Abstract
The formation of cataracts is associated with the accumulation of protein aggregates in the ocular lens, suggesting that defective protein degradation plays a role in cataract pathogenesis. Accumulation of the p62 protein has recently been identified as a marker for impaired autophagy in a variety of tissues; however, little information exists on its expression in the ocular lens and in cataracts. In the present study we examined the expression of p62 in the mouse lens and compared its expression in wild-type lenses with that in lenses from knock-in mice with an arginine to glycine mutation in αB-crystallin (αB-R120G) that is known to cause human hereditary cataract. Immunohistochemical, immunoblotting, and transmission electron microscopic analyses of wild type and αB-R120G mutant mice were performed. To assess the effect of increased protein aggregation on autophagy, immunohistochemical staining was performed with an anti-p62 antibody, revealing the presence of p62-positive punctate staining in a band of denucleated cortical fiber cells. The number and size of p62 puncta were significantly greater in αB-R120G homozygous mutant lenses than in wild type and heterozygous mutant lenses. p62 staining was also abundant in lens epithelial cells and was concentrated around the nuclear membrane. Double-membraned structures similar to autophagosomes containing cellular cytoplasmic content were detected in lens epithelial cells by transmission electron microscopy. The autophagosomes in lens epithelial cells from αB-R120G homozygous mutant mice were larger than those in wild type mice. Double-membraned structures that are probably autophagosomes were also detected in cortical fiber cells and were more abundant in the αB-R120G homozygous mutant lens than the wild type lens. This study demonstrates p62 distribution as speckles in the lens fiber cells, altered levels of p62 expression, and the presence of autophagosomes in the ocular lens of αB-R120G mutant mice. We propose that autophagy is inhibited in the αB-R120G mutant lenses because of a defect in protein degradation after autophagosome formation. Further work is necessary to determine the relationship between αB-crystallin function, autophagy, and cataract formation.
Keywords: Autophagy, cataracts, αB-crystallin, protein degradation, p62 protein
1. Introduction
Aberrant protein aggregation is a characteristic feature of several age-associated diseases, and protein aggregation has been extensively studied in neurodegenerative diseases, cardiomyopathy, and cataracts. Disorders associated with increased protein aggregation exhibit accumulation of poly-ubiquitinated (polyUb)-positive aggregates, indicating alterations in the protein quality control system during aging and/or disease pathology (Sanbe et al. 2004; Tannous et al. 2008; Tannous et al. 2008). Approaches aimed at decreasing protein aggregate accumulation by enhancing their degradation and clearance are therefore likely to be of significant clinical interest.
Cellular protein aggregation in cataracts has been associated with long-term exposure to UV radiation, stress conditions (such as oxidative stress), and mutations in specific genes (Bloemendal et al. 2004). The formation of light-scattering aggregates in hereditary cataracts is caused by mutations in crystallin genes (Andley et al. 2008). Mouse models for crystallin mutations have recently been employed to study the mechanism of lens protein aggregation and the metabolic pathways that are altered during cataractogenesis (Andley 2006; Andley 2007; Xi et al. 2008; Huang et al. 2009; Andley et al. 2011). The αB-crystallin R120G mutation (αB-R120G) causes desminrelated myopathy (DRM) and cataracts (Vicart et al. 1998) and has also received significant attention in the field of cardiomyopathy (Sanbe et al. 2005). Recently, our laboratory generated a knock-in mouse model to investigate the mechanism of hereditary cataract formation and desmin-related myopathy caused by the R120G αB-crystallin mutation (Andley et al. 2011). This model for hereditary cataract exhibits increased protein aggregation and formation of lens opacities (Andley et al. 2011). Using this model, we demonstrated that the molecular weight of lens α-crystallin increases with age and gene dosage. In addition, αB-crystallin in the knock-in mice becomes increasingly insoluble and associates more strongly with intermediate filament proteins vimentin and desmin in lens and muscle cells, respectively. We also observed a decrease in the grip strength of mutant mice. αB-crystallin is a member of the small heat shock protein family that possesses a chaperone-like function. Several laboratories have shown that αB-R120G crystallin assumes an abnormal conformation in vitro and has reduced chaperone activity (Bova et al. 1999; Perng et al. 1999). Although the increase in protein aggregation in αB-R120G mutant mouse lenses suggests a defect in protein degradation, protein degradation mechanisms have not been studied in this mouse model.
Two protein degradation mechanisms exist in eukaryotic cells: the proteasome, which degrades proteins by the ubiquitin-proteasome pathway, and autophagy, which involves the lysosomal degradation machinery. The ubiquitin-proteasome pathway recognizes and selectively degrades oxidatively damaged proteins, and in the lens this pathway has been shown to recognize selectively truncated αA-crystallin, deamidated βB2-crystallin, and oxidatively-modified proteins (He et al. 2008). Autophagy is a multi-step protein degradation mechanism that is regulated by several ATG proteins and is distinct from the ubiquitin-proteasome pathway. During autophagy, protein substrates targeted for degradation are first packaged into inclusion bodies and engulfed by a double-layered membrane structure known as the autophagosome. The autophagosome moves along microtubules to fuse with lysosomes, and the protein content is degraded by lysosomal proteases (Cao and Klionsky 2008).
Autophagy is critical for survival during periods of increased nutrient requirements, and can also be regulated by non-metabolic factors such as oxidative stress and accumulation of aggregated proteins (Lee et al. 2012). Loss of autophagy function has been suggested as a cause of accumulation of misfolded proteins, cytoplasmic protein aggregation, and even cardiac disease (Rubinsztein 2006; Williams et al. 2006). Enhancement of autophagy by rapamycin improves cardiac function and reduces desmin aggregate formation in LMNA-deficient mice (Ramos et al. 2012). Cardiomyocytes expressing mutant R120G αB-crystallin show reduced autophagic flux, and coexpression of Atg7, a regulator of autophagic function in cardiomyocytes expressing αB-R120G protein, reduces protein aggregate content and cardiomyocyte toxicity (Pattison et al. 2011).
p62 is an ubiquitin-binding scaffold protein that colocalizes with ubiquitinated protein aggregates in many protein aggregation diseases associated with neurodegeneration and myofibrillar myopathies (Kuusisto et al. 2001; Son et al. 2012). Because p62 is specifically degraded by autophagy (Bjorkoy et al. 2005), it accumulates in the cells and tissues of autophagy-deficient mice (Wang et al. 2006; Komatsu et al. 2007; Nakai et al. 2007) and this accumulation of p62 has been used as a marker for inhibition of autophagy and defects in autophagic degradation (Bjorkoy et al. 2005; Mizushima and Yoshimori 2007; Settembre et al. 2008). The p62 protein contains domains essential for interaction with the autophagic machinery and interaction with signaling pathways involved in antioxidant responses, inflammation, and metabolism (Lamark et al. 2003; Wilson et al. 2003). p62 self-oligomerizes via an N-terminal PB1 domain. This domain is also involved in binding of p62 to ERK1 and appears to be required for the autophagic degradation of p62. The p62 protein also interacts with ubiquitinated proteins via the C-terminal UBA domain. It has been proposed that p62 transfers ubiquitinated substrates to the proteasome and functions as a link between ubiquitinated proteins and the autophagic pathway to facilitate lysosomal degradation of these aggregates (Seibenhener et al. 2004; Pankiv et al. 2007). More recently, the accumulation of p62 aggregates has been considered a specific marker of autophagic flux inhibition at any point beyond autophagosome formation (Klionsky et al. 2012).
The p62 protein can directly bind microtubule-associated protein light chain 3 (LC3) via a specific C-terminal sequence domain (Pankiv et al. 2007). LC3 is an ubiquitin-like protein that exists in two forms, non-lipidated LC3-I and phosphatidyl ethanolamine-lipidated form LC3-II (Klionsky et al. 2008). LC3-II is bound to autophagosome membranes and can also be localized on phagophores. LC3-II levels have traditionally been used as a marker for autophagy; however, in mammalian systems, the in vivo levels of LC3-II can either increase during autophagy or decrease relative to LC3-I if LC3-II degradation via the lysosomal machinery is particularly rapid (Klionsky et al. 2008). Moreover, changes in the levels of LC3-II have been observed to be tissue- and cell environment-dependent (Mizushima et al. 2004; Tanida et al. 2005). Detectable levels of LC3-II are not sufficient evidence for autophagy without accurate quantification (Suzuki et al. 2001; Matsui et al. 2007). In skeletal muscle cells, investigators have shown that a more accurate measure of autophagy in vivo is the autophagic flux of LC3-II in the presence of lysomotropic agents (Ju et al. 2010).
