Abstract
Brain-derived neurotrophic factor (BDNF) is a critical activity-dependent modulator of gene expression, which can regulate both transcription and translation. Several functions of BDNF, including the induction of dendrite outgrowth and long-term synaptic plasticity, are known to depend, in particular, upon the ability of BDNF to regulate protein synthesis. Although BDNF modestly increases total neuronal protein synthesis, substantial evidence indicates that BDNF induces the translation of only a small subset of expressed mRNAs and demonstrates an extraordinary degree of transcript specificity. The mechanism by which BDNF selectively upregulates the translation of only a discrete group of mRNAs is of intrinsic importance to its trophic function in promoting neuronal growth and plasticity, and is the focus of this review.
1. Requirement for BDNF-regulated protein synthesis in synaptic plasticity, learning and memory
The neurotrophin BDNF is widely expressed in both the developing and mature mammalian brain where it serves as a crucial regulator of neuronal survival, growth, and activity-dependent synaptic plasticity. Many of the well-known pro-growth functions of BDNF rely upon its ability to enhance the production of ensembles of proteins that support neuronal growth and excitatory synaptic function. This review focuses on the physiological function of BDNF-regulated protein synthesis and how BDNF achieves the gene target specificity for pro-growth mRNAs that are required for enhanced synaptic function and plasticity. Multiple lines of evidence indicate that novel protein synthesis is required for long-lasting forms of synaptic plasticity associated with learning and memory. The importance of BDNF in supporting synaptic plasticity and memory formation stems, at least in part, from its capacity to regulate protein synthesis. BDNF has been shown to increase total cellular translation by signaling through its tropomyosin-related kinase B (TrkB) receptor leading to the activation of the PLCγ P13K, and the MAPK pathways. Multiple forms of BDNF-mediated plasticity depend upon the regulation of protein synthesis by BDNF. The enhancement of dendritic arborization in response to BDNF has been shown to require the regulation of translation (Jaworski et al., 2005). Likewise, BDNF-dependent structural plasticity of dendritic spines, specifically spine head enlargement, was also shown to depend upon protein synthesis in rat brain slices (Tanaka et al., 2008). Additional protein-synthesis dependent effects of BDNF include enhanced abundance of GluA1 association with the postsynaptic plasma membrane (Caldeira et al., 2007). BDNF also upregulates both the abundance and plasma membrane-association of NMDA receptor subunits NR1, NR2A, and NR2B in a protein synthesis-dependent manner, and this was shown to correlate with an increase in NMDA receptor activity (Caldeira et al., 2007). Multiple reports support a critical role for BDNF-regulated protein synthesis in in-vitro assays of long-term use-dependent synaptic plasticity. BDNF can enhance the induction of early phase LTP (E-LTP) and late-phase LTP (L-LTP) in hippocampal slices, and BDNF-mediated stimulation of de novo protein synthesis has been reported to be essential for the maintenance of L-LTP (Korte et al., 1995; Kang & Schuman, 1996; Patterson et al., 1996; Messaoudi et al., 2007; Tanaka et al., 2008). These findings are consistent with in-vivo studies showing that secretion of BDNF is crucial for the persistence of long-term memory storage in the hippocampus (Bekinschtein et al., 2007).
2. Effects of BDNF on global protein synthesis
BDNF has been shown to enhance total neuronal protein synthesis, as assessed by metabolic labeling studies, through activation of P13K-mTOR and MAPK cascades in both primary cortical neurons (Takei et al., 2001), as well as in isolated synaptic preparations (Takei et al., 2004). The association of eukaryotic initiation factor 4E (eIF4E) with the mRNA 5′ cap is the rate-limiting step for the induction of cap-dependent translation initiation. This association can be increased by phosphorylation of eIF4E as well as by phosphorylation of the eIF4E binding protein (eIF4E-BP), which releases eIF4E to participate in the mRNA translation complex; BDNF has been shown to promote translation initiation by enhancing both of these processes. Infusion of BDNF into the hippocampal dentate gyrus was shown to increase cap-dependent initiation through induction of MAPK-mediated eIF4E phosphorylation(Kanhema et al., 2006). Enhanced eIF4E phosphorylation as well as mTOR-mediated phosphorylation of eIF4E-BP were shown to mediate increased translation initiation by BDNF in cortical neurons (Takei et al., 2001). BDNF may also modulate translation at the step of elongation. BDNF was found to elicit a rapid and transient increase in eEF2 phosphorylation leading to arrest of elongation in the dentate gyrus but not in synaptoneurosomes, suggesting that regulation of elongation by BDNF could be restricted to non-synaptic sites (Kanhema et al., 2006). Phosphorylation of eEF2 and enhanced elongation via the mTOR cascade by BDNF has also been observed in cultured cortical neurons (Inamura et al., 2005). Contrasting evidence for BDNF-induced regulation of eEF2 phosphorylation suggests that the control of elongation by BDNF could be compartment-specific.
The capacity of BDNF to produce locally enhanced translation initiation through effects in neuronal processes and at synapses has been demonstrated by multiple approaches. BDNF increases phosphorylation of 4EBP1 via the mTOR pathway in neuronal dendrites (Takei et al., 2004; Kanhema et al., 2006). BDNF also enhances the phosphorylation of p70S6K in isolated synapses, which may promote the translation of 5′ terminal oligopyrimidine tract (TOP)-containing transcripts, which include mRNAs for ribosomal proteins and elongation factors (Takei et al., 2004). Additionally BDNF can stimulate translation initiation by causing the disassociation of eIF4E from the repressing CYFIP1-mRNA binding protein in isolated synapses (synaptoneurosomes)(Napoli et al., 2008). In non-neuronal systems, the subcellular redistribution of translation initiation and elongation factors, often in a cytoskeleton-dependent manner, has been revealed as a mechanism for the regulation of protein synthesis. Similarly, BDNF has been found to enhance the translocation of eIF4E into dendritic spines and to increase the association of eIF4E with synaptic mRNA granules through a mechanism blocked by actin disruption(Smart et al., 2003).
