Abstract
Platinum-based concurrent chemo-radiotherapy is considered a standard treatment approach for locoregionally advanced nasopharyngeal carcinoma (NPC). However, only a minority of patients benefit from this treatment regimen compared to radiotherapy alone. Identification of a set of molecular markers predicting sensitivity of platinum-based chemotherapy may contribute to personalized treatment of NPC patients for better clinical outcome with less toxicity. Previously, we generated a cisplain sensitive NPC cell line, S16, by clonal selection from CNE-2 cells and found that eIF3a is up-regulated and contributes to cisplatin sensitivity by down-regulating the synthesis of NER proteins. In this study, we conducted a gene expression profiling analysis and found three other genes, asparagine synthetase (ASNS), choriogonadotropin α subunit (CGA), and matrix metalloproteinase 19 (MMP19), that are up-regulated in the cisplatin-sensitive S16 cells compared with the CNE-2 cells. However, only ASNS and MMP19, but not CGA, contributes to cisplatin sensitivity by potentiating cisplatin-induced DNA damage and apoptosis. Thus, ASNS and MMP19, along with eIF3a, are sensitivity factors for cisplatin treatment and may serve as potential candidate molecular markers for predicting cisplatin sensitivity of advanced nasopharyngeal carcinoma.
Introduction
Nasopharyngeal carcinoma (NPC) is generally a rare malignancy in most part of the world. It, however, has a high incidence in a few well-defined populations, including natives of southern China, Southeast Asia, the Arctic, and the Middle East/North Africa (1–3). Concurrent chemoradiotherapy is considered as a standard treatment approach for locoregionally advanced NPC and platinum-based regimen is thought to be one of the best protocols by meta-analysis (4, 5). However, meta-analysis of individual patient data from eight randomized trials containing 1753 patients showed that, compared to radiotherapy alone, cisplatin-based concurrent chemoradiotherapy improved 5-year disease-free survival by only 10% (52% vs. 42%) in locaoregionally advanced NPC (4). Furthermore, many NPC patients do not benefit but suffer from side effects of the additional chemotherapy. These findings suggest that identifying patients who potentially do not benefit from concurrent chemotherapy may be helpful to personalize treatment strategies for better clinical outcome with less toxicity. Thus, it is imperative to identify molecular markers predicting sensitivity and responses of platinum-based chemotherapy of NPC patients for better clinical outcome with less toxicity.
To this end, we have established a cisplatin sensitive human NPC cell line S16 from CNE-2 cells using clonal selection and limited dilution and identified eIF3a as a potential marker predicting platinum sensitivity in a recent study (6). The increased eIF3a expression in S16 cells appears to suppress the synthesis of DNA repair proteins which in turn leads to reduced DNA repair and increased cisplatin sensitivity. To determine if other genes are also potentially up-regulated in S16 cells and contribute to cisplatin sensitivity, we performed comparative gene expression profiling analysis between the cisplain sensitive S16 clone and its parental CNE-2 cells using microarray analysis, followed by confirmative real-time PCR analyses. Three genes, asparagine synthetase (ASNS), choriogonadotropin α subunit (CGA), and matrix metalloproteinase 19 (MMP19), were found to have significant changes in expression level between S16 and CNE-2 cells. However, only ASNS and MMP19 were found to contribute to cisplatin sensitivity of S16 cells by promoting cisplatin-induced DNA damage and apoptosis in S16 cells. Thus, ASNS and MMP19, along with eIF3a, are sensitivity factors for cisplatin treatment and may serve as candidate molecular markers predicting sensitivity and possibly clinical outcome of cisplatin-based chemotherapy for advanced NPC.
Materials and Methods
Materials
AmpliTaq Gold polymerase, Power SYBR® Green RNA-to-CT™ 1-Step Kit, Dulbecco's Modified Eagle Medium (DMEM), G418, Hoechst 33342, TRIzol reagent, Superscript™ II reverse transcriptase, and Lipofectamine™ 2000 were all from Applied Biosystems (Carlsbad, CA). Antibody against actin, HRP-conjugated anti-mouse or rabbit secondary antibodies, (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), and cis-dichlorodiammine platinum (II) (cisplatin) were from Sigma-Aldrich (St Louis, MO, USA). The enhanced chemiluminescence (ECL) system, Cy3-dCTP, and Cy5-dCTP were obtained from Amersham Pharmacia Biotech (Piscataway, NJ). RNeasy Mini Kit, siRNAs for CGA, ASNS (7), and MMP19 (8) (Supplemental Table S1) were purchased from or custom synthesized by QIAGEN (Valencia, CA, USA). Scrambled control siRNA (Silencer Negative Control #1 siRNA) was purchased from Ambion (Austin, TX, USA). Polyvinylidene difluoride (PVDF) membrane and concentrated protein assay dye reagents were from Bio-Rad (Hercules, CA). Restriction Endonucleases and T4 DNA Ligase were from New England Biolabs Inc. (Ipswich, MA). Anti-MMP19 (ab53146), ASNS (H00000440-B01), CGA (sc18224 or sc57185), p-H2AX (KAM-CC255), and cleaved PARP (19F4, #9546) antibodies were from Abcam (Cambridge, MA USA), ABNOVA (Taipei, Taiwan, China), Santa Cruz biotechnology Inc. (Santa Cruz, CA), Stressgen (Brussels, Belgium), and Cell Signaling Technology, Inc. (Danvers, MA), respectively. All other reagents were of molecular biology grade and obtained from Sigma-Aldrich or Fisher Scientific (Chicago, IL).
Cell lines and transfection
The cisplatin-sensitive cell clone S16 was selected and established from a poorly differentiated human NPC cell line CNE-2 (9) by clonal selection and limited dilution (6). Both S16 and CNE-2 cells were cultured and maintained in DMEM supplemented with 10% fetal bovine serum. The parental cell line CNE-2 has been kept in the Laboratory of Cancer Genetics, VARI, since 2000 and the S16 derivative was established in 2001. These cell lines were authenticated in 2003 by examining five STR sequences and one point mutation sequence of the p53 gene.
For transient transfection, cells were seeded in 10-cm dishes at 1 × 106 per dish and cultured for 24 hrs followed by transfection with siRNAs (or plasmids using Lipofectamine™ 2000 according to manufacture's instructions. Two siRNAs for each target gene (Supplemental Table S1) were used as a mixture at the ratio of 1:1. The cells were cultured for additional 24 hrs in the standard media and were harvested for further analysis. To generate stable pool of CNE-2 cells with over-expression of ectopic ASNS or MMP19, the standard media were replaced with media containing G418 24 hours after transfection and cultured continuously in the media containing 750 µg/mL of G418 for 3 weeks and pools of selected cells were maintained in media containing 200 µg/mL of G418.
Engineering of ASNS and MMP19 ectopic over-expression constructs
cDNA encoding ASNS and MMP19 was cloned by reverse transcription from isolated mRNAs using Superscript II reverse transcriptase and PCR with specific primers shown in Supplemental Table S2. The PCR products were then cloned into pcDNA3.1(+) plasmid as previously described (10) and verified by double strand DNA sequencing.
cDNA microarray
Parental CNE-2 and cisplatin-sensitive subclone S16 cells were seeded in 15-cm dishes at 3 × 106 per dish. After 24 h of culture, cells were treated with cisplatin at a final concentration of 8.67 µM (IC50 of CNE-2 cells) for 0, 2 or 8 h respectively, followed by total RNA extraction by using TRIzol reagent. The methods for cDNA microarray production, hybridizations, and data normalization using GenePix Pro 3.0 software were reported previously (11–13) with slight modification. Briefly, a total of 33,099 cDNA clones of the Sequence Validated Human cDNA library (ResGen, Invitrogen Corporation) were amplified and robotically printed onto two sets of polylysine-coated microarray slides, each with 19,159 or 13,940 cDNAs, respectively for the two sub-arrays. Fifty micrograms of total RNA from each side of comparison were reverse transcribed with oligo(dT)12–18 primer and Superscript II in the presence of either Cy3-dCTP or Cy5-dCTP. The Cy3- and Cy5-labeled cDNA probes were mixed and hybridized to the microarray slides. The slides were then scanned by using a confocal fluorescent scanner equipped with lasers operating at 532- and 635-nm wavelengths (Scan Array Lite, GSI Lumonics, Billerica, CA). After subtraction of the background intensity, the ratio of the net fluorescence from Cy5-specific channel and the net fluorescence from Cy3-specific channel were calculated for each cDNA spot and represented the expressions of the RNA in the Cy5-labelled sample relative to the expressions in the Cy3-labelled sample. Each experiment was repeated with a switch in fluorescent labels to account for dye effects.