Although there is strong evidence that a defect in autophagy may be involved in the development of diseases associated with aberrant protein aggregation very little information exists on the role of autophagy in hereditary cataract development. An important study recently reported that mutations of the autophagy gene FYCO1 are associated with autosomal recessive cataracts in 12 Pakistani families, suggesting that autophagy plays an important role in hereditary cataractogenesis (Chen et al. 2011). Further studies have demonstrated the expression of a large number of autophagy-related genes, including those encoding LC3 and p62, in the human lens, and serum starvation, which induces autophagy, results in an increased number of LC3B-positive punctate bodies in cultures of the human lens epithelial cell line HLE B-3 (Brennan et al. 2012). Lenses of mice carrying a deletion of the gene for the autophagy-inducer ATG5 exhibit normal organelle degradation (Matsui et al. 2006); however, more recent work demonstrates that autophagy can occur by ATG5/7-dependent and independent mechanisms (Nishida et al. 2009). In the eye, p62 has been shown to be localized in human retinal pigment epithelial (RPE) cells (Viiri et al. 2010). However, the relationship between increased protein aggregation in cataracts and lens autophagy has not been investigated.
In the present study, we investigated the expression of p62 and LC3-II and the presence of autophagosomes in the lenses of wild type and αB-R120G mutant mice. We demonstrate inhibition of autophagy in αB-R120G mutant mouse lenses as shown by an increase in p62 puncta in lens fiber cells. In addition, the autophagosomes were significantly larger in the lens epithelial layer of αB-R120G homozygous mutant mice than in wild type or heterozygous mice, suggesting a defect in protein degradation after formation of autophagosomes in the mutant lenses.
2. Materials and Methods
2.1 Animals and lenses
AlphaB-R120G knock-in mice expressing an A to G point mutation in codon 120 of the mouse αB-crystallin gene cryab, which results in the substitution of arginine 120 with glycine (R120G), were generated as described previously (Andley et al. 2011). Briefly, a knock-in plasmid was generated by cloning the 5’ and 3’ arms of the mouse αB-crystallin gene, followed by mutation and homologous recombination in mouse embryonic stem cells, which were implanted into mice to generate the αB-R120G knock-in mice. Heterozygous αB-R120G mutant mice expressed one copy of the mutant gene while the second copy was wild type. Homozygous mice containing two copies of the mutant gene were generated by interbreeding heterozygous mice. Wild type littermates were used as controls. Mice used in this study were from the 129 Sv strain backcrossed with C57BL6J strain by speed congenics and were on an approximately 90% C57 background. PCR-based screening was used to routinely genotype the mice. One- to six-month-old mice were used in these studies. The animals were maintained at Washington University School of Medicine in the Division of Comparative Medicine by trained veterinary staff. All protocols and procedures involving mice were approved by the Washington University Animal Studies Committee and performed by Mouse Genetics Core. Mice were euthanized by CO2 inhalation and lenses were immediately dissected and used for analysis.
2.2 Immunoblot analysis
Lenses were dissected and homogenized in 1 ml ice-cold RIPA buffer (150 mM NaCl, 10 mM Tris-HCL, pH 7.2, 0.1 % triton X-100, 1% sodium deoxycholate and 5 mM EDTA) containing protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO) with glass grinding tubes resting in an ice-water bath. The lens homogenates were centrifuged at 10,000 × g for 30 min at 4°C. The protein concentration of th e supernatant was determined using the BioRad protein assay kit (500-0001) with bovine serum albumin as the standard. The supernatant was solubilized further in NuPage LDS sample buffer (NP00007), boiled for 5 min, and separated by SDS-PAGE using 12% (for p62) or 18% gels (for LC3) and 100 µg of protein was loaded in each lane. Proteins were transferred to a PVDF membrane (162–1077, BioRad). After blocking with 10 ml of Odyssey blocking buffer (927–40000, LiCor), membranes were incubated with 1:1000 dilution of a polyclonal anti-LC3B antibody (L7543, Sigma-Aldrich) or 1:500 dilution of monoclonal antibody to p62 (P0067, Sigma-Aldrich) in 10 ml blocking buffer, and a 1:100 dilution of anti-β-actin antibody (A1978, Sigma-Aldrich). Protein bands were visualized in an Odyssey protein gel documentation system equipped with a software program that allowed simultaneous detection of p62 (or LC3) and actin. Bands were quantified using Odyssey software.
2.3 Transmission electron microscopy
For ultrastructural analysis, lenses were fixed in 2% paraformaldehyde/2.5% glutaraldehyde (Polysciences Inc., Warrington, PA) in 100 mM phosphate buffer, pH 7.2 for 1 hr at room temperature. Fixed lenses were washed in phosphate buffer and postfixed in 1% osmium tetroxide (Polysciences Inc., Warrington, PA) for 1 hr. Lenses were rinsed extensively in deionized water prior to en bloc staining with 1% aqueous uranyl acetate (Ted Pella Inc., Redding, CA) for 1 hr. Following several rinses in deionized water, lenses were dehydrated in a graded series of ethanol and embedded in Eponate 12 resin (Ted Pella Inc.). Sections of 90-nm thickness were cut with a Leica Ultracut UCT ultramicrotome (Leica Microsystems Inc., Bannockburn, IL), stained with uranyl acetate and lead citrate, and viewed on a JEOL 1200 EX transmission electron microscope (JEOL USA Inc., Peabody, MA). Twenty serial sections from four different areas of the tissue were analyzed. These studies were performed by Dr. Wandy Beatty of the Department of Molecular Microbiology, Washington University School of Medicine. To determine the area of the autophagosomes, lens fiber cell autophagosomes were examined in the cortical fibers adjacent to the boundary of the organelle-free zone in the equatorial region and the diameter of the autophagosomes was recorded from the images. To systematically count lens fiber cell autophagosomes, 5–6 serial sections from two different areas separated by 40–50 µm were placed on a single grid. Two different grids were examined.
2.4 Immunofluorescence and microscopy
Immunohistochemistry was conducted on mid-sagittal lens sections to analyze p62 and LC3 distribution in the lens. Mouse eyes were fixed in 4% paraformaldehyde, embedded in paraffin, and cross-sectioned into 3-µm sections for examination of lens cells as described previously (Andley 2009; Andley et al. 2011). Tissue sections were labeled with anti-human p62 monoclonal antibody (1 µg/ml) and Alexa568-labeled conjugated goat anti-mouse IgG, fluorescein phalloidin, and DRAQ5 (Invitrogen) and visualized using a Zeiss 510 confocal microscope (Carl Zeiss, Jena, Germany). A monoclonal antibody to β-actin was added to stain the actin cytoskeleton. Sections were also labeled with a 1:100 dilution of anti-LC3 polyclonal antibody (Sigma, L7543) and Alexa568-conjugated goat anti rabbit IgG (Invitrogen). Nuclei were counterstained with DRAQ5 and sections were visualized using a Zeiss 510 confocal microscope. Control sections were processed without adding the primary antibody.
2.5 Analysis of p62 puncta in lens sections
To systemically count punctate labels in the images of lens sections, ×40 magnification images were analyzed using the Image J program (http://rsb.info.nih.gov/ij). The p62 channel was duplicated and a Gaussian Blur (radius 30.0) was applied to eliminate background and discern the space between adjacent punctate labels. The image was set to binary with a threshold of 30 and 255. Five 400 × 400 (4.95 µm × 43.95 µm) boxes were drawn at random within the band of the punctate labels. The punctate bodies were counted and analyzed using the Analyze particle function of Image J. Results were compiled for total area per sample, counts per sample, average size, and fractional coverage. This protocol has been adapted from the Swiss Federal Institute of Technology, Zurich, Switzerland (http://www.microscopy.ethz.ch/downloads/particlesize.pdf).
2.6 Treatment of mice with rapamycin in vivo
Wild type or αB-R120G heterozygous mice (3–6 months old) were injected with either vehicle (5% Tween 80, 5% PEG 400, 4% ethanol), or rapamycin conjugated with vehicle (2.5 mg/ml) for 6 days. Two mice were used per treatment. Each mouse was weighed at the beginning of the experiment. Mice were injected with each compound according to weight (4 µl per gram). On the last day (day 7) mice were not injected, but were euthanized. The eyes were removed and placed in PBS at 4°C until lens removal. Four lenses were removed and combined per treatment. Lenses were homogenized in 400 µl RIPA buffer (150 mM NaCl, 10 mM Tris-HCl, 0.1% Triton X-100, 1% sodium deoxycholate, 5 mM EDTA, pH 7.2) containing a protease inhibitor cocktail. Samples were placed on ice, centrifuged for 10 min at 10,000 rpm, and analyzed by western blotting using standard protocols.