3. BDNF regulates the translation of a subset of neuronal mRNAs
While the capacity of BDNF to modestly enhance global cellular translation has been readily appreciated, accumulated evidence has also revealed that only select proteins are increased in response to BDNF. The current consensus is that the BDNF-mediated increase in total translation, which can be measured by approaches such as metabolic label incorporation, derives from the enhanced translation of a minority of available transcripts. Multiple investigations examining small numbers of transcripts suggested that BDNF could induce the translation of plasticity-related proteins (such as CaMKIIα, Staufen, Arc, Homer2, NR1, GluA1) while leaving the levels of other proteins unaffected (Aakalu et al., 2001; Yin et al., 2002; Ying et al., 2002; Kelleher et al., 2004; Takei et al., 2004; Kanhema et al., 2006; Jourdi et al., 2009). High-throughput approaches examining gene target selectivity on a more global scale have helped to elucidate the truly impressive extent of gene target specificity in BDNF-regulated protein synthesis. 2D electrophoresis of radiolabeled proteins from isolated synapses demonstrated that only specific proteins were enhanced by BDNF, while most proteins show no change, and a subset of proteins were decreased in response to BDNF (Yin et al., 2002). In 2004, Schratt and colleagues provided compelling evidence for the transcript selectivity of BDNF-induced protein synthesis using polysome profiling. In this study conducted in cortical neurons, BDNF was shown to induce a transcription-independent recruitment of a specific subset of mRNAs, less than 4 % of the total expressed transcripts, to polysomes (Schratt et al., 2004). This selective regulation was sensitive to inhibition of mTOR. Intriguingly, BDNF not only upregulates translation of a discrete group of mRNAs, but in some cases BDNF mediates the transcription-independent downregulation of specific transcripts and their corresponding proteins, such as the potassium co-transporter KCC2 and potassium channel Kv1.1 (Rivera et al., 2002; Raab-Graham et al., 2006). Using multidimensional protein identification technology (MudPIT) to analyze several thousand proteins, the selective downregulation of significant numbers of proteins by BDNF, in addition to upregulated proteins, was also observed in isolated cortical synapses (synaptoneurosomes) (Liao et al., 2007). Components of the translational machinery, including translation factors, ribonucleoproteins, ribosomal proteins, as well as proteins known to regulate synaptic function and dendritic spine morphology were among the protein classes upregulated by 30 minutes of BDNF treatment (Liao et al., 2007).
These investigations illustrate not only the effects of BDNF in promoting the synthesis of many proteins that support neuronal growth and synaptic plasticity, but also collectively underscore the high degree of selectivity in the regulation of target transcripts by BDNF. In the context of BDNF’s known roles in the brain, it is perhaps not surprising that this growth factor would need to selectively regulate only certain proteins in order to achieve a net pro-growth or pro-plasticity function. Nonetheless, this striking specificity for both up- and down-regulated gene targets is a feature of BDNF control of protein synthesis that has sometimes been overlooked in favor of the simplified understanding that BDNF enhances total cellular protein synthesis.
4. Post-transcriptional regulatory mechanisms underlying translational specificity of BDNF
4.1 RNA-binding proteins and microRNAs
How does BDNF selectively regulate the translation of only a discrete group of mRNAs? In addition to acting on initiation factors of protein synthesis machinery, BDNF also modulates several more selective post-transcriptional regulators of gene expression. Multiple modes of exerting transcript selectivity in the post-transcriptional regulation of gene expression have been described, including cis-regulatory elements such as internal ribosome entry sites (IRES) and cytoplasmic polyadenylation elements (CPEs), and trans-regulatory factors such as RNA-binding proteins, and microRNAs (miRNAs). RNA-binding proteins have been shown to modulate the synthesis of several plasticity-related proteins that are also targets of BDNF. The Cytoplasmic Polyadenylation Element Binding protein (CPEB) (Huang et al., 2002; Huang et al., 2003) and the Fragile-X Mental Retardation Protein (FMRP) (Vanderklish & Edelman, 2005; Zalfa et al., 2006) function in both localizing and regulating translation of mRNAs. Cytoplasmic polyadenylation, and likewise translation, can be regulated in mRNAs containing sequence-specific binding sites for CPEB (CPE sites). CPEB is phosphorylated in an activity-dependent manner which enhances translation initiation by promoting the recruitment of poly(A) polymerase and causing the dissociation of eIF4E from an inhibitory protein, Maskin. BDNF may promote the activity-dependent polyadenylation of some CPE-containing neuronal RNAs (Wu et al., 1998), and possibly other RNAs regulated by CPEB in an apparently CPE sequence-independent manner(Du & Richter, 2005). The translation of mRNAs interacting with the RNA-binding protein Fragile X mental retardation protein (FMRP) (Brown et al., 2001; Miyashiro et al., 2003) may also undergo BDNF-dependent regulation. BDNF has been shown to downregulate FMR1 mRNA expression in cultured hippocampal neurons as well as to decrease FMRP protein levels in the hippocampus of transgenic mice overexpressing TrkB receptors in vivo (Castren et al., 2002). In addition, BDNF treatment has been reported to post-translationally regulate FMRP by activating calcineurin-mediated FMRP dephosphorylation in hippocampal neurons(Wang et al., 2012). Dephosphorylation of FMRP has been suggested to promote mRNA translation (Ceman et al., 2003; Narayanan et al., 2007). By decreasing FMRP levels and promoting dephosphorylation of FMRP, BDNF may regulate the translation of a subset of plasticity-related genes. BDNF treatment has been shown to elicit an increase in protein synthesis from several mRNAs, CaMKIIα, Arc, Map1B, and APP, that are known to be targets of FMRP(Napoli et al., 2008). FMRP can modulate translation of mRNAs through interaction with the cytoplasmic FMRP interacting protein 1 (CYFIP1), which binds to eIF4E and forms a complex with FMRP-target mRNAs(Napoli et al., 2008). BDNF stimulation in cultured primary hippocampal neurons and cortical synaptoneurosomes was shown to decrease co-immunoprecipitation of CYFIP1 and eIF4E leading to activation of target mRNA translation(Napoli et al., 2008).