For two experiments with Cy3 and Cy5 swapped for each cDNA clones on the chip, the final relative expression (ratio) from these repeated experiments were calculated to be their geometric mean of the two expression ratios (ratio from the dye swapped chip will be inversed before taking the mean). If one of the ratios is missing, the non-missing ratio will be used instead of the geometric mean. If both ratios were missing, the final expression ratio would be absent. Probes were filtered according to the following schema. First, any cDNAs with more than 3 missing expression ratios were removed due to quality concern. Second, cDNAs with poor annotations were filtered and eliminated. After these filtering operations, the data set had ~13300 cDNAs remaining for further analyses. The microarray data have been deposited into GEO database (accession # GSE49813).
Real-time RT-PCR and Western blot analysis
Real time RT-PCR analysis was performed as previously described (6). Briefly, total RNAs were extracted using RNeasy Mini Kit and subjected to real time RT-PCR using Power SYBR® Green RNA-to-CT™ 1-Step Kit using primers shown in Supplemental Table S2. The threshold cycle (Ct) values were determined and normalized against that of GAPDH internal control. The relative mRNA levels were shown as the value of 2ΔCt normalized to the control group.
Western blot analysis was performed as described previously (14). Briefly, cell pellets were lysed with TNN-SDS buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 50 mM NaF, 1 mM sodium orthovanadate, 1 mM DTT, 2 mM PMSF, 0.5% NP-40, and 0.1% SDS) at 4 °C for 30 min followed by centrifugation (10,000 g for 10 min at 4 °C) to remove insoluble materials, and protein concentration in supernatants were measured using Protein Assay kit. Proteins were then separated by 10% SDS-polyacrylamide gel electrophoresis and transferred to PVDF membranes. The blots were then probed with specific primary antibody, followed by reaction with HRP-conjugated secondary antibody, and signals were enhanced by ECL detection system and captured using x-ray film.
Cell survival and apoptosis assays
Cell survival was determined using MTT assays as previously described (6, 15, 16). Briefly, cells were inoculated in 96-well plates at a density of 2000 cells/well and incubated for 24 hrs before treated with cisplatin for 72 hrs. Viable cells were then stained with MTT followed by determination of OD570 nm with a reference wavelength at 630 nm. The data were analyzed using GraphPad Prism4 software (GraphPad Software, La Jolla, CA) to obtain IC50.
Apoptosis was analyzed by staining of nuclei using Hoechst 33342 dye as we previously described (6). Briefly, both floating and adherent cells were harvested, and cells were washed twice in ice-cold PBS. Collected cells were adjusted to the density of 1×106 cells/mL in PBS with 1% FBS and stained with 5 µM of Hoechst 33342 at 37°C for 10 min. The stained cells were mounted onto a polylysine-coated slide and examined under a fluorescent microscope immediately. For each measurement, a total of 300–400 nuclei from 5~8 randomly chosen fields were examined. High blue fluorescent indicates apoptotic cells, low blue indicates live cells. Apoptosis was expressed as a percentage of the total number of nuclei examined. Alternatively, apoptosis was analyzed using Cell Death Detection ELISA kit (Roche, Indianapolis, IN) or by determining PARP cleavage using Western blot as we previously described (6). The experiments were repeated three times, and the results expressed as means and standard deviations.
Results
Identification of ASNS, CGA, and MMP19 with increased expression in cisplatin-sensitive S16 cells
Recently, we generated a cisplatin-sensitive clone S16 from a parental human nasopharyngeal carcinoma cell line CNE-2, which has not been selected against cisplatin and, thus, not cisplatin resistant, by clonal selection and limited dilution and it has been shown to be derived authentically from CNE-2 cells (6). S16 is about 3-fold more sensitive to cisplatin than the parental CNE-2 cells and eIF3a over-expression in S16 cells has been shown to contribute to this sensitivity by suppressing synthesis of nucleotide excision repair (NER) proteins (6).
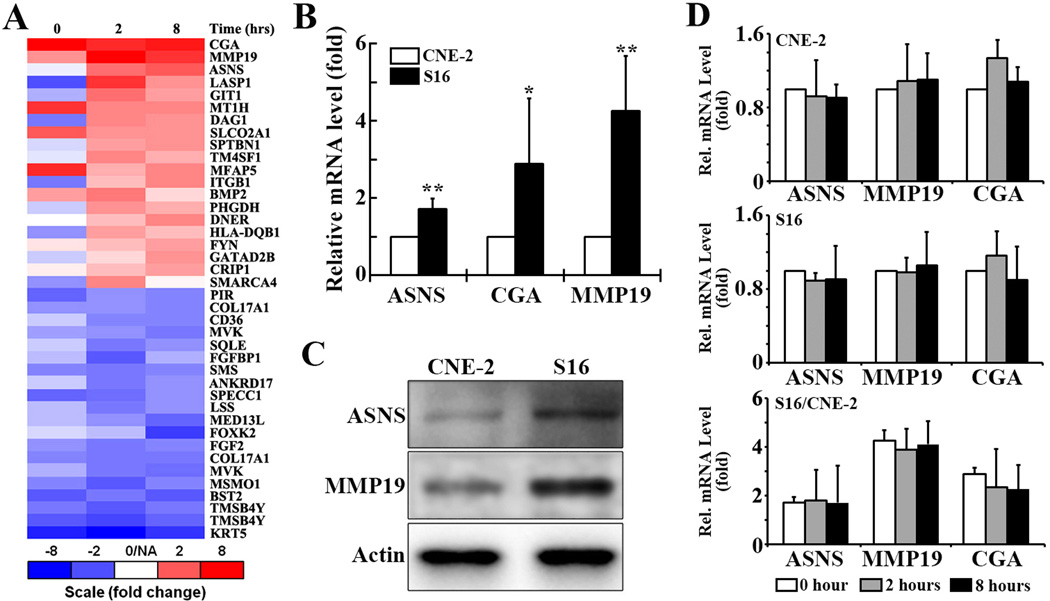
The genes with elevated expression such as eIF3a (6) in the cisplatin sensitive S16 cells or after cisplatin treatment were thought to be sensitivity factor for cisplatin treatment. Identifications of these sensitivity factors can broaden our knowledge on how chemotherapy agents kill cancer cells. Moreover, amplifying the functions of these sensitvity factors may help sensitize the resistant cells upon chemotherapy and, thus overcome drug resistance and enhance chemotherapy effect, which will have a potential broader clinical application. With this in mind and to determine what other genes are also potentially up-regulated in expression and may serve as contributors to cisplatin sensitivity of S16 cells, we performed a comparative mRNA expression profiling analysis between S16 and CNE-2 cells treated with or without cisplatin. We identified three genes (asparagine synthetase [ASNS], glycoprotein hormone alpha polypeptide [CGA], and matrix metallopeptidase 19 [MMP19]) that are most up-regulated in S16 cells compared to the parental CNE-2 cells with or without cisplatin treatment (Fig. 1A).