3. Results
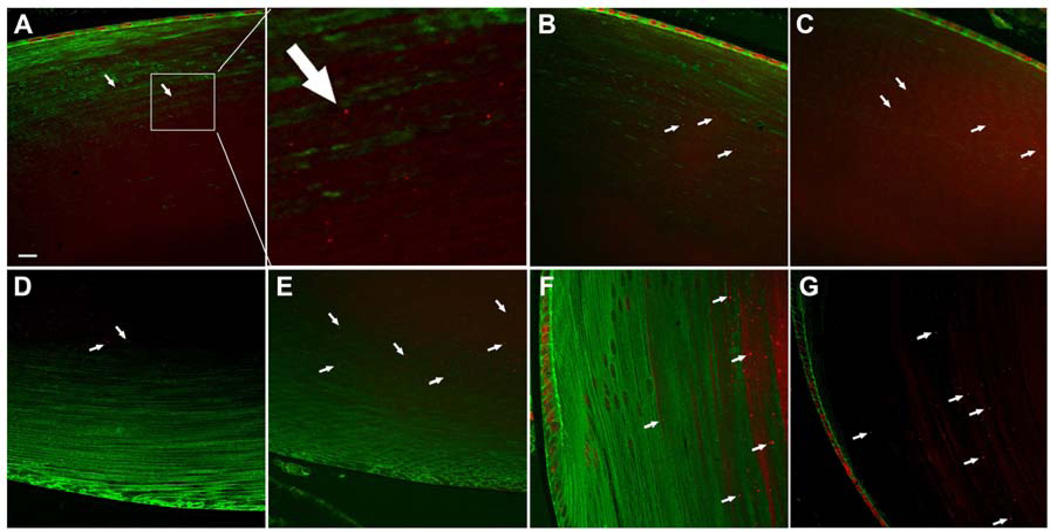
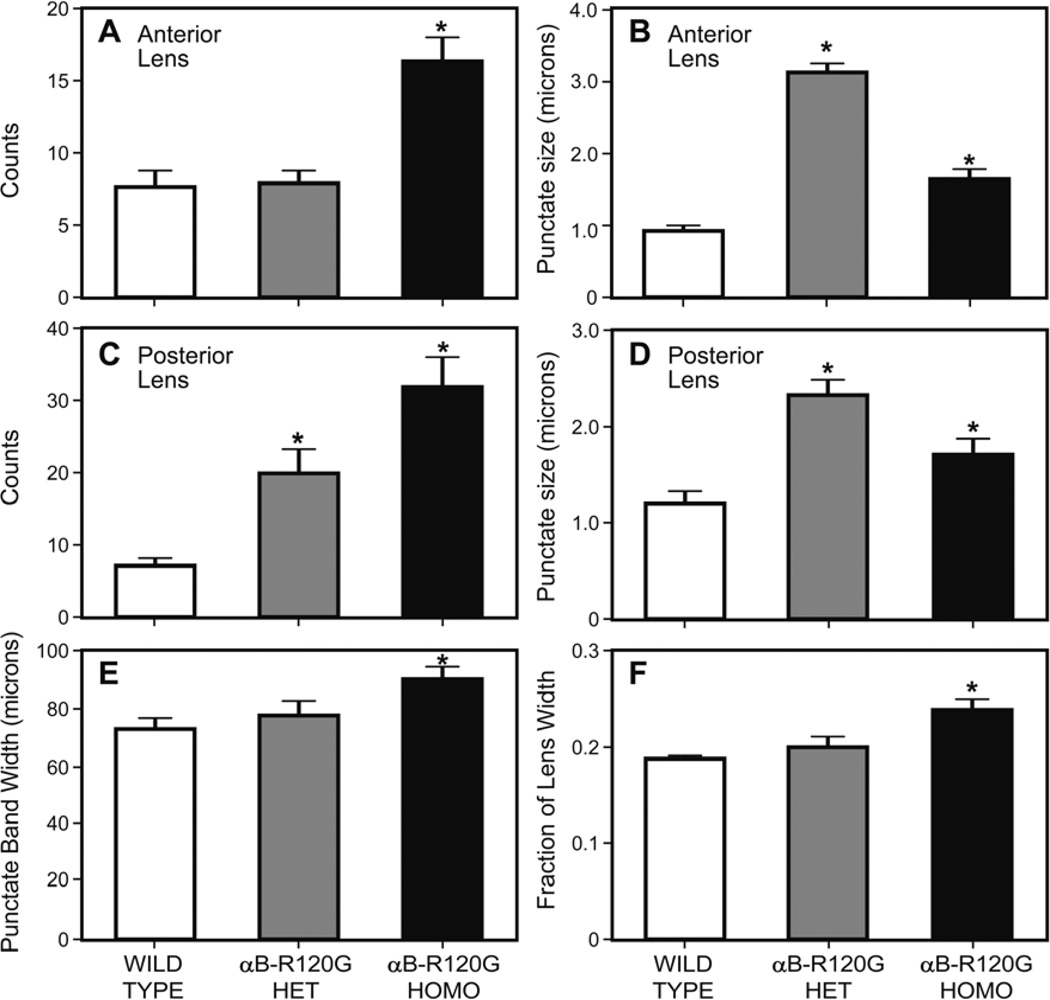
In αB-R120G knock-in mice, lens proteins aggregate into high molecular weight species that scatter light (Andley et al. 2011). To gain insight into the role of autophagy in αB-R120G knock-in mice, we investigated a key player of the autophagy pathway that is involved in degradation of polyUb-proteins – the ubiquitin-binding protein p62 that controls the sequestration of polyUb-proteins into inclusion bodies for autophagic degradation and is a well-established marker for autophagy. We examined the distribution of p62 in mouse lenses by immunohistochemistry and confocal microscopy (Figure 1). p62 expressed in lens epithelial cell nuclei was strongly localized on the nuclear membrane, as well as in a prominent band containing speckles in the inner cortical fibers of the organelle-free zone (Figure 1). Further analysis of p62-labeled speckles demonstrated that the number of p62 speckles was significantly higher in αB-R120G homozygous lenses than in heterozygous and wild type lenses (Figure 2A). In addition, the size of the speckles was greater in heterozygous and homozygous lenses compared with wild type (Figure 2B). The number and size of speckles in posterior fiber cells was increased in αB-R120G heterozygous and homozygous mutant lenses compared with the wild type (Figure 2C and D, respectively). The width of the band of speckles increased in the homozygous mutant lenses (Figure 2E), and the fractional lens diameter occupied by the p62 speckles also increased in these lenses (Figure 2F).
Figure 1.
Immunohistochemical analysis of p62 distribution in mouse lenses. p62 was localized to the lens epithelial cells and a band of speckles in lens fiber cells (arrows). (A–C) p62 labeling in the anterior region: (A) Wild type lens. The boxed area enclosed in the white square is enlarged in the second image of the top panel. (B) αB-R120G heterozygous mutant lens. (C) αB-R120G homozygous mutant lens. (D, E) p62 labeling in the posterior region: (D) αB-R120G heterozygous mutant lens. (E) αB-R120G homozygous mutant lens. (F, G) p62 labeling in the equatorial region: (F) αB-R120G heterozygous mutant lens. (G) αB-R120G homozygous mutant lens. Red, p62; Green, β-actin. Bar = 16 µm (A–F) and 10 µm (G).
Figure 2.
Quantification of p62 speckles in wild type and αB-R120G mutant lenses. (A) Number of p62 speckles in anterior lens fiber cells. Note the increase in the number of p62-positive puncta in the αB-R120G homozygous lenses. n=5. Asterisks indicate statistical significance (WT vs. Homozygous, p<0.0001). (B) Size of p62-positive puncta in anterior lens fiber cells. Note the increase in size of the puncta in αB-R120G homozygous and heterozygous mutant lenses compared with the wild type lenses (WT vs. Heterozygous p< 0.0001; WT vs. Homozygous p<0.05). (C) Number of p62 speckles in posterior lens fiber cells. Note the increase in the number of p62-positive puncta in the αB-R120G homozygous lenses (WT vs. Heterozygous p<0.05; WT vs. Homozygous p<0.05). (D) Size of p62-positive puncta in posterior lens fiber cells. Note the increase in size of the puncta in αB-R120G homozygous lenses compared with the wild type lenses. (E) Width of punctate/speckles band in wild type and αB-R120G mutant lenses (WT vs. Homozygous p<0.05). (F) Fraction of the lens equatorial diameter in wild type, αB-R120G homozygous, and αB-R120G heterozygous lenses (WT vs. Homozygous p<0.05).
