patients with cancer was significantly higher in those with an RR (HR, 0.42; 95% CI, 0.26 to 0.68; P = .0003) or RC donor (HR, 0.48; 95% CI, 0.28 to 0.82; P = .0075) than in those with a CC donor (Table 2). There was no statistical difference in the cumulative incidence of progression between the patients who received an RR graft or an RC graft (Fig 2A). When the analysis was limited to the 231 patients with hematologic malignancies, the difference in progression rate was still significant statistically (PRR,RC/CC =.01; Fig 2B). There was no significant difference in the risk of disease progression between AML and ALL patients among all three donor KIR2DL1 allelic groups (all P ≥ .54; Fig 2C). After adjustment for other risk factors, the PFS remained significantly higher in patients with an RR (P = .0001) or RC donor (P = .0049) than in those with a CC donor (Table 3).
Fig 2.
Cumulative incidence of disease progression after transplantations stratified by donor KIR2DL1 functional allelic groups. Shown are the cumulative incidence (CI) of disease progression or relapse in (A) 256 patients with malignancy, (B) 231 patients with hematologic malignancy, and (C) patients with acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML) displayed separately after receiving a KIR2DL1-R245 homozygous (RR), KIR2DL1-R245/C245 heterozygous (RC), or KIR2DL1-C245 homozygous (CC) graft. Deaths unrelated to disease were considered competing events. ALL:RR, patients with ALL who received an RR donor graft; ALL:RC, patients with ALL who received an RC donor graft; ALL:CC, patients with ALL who received a CC donor graft; AML:RR, patients with AML who received an RR donor graft; AML:RC, patients with AML who received an RC donor graft; AML:CC, patients with AML who received a CC donor graft; PRR,RC/CC, P value when comparing RR and RC groups with CC group; PRR/RC, P value between the RR and RC groups; PRR, PRC, and PCC indicate the P values when comparing ALL and AML patients who received a similar RR, RC, or CC donor graft.
GvHD
There was no significant correlation between grade II to IV acute GvHD in the transplantation recipients and the donor KIR2DL1 polymorphism (PRR/CC =.78; PRC/CC =.8; Table 2). Of the patients who received an RR graft, 25.1% had grade II to IV GvHD. The corresponding rates were 31.6% in the RC and 27.3% in the CC groups.
Effects of Receptor-Ligand Mismatch
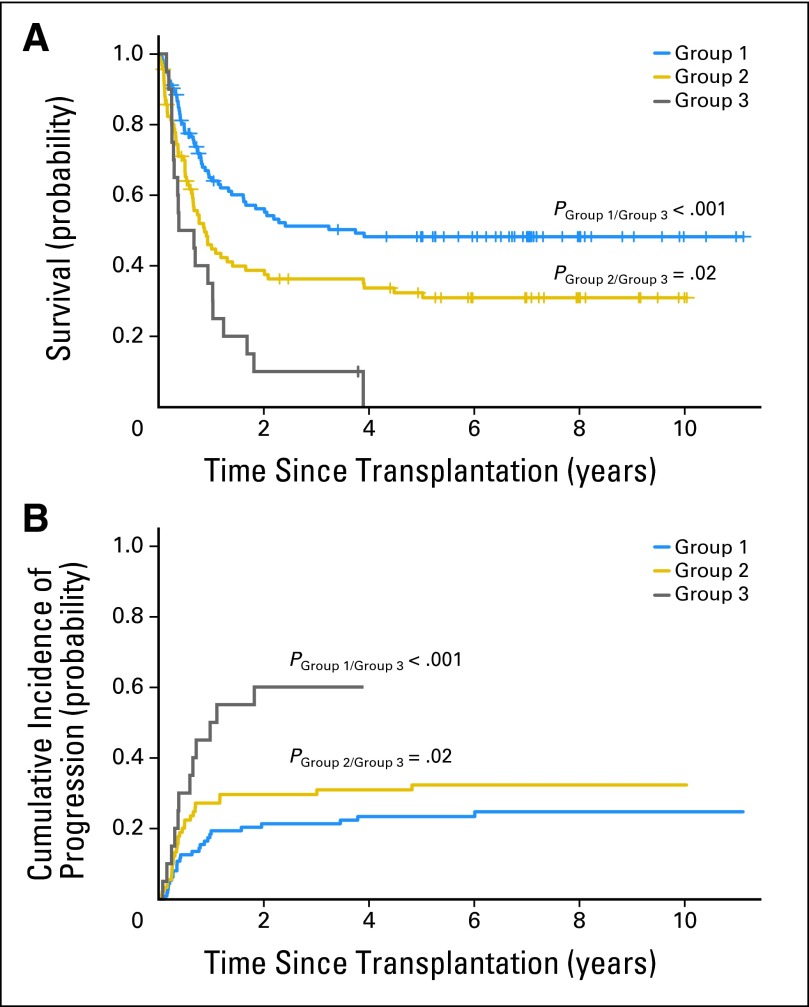
We then examined the effect of HLA-C receptor-ligand mismatch in the context of our KIR2DL1 polymorphism analysis. Among the 231 patients with hematologic malignancies, those with a donor who was RR or RC in KIR2DL1 and was HLA-C receptor-ligand mismatched had the best 5-year survival rates (48%) when compared with those with a donor who was RR or RC in KIR2DL1 but was not HLA-C receptor-ligand mismatched (5-year survival rate, 31%) or those with a CC donor with or without receptor-ligand mismatch (5-year survival rate, 0%; Fig 3A). The cumulative incidence of diseases progression was the lowest in the group with donors who were RR or RC in KIR2DL1 and were HLA-C receptor-ligand mismatched (Fig 3B).