Figure 1. Analysis of ASNS, CGA, and MMP19 expression.
A. Microarray analysis. Gene expression profiling using microarray analysis was performed on the S16 and CNE-2 cells treated with or without cisplatin for different times. The 20 most up-regulated and 20 most down-regulated gene sequences in S16 relative to CNE-2 cells are plotted and ranked by the averaged values of the ratio from the treatments at 2 and 8 hour time points. Some genes were detected by two different probes binding to two different transcriptional regions. B. Quantitative RT-PCR analysis the ASNS, CGA, and MMP19 mRNA level in S16 cells relative to that in CNE-2 cells (* p<0.05, ** p<0.01, two dialed t-test). C. Western blot analysis of ASNS and MMP19 proteins in CNE-2 and S16 cells. Actin was used as a loading control. D. Quantitative RT-PCR analysis of mRNA levels of ASNS, CGA, and MMP19 in S16 and CNE-2 cells following treatment with 8.7 µM cisplatin for 0, 2, or 8 hrs. Panel C shows the relative ration of ASNS, MMP19, and CGA mRNAs in S16 and CNE-2 cells.
Our preliminary findings in gene expression profiling were further validated by using a real time RT-PCR analysis, which was a more accurate and reliable approach (Fig. 1B), followed by immunoblotting analyses (Fig. 1C). The constitutive expression levels of ASNS and MMP19 were confirmed to be elevated in S16 cells compared with CNE-2 cells (Fig. 1B–1C). CGA, however, is undetectable in either S16 or CNE-2 cells using Western blot analysis (data not shown) although its mRNA level is increased in S16 cells as determined by real time RT-PCR analysis (Fig. 1B). To determine if the expression of these genes changes in response to cisplatin treatment, we next performed real time RT-PCR analysis of both S16 and CNE-2 cells following a 2- and 8-hr treatment with cisplatin. As shown in Fig. 1D, none of these three genes experiences any change in their expression in either S16 or CNE-2 cells. However, the ratio between the two cells following cisplatin treatment remains the same as the untreated cells, consistent with the increased constitutive expression of these genes in S16 relative to CNE-2 cells.
Knockdown of ASNS and MMP19, but not CGA, reduces S16 sensitivity to cisplatin
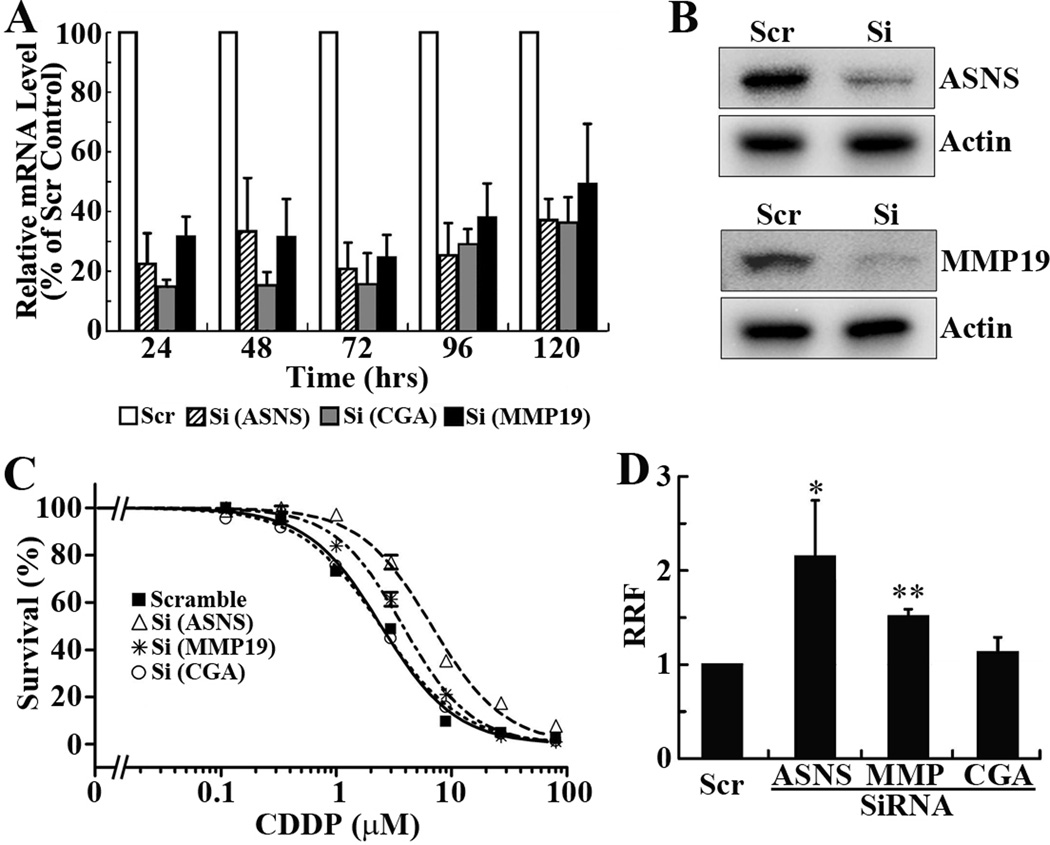
To determine whether the up-regulated expression of ASNS, MMP19, and CGA in S16 cells possibly contributes to the increased cisplatin sensitivity, S16 cells were transiently transfected with siRNAs targeting ASNS, CGA, and MMP19 individually followed by cisplatin treatment and MTT assay. Fig. 2A shows that the mRNA level of all three genes is effectively knocked down by their respective siRNAs and the knockdown lasts for at least 5 days. The knockdown of ASNS and MMP19 at 72 hrs following siRNA transfection was also confirmed using Western blot analysis (Fig. 2B). Fig. 2C shows that the cisplatin sensitivity of S16 cells is effectively reduced by knocking down ASNS and MMP19 expression. The relative resistance factor (RRF) is increased by 1.5–2 fold (Fig. 2D). However, CGA knockdown dis not appear to have any significant effect on cisplatin sensitivity of S16 cells (Fig. 2C and 2D). The lack of effect of CGA expression on cisplatin sensitivity is consistent with the observation of unable to detect CGA by Western blot. Thus, the up-regulated expressions of ASNS and MMP19, but not CGA, in S16 cells likely contribute to the increased cisplatin sensitivity compared to its parental CNE-2 cells.
Figure 2. Knockdown of ASNS and MMP19, but not CGA, increases cisplatin resistance.
A. Quantitative RT-PCR analysis of mRNA levels of ASNS, CGA, and MMP19 at different times following transfection with their respective siRNAs or a scrambled (Scr) control siRNA. GAPDH was used as an internal standard. B. Western blot analysis of ASNS and MMP19 at 5 days following transfection with their respective siRNAs or scrambled control siRNA. Actin was used as a loading control. C. Effect of ASNS, CGA, and MMP19 knockdown on cisplatin sensitivity. Twenty-four hrs following siRNA-transfection, S16 cells were seeded into 96-well plate and subjected to cisplatin treatment and MTT assay. D. Relative resistance factor. IC50 determined from dose response curves as shown in panel C and was used to calculate relative resistance factor as described in Materials and Methods. The data shown are from 4–5 independent experiments (* p<0.05, ** p<0.01, two tailed t-test).