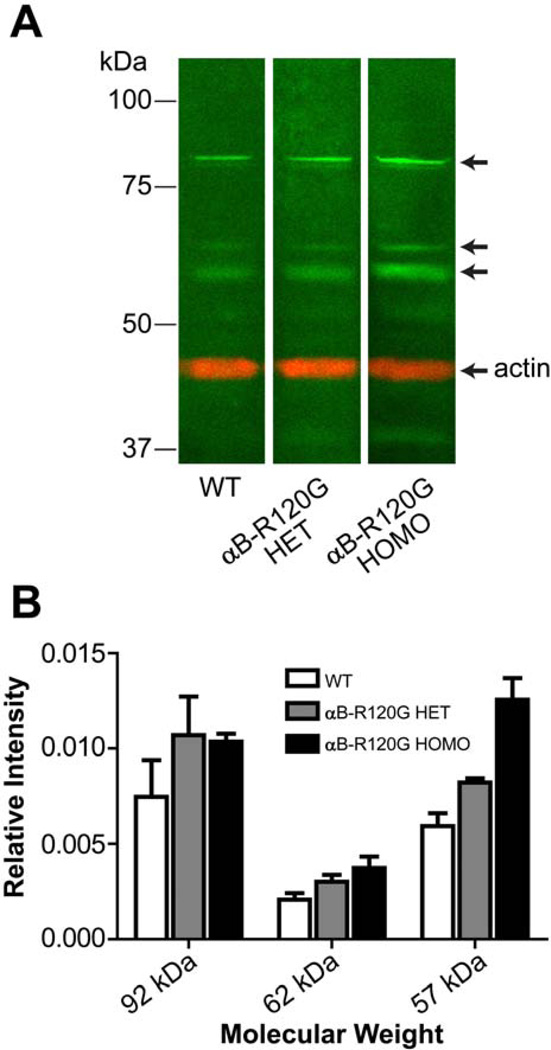
Expression of p62 was also analyzed by immunoblot analysis. The antibody to p62 recognizes a number of protein bands (Figure 3): in addition to the band at 62 kDa, an additional prominent band was detected at 57 kDa and a doublet at approximately 92 kDa was also observed in all adult lenses. The intensity of the bands at 57 and 62 kDa was statistically significantly higher (p<0.05) in αB-R120G mutant lenses compared with wild type lenses. The 92-kDa band may be a crosslinking product of p62 with other proteins (Bjorkoy et al. 2005; Pankiv et al. 2007).
Figure 3.
Immunoblot analysis of p62 in mouse lenses. (A) Wild type, αB-R120G heterozygous, and homozygous mutant lens proteins were analyzed by SDS-PAGE and immunoblotting using antibodies specific for p62 (green) and actin (red). Three prominent bands (green) that interact with the p62 antibody are indicated by arrows. Top, middle, and third arrows indicate the approximately 92, 62, and 57 kDa bands, respectively. The position of actin on the blot is also indicated by an arrow. (B) Bar graph showing an increase in bands that interact with the p62 antibody in mutant lenses. Data are the mean of four independent sets of lenses, error bars show standard deviation.
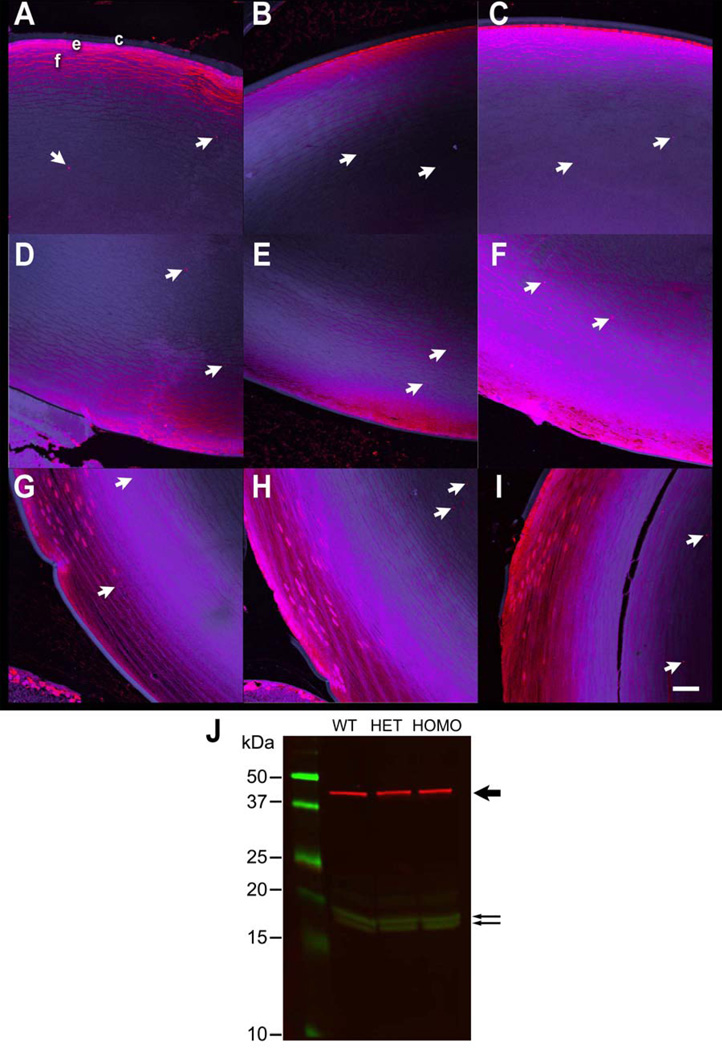
During autophagy, the microtubule-associated protein light chain 3 (LC3-I) protein is converted to LC3-II, an autophagosome-associated protein. We examined the distribution of LC3 in mouse lenses by immunochemical staining of lens sections and confocal microscopy. The LC3 antibody stained the epithelial cell layer, anterior lens fiber cells, and appeared as puncta in the inner cortical fiber cells (Figure 4A–I). The LC3 puncta increased in the posterior lens fibers cells of αB-R120G homozygous mutant mice (Figure 4G–I). LC3 expression in mouse lenses was examined by immunoblot analysis with an LC3B antibody that recognizes both LC3-I and LC3-II (Ju et al. 2010). Immunoblot analysis suggested a slight increase in LC3-II levels in αB-R120G heterozygous and homozygous mutant lenses compared with the wild type lenses (Figure 4J).
Figure 4.
(A–I) Detection of LC3 in mouse lenses by immunofluorescence analysis. (A–C) Anterior region: (A) Wild type lens; (B) αB-R120G heterozygous mutant lens; (C) αB-R120G homozygous mutant lens. (D–F) Posterior region: (D) Wild type lens; (E) αB-R120G heterozygous mutant lens; (F) αB-R120G homozygous mutant lens. (G–I) Equatorial region: (G) Wild type lens; (H) αB-R120G heterozygous mutant lens; (I) αB-R120G homozygous mutant lens. Red, LC3; Blue, DRAQ 5. c, lens capsule; e, lens epithelium; f, lens fibers. The intensity of the blue fluorescence has been enhanced in these images to clearly visualize the lens capsule. Scale bar, 60 µm. (J) Immunoblot analysis of LC3-1, LC3-II and F-actin in mouse lenses. The molecular weight marker proteins are in the left lane, and the size of the markers is indicated. The bold arrow on the right indicates the F-actin band (red). The lower two arrows indicate the LC3-I and LC3-II bands, respectively. WT, Wild type lens; HET, αB-R120G heterozygous mutant lens; HOMO, αB-R120G homozygous mutant lens.
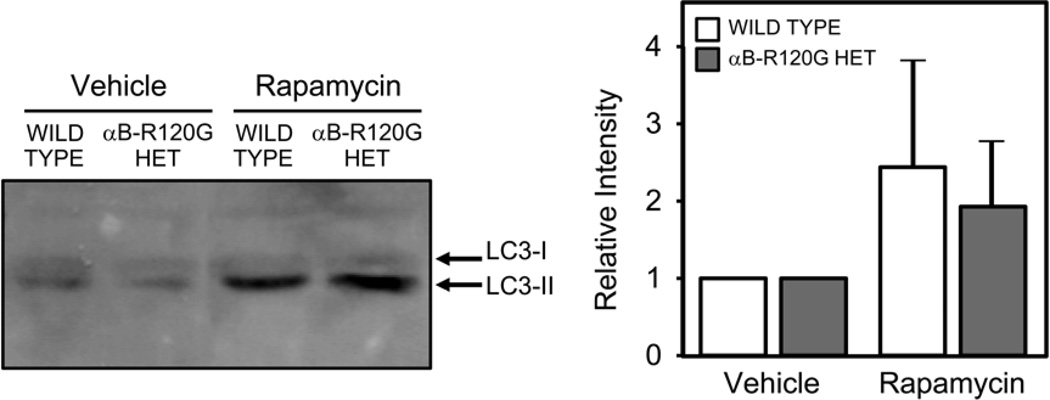
To confirm the effect of autophagy stimulation on LC3 levels in mouse lenses, mice were treated with vehicle or rapamycin, as a positive control for autophagy induction, and the lenses were analyzed by immunoblotting with antibodies specific to LC3. Treatment with rapamycin increased expression of LC3-II in both wild type and heterozygous mutant lenses (Figure 5), supporting the conclusion that the LC3-II responses observed in our study reflect autophagic activity.