Fig 3.
Survival and cumulative incidence (CI) of disease progression stratified by donor KIR2DL1 allelic groups and the presence of HLA-C receptor-ligand mismatch. Shown are the (A) overall survival probability and (B) CI of disease progression or relapse among different groups of patients with hematologic malignancy. Group 1, patients with a KIR2DL1-R245 homozygous (RR) or KIR2DL1-R245/C245 heterozygous (RC) donor and HLA-C receptor-ligand mismatch; group 2, patients with a KIR2DL1 RR or RC donor but not HLA-C receptor-ligand mismatched; group 3, patients who received a KIR2DL1-C245 homozygous (CC) graft with or without HLA-C receptor-ligand mismatch. Pgroup 1/group 3, P value between group 1 and group 3; Pgroup 2/group 3, P value between group 2 and group 3.
DISCUSSION
We found that patients who received an HSCT graft from donors with KIR2DL1-R245 had better overall survival and progression-free survival than those who received HSCT from donors with KIR2DL1-C245 alleles only. The outcome of HSCT was also dependent on other known factors, including primary disease, disease status, conditioning, and T-cell depletion. After adjusting for these prognostic factors, our analysis showed that the probability of patient survival was still much higher if the donor was KIR2DL1-R245-positive. The effect of donor KIR2DL1 alleles seemed to be comparable in various subset analyses in Figure 1 and Appendix Figure A1; however, the numbers of patients in some subsets were small and similar investigations in other patient cohorts will be necessary to assess the generalizability of our findings. Previous studies, for example, have shown that KIR effect on relapse was only observed in adult patients with AML but not with ALL.7 Several pediatric studies, however, did not observe such a distinction between AML and ALL.18,23,33–35 In our study, the effect of KIR2DL1 allelic polymorphism on the patterns of survival estimates was similar in patients with childhood AML or ALL among all allele groups.
The better survival rates and cancer control in patients receiving grafts from donors with KIR2DL1-R245 alleles may be related to the strength of KIR signaling. We have recently shown that KIR2DL1-R245 alleles are functionally stronger than KIR2DL1-C245 alleles.28 It has been documented in several studies that NK cells lacking inhibitory receptor-mediated NK cell licensing or arming are hyporesponsive against target cells.16,36 In keeping with these observations, we previously found that NK cells expressing KIR2DL1-C245 have significantly less degranulation and interferon gamma (IFN-γ) secretion than those with R245-containing alleles after coculture with identical target cells.28,29 Yawata et al37 demonstrated that enhancement of IFN-γ response to missing-self mediated by HLA-C2 in normal donors was less in KIR2DL1*004 (KIR2DL1-C245) NK cells than in those with other KIR alleles. Taken together, these studies suggested that the functionally stronger KIR2DL1-R245 alleles are apparently more potent than the weaker KIR2DL1-C245 alleles in licensing NK cells, resulting in better cancer control and patient survival after HSCT.