Asparagine supplementation rescues growth inhibition but not cisplatin resistance induced by ASNS gene knockdown
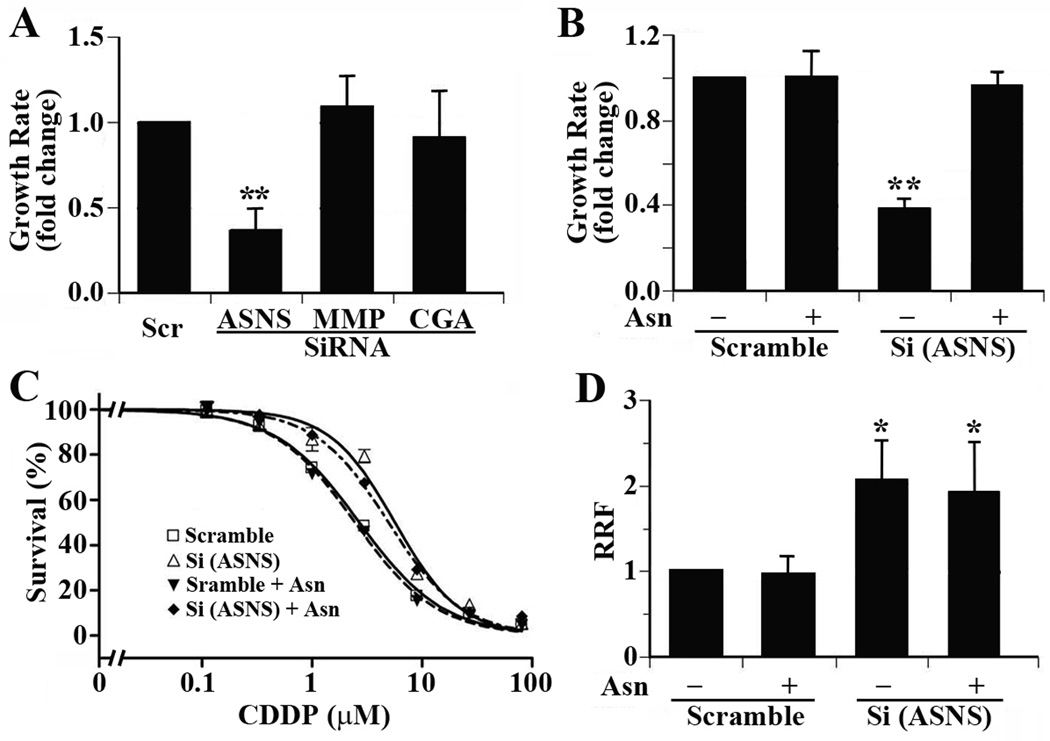
The effect of ASNS and MMP19 knockdown on cisplain sensitivity could be due to their effect on the proliferation of S16 cells. To test this possibility, we examined the effect of ASNS and MMP19 knockdown on cell proliferation. As shown in Fig. 3A, ASNS knockdown significantly slows down cell proliferation, while CGA and MMP19 knockdown has no effect on S16 growth. Thus, the effect of MMP19 knockdown on cisplatin sensitivity is unlikely due to its effect on S16 cell proliferation. Considering that ASNS is responsible for the synthesis of asparagine (Asn), a non-essential amino acid which is absent in culture media, it is reasonable to assume that reduced synthesis of Asn by ASNS knockdown may reduce cell growth. To determine if the reduced cisplatin sensitivity by ASNS knockdown is due to decreased cell proliferation, we supplemented 0.4 mM of Asn into the culture media and tested if ASNS knockdown still affects S16 cell proliferation and cisplatin sensitivity. As shown in Fig. 3B, Asn supplementation restored the growth rate of S16 cells transfected with ASNS siRNA. However, Asn supplementation had no significant effect on the decrease in cisplatin sensitivity induced by ASNS knockdown (Fig. 3C and 3D). Thus, the reduced sensitivity to cisplatin in S16 cells by ASNS knockdown unlikely results from the reduced growth rates due to inadequate Asn synthesis. This observation also suggests that the effect of ASNS on cellular sensitivity to cisplatin is not due to its end product, Asn.
Figure 3. Effect of proliferation on cisplatin sensitivity.
A. Effect of ASNS, CGA, and MMP19 knockdown on S16 cell proliferation. B. Effect of Asn supplementation on ASNS knockdown-induced cell proliferation inhibition. C. Effect of Asn supplementation on ASNS knockdown-induced cisplatin resistance. D. Effect of Asn supplementation on relative resistance factor derived from three independent experiments. (* p<0.05, ** p<0.01, two tailed t-test)
Ectopic overexpression of ASNS or MMP19 increases cisplatin sensitivity in CNE-2 cells
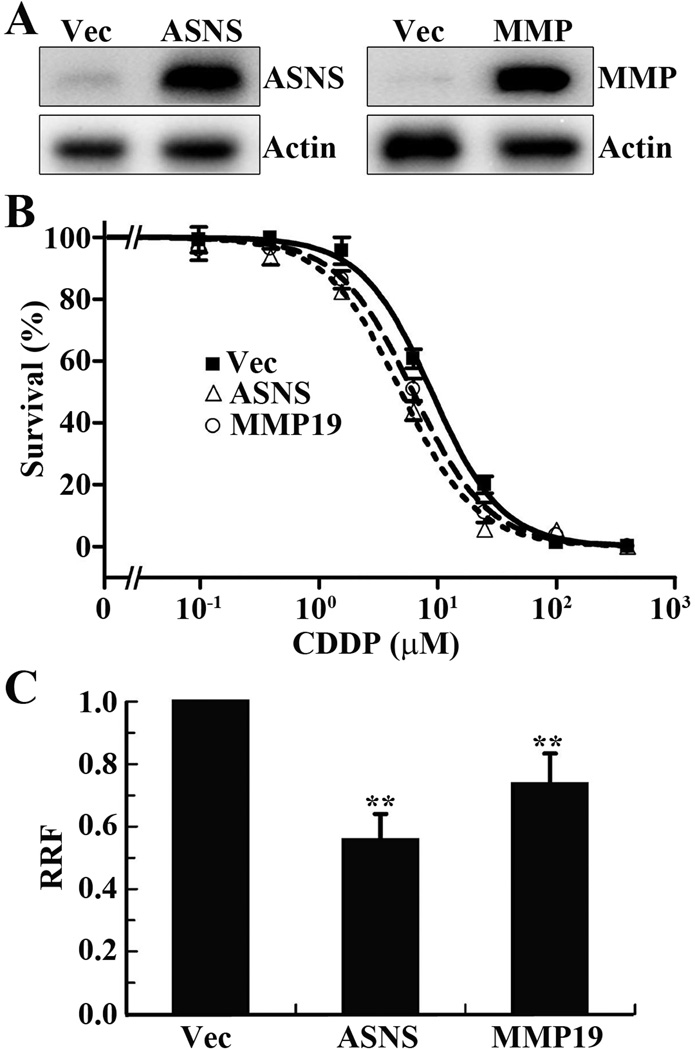
To confirm the above conclusion, we next performed a reverse experiment by over-expressing ectopic ASNS and MMP19 individually in the parental CNE-2 cells and determined if the increased ASNS and MMP19 expression would increase cisplatin sensitivity. For this purpose, a pool of stable CNE-2 cells transfected with ASNS or MMP19 cDNA for ectopic over-expression of ASNS and MMP19, respectively, was selected and subjected to Western blot analysis and MTT assay. Fig. 4A shows the stable over-expression of ASNS and MMP19 proteins in these cells transfected with ASNS and MMP19 cDNA compared with vector-transfected control cells. MTT assay shows that ASNS and MMP19 over-expression significantly increases cisplatin sensitivity by reducing RRF by 30–50% compared to the vector-transfected control cells (Fig. 4B and 4C). Together with the data shown above, we conclude that ASNS and MMP19 over-expression likely contributes to the increased cisplatin sensitivity of S16 cells.