Figure 5.
Effect of rapamycin on autophagy in lenses in vivo. Mice were treated with rapamycin or vehicle as described in Methods. The lenses were homogenized and analyzed by immunoblotting with antibodies to LC3 and actin. Four lenses were combined and analyzed under each condition. Note the increase in the LC3-II band in rapamycin-treated lenses. Bar graphs show the quantitative changes in LC3-II in three independent experiments.
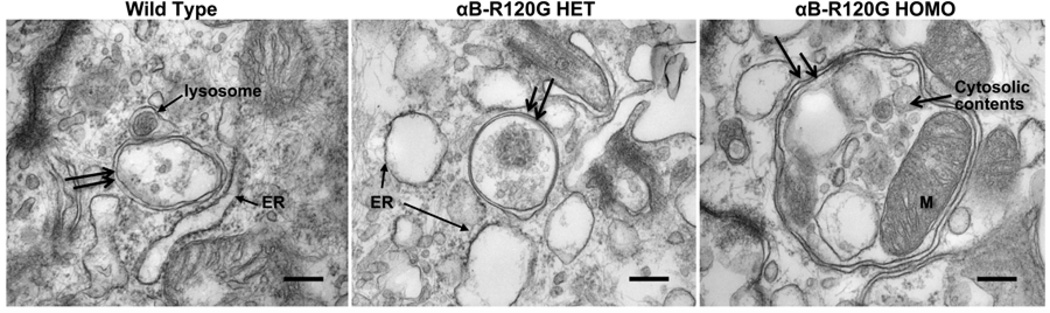
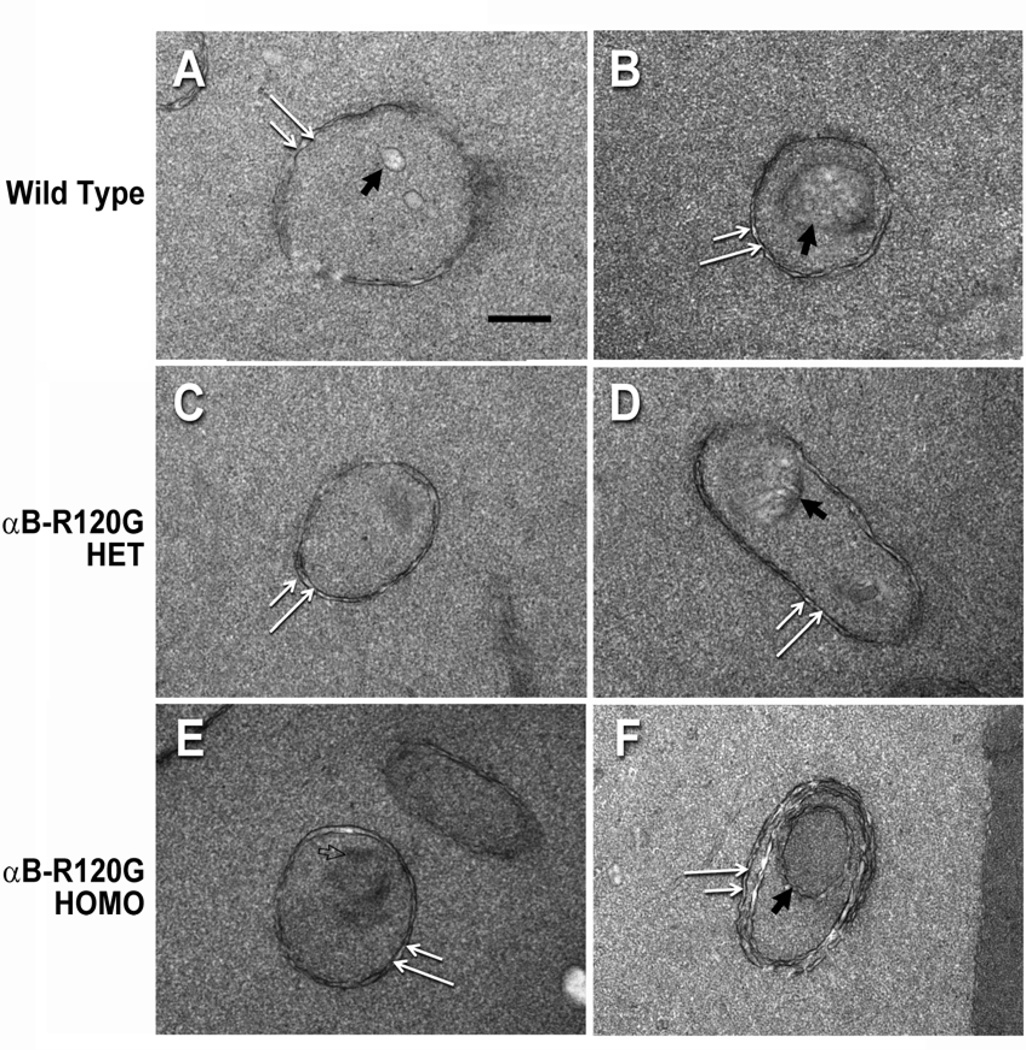
To examine the effect of the αB-R120G mutation on autophagosome formation we examined the lens epithelial and cortical fiber cell layers of these mice by transmission electron microscopy. Double-membraned structures were considered to be autophagosomes based on the following criteria: the structures were vesicles with double membranes, had clearly recognizable cytoplasmic content (intact or degraded), were between 0.3 and 2 microns in diameter, and were not multilamellar bodies. These structures were observed in all genotypes examined. In addition, lens epithelial cells of αB-R120G knock-in mice had larger than normal autophagosomes, and many of these contained mitochondria. These autophagosomes were in close proximity to the lysosomes, and appeared to contain swollen endoplasmic reticulum (Figure 6). The area of the autophagosomes was 2.5-fold larger in lens epithelial cells of the αB-R120G homozygous mutant than in wild type or the heterozygous mutant. The area of the autophagosomes in the lens epithelial cell layer of the wild type and αB-R120G heterozygous mutant lenses was 0.41± 0.25 µm2 (n = 11) and 0.38 ± 0.19 µm2 (n = 11) respectively, and increased to 1.06 ± 0.52 µm2 (n = 9) in the αB-R120G homozygous mutant lens (p= 0.001, wild type versus homozygous mutant). We next examined cortical lens fiber cells at the boundary of the organelle-free zone and observed double-membraned structures containing degraded cellular material that were presumed to be autophagosomes (Figure 7A, B, and D). The density of the cytoplasmic material inside the double-membraned structures was different from that outside (Figure 7C, E), suggesting that they are not ball-and-socket junctions or protrusions. In addition, these structures did not contain gap junctions typical of ball-and socket junctions (Biswas et al. 2010). One of the double-membraned structures (Figure 7F) had three pairs of membranes, suggesting that it was an early-stage autophagosome in which sorting of membranes had not occurred. The lens fiber cell autophagosomes were similar in size in the three genotypes. To assess the number of autophagosomes in the lens fiber cells while maintaining the same electron microscopic analysis time for the three genotypes, each genotype was examined for 90 minutes. This resulted in the sampling of three autophagosomes each in the wild type and αB-R120G heterozygous mutant lenses, and nine autophagosomes in the αB-R120G homozygous mutant lenses, suggesting that the number of autophagosomes was higher in the homozygous mutant lenses.
Figure 6.
Electron micrographs of autophagosomes in the lens epithelial layer of mouse lenses. (WT) Wild type lens, (αB-R120G HET) αB-R120G heterozygous mutant lens, (αB-R120G HOMO) αB-R120G homozygous mutant lens. Note that double-membraned structures (double black arrows) resembling autophagosomes that contained cytoplasmic material and/or organelles were detected in each genotype. The double membranes are visible as two parallel membranes that have a similar structure, but are not related or linked. Note also the proximity of autophagosomes to the ER and lysosomes. The ER appears swollen in the image shown for the αB-R120G heterozygous sample. The area of the autophagosomes was 2.5-fold larger in lens cells of αB-R120G homozygous mutant (n = 9) than in the wild type (n = 11) or heterozygous mutant (n = 11); p = 0.001. Scale bar = 250 nm.
Figure 7.