KIR2DL1 is present in the centromeric motif of typical group A KIR haplotypes but in only approximately half of the group B haplotypes.38,39 An alternative or complementary explanation of our findings is the association of KIR2DL1-R245 with the Cen-A–containing haplotypes and KIR2DL1-C245 with the Cen-B haplotypes. For instance, the common alleles KIR2DL1*003 and KIR2DL1*002 in Cen-A haplotypes are KIR2DL1-R245.38–40 Our findings of uniformly poor outcomes with KIR2DL1-C245 homozygous donors, however, cannot be explained entirely by the association with the Cen-B KIR haplotypes, because some Cen-B–associated KIR2DL1 alleles are KIR2DL1-C245 (eg, KIR2DL1*004 and *007), whereas others are KIR2DL1-R245 (eg, KIR2DL1*010).41 The frequency of these two groups of Cen-B–associated KIR2DL1 alleles was similar (approximately 12%) in our previous study.28 Furthermore, some KIR2DL1-C245 alleles (eg, KIR2DL1*006) are not associated with particular Cen-A or Cen-B regions.41 In some previous studies, donor Cen-B was a favorable factor in HSCTs,1,42,43 whereas others observed a worse outcome.44–46 In our study, Cen-A/B haplotypes were not associated statistically with overall survival or PFS (all P ≥ .15).
The level of transcripts and the proportion of NK cells expressing KIR2DL1 are dependent on the gene copy number.27,47 Thus, we hypothesized that recipients with an RR donor might have better outcome than those with an RC donor. Although transplantations from an RC donor was associated with a 15% higher risk in PFS analyses compared with those from an RR donor (HR, 1.15; 95% CI, 0.79 to 1.64; P = .48), the difference was not statistically significant, in part because the effect size was relatively small (compared with a 2.5-fold difference in HR between RR and CC), and the number of HSCTs in this study is limited. Nevertheless, analyses of larger cohorts of patients are warranted, as NK cells from RC donors have an intermediate cytolytic phenotype in vitro when compared with RR or CC homozygous NK cells.29
Leukemic relapse and patient survival after HSCT can also be predicted by the mismatches between donor inhibitory KIRs and recipient ligands.7–9 Based on the Memphis receptor-ligand model,9,18 patients who do not have a ligand for one or more inhibitory KIRs of the donor are predicted to have a low relapse rate and favorable survival probability. Our new findings support this model and show that receptor-ligand mismatch is also prognostic in the context of KIR2DL1 functional allele polymorphism. Patients with HLA-C receptor-ligand mismatch had better survival rates and leukemic control than those without a mismatch after receiving a donor graft with the strong KIR2DL1-R245 functional allele. In contrast, patients who received a donor graft with the weak KIR2DL1-C245 allele had poor survival, regardless of the presence or absence of receptor-ligand mismatch, in agreement with our prior laboratory finding that KIR2DL1-C245 NK cells are hyporeactive even in the setting of missing ligands.28,29
In the field of donor HLA typing, we have witnessed the progression from low-resolution serology typing, to intermediate-resolution antigen typing, and finally to high-resolution allele typing. The advance in donor KIR typing is reminiscent of that in HLA typing. KIR-mediated alloreactivity was first predicted based on the donor HLA ligand repertoire.5,13 KIR genotyping was then used to directly measure the donor gene content.18,48 A KIR expression assay was subsequently implemented to select the donor with the largest number of receptor-ligand mismatched NK cells.49 Now, based on the findings of our study, we propose to include functional allele typing in the selection of HSCT donors. This extension should not slow down donor work-up, because HLA and KIR typing can be performed using the same blood sample collected from potential donors during routine HLA matching. Functional KIR allele typing and KIR ligand typing can be performed simultaneously and can be completed in one day using the SNP assays, which are much cheaper and faster than existing high-resolution typing.29 Importantly, this high-throughput SNP assay provides readouts that are informative in predicting NK cell activity and patient outcome in HSCT. Because the amount of DNA required is small (100 ng), KIR allele and ligand typing should also be feasible before or after cord blood units are banked.
Acknowledgment
We thank David Galloway for scientific editing.
Appendix
Fig A1.
Overall survival stratified by donor KIR2DL1 functional allelic groups. (A) Survival of patients who received a graft from a haploidentical (H), sibling (S), or unrelated (U) donor. (B) Survival of patients who received a T cell depleted (yes) or replete (no) graft. (C) Survival of patients who received a myeloablative (yes) or nonmyeloablative (no) conditioning. CC, KIR2DL1-C245 homozygous graft; PRR, PRC, and PCC, P values when comparing patients who received a similar RR, RC, or CC donor graft, respectively; P×RR, RC/CC, P value when comparing the RR and RC groups with the CC group in the corresponding subset; RC, KIR2DL1-R245/C245 heterozygous graft; KIR2DL1-R245 homozygous graft.