Figure 4. Ectopic overexpression of ASNS or MMP19 increases cisplatin-sensitivity.
A. Western blot analysis of stable CNE-2 cells with over-expression of ectopic ASNS and MMP19 or transfected with vector (Vec) control. Actin was used as a loading control. B. Dose response of the stable cells to cisplatin treatment as determined using MTT assay. C. Relative resistance factor (RRF). IC50 determined from dose response curves as shown in panel B and was used to calculate RRF as described in Materials and Methods. The data shown are from 3 independent experiments (** p<0.01, two tailed t-test).
ASNS and MMP19 over-expression potentiates cisplatin-induced DNA damage and apoptosis
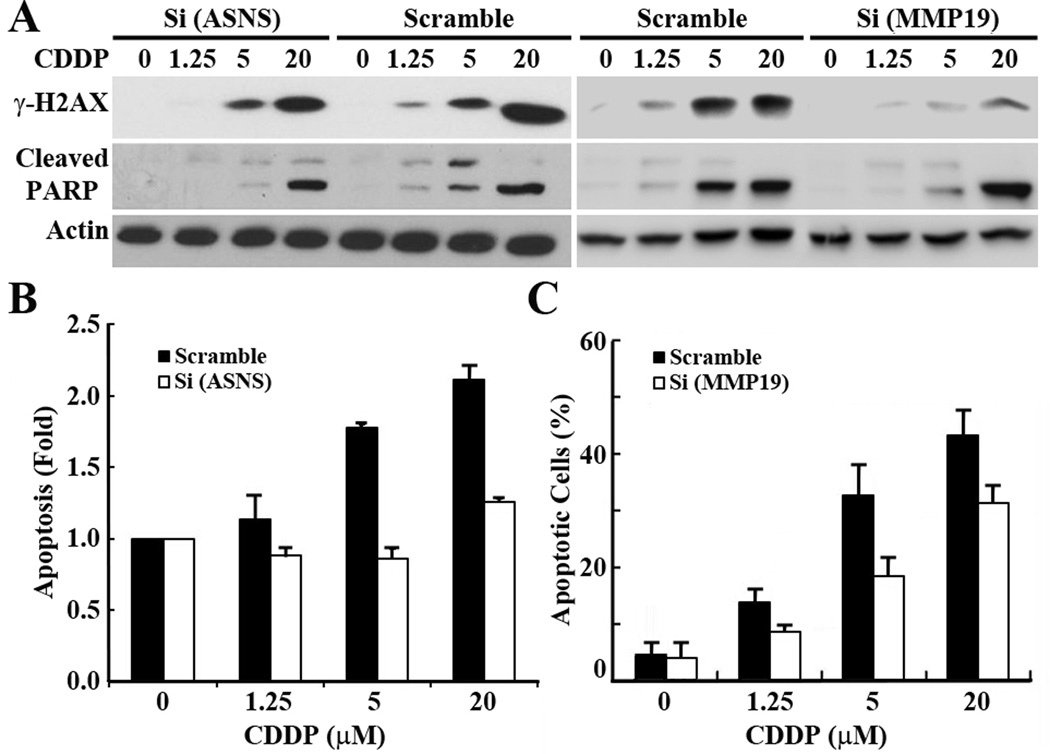
Cisplatin is known to exert its cytotoxicity by inducing DNA damage and apoptosis. To determine if ASNS and MMP19 over-expression in S16 cells possibly potentiates S16 cells to cisplatin-induced DNA damage and apoptosis, we first examined the effect of knocking down ASNS or MMP19 on cisplatin-induced DNA damage and apoptosis in S16 cells by evaluating the level of phosphorylated H2AX (γH2AX), a marker for DNA damage (17), and cleaved 85-kDa poly(ADP-ribose) polymerase (cPARP), a marker for early apoptosis (18) following cisplatin treatment. As shown in Fig. 5A, the level of γ-H2AX and cleaved PARP increased along with the escalating doses of cisplatin in the control S16 cells transfected with scrambled siRNA, indicating that more DNA damage and apoptosis may be induced by higher concentrations of cisplatin. However, the cisplatin-induced increase in γ-H2AX and cleaved PARP is attenuated by knocking down ASNS and MMP19, suggesting that both ASNS and MMP19 knockdown protects S16 cells against cisplatin-induced DNA damage and apoptosis. It is noteworthy that ASNS and MMP19 knockdown provided limited protection of cells against cisplatin-induced apoptosis with high concentration (20 µM) of cisplatin. The effect of ASNS and MMP19 knockdown on cisplatin-induced apoptosis was also examined using cell death detection ELISA and Hoechest 33342 staining of disintegrated nuclei, respectively, as previously described (6). As shown in Fig. 5B–5C, significantly less apoptosis was detected in the ASNS and MMP19 knockdown cells compared with the control cells transfected with scrambled siRNAs. Thus, up-regulated ASNS and MMP19 expression in S16 cells potentiates cisplatin-induced DNA damage and apoptosis.
Figure 5. Effect of MMP19 and ASNS knockdown on cisplatin-induced DNA damage and apoptosis.
S16 cells were transiently transfected with siRNAs against ASNS and MMP19 or scrambled control siRNA followed by treatment with cisplatin at different doses and analysis of DNA damage by detection of γ-H2AX and apoptosis by detection of cleaved PARP on Western blot (A), by staining of disintegrated nuclei (C), or by determination of cell death using ELISA (B). Actin was used as a loading control for Western blot analysis. The apoptosis assay and detection of γ-H2AX and cleaved PARP (cPARP) for ASNS knockdown cells were performed in the condition with supplementation of 0.4 mM Asn.