Recent evidence indicates that miRNAs may play an important role in the capacity of BDNF to selectively regulate specific mRNA targets. miRNAs are small 22–24 nucleotide non-coding endogenous RNAs that regulate post-transcriptional translational by binding to partially complementary sites in target mRNAs. Perfect complementarity of a miRNA seed sequence (miRNA nucleotides 2–8) for the mRNA has been shown to be a strong predictor of miRNA binding and functional regulation of a given mRNA target (Grimson et al., 2007; Nielsen et al., 2007; Bartel, 2009). Binding of an mRNA by a miRNA can lead to translational repression of the mRNA which may be accompanied by degradation of the target mRNA.
Hundreds of miRNAs are expressed in the mammalian brain and there is strong evidence that brain-specific miRNAs are crucial for neural development and synaptic plasticity(Schratt et al., 2006; Bonev et al., 2011). An initial report implicating miRNAs in the regulation of protein synthesis by BDNF involved the brain-specific miRNA, miR-134. miR-134 was found to negatively regulate dendritic spine size by inhibiting the translation of an mRNA encoding Limk1(Schratt et al., 2006). Limk1 mRNA undergoes enhanced translation in response to BDNF (Schratt et al., 2004), and loss of Limk1 protein produces deficits in spatial learning and hippocampal LTP(Meng et al., 2002). BDNF was shown to relieve translational suppression of Limk1 by miR-134 and to permit increased protein synthesis of synaptic Limk1 and spine growth(Schratt et al., 2006); the mechanism by which BDNF relieved miR-134 dependent suppression was not elucidated. This study highlighted the concept that BDNF could regulate gene expression through altering the function of a specific miRNA to relieve suppression.
While this review focuses on BDNF-mediated post-transcriptional regulation of specificity in protein synthesis, it should be briefly noted that BDNF has also been reported to regulate the transcription of miRNA precursors. A BDNF-mediated increase in pre-miR-132 levels in cultured cortical neurons, for example, was shown to peak 2 – 4 hours after BDNF exposure and could be inhibited by expression of a dominant negative mutant (A-CREB) based on the transcription factor CREB (cAMP response element binding protein), as well as by inhibition of the MAPK pathway(Vo et al., 2005; Wayman et al., 2008; Kawashima et al., 2010). miR-132 has been shown to be necessary and sufficient to promote neurite outgrowth in primary cortical neurons (Vo et al., 2005), as well as regulating dendrite morphogenesis in organotypic hippocampal slices(Wayman et al., 2008). mRNA encoding p250GAP, a member of the Rac/Rho family of GAPs, was identified as one putative target of miR-132 in the regulation of structural plasticity (Wayman et al., 2008).
Additional evidence for reversal of miRNA-mediated silencing in neurons came from Ashraf and colleagues’ demonstration that loss of an RNA-induced silencing complex (RISC) component, Armitage, released translational repression of a synaptic mRNA, CaMKII, in drosophila (Ashraf et al., 2006). MOV10, the mammalian homolog of Armitage, was subsequently shown to undergo activity-dependent proteasomal degradation that leads to relief in translational silencing of several mRNAs (Banerjee et al., 2009). While MOV10 degradation was never explicitly linked to BDNF, these studies drew attention to the concept that reversal of miRNA-mediated silencing could present a mechanism for the regulation of synaptic protein synthesis important for neuronal plasticity and memory formation. MOV10 degradation was found to require NMDA receptor activation, and many studies have positively linked BDNF to NMDA signaling. BDNF increases mRNA and protein levels of NMDA receptor subunits NR1, NR2A, and NR2B(Caldeira et al., 2007), enhances the phosphorylation of NR1 and NR2B in hippocampal and cortical neurons(Lin et al., 1998) and contributes to increased glutamatergic transmission through NMDAR channels(Levine et al., 1998; Levine & Kolb, 2000; Alder et al., 2005). In contrast to modulation of miR-134, the regulation of Armitage or MOV10 might be expected to produce more ‘global’, rather than miRNA-specific, changes in relief of mi-RNA-mediated repression. While the effects of loss of MOV10 were not assayed in a high-throughput manner, a candidate-based screen did reveal multiple candidate mRNAs, including CaMKIIα and Lypla1, that underwent enhanced translation upon loss of MOV10(Banerjee et al., 2009).