A gallery of electron micrographs of lens fiber cells in the organelle-free zone of wild type, αB-R120G heterozygous mutant, and αB-R120G homozygous mutant lenses. Double-membraned structures that are presumed to be autophagosomes were detected in each genotype (double white arrows). (A, B): Wild type lens; (C, D): αB-R120G heterozygous mutant lens; (E, F): αB-R120G homozygous mutant lens. Note that the structures in (A–E) are enclosed by a pair of membranes, and surrounded by a region of the cytoplasm. Note also that some of these structures contain distinct cellular material (filled black arrows). In addition to the conventional double-membrane autophagosomes, a multilamellar structure is observed in image (F) that has an inner core of double membranes surrounded by a middle pair of membranes, which is in turn surrounded by an outermost membrane pair. Each of the membranes has undulations. The open arrow in panel E indicates a region of darker density than the surrounding cytoplasm, suggesting that the double-membraned structure is not a ball-and-socket junction. Scale bar = 250 nm.
4. Discussion
In this study we analyzed the expression of autophagy markers p62 and LC3II and autophagosome formation in the αB-R120G knock-in mouse model of cataract formation in which mutant αB-crystallin is expressed in all cells under the influence of the endogenous promoter (Andley et al. 2011). In this model, crystallins become increasingly aggregated and αB-crystallin becomes water-insoluble concomitant with the formation of lens opacities (Andley et al. 2011). The p62 protein is specifically degraded by autophagy; therefore its accumulation is used as a marker for defective autophagic degradation. The results of the current study indicate that the p62 protein is expressed in the nuclear membrane of lens epithelial cells, and as speckles in a band of cortical fiber cells. The accumulation of p62 in the speckles and the size of these speckles were increased in the homozygous mutant lens fiber cells compared with heterozygous mutant and wild type cells. The width of the band of speckles was also increased in the mutant lenses. The LC3II protein binds to the autophagosome membrane. p62 interacts with LC3 and is present in complexes with ubiquitinated protein complexes and LC3. Mutations in the LC3 binding site of p62 cause the formation of inclusion bodies similar to those observed during autophagy deficiency (Seibenhener et al. 2004; Mizushima 2007; Pankiv et al. 2007). In the present study LC3 was detected in epithelial and anterior cortical fiber cells, and as punctate bodies in inner cortical fiber cells of adult mouse lenses. Punctate bodies containing GFP-LC3 have been reported in adult mouse lens epithelial and embryonic lens fiber cell layers (Mizushima et al. 2004; Matsui et al. 2006). In newborn lenses LC3 distribution has been reported as diffuse staining in the epithelial and central fiber cells (Brennan et al. 2012). Our immunofluorescence data are in agreement with these reports. Immunoblot analysis also revealed minor increases in the level of LC3-II in the αB-R120G mutant lenses. It should be noted that LC3 puncta can also be induced by detergents (Ciechomska and Tolkovsky, 2007). However, as we used the same conditions to treat the wild type and mutant lenses it is unlikely that the increase in puncta observed in this study is due to the presence of detergent in our homogenizing buffer. The expression patterns of p62 and LC3II in the homozygous mutant lenses, combined with the increase in the size of autophagosomes observed by TEM, suggest impairment of autophagic degradation rather than impaired formation of the autophagosome. As a positive control we confirmed that rapamycin induced autophagy with increased expression of LC3II and p62 in the αB-R120G mutant lens in vivo.
The αB-crystallin-R120G mutant protein forms high-molecular weight protein aggregates in lens and muscle cells in the αB-R120G knock-in mice (Andley et al. 2011). Previous studies also demonstrated an increase in total ubiquitinated protein in the insoluble fractions of muscle cells. Since misfolded protein aggregates are degraded by autophagy, the persistence of these aggregates in the αB-R120G mutant lenses suggests impaired autophagic clearance. Protein aggregate formation can occur either when autophagy is inhibited or when the capacity for degradation is exceeded by the formation of protein aggregates (Korolchuk et al. 2009; Korolchuk et al. 2010). It has also been proposed that the accumulation of misfolded proteins and protein aggregates may itself impair autophagy (Rubinsztein 2006; Williams et al. 2006). Investigators have shown that the αB-R120G mutation diminishes the chaperone activity of the protein in vitro (Bova et al. 1999). Further studies are necessary to determine if the observed changes in autophagy-related proteins are associated with impaired chaperone activity of the αB-R120G or an increase in protein aggregate size and impaired interaction with cytoskeletal proteins (Andley et al. 2011).
A novel observation in the current study was that p62 speckles were not expressed in the outermost cortical fiber cells but rather were present as a diffuse layer of cells bordering the organelle-free zone in the mid-cortical regions of the lens. This distribution pattern coincides with the sites of cleavage of a number of proteins in the lens. For example, in this region of the lens proteolytic degradation by the ubiquitin-proteasome pathway is active (Bassnett 2009); the membrane intrinsic protein MIP26 is cleaved to its maturation product (Gutierrez et al. 2011); and calpain-cleaved spectrin has been detected (De Maria et al. 2009). The lens-specific protein lengsin is also expressed in the transition zone as the maturing fiber cells lose their organelles (Wyatt et al. 2008). As components of nuclei and other organelles are degraded, DNAse-IIβ is activated in the fiber cells adjacent to the border of the organelle-free zone (De Maria and Bassnett 2007). The increase in the number and size of the p62-positive speckles in a band of fiber cells near the border of the organelle-free zone suggests that the degradation of p62 is inhibited in the αB-R120G mutant lenses. Clarifying the nature and composition of these p62 speckles and their relationship with αB-R120G aggregates is necessary for a better understanding of the quality control mechanisms that are impaired in the pathology of cataracts.
Autophagosomes are double-membraned vesicles in which protein aggregates or cytosol are trapped during autophagy and delivered to lysosomes for degradation. Protein aggregates are degraded by engulfment in autophagosomes followed by activity of lysosomal proteases (Komatsu et al. 2007). Autophagosomes are best characterized by electron microscopic examination of individual cells (Gao et al. 2010). However, the study of lens autophagosomes is a new area of investigation and there are no reports of TEM analysis of autophagosomes in the lens. The high-magnification TEM images of autophagosomes in the lens epithelium shown here are the first demonstration of these structures in intact lens tissue, although vacuoles labeled with GFP-LC3 have been described in lens epithelium at the light microscopy level (Mizushima et al. 2004; Matsui et al. 2006). Increased autophagosome size may suggest failure in fusion of autophagosomes with lysosomes as a result of lack of transport of the autophagosome to the lysosome. Since αB-crystallin interacts with cytoskeletal structures, mutant αB-crystallin expression may impair autophagosomal transport in the mutant knock-in lenses and impair autophagy (Perng et al. 1999; Perng et al. 2004; Xi et al. 2006; Andley et al. 2011). It remains to be tested whether the increase in autophagosomal size is the result of accumulation of mutant αB-crystallin in the autophagosomes. Investigators have shown that cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease (Martinez-Vicente et al. 2010). Autophagic vacuoles form at a normal or accelerated rate in cell culture models for Huntington’s disease (HD) and are adequately eliminated by the lysosomes, but they fail to efficiently trap cytosolic cargo in their lumen and inefficient engulfment of cytosolic components by autophagosomes is responsible for their accumulation inside HD cells (Martinez-Vicente et al. 2010). Autophagosome formation and the sequestration of organelles within the autophagosome appear to occur normally in the αB-R120G mutant lenses (Figure 6) but protein degradation by lysosomes may be defective. Our electron microscopic examination of lenses showed swelling of the endoplasmic reticulum (ER) in the αB-R120G mutant lens epithelial cells. Such ER swelling has been reported in regions where autophagy is inhibited (Ding et al. 2007; Ding et al. 2007; Nishiyama et al. 2007). The swollen ER observed in our study may be further evidence of upregulation of autophagy without complete lysosomal degradation, leading to accumulation of aggregated protein in mutant mice. We propose that the αB-R120G mutation leads to a block in autophagy due to a defect in protein degradation required for the completion of autophagy.
We believe that the double-membraned structures in lens cortical fiber cells adjacent to the boundary of the organelle-free zone are autophagosomes (Figure 7). These structures in lens fiber cells are similar to reported autophagosomes and appear to contain cellular debris, a known characteristic of autophagosomes (Xie and Klionsky 2007). Furthermore, the absence of gap junctions in these structures suggests that they are not ball-and-socket junctions (Biswas et al. 2010). These double-membraned structures are also unlikely to be protrusions between lens fiber cells since protrusions are associated with adherens junctions (Biswas et al. 2010). An accumulation of autophagosomes has been associated with autophagic inhibition in rat hepatocytes (Seglen et al. 1996). In Chinese hamster ovary cell cultures, a vinblastine-induced increase in autophagic vacuole size has also been linked with inhibition of autophagy (Munafo and Colombo 2001). Taken together, these studies support the concept that the increase in autophagosome size and in the size and number of p62 speckles observed in our current work is due to inhibition of autophagy in the αB-R120G mutant lenses. Further studies will be necessary to determine whether this increase in p62 speckles and number or size of autophagosomes is due to a lack of autophagosomal fusion with lysosomes and defective protein degradation.