ASNS and MMP19 down-regulate the expression of NER and survival genes
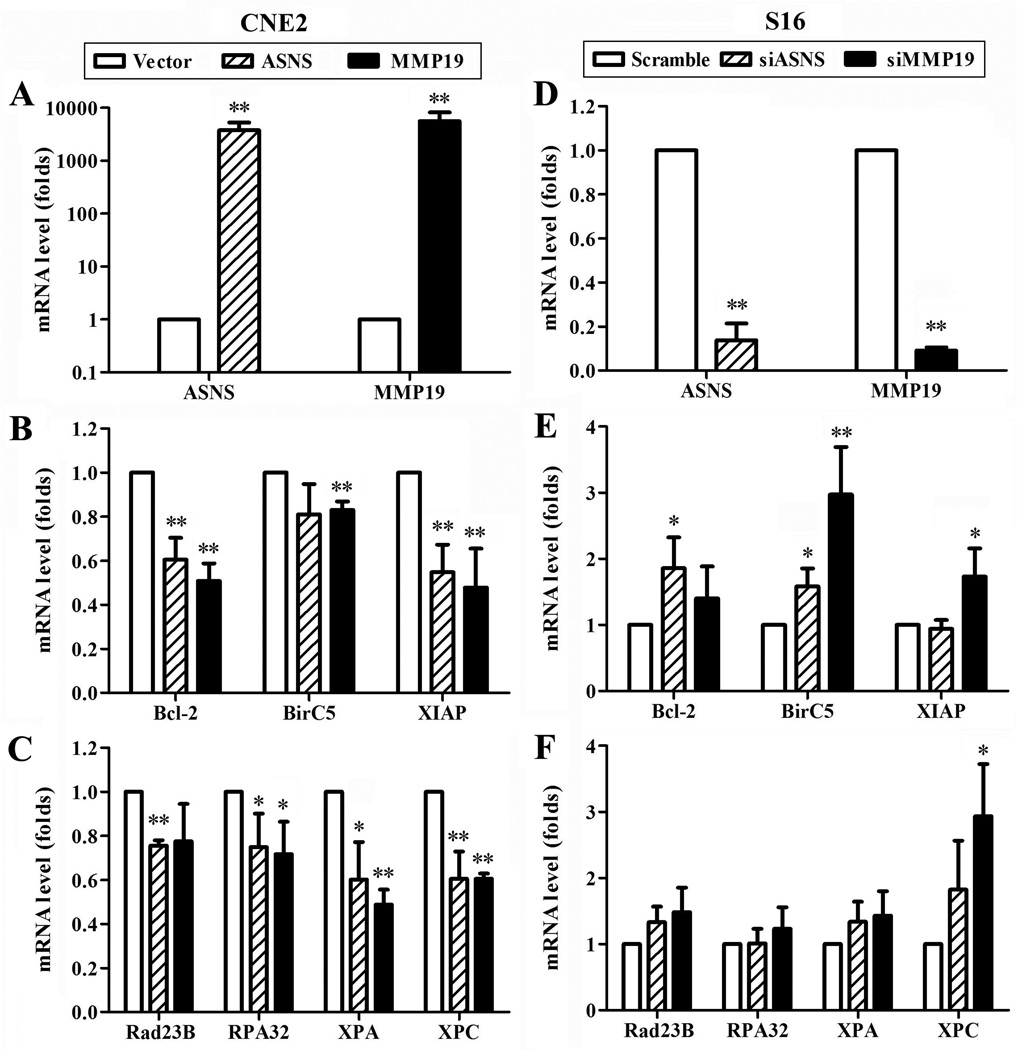
Considering that NER is the major pathway to repair cisplatin-induced DNA damages and that cisplatin-induced apoptosis can be abrogated by up-regulating survival factors, we next tested potential effects of ASNS or MMP19 on the expression of representative NER and survival genes by ectopic over-expressing or knocking down ASNS and MMP19 using real time RT-PCR. As shown in Fig. 6, ectopic over-expression of ASNS or MMP19 in CNE-2 cells significantly reduces the mRNA level of survival genes Bcl-2, XIAP, and BirC5 and NER genes Rad23B, RPA32, XPA, and XPC while knocking down ASNS or MMP19 expression in S16 cells increases the expression of these genes. These data suggest that ASNS and MMP19 over-expression may potentiate cisplatin-induced DNA damage and apoptosis by inhibiting the expression of NER and survival genes in nasopharyngeal carcinoma.
Figure 6. ASNS and MMP19 down-regulate the expression of survival genes and nucleotide excision repair genes.
CNE-2 (A–C) and S16 (D–F) cells were transiently transfected with cDNA and siRNAs, respectively, followed by real time RT-PCR analysis at 48 hrs after transfection for the mRNA levels of ASNS and MMP19 (A and D), survival genes Bcl-2, BirC5, and XIAP (B and E), and nucleotide excision repair genes (C and F). GAPDH was used as an internal control. The data of three independent experiments were presented as average ± SD. (*p<0.05; **p<0.01).
Discussion
Cisplatin is a widely used anticancer agent for the treatment of advanced NPC as well as a variety of other human cancers including that of lung, cervix, head and neck, colorectal, bladder, testis, and ovary (19–21). However, the efficacy of cisplatin is always limited by resistance from cancer cells and its toxicities to normal organs (22–24). Previously, various possible mechanisms of cisplatin resistance have been identified using cisplatin-selected resistant cell lines and these mechanisms include inactivation of cisplatin by glutathione, metallothionein or other sulphur-containing molecules; increased repair of cisplatin adducts; reduced cisplatin accumulation by changing the profile of uptake/efflux; increased cisplatin adducts tolerance and failure of apoptotic pathways (22, 24–31). In the present study, we performed a comparative gene profiling analysis of a cisplatin sensitive S16 cells derived from human NPC cell line CNE-2 using a different approach from commonly used drug selection but by clonal selection and limited dilution. We found that the expression of ASNS, CGA, and MMP19 in S16 cells was up-regulated compared with its parental CNE-2 cells. This finding was validated using real-time RT-PCR. However, only the increased level of ASNS and MMP19 could be validated at protein level using Western blot analysis. CGA protein could not be detected by Western blot, suggesting that the increased mRNA level of CGA in S16 cells did not result in detectable level of cellular CGA protein. The inability to detect cellular CGA could also be due to the fact that CGA is a secretory protein and it does not accumulate well in cells. Nevertheless, the observation that knocking down CGA has no effect on cellular response to cisplatin suggests that the increased CGA mRNA level in S16 cells does not contribute to the cisplatin sensitivity of this cell.
On the other hand, ASNS or MMP19 over-expression in S16 cells likely contribute to the increased cisplatin sensitivity of S16 cells compared to the parental CNE-2 cells. While knocking down ASNS and MMP19 expression reduces cisplatin sensitivity of S16 cells, ectopic over-expression of ASNS and MMP19 in the parental CNE-2 cells increases cisplatin sensitivity. We also found that knocking down ASNS and MMP19 expression in S16 cells reduces the level of cisplatin-induced DNA damage and apoptosis, suggesting that the increased ASNS and MMP19 level in S16 cells may attenuate cellular defense against cisplatin-induced DNA damage and apoptosis. Because ASNS and MMP19 inhibit the expression of NER and survival genes, it is likely that ASNS and MMP19 expression may also associates with other anticancer drug treatments and cellular response to these therapeutics.
It is noteworthy that the change in cisplatin sensitivity due to changes in ASNS and MMP19 is small but significant. Whether this small change in cisplatin sensitivity due to different levels of a single gene is relevant in clinical response is not yet known. However, clinical outcome is likely influenced by changes in multiple genes and it could be a result of additive effects of multiple genes. Our findings that multiple genes have altered expression in the S16 cells as demonstrated in this and a previous study (6) corroborate with this possibility.
ASNS is an enzyme responsible for catalyzing glutamine- and ATP-dependent conversion of aspartic acid to Asn, a known non-essential amino acid for mammalian cells (32). Some cancer cells such as T-cell leukemia are known to lack this enzyme and are dependent on external Asn and, thus, L-asparaginase has been used to treat theses cancers (33–35). However, how increased ASNS expression sensitizes NPC cells to cisplatin treatment is currently unknown. While cells with higher levels of ASNS may grow faster than cells with low level of ASNS possibly due to different level of Asn, the growth rate does not appear to be the cause of cisplatin sensitivity of S16 cells. Supplementation of Asn in culture media had no effect on survival of S16 cells with ASNS knockdown in the presence of cisplatin although it restored the growth rate of this cell line. This observation is consistent with previous studies in which supplementation of L-asparaginase to culture media did not affect cisplatin sensitivity of pancreatic cancer cells (36). Thus, ASNS may have additional function, which contributes to cellular sensitivity to cisplatin-induced DNA damage.
Previously, it has been reported that enhanced ASNS expression protects MiaPaCa-2 pancreatic cancer cells from apoptosis induced by cisplatin under glucose-deprived condition but not under normal condition (36). It appears that glucose deprivation induces ASNS over-expression in MiaPaCa-2 pancreatic cancer cells, which may suppress cisplatin-induced activation of JNK/SAPK and, thus, inhibiting cisplatin-induced cell death. This observation is different from our study in which ASNS up-regulation sensitizes cellular response to cisplatin treatment and potentiates cisplatin-induced cell death. The cause for this difference is currently unknown. However, it is possible that ASNS in different cancers (nasopharyngeal carcinoma vs pancreatic cancer) may play different roles in cellular sensitivity to cisplatin. It is also possible that glucose deprivation may modify ASNS function and make it a cellular defense mechanism to DNA damage-induced stress. Nevertheless, the findings from both studies consistently demonstrated that ASNS expression plays a role in cellular response to cisplatin treatment although further work is needed to delineate the differences of these studies and the detailed molecular mechanism of ASNS action in response to cisplatin treatment. Furthermore, the finding that ASNS over-expression in MiaPaCa-2 cells under glucose deprivation conditions causes resistance only to platinum drugs but not to etopside, paclitaxel, gemcitabine, and 5-FU (36), suggests that ASNS may affect cellular nucleotide excision repair (NER) system as the DNA damage induced by platinum-derived anti-cancer drugs are mainly repaired by NER (6). This is supported by our finding that cisplatin-induced production of γ-H2AX, an indicator of DNA damage, is attenuated by ASNS knockdown in S16 cells.