4.2 Regulation of miRNA biogenesis
Since the discovery of miRNAs twenty years ago(Lee et al., 1993), nuclear and cytoplasmic steps have been elucidated which are necessary for the processing of miRNA precursors to mature functional miRNAs. Regulatory mechanisms that impact and control the biogenesis of miRNAs at these steps continue to be revealed. Given that miRNAs are predicted to regulate more than 60% of mRNAs, the control of cellular miRNA composition holds significant potential for post-transcriptional gene regulation. Recent work from our laboratory revealed that mRNA transcript selectivity in protein synthesis mediated by BDNF can be achieved through control of miRNA biogenesis (Huang et al., 2012). BDNF rapidly enhanced the protein levels of the RNAse III family endoribonuclease, Dicer, which processes pre-miRNA to mature miRNA duplexes. The rapid elevation of Dicer protein levels by BDNF were shown to occur independently of new transcription. BDNF-mediated enhancement of miRNA processing by Dicer led to a post-transcriptional increase in the levels of many miRNAs. However, BDNF was also observed to decrease production of a select group of miRNAs comprised largely of Let-7 miRNA family members. Investigations from multiple laboratories have elucidated the pathways by which Let-7 miRNAs can be subject to post-transcriptional regulation by Lin28, a conserved RNA-binding protein and pluripotency factor. Lin28 negatively regulates the production of Let-7 miRNAs by binding to “GGAG” motif in the terminal loop of Let-7 precursor miRNAs to block Dicer processing and facilitate degradation of the Let-7 precursor miRNA. BDNF was found to rapidly and post-transcriptionally upregulate Lin28 protein levels in mature hippocampal neurons, mediating a subsequent selective decline in the levels of mature Let-7 miRNA levels (Huang et al., 2012). Collectively, the positive Dicer-mediated and negative Lin28-mediated effects of BDNF on miRNA biogenesis allow BDNF to shape the neuronal miRNA pool and differentially control miRNA-mediated repression of distinct transcripts. Elevated production of mature miRNAs, by the BDNF-induced Dicer elevation, targets a greater number of mRNAs for translational repression and can explain why many mRNAs do not undergo enhanced translation in response to BDNF and how levels of some proteins are decreased in response to BDNF (Rivera et al., 2004; Raab-Graham et al., 2006). In contrast, the simultaneous increase in Lin28 levels by BDNF reduces neuronal levels of Lin28-targeted miRNAs, mainly Let-7 family miRNAs, which permits Let-7 targeted mRNAs to escape translational repression and selectively undergo increased translation in response to BDNF. Lin28 confers specificity to BDNF-regulation of translation and may be viewed as a ‘selector’ molecule that excludes certain pre-miRNAs from being processed to mature miRNAs. In the absence of Lin28, BDNF is unable to upregulate the translation of the many growth-promoting targets that harbor Let-7 miRNA sites (Huang et al., 2012) and that are associated with the important role of BDNF in neuronal plasticity and cognition.
4.3 The Lin28 / Let-7 Axis in post-transcriptional control of pro-growth gene expression programs
There is considerable precedence for a prominent role of Let-7 family miRNAs in the repression of genes controlling growth, proliferation, and pluripotency. In mammals, the cellular abundance of mature Let-7 miRNAs can be controlled, at least in part, in a post-transcriptional manner by Lin28 (Newman et al., 2008; Viswanathan et al., 2008) Lin28 was first discovered in C. elegans as a heterochronic gene and regulator of developmental timing (Ambros & Horvitz, 1984; Moss et al., 1997). Recent evidence from genome-wide association studies (GWAS) indicates that Lin28 may retain its heterochronic function in humans and play a critical role in coordinating growth and development. Genetic variation near and within Lin28 (LIN28B loci) has been correlated with human age at onset of puberty and height (Ong et al., 2009; Perry et al., 2009). Work from the Daley lab further substantiated these findings with transgenic mice overexpressing Lin28a, which exhibited increased body size and delayed onset of puberty. Increased glucose metabolism and insulin sensitivity, phenotypes that recapitulated human developmental traits identified in GWAS studies, were also observed in mice overexpressing Lin28 (Lettre et al., 2008; He et al., 2009; Ong et al., 2009; Sulem et al., 2009; Zhu et al., 2010). Subsequently, Zhu and colleagues showed that Lin28 regulates mammalian glucose metabolism in an mTOR-dependent manner in part through derepression of metabolic genes, including INSR, IGF1R, and IRS2, which are targets of Let-7 miRNAs (Zhu et al., 2011).
While Let-7-independent functions for Lin28 are known, the role of Lin28 in managing growth and metabolism is most well-established through its regulation of the Let-7 family miRNAs that can coordinately repress multiple pro-growth genes. As a consequence of these broad pro-growth effects, Let-7 family miRNAs are often classified as tumor suppressor genes. Lin28, which blocks Let-7 miRNA production, is believed to be overexpressed in up to 15 % of all cancers. Dysregulated reduced expression of Let-7 miRNAs is frequently found in lung cancer cell lines and patient tumor samples, and Let-7 miRNA overexpression has been used to inhibit proliferation of lung cancer cells (Takamizawa et al., 2004). Other cancers exhibiting low levels of Let-7 miRNAs include chronic myelogenous leukemia, B cell lymphoma, breast, colon, esophageal, ovarian, liver, kidney, prostate cancer. Several studies have identified oncogenes including Ras, Myc, and HMGA2, as Let-7 targets providing molecular insight into how aberrant reductions in Let-7 miRNAs could contribute to tumorigenesis(Johnson et al., 2005; Johnson et al., 2007; Shell et al., 2007; Kumar et al., 2008); Yu, 2007 #1166}. In healthy tissue, a major role of the Let-7 miRNA family is in the control of target genes that regulate cellular proliferation and differentiation, as required for proper development. Let-7 miRNA levels are progressively elevated during development, across many species, and function to promote differentiation and represses self-renewal by decreasing the expression of target genes that induce proliferation and inhibit differentiation such as c-Myc, Pax6, Ascl1, and the transcription factors HBL-1 and DAF-12 (Abrahante et al., 2003; Lin et al., 2003; Grosshans et al., 2005; Sampson et al., 2007; Ramachandran et al., 2010) Let-7 miRNA levels are low or absent in a variety of stem cell or progenitor cell populations of normal tissue (Thomson et al., 2004; Wulczyn et al., 2007). High Let-7 expression is detected late in embryonic development and in adult tissues (Sempere et al., 2004; Thomson et al., 2004). It is now well-established that Let-7 functions as a key developmental timing switch in the transition from stem cell to differentiated cell fate across multiple organisms from C. elegans to vertebrates.