Electron microscopic analyses of lenses from human and rat lenses have also revealed the presence of multilamellar bodies (MLBs) that show an increased abundance in human cataracts (Gilliland et al. 2004; Marsili et al. 2004). These MLBs are generally spherical but can have other shapes (Gilliland et al. 2004). The size of the MLBs is similar to the size of the structures that we consider to be autophagosomes in our current study. Interestingly, earlier studies on lung epithelial cell cultures demonstrated that autophagy is specifically involved in MLB formation (Hariri et al. 2000).
p62 is a scaffold protein involved in cell signaling, receptor internalization, and protein turnover. It is known to be a regulator of transcription of NF-κB (Moscat et al. 2007). p62 is normally found in cytosolic cellular speckles or inclusion bodies that contain ubiquitinated protein aggregates. These ubiquitinated protein aggregates are p62-positive in several neurodegenerative diseases and the size of these p62 speckles is typically increased in diseases associated with protein aggregation (Lee et al. 2012). Recently, p62 and LC3 immunohistochemistry has been utilized in the diagnosis of autophagic vacuolar myopathies (Lee et al. 2012). p62 and LC3 exhibit punctate staining in the muscle of patients treated with drugs to induce myopathy but not in normal controls. These methods have been proposed as a tissue diagnostic marker for autophagic vacuolar myopathy. Our study indicates that p62 speckles in the lens may be an excellent marker of autophagic clearance in the lens. Drugs that can increase autophagy have been used to rescue cardiac and skeletal muscle function in animal models of cardiomyopathy (Choi et al. 2012; Ramos et al. 2012). Activated mTOR inhibits autophagy, and inhibition of the mTOR pathway in the heart caused a reduction in p62 levels, indicating increased autophagic degradation. These results suggest that characterization of drugs that inhibit the mTOR pathway in the lens could open new avenues for the development of therapies targeting p62 in cataract.
In summary, the increased p62 immunolabeling in lens fiber cells, the increased size of the p62 speckles, and the increased size of the autophagosomes in αB-R120G mutant lenses are consistent with impairment of autophagy and suggest ineffective autophagic clearance. Based on these results, it would be interesting to identify drugs that enhance lysosomal clearance. Such studies may help elucidate the pathogenesis of cataract formation since preservation of autophagic activity is associated with a reduction in intracellular accumulation of damaged proteins and enhanced protein homeostasis (Salminen and Kaarniranta 2009). It would also be interesting to investigate whether autophagy is inhibited in other models of cataract in which high molecular weight crystallin aggregates are formed. Age-related cataracts are thought to have a genetic component (Heiba et al. 1995; Hammond et al. 2000; Shiels and Hejtmancik 2007). A wealth of literature indicates that in many human age-related cataracts, lens crystallins aggregate into high molecular weight species that scatter light (Carver et al. 1996; Bloemendal et al. 2004; Andley 2007; Sharma and Santhoshkumar 2009). It has also been suggested that the results of studies on the mechanisms of hereditary cataracts may have implications for understanding age-related cataracts (Moore 2004). Thus, the αB-R120G mouse model can be used in future studies to explore avenues to decrease protein aggregation in the lens.
Highlights.
Strong expression of autophagy marker p62 in mouse lens epithelial and fiber cells
Upregulation of p62 in hereditary cataract model caused by the αB-crystallin R120G mutation
Increase in LC3-II and autopohagosome size in hereditary cataract model
Findings suggest dysfunctional autophagy in hereditary cataract model
Acknowledgements
This work is supported by the National Institutes of Health (NIH) grants EY05681 (UPA), Core Grant EY02687 to the Department of Ophthalmology and Visual Sciences, and a Research to Prevent Blindness unrestricted grant to the Department of Ophthalmology and Visual Sciences. Dr. Conrad C. Weihl (Department of Neurology, Washington University School of Medicine) was supported by the Muscular Dystrophy Association. We thank Belinda McMahan (Department of Ophthalmology and Visual Sciences, Washington University School of Medicine) for immunohistochemical staining and Dr. Wandy Beatty (Department of Molecular Microbiology, Washington University School of Medicine) for electron microscopy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andley UP. Crystallins and hereditary cataracts: molecular mechanisms and potential for therapy. Expert Rev Mol Med. 2006;8(25):1–19. doi: 10.1017/S1462399406000111. [DOI] [PubMed] [Google Scholar]
- Andley UP. Crystallins in the eye: Function and pathology. Prog Retin Eye Res. 2007;26(1):78–98. doi: 10.1016/j.preteyeres.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Andley UP. AlphaA-crystallin R49Cneo mutation influences the architecture of lens fiber cell membranes and causes posterior and nuclear cataracts in mice. BMC Ophthalmol. 2009;9:4. doi: 10.1186/1471-2415-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andley UP, Hamilton PD, et al. Mechanism of insolubilization by a single-point mutation in alphaA-crystallin linked with hereditary human cataracts. Biochemistry. 2008;47(36):9697–9706. doi: 10.1021/bi800594t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andley UP, Hamilton PD, et al. A knock-in mouse model for the R120G mutation of alphaB-crystallin recapitulates human hereditary myopathy and cataracts. PLoS ONE. 2011;6(3):e17671. doi: 10.1371/journal.pone.0017671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassnett S. On the mechanism of organelle degradation in the vertebrate lens. Exp Eye Res. 2009;88(2):133–139. doi: 10.1016/j.exer.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas SK, Lee JE, et al. Gap junctions are selectively associated with interlocking ball-and-sockets but not protrusions in the lens. Mol Vis. 2010;16:2328–2341. [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171(4):603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloemendal H, de Jong W, et al. Ageing and vision: structure, stability and function of lens crystallins. Prog Biophys Mol Biol. 2004;86(3):407–485. doi: 10.1016/j.pbiomolbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Bova MP, Yaron O, et al. Mutation R120G in alphaB-crystallin, which is linked to a desmin-related myopathy, results in an irregular structure and defective chaperone-like function. Proc Natl Acad Sci U S A. 1999;96(11):6137–6142. doi: 10.1073/pnas.96.11.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan LA, Kantorow WL, et al. Spatial expression patterns of autophagy genes in the eye lens and induction of autophagy in lens cells. Mol Vis. 2012;18:1773–1786. [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Klionsky DJ. New insights into autophagy using a multiple knockout strain. Autophagy. 2008;4(8):1073–1075. doi: 10.4161/auto.6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver JA, Nicholls KA, et al. Age-related changes in bovine alpha-crystallin and high-molecular-weight protein. Exp Eye Res. 1996;63(6):639–647. doi: 10.1006/exer.1996.0158. [DOI] [PubMed] [Google Scholar]
- Chen J, Ma Z, et al. Mutations in FYCO1 cause autosomal-recessive congenital cataracts. Am J Hum Genet. 2011;88(6):827–838. doi: 10.1016/j.ajhg.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JC, Muchir A, et al. Temsirolimus activates autophagy and ameliorates cardiomyopathy caused by lamin a/c gene mutation. Sci Transl Med. 2012;4(144):144ra102. doi: 10.1126/scitranslmed.3003875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechomska IA, Tolkovsky AM. Non-autophagic GFP-LC3 puncta induced by saponin and other detergents. Autophagy. 2007;3:586–590. doi: 10.4161/auto.4843. [DOI] [PubMed] [Google Scholar]
- De Maria A, Bassnett S. DNase IIbeta distribution and activity in the mouse lens. Invest Ophthalmol Vis Sci. 2007;48(12):5638–5646. doi: 10.1167/iovs.07-0782. [DOI] [PubMed] [Google Scholar]
- De Maria A, Shi Y, et al. Calpain expression and activity during lens fiber cell differentiation. J Biol Chem. 2009;284(20):13542–13550. doi: 10.1074/jbc.M900561200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WX, Ni HM, et al. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem. 2007;282(7):4702–4710. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- Ding WX, Ni HM, et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171(2):513–524. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Kang JH, et al. Biochemical isolation and characterization of the tubulovesicular LC3-positive autophagosomal compartment. J Biol Chem. 2010;285(2):1371–1383. doi: 10.1074/jbc.M109.054197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland KO, Freel CD, et al. Distribution, spherical structure and predicted Mie scattering of multilamellar bodies in human age-related nuclear cataracts. Exp Eye Res. 2004;79(4):563–576. doi: 10.1016/j.exer.2004.05.017. [DOI] [PubMed] [Google Scholar]
- Gutierrez DB, Garland D, et al. Spatial analysis of human lens aquaporin-0 post-translational modifications by MALDI mass spectrometry tissue profiling. Exp Eye Res. 2011;93(6):912–920. doi: 10.1016/j.exer.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond CJ, Snieder H, et al. Genetic and environmental factors in age-related nuclear cataracts in monozygotic and dizygotic twins. N Engl J Med. 2000;342(24):1786–1790. doi: 10.1056/NEJM200006153422404. [DOI] [PubMed] [Google Scholar]