MMP19 belongs to the multi-protein family of zinc-binding matrix metalloproteinase (37, 38). While most MMPs are expressed under conditions involving extensive connective tissue remodeling (39) or neoplastic progression (40), MMP19 is mostly expressed in adult human normal placenta, lung, pancreas, ovary, spleen, and intestine (37). It, however, has been reported that MMP19 expression is down-regulated in NPC cell lines and NPC tissues as compared to normal control, lymphohyperplasia and adenoid tissues (41, 42) and that MMP19 may have tumor suppressor function in NPC (42). On the other hand, MMP19 expression has been found to increase in oropharyngeal squamous cell carcinoma, cutaneous melanomas and astroglial tumors, and promotes the invasions of glioma, melanoma and breast cancer cells (8, 43–45).
The findings that MMP19 is up-regulated in a cisplatin-sensitive cells and it contributes to cisplatin sensitivity is unprecedented. Although its mechanism of action in cisplatin response is currently unknown, the finding that MMP19 knockdown reduces cisplatin-induced production of γ-H2AX suggests that MMP19 up-regulation may attenuate NER activity to repair cisplatin-induced DNA damage, similar as ASNS. Previously, we have shown that eIF3a expression is also up-regulated in S16 cells and eIF3a suppresses the synthesis of NER proteins, resulting reduced NER activity and, thus, increased sensitivity to cisplatin (6). Both MMP19 and ASNS may work similarly as eIF3a to suppress NER in S16 cells. Indeed, we found that MMP19 and ASNS over-expression suppresses the expression of NER and survival genes although it is yet to be determined how MMP19 and ASNS genes regulate the expression of these downstream target genes.
Although no studies have been reported regarding MMP19 in cisplatin response, it has been shown recently that MMP7 and MMP13 expression positively correlates with cisplatin resistance in human head and neck cancer cell lines (46) although it is unknown if their higher expression level contributes to cisplatin resistance. Once again, this finding of the association study is in disagreement with our study although different MMPs were studied. However, the difference is not unexpected as MMP19 is considered a tumor suppressor gene for NPC but a tumor and metastasis promoting gene for other cancers (see discussion above)..
In summary, we found that the expression of ASNS, CGA and MMP19 genes is up-regulated at mRNA level in a cisplatin-sensitive clone S16 derived from human NPC cell line CNE-2. However, the changes could be validated at the protein level only for ASNS and MMP19. Similarly, only the up-regulated expression of ASNS and MMP19 but not CGA appears to contribute to the increased cisplatin sensitivity of S16 cells possibly by reducing cellular capacity to repair cisplain-induced DNA damage and, thus, reduced tolerance to cisplatin-induced apoptosis. Based this and our previous study (6), ASNS, MMP19, and eIF3a together all contributes to increased cisplatin sensitivity of S16 cells compared with its parental CNE-2 cells possibly by reducing NER repair activity and cisplatin-induced apoptosis.
Supplementary Material
Acknowledgments
Grant Support
This work was supported in part by grants from the National Institutes of Health R01CA94961 (J.T. Zhang), and National Natural Science Foundation of China (Grant No. 81071822 (R.Y. Liu), 81030043 (C.N. Qian), and 81272340 (C.N. Qian)).
Abbreviations
- NPC
nasopharyngeal carcinoma
- ASNS
asparagine synthetase
- CGA
choriogonadotropin α subunit
- MMP19
matrix metalloproteinase 19
- cisplatin
cis-Dichlorodiammine platinum(II)
- Asn
asparagine
Footnotes
All authors declare no conflict of interest for the studies reported in this manuscript.
References
- 1.Adham M, Kurniawan AN, Muhtadi AI, Roezin A, Hermani B, Gondhowiardjo S, et al. Nasopharyngeal carcinoma in Indonesia: epidemiology, incidence, signs, and symptoms at presentation. Chinese journal of cancer. 2012;31:185–196. doi: 10.5732/cjc.011.10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao SM, Simons MJ, Qian CN. The prevalence and prevention of nasopharyngeal carcinoma in China. Chinese journal of cancer. 2011;30:114–119. doi: 10.5732/cjc.010.10377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wee JT, Ha TC, Loong SL, Qian CN. Is nasopharyngeal cancer really a "Cantonese cancer"? Chinese journal of cancer. 2010;29:517–526. doi: 10.5732/cjc.009.10329. [DOI] [PubMed] [Google Scholar]
- 4.Baujat B, Audry H, Bourhis J, Chan AT, Onat H, Chua DT, et al. Chemotherapy in locally advanced nasopharyngeal carcinoma: an individual patient data meta-analysis of eight randomized trials and 1753 patients. Int J Radiat Oncol Biol Phys. 2006;64:47–56. doi: 10.1016/j.ijrobp.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 5.Thephamongkhol K, Zhou J, Browman G, Wong R, Chan J, Zeng Y, et al. Chemo-radiotherapy versus radiotherapy alone for nasopharyngeal carcinoma: A meta-analysis of 78 randomized controlled trials (RCTs) from English and non-English databases. J Clin Oncol. 2004;22:493s. [Google Scholar]
- 6.Liu RY, Dong Z, Liu J, Yin JY, Zhou L, Wu X, et al. Role of eIF3a in regulating cisplatin sensitivity and in translational control of nucleotide excision repair of nasopharyngeal carcinoma. Oncogene. 2011;30:4814–4823. doi: 10.1038/onc.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lorenzi PL, Reinhold WC, Rudelius M, Gunsior M, Shankavaram U, Bussey KJ, et al. Asparagine synthetase as a causal, predictive biomarker for L-asparaginase activity in ovarian cancer cells. Mol Cancer Ther. 2006;5:2613–2623. doi: 10.1158/1535-7163.MCT-06-0447. [DOI] [PubMed] [Google Scholar]
- 8.Hegedus L, Cho H, Xie X, Eliceiri GL. Additional MDA-MB-231 breast cancer cell matrix metalloproteinases promote invasiveness. J Cell Physiol. 2008;216:480–485. doi: 10.1002/jcp.21417. [DOI] [PubMed] [Google Scholar]
- 9.Sizhong Z, Xiukung G, Yi Z. Cytogenetic studies on an epithelial cell line derived from poorly differentiated nasopharyngeal carcinoma. Int J Cancer. 1983;31:587–590. doi: 10.1002/ijc.2910310509. [DOI] [PubMed] [Google Scholar]
- 10.Yang Y, Chen Q, Zhang JT. Structural and functional consequences of mutating cysteine residues in the amino terminus of human multidrug resistance-associated protein 1. J Biol Chem. 2002;277:44268–44277. doi: 10.1074/jbc.M207003200. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi M, Rhodes DR, Furge KA, Kanayama H, Kagawa S, Haab BB, et al. Gene expression profiling of clear cell renal cell carcinoma: gene identification and prognostic classification. Proc Natl Acad Sci U S A. 2001;98:9754–9759. doi: 10.1073/pnas.171209998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray SG, Qian CN, Furge K, Guo X, Teh BT. Microarray profiling of the effects of histone deacetylase inhibitors on gene expression in cancer cell lines. Int J Oncol. 2004;24:773–795. doi: 10.3892/ijo.24.4.773. [DOI] [PubMed] [Google Scholar]