Why does the reduction in Let-7 family miRNAs by BDNF have a profound effect on the translation of neuronal mRNAs containing binding sites for Let-7 family miRNAs? This is an interesting consideration, particularly given that miRNAs are sometimes viewed as mediators that serve only to fine-tune gene expression. There are several reasons why targeting Let-7 miRNA levels may be particularly effective in allowing BDNF to promote the synthesis of an ensemble of neuronal proteins with roles in growth and synaptic plasticity. Several high-throughput studies have been conducted to quantitatively assess brain region-specific miRNA expression patterns, and by multiple approaches the Let-7 family of miRNAs appears to be highly abundant in the adult mammalian brain. A miRNA profiling study using deep sequencing (Illumina Genome analyzer) of bilateral rat hippocampal CA3 regions reported that the Let-7 family of miRNAs represent nearly 50% of small RNA sequences(Shinohara et al., 2011). In the adult mouse frontal cortex and hippocampus, quantitative comparative analysis of miRNA expression using methods of both miRNA-Seq (Illumina Genome analyzer) and miRNA microarray (Affymetrix) found that the Let-7 family of miRNAs was collectively by far the most abundant class of miRNAs in both the hippocampus (59%) and frontal cortex (47%)(Juhila et al., 2011). Remarkably, individual members of the Let-7 family represented 7 of the 15 most abundant miRNAs in the hippocampus(Juhila et al., 2011). These studies exemplify why mRNAs containing seed-matched sites for Let-7 miRNAs might be anticipated to be repressed by these miRNAs under basal conditions, and to undergo significantly enhanced translation when a BDNF stimulus leads to relief from miRNA-mediated repression by a substantial and selective decline in the highly abundant Let-7 family miRNAs.
5. Concluding Remarks
While a foundation of previous research has illuminated the mechanisms of bulk regulation of protein synthesis by BDNF, this review has focused on more recent efforts to understand how selectivity of BDNF-induced protein synthesis is achieved. The physiological effects of BDNF include neuronal growth, survival, and plasticity suggesting that BDNF must selectively activate the translation of pro-growth or plasticity-related mRNAs, rather than exert nonspecific effects on gene expression. New insights into the effects of BDNF on post-transcriptional regulators, including RNA-binding proteins and miRNAs, has shed light on mechanisms that contribute to the specificity of BDNF translational control. Future work directed towards the elucidation of the signaling pathways responsible for the regulation of RNA-binding proteins and miRNAs by BDNF will help establish vital regulatory points that may be essential to the physiological impact of BDNF on brain function.
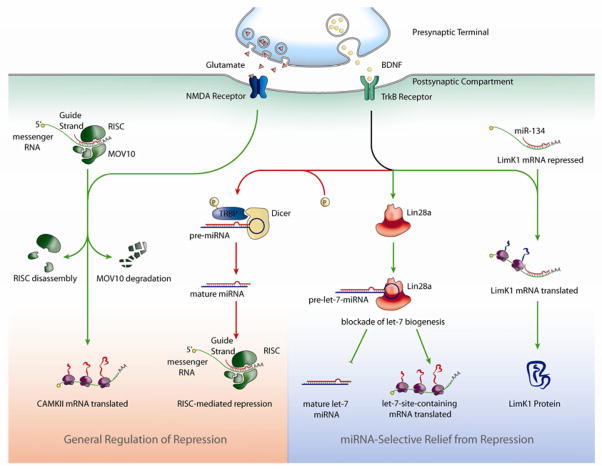
Figure 1. BDNF exerts post-transcriptional control of protein synthesis through regulation of miRNA-dependent repression.
miRNA-mediated repression can be regulated by BDNF to provide general (left panel) or miRNA-selective (right panel) control of translation. miRNA-selective effects of BDNF on the efficacy of repression (miR-134) (Schratt et al., 2006)or abundance (Let-7 family miRNAs; (Huang et al., 2012)) for particular miRNAs, can reduce repression by particular miRNAs and impart transcript specificity to the induction of protein synthesis by BDNF. BDNF stimulation upregulates Lin28, an RNA binding protein that can bind precursors of the Let-7 miRNA family (Let-7 pre-miRNA), preventing them from being processed by the Dicer-TRBP machinery. The resulting diminished levels of mature Let-7 miRNAs relieve repression of mRNAs with Let-7 binding sites and permit their translation. BDNF can also regulate miRNA-mediated repression in a global manner that is not selective for a particular miRNA family (left panel). By facilitating the phosphorylation of TRBP, BDNF stabilizes and enhances levels of the Dicer-TRBP complex that processes pre-miRNA into mature miRNA. This leads to a general rapid increase in miRNA levels and, in turn, increased miRNA-mediated repression of many transcripts. BDNF also modulates NMDA-receptor dependent signaling which has been shown to produce general effects on RISC-dependent mRNA repression. NMDA-dependent signaling induces proteasome-dependent degradation of MOV10, a component of RISC, leading to a loss of RISC function and freeing repressed mRNA to enable translation (as was demonstrated for the CaMKIIα transcript).