Footnotes
See accompanying editorial on page 3742
Supported in part by research Grant No. CA-21765 from the National Institutes of Health, the Assisi Foundation of Memphis, and the American Lebanese Syrian Associated Charities.
R.B. and W.L. hold the patent for the single nucleotide polymorphism assay used for killer-cell immunoglobulin-like receptors allele typing.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Rafijul Bari, Guolian Kang, Wing Leung
Collection and assembly of data: Rafijul Bari, Erin Sullivan, Victoria Turner, Kwan Gan
Data analysis and interpretation: Rafijul Bari, Piya Rujkijyanont, Guolian Kang, Wing Leung
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Cooley S, Weisdorf DJ, Guethlein LA, et al. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood. 2010;116:2411–2419. doi: 10.1182/blood-2010-05-283051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen DF, Prasad VK, Broadwater G, et al. Differential impact of inhibitory and activating Killer Ig-Like Receptors (KIR) on high-risk patients with myeloid and lymphoid malignancies undergoing reduced intensity transplantation from haploidentical related donors. Bone Marrow Transplant. 2012;47:817–823. doi: 10.1038/bmt.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venstrom JM, Pittari G, Gooley TA, et al. HLA-C-dependent prevention of leukemia relapse by donor activating KIR2DS1. N Engl J Med. 2012;367:805–816. doi: 10.1056/NEJMoa1200503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malmberg KJ, Michaëlsson J, Parham P, et al. Killer cell immunoglobulin-like receptor workshop: Insights into evolution, genetics, function, and translation. Immunity. 2011;35:653–657. doi: 10.1016/j.immuni.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Ruggeri L, Capanni M, Urbani E, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295:2097–2100. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]
- 6.Olson JA, Leveson-Gower DB, Gill S, et al. NK cells mediate reduction of GVHD by inhibiting activated, alloreactive T cells while retaining GVT effects. Blood. 2010;115:4293–4301. doi: 10.1182/blood-2009-05-222190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Velardi A, Ruggeri L, Mancusi A. Killer-cell immunoglobulin-like receptors reactivity and outcome of stem cell transplant. Curr Opin Hematol. 2012;19:319–323. doi: 10.1097/MOH.0b013e32835423c3. [DOI] [PubMed] [Google Scholar]
- 8.Moretta L, Locatelli F, Pende D, et al. Killer Ig-like receptor-mediated control of natural killer cell alloreactivity in haploidentical hematopoietic stem cell transplantation. Blood. 2011;117:764–771. doi: 10.1182/blood-2010-08-264085. [DOI] [PubMed] [Google Scholar]
- 9.Leung W. Use of NK cell activity in cure by transplant. Br J Haematol. 2011;155:14–29. doi: 10.1111/j.1365-2141.2011.08823.x. [DOI] [PubMed] [Google Scholar]
- 10.Hsu KC, Chida S, Geraghty DE, et al. The killer cell immunoglobulin-like receptor (KIR) genomic region: Gene-order, haplotypes and allelic polymorphism. Immunol Rev. 2002;190:40–52. doi: 10.1034/j.1600-065x.2002.19004.x. [DOI] [PubMed] [Google Scholar]
- 11.Parham P, Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat Rev Immunol. 2013;13:133–144. doi: 10.1038/nri3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Single RM, Martin MP, Gao X, et al. Global diversity and evidence for coevolution of KIR and HLA. Nat Genet. 2007;39:1114–1119. doi: 10.1038/ng2077. [DOI] [PubMed] [Google Scholar]
- 13.Ruggeri L, Capanni M, Casucci M, et al. Role of natural killer cell alloreactivity in HLA-mismatched hematopoietic stem cell transplantation. Blood. 1999;94:333–339. [PubMed] [Google Scholar]
- 14.Giebel S, Locatelli F, Wojnar J, et al. Homozygosity for human leucocyte antigen-C ligands of KIR2DL1 is associated with increased risk of relapse after human leucocyte antigen-C-matched unrelated donor haematopoietic stem cell transplantation. Br J Haematol. 2005;131:483–486. doi: 10.1111/j.1365-2141.2005.05797.x. [DOI] [PubMed] [Google Scholar]
- 15.Hou LH, Steiner NK, Chen M, et al. KIR2DL1 allelic diversity: Four new alleles characterized in a bone marrow transplant population and three families. Tissue Antigens. 2007;69:250–254. doi: 10.1111/j.1399-0039.2006.00793.x. [DOI] [PubMed] [Google Scholar]