- 13.Guo X, Lui WO, Qian CN, Chen JD, Gray SG, Rhodes D, et al. Identifying cancer-related genes in nasopharyngeal carcinoma cell lines using DNA and mRNA expression profiling analyses. Int J Oncol. 2002;21:1197–1204. [PubMed] [Google Scholar]
- 14.Dong Z, Liu Z, Cui P, Pincheira R, Yang Y, Liu J, et al. Role of eIF3a in regulating cell cycle progression. Exp Cell Res. 2009;315:1889–1894. doi: 10.1016/j.yexcr.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 15.Peng H, Dong Z, Qi J, Yang Y, Liu Y, Li Z, et al. A Novel Two Mode-Acting Inhibitor of ABCG2-Mediated Multidrug Transport and Resistance in Cancer Chemotherapy. PLoS ONE. 2009;4:e5676. doi: 10.1371/journal.pone.0005676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu H, Liu Y, Zhang JT. A new mechanism of drug resistance in breast cancer cells: fatty acid synthase overexpression-mediated palmitate overproduction. Mol Cancer Ther. 2008;7:263–270. doi: 10.1158/1535-7163.MCT-07-0445. [DOI] [PubMed] [Google Scholar]
- 17.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 18.Kaufmann SH, Desnoyers S, Ottaviano Y, Davidson NE, Poirier GG. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 1993;53:3976–3985. [PubMed] [Google Scholar]
- 19.Kong L, Zhang YW, Hu CS, Guo Y. Neoadjuvant chemotherapy followed by concurrent chemoradiation for locally advanced nasopharyngeal carcinoma. Chinese journal of cancer. 2010;29:551–555. doi: 10.5732/cjc.009.10518. [DOI] [PubMed] [Google Scholar]
- 20.Desoize B, Madoulet C. Particular aspects of platinum compounds used at present in cancer treatment. Crit Rev Oncol Hematol. 2002;42:317–325. doi: 10.1016/s1040-8428(01)00219-0. [DOI] [PubMed] [Google Scholar]
- 21.Lebwohl D, Canetta R. Clinical development of platinum complexes in cancer therapy: an historical perspective and an update. Eur J Cancer. 1998;34:1522–1534. doi: 10.1016/s0959-8049(98)00224-x. [DOI] [PubMed] [Google Scholar]
- 22.Martin LP, Hamilton TC, Schilder RJ. Platinum resistance: the role of DNA repair pathways. Clin Cancer Res. 2008;14:1291–1295. doi: 10.1158/1078-0432.CCR-07-2238. [DOI] [PubMed] [Google Scholar]
- 23.Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007;33:9–23. doi: 10.1016/j.ctrv.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 25.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 26.Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shah NP. Bench to bedside: BRCA: from therapeutic target to therapeutic shield. Nat Med. 2008;14:495–496. doi: 10.1038/nm0508-495. [DOI] [PubMed] [Google Scholar]
- 28.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 29.Howell SB, Safaei R, Larson CA, Sailor MJ. Copper transporters and the cellular pharmacology of the platinum-containing cancer drugs. Mol Pharmacol. 2010;77:887–894. doi: 10.1124/mol.109.063172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo MT, Chen HH, Song IS, Savaraj N, Ishikawa T. The roles of copper transporters in cisplatin resistance. Cancer Metastasis Rev. 2007;26:71–83. doi: 10.1007/s10555-007-9045-3. [DOI] [PubMed] [Google Scholar]
- 31.Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 32.Richards NG, Schuster SM. Mechanistic issues in asparagine synthetase catalysis. Adv Enzymol Relat Areas Mol Biol. 1998;72:145–198. doi: 10.1002/9780470123188.ch5. [DOI] [PubMed] [Google Scholar]
- 33.Pieters R, Hunger SP, Boos J, Rizzari C, Silverman L, Baruchel A, et al. L-asparaginase treatment in acute lymphoblastic leukemia: a focus on Erwinia asparaginase. Cancer. 2011;117:238–249. doi: 10.1002/cncr.25489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avramis VI, Tiwari PN. Asparaginase (native ASNase or pegylated ASNase) in the treatment of acute lymphoblastic leukemia. Int J Nanomedicine. 2006;1:241–254. [PMC free article] [PubMed] [Google Scholar]
- 35.Muller HJ, Boos J. Use of L-asparaginase in childhood ALL. Crit Rev Oncol Hematol. 1998;28:97–113. doi: 10.1016/s1040-8428(98)00015-8. [DOI] [PubMed] [Google Scholar]
- 36.Cui H, Darmanin S, Natsuisaka M, Kondo T, Asaka M, Shindoh M, et al. Enhanced expression of asparagine synthetase under glucose-deprived conditions protects pancreatic cancer cells from apoptosis induced by glucose deprivation and cisplatin. Cancer Res. 2007;67:3345–3355. doi: 10.1158/0008-5472.CAN-06-2519. [DOI] [PubMed] [Google Scholar]
- 37.Pendas AM, Knauper V, Puente XS, Llano E, Mattei MG, Apte S, et al. Identification and characterization of a novel human matrix metalloproteinase with unique structural characteristics, chromosomal location, and tissue distribution. J Biol Chem. 1997;272:4281–4286. doi: 10.1074/jbc.272.7.4281. [DOI] [PubMed] [Google Scholar]
- 38.Mueller MS, Mauch S, Sedlacek R. Structure of the human MMP-19 gene. Gene. 2000;252:27–37. doi: 10.1016/s0378-1119(00)00236-5. [DOI] [PubMed] [Google Scholar]
- 39.Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8:221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu J, Chua HH, Chen SY, Chen JY, Tsai CH. Regulation of matrix metalloproteinase-1 by Epstein-Barr virus proteins. Cancer Res. 2003;63:256–262. [PubMed] [Google Scholar]
- 42.Chan KC, Ko JM, Lung HL, Sedlacek R, Zhang ZF, Luo DZ, et al. Catalytic activity of Matrix metalloproteinase-19 is essential for tumor suppressor and anti-angiogenic activities in nasopharyngeal carcinoma. Int J Cancer. 2011;129:1826–1837. doi: 10.1002/ijc.25855. [DOI] [PubMed] [Google Scholar]
- 43.Velinov N, Aebersold D, Haeni N, Hlushchuk R, Mishev G, Weinstein F, et al. Matrix metalloproteinase-19 is a predictive marker for tumor invasiveness in patients with oropharyngeal squamous cell carcinoma. Int J Biol Markers. 2007;22:265–273. [PubMed] [Google Scholar]
- 44.Muller M, Beck IM, Gadesmann J, Karschuk N, Paschen A, Proksch E, et al. MMP19 is upregulated during melanoma progression and increases invasion of melanoma cells. Mod Pathol. 2010;23:511–521. doi: 10.1038/modpathol.2009.183. [DOI] [PubMed] [Google Scholar]
- 45.Lettau I, Hattermann K, Held-Feindt J, Brauer R, Sedlacek R, Mentlein R. Matrix metalloproteinase-19 is highly expressed in astroglial tumors and promotes invasion of glioma cells. J Neuropathol Exp Neurol. 2010;69:215–223. doi: 10.1097/NEN.0b013e3181ce9f67. [DOI] [PubMed] [Google Scholar]
- 46.Ansell A, Jerhammar F, Ceder R, Grafstrom R, Grenman R, Roberg K. Matrix metalloproteinase-7 and -13 expression associate to cisplatin resistance in head and neck cancer cell lines. Oral Oncol. 2009;45:866–871. doi: 10.1016/j.oraloncology.2009.02.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.