Highlights.
BDNF selectively regulates the translation of a subset of target genes
BDNF achieves target specificity by post-transcriptional regulation of translation
Target specificity requires BDNF regulation of miRNA biogenesis and mRNA repression
Acknowledgments
This work was supported by grants from the National Institute of Mental Health (MH098016 to M.K.M. and MH098634 to C.R.R.) and the Braude Foundation (to M.K.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Developmental cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- Alder J, Thakker-Varia S, Crozier RA, Shaheen A, Plummer MR, Black IB. Early presynaptic and late postsynaptic components contribute independently to brain-derived neurotrophic factor-induced synaptic plasticity. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2005;25:3080–3085. doi: 10.1523/JNEUROSCI.2970-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Neveu P, Kosik KS. A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron. 2009;64:871–884. doi: 10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekinschtein P, Cammarota M, Igaz LM, Bevilaqua LR, Izquierdo I, Medina JH. Persistence of long-term memory storage requires a late protein synthesis- and BDNF- dependent phase in the hippocampus. Neuron. 2007;53:261–277. doi: 10.1016/j.neuron.2006.11.025. [DOI] [PubMed] [Google Scholar]
- Bonev B, Pisco A, Papalopulu N. MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Developmental cell. 2011;20:19–32. doi: 10.1016/j.devcel.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Caldeira MV, Melo CV, Pereira DB, Carvalho R, Correia SS, Backos DS, Carvalho AL, Esteban JA, Duarte CB. Brain-derived neurotrophic factor regulates the expression and synaptic delivery of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor subunits in hippocampal neurons. The Journal of biological chemistry. 2007;282:12619–12628. doi: 10.1074/jbc.M700607200. [DOI] [PubMed] [Google Scholar]
- Castren M, Lampinen KE, Miettinen R, Koponen E, Sipola I, Bakker CE, Oostra BA, Castren E. BDNF regulates the expression of fragile X mental retardation protein mRNA in the hippocampus. Neurobiology of disease. 2002;11:221–229. doi: 10.1006/nbdi.2002.0544. [DOI] [PubMed] [Google Scholar]
- Ceman S, O’Donnell WT, Reed M, Patton S, Pohl J, Warren ST. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Human molecular genetics. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- Du L, Richter JD. Activity-dependent polyadenylation in neurons. RNA. 2005;11:1340–1347. doi: 10.1261/rna.2870505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Molecular cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Developmental cell. 2005;8:321–330. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- He C, Kraft P, Chen C, Buring JE, Pare G, Hankinson SE, Chanock SJ, Ridker PM, Hunter DJ, Chasman DI. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nature genetics. 2009;41:724–728. doi: 10.1038/ng.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Carson JH, Barbarese E, Richter JD. Facilitation of dendritic mRNA transport by CPEB. Genes & development. 2003;17:638–653. doi: 10.1101/gad.1053003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Jung MY, Sarkissian M, Richter JD. N-methyl-D-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and alpha CaMKII mRNA polyadenylation at synapses. The EMBO journal. 2002;21:2139–2148. doi: 10.1093/emboj/21.9.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YW, Ruiz CR, Eyler EC, Lin K, Meffert MK. Dual regulation of miRNA biogenesis generates target specificity in neurotrophin-induced protein synthesis. Cell. 2012;148:933–946. doi: 10.1016/j.cell.2012.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamura N, Nawa H, Takei N. Enhancement of translation elongation in neurons by brain-derived neurotrophic factor: implications for mammalian target of rapamycin signaling. J Neurochem. 2005;95:1438–1445. doi: 10.1111/j.1471-4159.2005.03466.x. [DOI] [PubMed] [Google Scholar]
- Jaworski J, Spangler S, Seeburg DP, Hoogenraad CC, Sheng M. Control of dendritic arborization by the phosphoinositide-3′-kinase-Akt-mammalian target of rapamycin pathway. J Neurosci. 2005;25:11300–11312. doi: 10.1523/JNEUROSCI.2270-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, Brown D, Slack FJ. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer research. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Jourdi H, Hsu YT, Zhou M, Qin Q, Bi X, Baudry M. Positive AMPA receptor modulation rapidly stimulates BDNF release and increases dendritic mRNA translation. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2009;29:8688–8697. doi: 10.1523/JNEUROSCI.6078-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhila J, Sipila T, Icay K, Nicorici D, Ellonen P, Kallio A, Korpelainen E, Greco D, Hovatta I. MicroRNA expression profiling reveals miRNA families regulating specific biological pathways in mouse frontal cortex and hippocampus. PLoS ONE. 2011;6:e21495. doi: 10.1371/journal.pone.0021495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- Kanhema T, Dagestad G, Panja D, Tiron A, Messaoudi E, Havik B, Ying SW, Nairn AC, Sonenberg N, Bramham CR. Dual regulation of translation initiation and peptide chain elongation during BDNF-induced LTP in vivo: evidence for compartment-specific translation control. Journal of neurochemistry. 2006;99:1328–1337. doi: 10.1111/j.1471-4159.2006.04158.x. [DOI] [PubMed] [Google Scholar]