- 16.Anfossi N, André P, Guia S, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 17.Parham P. Taking license with natural killer cell maturation and repertoire development. Immunol Rev. 2006;214:155–160. doi: 10.1111/j.1600-065X.2006.00462.x. [DOI] [PubMed] [Google Scholar]
- 18.Leung W, Iyengar R, Turner V, et al. Determinants of antileukemia effects of allogeneic NK cells. J Immunol. 2004;172:644–650. doi: 10.4049/jimmunol.172.1.644. [DOI] [PubMed] [Google Scholar]
- 19.Hsu KC, Keever-Taylor CA, Wilton A, et al. Improved outcome in HLA-identical sibling hematopoietic stem-cell transplantation for acute myelogenous leukemia predicted by KIR and HLA genotypes. Blood. 2005;105:4878–4884. doi: 10.1182/blood-2004-12-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller JS, Cooley S, Parham P, et al. Missing KIR ligands are associated with less relapse and increased graft-versus-host disease (GVHD) following unrelated donor allogeneic HCT. Blood. 2007;109:5058–5061. doi: 10.1182/blood-2007-01-065383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu J, Venstrom JM, Liu XR, et al. Breaking tolerance to self, circulating natural killer cells expressing inhibitory KIR for non-self HLA exhibit effector function after T cell-depleted allogeneic hematopoietic cell transplantation. Blood. 2009;113:3875–3884. doi: 10.1182/blood-2008-09-177055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu KC, Gooley T, Malkki M, et al. KIR ligands and prediction of relapse after unrelated donor hematopoietic cell transplantation for hematologic malignancy. Biol Blood Marrow Transplant. 2006;12:828–836. doi: 10.1016/j.bbmt.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 23.Scquizzato E, Zambello R, Teramo A, et al. KIR/HLA-I mismatching and risk of relapse in paediatric patients undergoing non-haploidentical allogeneic haematopoietic stem cell transplantation. Pediatr Transplant. 2011;15:198–204. doi: 10.1111/j.1399-3046.2010.01447.x. [DOI] [PubMed] [Google Scholar]
- 24.Pando MJ, Gardiner CM, Gleimer M, et al. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J Immunol. 2003;171:6640–6649. doi: 10.4049/jimmunol.171.12.6640. [DOI] [PubMed] [Google Scholar]
- 25.Steiner NK, Dakshanamurthy S, VandenBussche CJ, et al. Extracellular domain alterations impact surface expression of stimulatory natural killer cell receptor KIR2DS5. Immunogenetics. 2008;60:655–667. doi: 10.1007/s00251-008-0322-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodridge JP, Lathbury LJ, Steiner NK, et al. Three common alleles of KIR2DL4 (CD158d) encode constitutively expressed, inducible and secreted receptors in NK cells. Eur J Immunol. 2007;37:199–211. doi: 10.1002/eji.200636316. [DOI] [PubMed] [Google Scholar]
- 27.Yawata M, Yawata N, Draghi M, et al. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bari R, Bell T, Leung WH, et al. Significant functional heterogeneity among KIR2DL1 alleles and a pivotal role of arginine 245. Blood. 2009;114:5182–5190. doi: 10.1182/blood-2009-07-231977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bari R, Leung M, Turner VE, et al. Molecular determinant-based typing of KIR alleles and KIR ligands. Clin Immunol. 2011;138:274–281. doi: 10.1016/j.clim.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leung W, Campana D, Yang J, et al. High success rate of hematopoietic cell transplantation regardless of donor source in children with very high-risk leukemia. Blood. 2011;118:223–230. doi: 10.1182/blood-2011-01-333070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leung WH, Turner V, Richardson SL, et al. Effect of HLA class I or class II incompatibility in pediatric marrow transplantation from unrelated and related donors. Hum Immunol. 2001;62:399–407. doi: 10.1016/s0198-8859(01)00220-8. [DOI] [PubMed] [Google Scholar]