- Hariri M, Millane G, et al. Biogenesis of multilamellar bodies via autophagy. Mol Biol Cell. 2000;11(1):255–268. doi: 10.1091/mbc.11.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Baba M, et al. Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy. Mol Biol Cell. 2008;19(12):5506–5516. doi: 10.1091/mbc.E08-05-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiba IM, Elston RC, et al. Evidence for a major gene for cortical cataract. Invest Ophthalmol Vis Sci. 1995;36(1):227–235. [PubMed] [Google Scholar]
- Huang Q, Ding L, et al. Mechanism of cataract formation in alphaA-crystallin Y118D mutation. Invest Ophthalmol Vis Sci. 2009;50(6):2919–2926. doi: 10.1167/iovs.08-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju JS, Varadhachary AS, et al. Quantitation of "autophagic flux" in mature skeletal muscle. Autophagy. 2010;6(7):929–935. doi: 10.4161/auto.6.7.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4(2):151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131(6):1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Mansilla A, et al. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33(4):517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk VI, Menzies FM, et al. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010;584(7):1393–1398. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- Kuusisto E, Salminen A, et al. Ubiquitin-binding protein p62 is present in neuronal and glial inclusions in human tauopathies and synucleinopathies. Neuroreport. 2001;12(10):2085–2090. doi: 10.1097/00001756-200107200-00009. [DOI] [PubMed] [Google Scholar]
- Lamark T, Perander M, et al. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J Biol Chem. 2003;278(36):34568–34581. doi: 10.1074/jbc.M303221200. [DOI] [PubMed] [Google Scholar]
- Lee HS, Daniels BH, et al. Clinical utility of LC3 and p62 immunohistochemistry in diagnosis of drug-induced autophagic vacuolar myopathies: a case-control study. PLoS ONE. 2012;7(4):e36221. doi: 10.1371/journal.pone.0036221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsili S, Salganik RI, et al. Cataract formation in a strain of rats selected for high oxidative stress. Exp Eye Res. 2004;79(5):595–612. doi: 10.1016/j.exer.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci. 2010;13(5):567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M, Yamamoto A, et al. Organelle degradation during the lens and erythroid differentiation is independent of autophagy. Biochem Biophys Res Commun. 2006;339(2):485–489. doi: 10.1016/j.bbrc.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Matsui Y, Takagi H, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100(6):914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21(22):2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, et al. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15(3):1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3(6):542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- Moore AT. Understanding the molecular genetics of congenital cataract may have wider implications for age related cataract. Br J Ophthalmol. 2004;88(1):2–3. doi: 10.1136/bjo.88.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J, Diaz-Meco MT, et al. Signal integration and diversification through the p62 scaffold protein. Trends Biochem Sci. 2007;32(2):95–100. doi: 10.1016/j.tibs.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Munafo DB, Colombo MI. A novel assay to study autophagy: regulation of autophagosome vacuole size by amino acid deprivation. J Cell Sci. 2001;114(Pt 20):3619–3629. doi: 10.1242/jcs.114.20.3619. [DOI] [PubMed] [Google Scholar]
- Nakai A, Yamaguchi O, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13(5):619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- Nishida Y, Arakawa S, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461(7264):654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- Nishiyama J, Miura E, et al. Aberrant membranes and double-membrane structures accumulate in the axons of Atg5-null Purkinje cells before neuronal death. Autophagy. 2007;3(6):591–596. doi: 10.4161/auto.4964. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Pattison JS, Osinska H, et al. Atg7 induces basal autophagy and rescues autophagic deficiency in CryABR120G cardiomyocytes. Circ Res. 2011;109(2):151–160. doi: 10.1161/CIRCRESAHA.110.237339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng MD, Muchowski PJ, et al. The cardiomyopathy and lens cataract mutation in alphaB-crystallin alters its protein structure, chaperone activity, and interaction with intermediate filaments in vitro. J Biol Chem. 1999;274(47):33235–33243. doi: 10.1074/jbc.274.47.33235. [DOI] [PubMed] [Google Scholar]
- Perng MD, Wen SF, et al. Desmin aggregate formation by R120G alphaBcrystallin is caused by altered filament interactions and is dependent upon network status in cells. Mol Biol Cell. 2004;15(5):2335–2346. doi: 10.1091/mbc.E03-12-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos FJ, Chen SC, et al. Rapamycin reverses elevated mTORC1 signaling in lamin A/C-deficient mice, rescues cardiac and skeletal muscle function, and extends survival. Sci Transl Med. 2012;4(144):144ra103. doi: 10.1126/scitranslmed.3003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443(7113):780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Salminen A, Kaarniranta K. Regulation of the aging process by autophagy. Trends Mol Med. 2009;15(5):217–224. doi: 10.1016/j.molmed.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Sanbe A, Osinska H, et al. Desmin-related cardiomyopathy in transgenic mice: a cardiac amyloidosis. Proc Natl Acad Sci U S A. 2004;101(27):10132–10136. doi: 10.1073/pnas.0401900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanbe A, Osinska H, et al. Reversal of amyloid-induced heart disease in desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2005;102(38):13592–13597. doi: 10.1073/pnas.0503324102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen PO, Berg TO, et al. Structural aspects of autophagy. Adv Exp Med Biol. 1996;389:103–111. doi: 10.1007/978-1-4613-0335-0_12. [DOI] [PubMed] [Google Scholar]
- Seibenhener ML, Babu JR, et al. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24(18):8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Fraldi A, et al. A block of autophagy in lysosomal storage disorders. Hum Mol Genet. 2008;17(1):119–129. doi: 10.1093/hmg/ddm289. [DOI] [PubMed] [Google Scholar]
- Sharma KK, Santhoshkumar P. Lens aging: effects of crystallins. Biochim Biophys Acta. 2009;1790(10):1095–1108. doi: 10.1016/j.bbagen.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiels A, Hejtmancik JF. Genetic origins of cataract. Arch Ophthalmol. 2007;125(2):165–173. doi: 10.1001/archopht.125.2.165. [DOI] [PubMed] [Google Scholar]
- Son JH, Shim JH, et al. Neuronal autophagy and neurodegenerative diseases. Exp Mol Med. 2012;44(2):89–98. doi: 10.3858/emm.2012.44.2.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, et al. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. Embo J. 2001;20(21):5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Minematsu-Ikeguchi N, et al. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1(2):84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- Tannous P, Zhu H, et al. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2008;105(28):9745–9750. doi: 10.1073/pnas.0706802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannous P, Zhu H, et al. Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy. Circulation. 2008;117(24):3070–3078. doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicart P, Caron A, et al. A missense mutation in the alphaB-crystallin chaperone gene causes a desmin-related myopathy. Nat Genet. 1998;20(1):92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- Viiri J, Hyttinen JM, et al. p62/sequestosome 1 as a regulator of proteasome inhibitor-induced autophagy in human retinal pigment epithelial cells. Mol Vis. 2010;16:1399–1414. [PMC free article] [PubMed] [Google Scholar]
- Wang QJ, Ding Y, et al. Induction of autophagy in axonal dystrophy and degeneration. J Neurosci. 2006;26(31):8057–8068. doi: 10.1523/JNEUROSCI.2261-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A, Jahreiss L, et al. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr Top Dev Biol. 2006;76:89–101. doi: 10.1016/S0070-2153(06)76003-3. [DOI] [PubMed] [Google Scholar]
- Wilson MI, Gill DJ, et al. PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol Cell. 2003;12(1):39–50. doi: 10.1016/s1097-2765(03)00246-6. [DOI] [PubMed] [Google Scholar]
- Wyatt K, Gao C, et al. A role for lengsin, a recruited enzyme, in terminal differentiation in the vertebrate lens. J Biol Chem. 2008;283(10):6607–6615. doi: 10.1074/jbc.M709144200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi JH, Bai F, et al. Mechanism of small heat shock protein function in vivo: a knock-in mouse model demonstrates that the R49C mutation in alpha A-crystallin enhances protein insolubility and cell death. J Biol Chem. 2008;283(9):5801–5814. doi: 10.1074/jbc.M708704200. [DOI] [PubMed] [Google Scholar]
- Xi JH, Bai F, et al. Alpha-crystallin expression affects microtubule assembly and prevents their aggregation. Faseb J. 2006;20(7):846–857. doi: 10.1096/fj.05-5532com. [DOI] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9(10):1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]