- Kawashima H, Numakawa T, Kumamaru E, Adachi N, Mizuno H, Ninomiya M, Kunugi H, Hashido K. Glucocorticoid attenuates brain-derived neurotrophic factor-dependent upregulation of glutamate receptors via the suppression of microRNA-132 expression. Neuroscience. 2010;165:1301–1311. doi: 10.1016/j.neuroscience.2009.11.057. [DOI] [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Govindarajan A, Jung HY, Kang H, Tonegawa S. Translational control by MAPK signaling in long-term synaptic plasticity and memory. Cell. 2004;116:467–479. doi: 10.1016/s0092-8674(04)00115-1. [DOI] [PubMed] [Google Scholar]
- Korte M, Carroll P, Wolf E, Brem G, Thoenen H, Bonhoeffer T. Hippocampal long-term potentiation is impaired in mice lacking brain- derived neurotrophic factor. Proc Natl Acad Sci U S A. 1995;92:8856–8860. doi: 10.1073/pnas.92.19.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, Sanna S, Eyheramendy S, Voight BF, Butler JL, Guiducci C, Illig T, Hackett R, Heid IM, Jacobs KB, Lyssenko V, Uda M, Boehnke M, Chanock SJ, Groop LC, Hu FB, Isomaa B, Kraft P, Peltonen L, Salomaa V, Schlessinger D, Hunter DJ, Hayes RB, Abecasis GR, Wichmann HE, Mohlke KL, Hirschhorn JN. Identification of ten loci associated with height highlights new biological pathways in human growth. Nature genetics. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ES, Crozier RA, Black IB, Plummer MR. Brain-derived neurotrophic factor modulates hippocampal synaptic transmission by increasing N-methyl-D-aspartic acid receptor activity. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:10235–10239. doi: 10.1073/pnas.95.17.10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ES, Kolb JE. Brain-derived neurotrophic factor increases activity of NR2B-containing N-methyl-D-aspartate receptors in excised patches from hippocampal neurons. Journal of neuroscience research. 2000;62:357–362. doi: 10.1002/1097-4547(20001101)62:3<357::AID-JNR5>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Liao L, Pilotte J, Xu T, Wong CC, Edelman GM, Vanderklish P, Yates JR., 3rd BDNF induces widespread changes in synaptic protein content and up-regulates components of the translation machinery: an analysis using high-throughput proteomics. Journal of proteome research. 2007;6:1059–1071. doi: 10.1021/pr060358f. [DOI] [PubMed] [Google Scholar]
- Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Developmental cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- Lin SY, Wu K, Levine ES, Mount HT, Suen PC, Black IB. BDNF acutely increases tyrosine phosphorylation of the NMDA receptor subunit 2B in cortical and hippocampal postsynaptic densities. Brain research. Molecular brain research. 1998;55:20–27. doi: 10.1016/s0169-328x(97)00349-5. [DOI] [PubMed] [Google Scholar]
- Meng Y, Zhang Y, Tregoubov V, Janus C, Cruz L, Jackson M, Lu WY, MacDonald JF, Wang JY, Falls DL, Jia Z. Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron. 2002;35:121–133. doi: 10.1016/s0896-6273(02)00758-4. [DOI] [PubMed] [Google Scholar]
- Messaoudi E, Kanhema T, Soule J, Tiron A, Dagyte G, da Silva B, Bramham CR. Sustained Arc/Arg3.1 synthesis controls long-term potentiation consolidation through regulation of local actin polymerization in the dentate gyrus in vivo. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2007;27:10445–10455. doi: 10.1523/JNEUROSCI.2883-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro KY, Beckel-Mitchener A, Purk TP, Becker KG, Barret T, Liu L, Carbonetto S, Weiler IJ, Greenough WT, Eberwine J. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, Witke W, Costa-Mattioli M, Sonenberg N, Achsel T, Bagni C. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Narayanan U, Nalavadi V, Nakamoto M, Pallas DC, Ceman S, Bassell GJ, Warren ST. FMRP phosphorylation reveals an immediate-early signaling pathway triggered by group I mGluR and mediated by PP2A. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2007;27:14349–14357. doi: 10.1523/JNEUROSCI.2969-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. Rna. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen CB, Shomron N, Sandberg R, Hornstein E, Kitzman J, Burge CB. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–1910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong KK, Elks CE, Li S, Zhao JH, Luan J, Andersen LB, Bingham SA, Brage S, Smith GD, Ekelund U, Gillson CJ, Glaser B, Golding J, Hardy R, Khaw KT, Kuh D, Luben R, Marcus M, McGeehin MA, Ness AR, Northstone K, Ring SM, Rubin C, Sims MA, Song K, Strachan DP, Vollenweider P, Waeber G, Waterworth DM, Wong A, Deloukas P, Barroso I, Mooser V, Loos RJ, Wareham NJ. Genetic variation in LIN28B is associated with the timing of puberty. Nature genetics. 2009;41:729–733. doi: 10.1038/ng.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson SL, Abel T, Deuel TA, Martin KC, Rose JC, Kandel ER. Recombinant BDNF rescues deficits in basal synaptic transmission and hippocampal LTP in BDNF knockout mice. Neuron. 1996;16:1137–1145. doi: 10.1016/s0896-6273(00)80140-3. [DOI] [PubMed] [Google Scholar]
- Perry JR, Stolk L, Franceschini N, Lunetta KL, Zhai G, McArdle PF, Smith AV, Aspelund T, Bandinelli S, Boerwinkle E, Cherkas L, Eiriksdottir G, Estrada K, Ferrucci L, Folsom AR, Garcia M, Gudnason V, Hofman A, Karasik D, Kiel DP, Launer LJ, van Meurs J, Nalls MA, Rivadeneira F, Shuldiner AR, Singleton A, Soranzo N, Tanaka T, Visser JA, Weedon MN, Wilson SG, Zhuang V, Streeten EA, Harris TB, Murray A, Spector TD, Demerath EW, Uitterlinden AG, Murabito JM. Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nature genetics. 2009;41:648–650. doi: 10.1038/ng.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab-Graham KF, Haddick PC, Jan YN, Jan LY. Activity- and mTOR-dependent suppression of Kv1.1 channel mRNA translation in dendrites. Science. 2006;314:144–148. doi: 10.1126/science.1131693. [DOI] [PubMed] [Google Scholar]