- 32.Vilches C, Castaño J, Gómez-Lozano N, et al. Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens. 2007;70:415–422. doi: 10.1111/j.1399-0039.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 33.Pende D, Marcenaro S, Falco M, et al. Anti-leukemia activity of alloreactive NK cells in KIR ligand-mismatched haploidentical HSCT for pediatric patients: Evaluation of the functional role of activating KIR and redefinition of inhibitory KIR specificity. Blood. 2009;113:3119–3129. doi: 10.1182/blood-2008-06-164103. [DOI] [PubMed] [Google Scholar]
- 34.Triplett B, Handgretinger R, Pui CH, et al. KIR-incompatible hematopoietic-cell transplantation for poor prognosis infant acute lymphoblastic leukemia. Blood. 2006;107:1238–1239. doi: 10.1182/blood-2005-07-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernardo M, Zecca M, Giorgiani G, et al. Donor NK alloreactivity improves the outcome of children with haematological malignancies given transplantation of T-cell depleted peripheral blood haematopoietic stem cells from an HLA-disparate family donor. Bone Marrow Transplant. 2011;46(suppl):S16. abstr 0172. [Google Scholar]
- 36.Kim S, Poursine-Laurent J, Truscott SM, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 37.Yawata M, Yawata N, Draghi M, et al. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112:2369–2380. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pyo CW, Guethlein LA, Vu Q, et al. Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PLoS One. 2010;5:e15115. doi: 10.1371/journal.pone.0015115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parham P, Norman PJ, Abi-Rached L, et al. Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules. Philos Trans R Soc Lond B Biol Sci. 2012;367:800–811. doi: 10.1098/rstb.2011.0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meenagh A, Gonzalez A, Sleator C, et al. Investigation of killer cell immunoglobulin-like receptor gene diversity, KIR2DL1 and KIR2DS1. Tissue Antigens. 2008;72:383–391. doi: 10.1111/j.1399-0039.2008.01093.x. [DOI] [PubMed] [Google Scholar]
- 41.Chazara O, Xiong S, Moffett A. Maternal KIR and fetal HLA-C: A fine balance. J Leukoc Biol. 2011;90:703–716. doi: 10.1189/jlb.0511227. [DOI] [PubMed] [Google Scholar]
- 42.Verheyden S, Schots R, Duquet W, et al. A defined donor activating natural killer cell receptor genotype protects against leukemic relapse after related HLA-identical hematopoietic stem cell transplantation. Leukemia. 2005;19:1446–1451. doi: 10.1038/sj.leu.2403839. [DOI] [PubMed] [Google Scholar]
- 43.Cooley S, Trachtenberg E, Bergemann TL, et al. Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood. 2009;113:726–732. doi: 10.1182/blood-2008-07-171926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kröger N, Binder T, Zabelina T, et al. Low number of donor activating killer immunoglobulin-like receptors (KIR) genes but not KIR-ligand mismatch prevents relapse and improves disease-free survival in leukemia patients after in vivo T-cell depleted unrelated stem cell transplantation. Transplantation. 2006;82:1024–1030. doi: 10.1097/01.tp.0000235859.24513.43. [DOI] [PubMed] [Google Scholar]
- 45.Giebel S, Nowak I, Wojnar J, et al. Impact of activating killer immunoglobulin-like receptor genotype on outcome of unrelated donor-hematopoietic cell transplantation. Transplant Proc. 2006;38:287–291. doi: 10.1016/j.transproceed.2005.11.091. [DOI] [PubMed] [Google Scholar]
- 46.McQueen KL, Dorighi KM, Guethlein LA, et al. Donor-recipient combinations of group A and B KIR haplotypes and HLA class I ligand affect the outcome of HLA-matched, sibling donor hematopoietic cell transplantation. Hum Immunol. 2007;68:309–323. doi: 10.1016/j.humimm.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McErlean C, Gonzalez AA, Cunningham R, et al. Differential RNA expression of KIR alleles. Immunogenetics. 2010;62:431–440. doi: 10.1007/s00251-010-0449-9. [DOI] [PubMed] [Google Scholar]
- 48.Gagne K, Brizard G, Gueglio B, et al. Relevance of KIR gene polymorphisms in bone marrow transplantation outcome. Hum Immunol. 2002;63:271–280. doi: 10.1016/s0198-8859(02)00373-7. [DOI] [PubMed] [Google Scholar]
- 49.Leung W, Iyengar R, Triplett B, et al. Comparison of killer Ig-like receptor genotyping and phenotyping for selection of allogeneic blood stem cell donors. J Immunol. 2005;174:6540–6545. doi: 10.4049/jimmunol.174.10.6540. [DOI] [PubMed] [Google Scholar]