- Ramachandran R, Fausett BV, Goldman D. Ascl1a regulates Muller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nature cell biology. 2010;12:1101–1107. doi: 10.1038/ncb2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera C, Li H, Thomas-Crusells J, Lahtinen H, Viitanen T, Nanobashvili A, Kokaia Z, Airaksinen MS, Voipio J, Kaila K, Saarma M. BDNF-induced TrkB activation down-regulates the K+-Cl- cotransporter KCC2 and impairs neuronal Cl- extrusion. The Journal of cell biology. 2002;159:747–752. doi: 10.1083/jcb.200209011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera C, Voipio J, Thomas-Crusells J, Li H, Emri Z, Sipila S, Payne JA, Minichiello L, Saarma M, Kaila K. Mechanism of activity-dependent downregulation of the neuron-specific K-Cl cotransporter KCC2. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2004;24:4683–4691. doi: 10.1523/JNEUROSCI.5265-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer research. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Nigh EA, Chen WG, Hu L, Greenberg ME. BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway during neuronal development. J Neurosci. 2004;24:7366–7377. doi: 10.1523/JNEUROSCI.1739-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome biology. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA, Feig C, Lengyel E, Peter ME. Let-7 expression defines two differentiation stages of cancer. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11400–11405. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara Y, Yahagi K, Kawano M, Nishiyori H, Kawazu C, Suzuki N, Manabe R, Hirase H. miRNA profiling of bilateral rat hippocampal CA3 by deep sequencing. Biochemical and biophysical research communications. 2011;409:293–298. doi: 10.1016/j.bbrc.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Smart FM, Edelman GM, Vanderklish PW. BDNF induces translocation of initiation factor 4E to mRNA granules: evidence for a role of synaptic microfilaments and integrins. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:14403–14408. doi: 10.1073/pnas.2436349100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulem P, Gudbjartsson DF, Rafnar T, Holm H, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Alexandersen P, Feenstra B, Boyd HA, Aben KK, Verbeek AL, Roeleveld N, Jonasdottir A, Styrkarsdottir U, Steinthorsdottir V, Karason A, Stacey SN, Gudmundsson J, Jakobsdottir M, Thorleifsson G, Hardarson G, Gulcher J, Kong A, Kiemeney LA, Melbye M, Christiansen C, Tryggvadottir L, Thorsteinsdottir U, Stefansson K. Genome-wide association study identifies sequence variants on 6q21 associated with age at menarche. Nature genetics. 2009;41:734–738. doi: 10.1038/ng.383. [DOI] [PubMed] [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer research. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Takei N, Inamura N, Kawamura M, Namba H, Hara K, Yonezawa K, Nawa H. Brain-derived neurotrophic factor induces mammalian target of rapamycin-dependent local activation of translation machinery and protein synthesis in neuronal dendrites. J Neurosci. 2004;24:9760–9769. doi: 10.1523/JNEUROSCI.1427-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei N, Kawamura M, Hara K, Yonezawa K, Nawa H. Brain-derived neurotrophic factor enhances neuronal translation by activating multiple initiation processes: comparison with the effects of insulin. J Biol Chem. 2001;276:42818–42825. doi: 10.1074/jbc.M103237200. [DOI] [PubMed] [Google Scholar]
- Tanaka J, Horiike Y, Matsuzaki M, Miyazaki T, Ellis-Davies GC, Kasai H. Protein synthesis and neurotrophin-dependent structural plasticity of single dendritic spines. Science. 2008;319:1683–1687. doi: 10.1126/science.1152864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nature methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- Vanderklish PW, Edelman GM. Differential translation and fragile X syndrome. Genes Brain Behav. 2005;4:360–384. doi: 10.1111/j.1601-183X.2005.00134.x. [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhu JZ, Chang KT, Min KT. DSCR1 interacts with FMRP and is required for spine morphogenesis and local protein synthesis. The EMBO journal. 2012;31:3655–3666. doi: 10.1038/emboj.2012.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Wells D, Tay J, Mendis D, Abbott MA, Barnitt A, Quinlan E, Heynen A, Fallon JR, Richter JD. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of alpha-CaMKII mRNA at synapses. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2007;21:415–426. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- Yin Y, Edelman GM, Vanderklish PW. The brain-derived neurotrophic factor enhances synthesis of Arc in synaptoneurosomes. Proc Natl Acad Sci U S A. 2002;99:2368–2373. doi: 10.1073/pnas.042693699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying SW, Futter M, Rosenblum K, Webber MJ, Hunt SP, Bliss TV, Bramham CR. Brain-derived neurotrophic factor induces long-term potentiation in intact adult hippocampus: requirement for ERK activation coupled to CREB and upregulation of Arc synthesis. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2002;22:1532–1540. doi: 10.1523/JNEUROSCI.22-05-01532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa F, Achsel T, Bagni C. mRNPs, polysomes or granules: FMRP in neuronal protein synthesis. Current opinion in neurobiology. 2006;16:265–269. doi: 10.1016/j.conb.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Zhu H, Shah S, Shyh-Chang N, Shinoda G, Einhorn WS, Viswanathan SR, Takeuchi A, Grasemann C, Rinn JL, Lopez MF, Hirschhorn JN, Palmert MR, Daley GQ. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nature genetics. 2010;42:626–630. doi: 10.1038/ng.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, Urbach A, Thornton JE, Triboulet R, Gregory RI, Altshuler D, Daley GQ. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]