Abstract
The Electronic Medical Records and Genomics Network is a National Human Genome Research Institute–funded consortium engaged in the development of methods and best practices for using the electronic medical record as a tool for genomic research. Now in its sixth year and second funding cycle, and comprising nine research groups and a coordinating center, the network has played a major role in validating the concept that clinical data derived from electronic medical records can be used successfully for genomic research. Current work is advancing knowledge in multiple disciplines at the intersection of genomics and health-care informatics, particularly for electronic phenotyping, genome-wide association studies, genomic medicine implementation, and the ethical and regulatory issues associated with genomics research and returning results to study participants. Here, we describe the evolution, accomplishments, opportunities, and challenges of the network from its inception as a five-group consortium focused on genotype–phenotype associations for genomic discovery to its current form as a nine-group consortium pivoting toward the implementation of genomic medicine.
Genet Med 15 10, 761–771.
Keywords: collaborative research, electronic medical records, genetics and genomics, genome-wide association studies, personalized medicine
The Electronic Medical Records and Genomics (eMERGE) Network is a National Human Genome Research Institute (NHGRI)–funded consortium tasked with developing methods and best practices for the utilization of the electronic medical record (EMR) as a tool for genomic research. The eMERGE Network comprises nine geographically distinct groups (Figure 1), each with its own biorepository where DNA specimens are linked to phenotypic data contained within EMRs. The large number of study participants and considerable diversity of the network sites provide a unique opportunity to conduct cost-effective studies in genomic medicine. Longitudinal phenotypic data already contained within EMRs linked to each group's biorepository can be extracted and repurposed so that cases and controls for a large number of phenotypes can be collected efficiently and merged across eMERGE Network sites. These data can then be combined with genomic data for the discovery of genotype–phenotype associations, and these discoveries, once validated, may be introduced back into the EMR to augment clinical care (Figure 2).
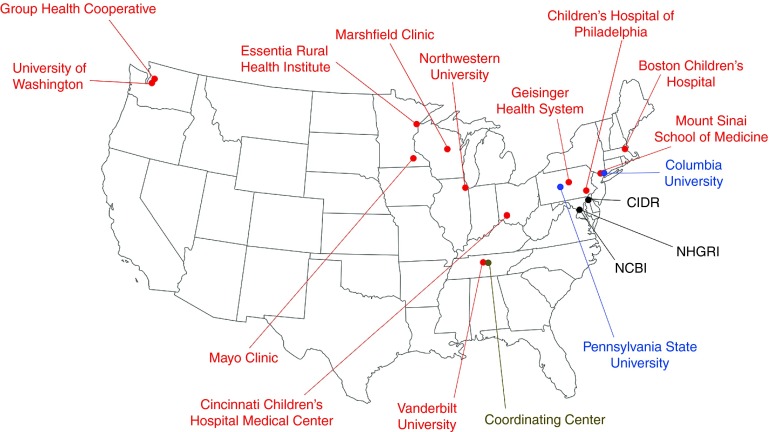
Figure 1.
Locations of member sites, affiliates, and support and service centers of the eMERGE Network. Red color indicates the nine members of eMERGE- II, gray color indicates the eMERGE Coordinating Center, blue color indicates an eMERGE affiliate or subcontract site, and black color indicates centers that provide services and support to eMERGE. eMERGE, Electronic Medical Records and Genomics.
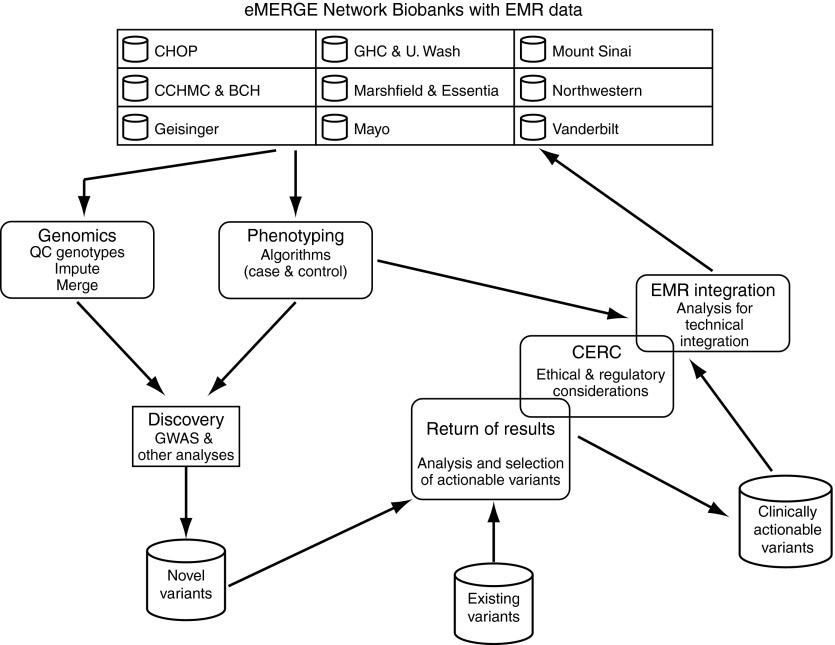
Figure 2.
Outline of the activities in the eMERGE Network. The main activities of the network and how they are integrated together are summarized. See text for details. eMERGE, Electronic Medical Records and Genomics; EMR, electronic medical record; GWAS, genome-wide association studies.
Now in its sixth year and second funding cycle, the network continues to make advances in multiple disciplines related to the fields of genomics and health-care informatics. Locations of the nine research groups, their affiliated sites, a coordinating center, and the services and support centers constituting the current eMERGE Network are shown in Figure 1. Outlines of the activities of the eMERGE Network are shown in Figure 2, and the organizational structure of the network is represented in Figure 3. Details of the biorepositories, EMR systems, and genotyping projects are summarized in Table 1, and goals of the projects at each eMERGE site are listed in Supplementary Table S1 online. The primary and secondary phenotypes selected by each site are summarized in Supplementary Table S2 online. Additional site and project descriptions were authored by each site and are presented in the Supplementary Materials online. In the following sections, we describe the evolution of the network in the context of the rapidly changing landscape of genomic medicine.
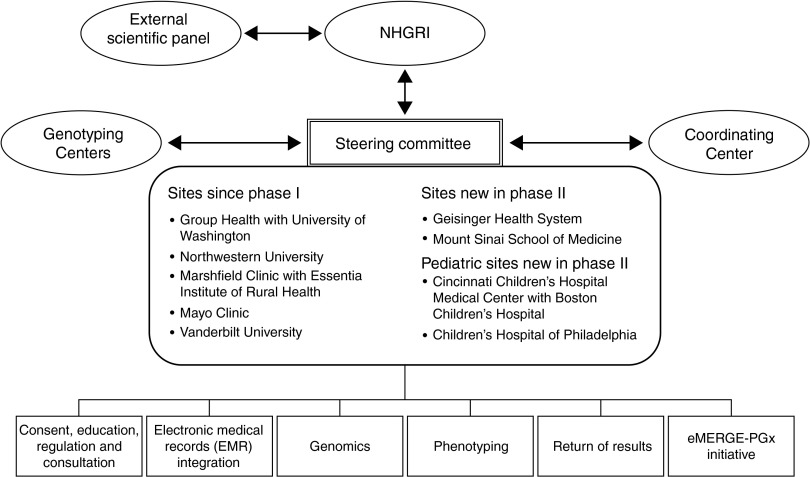
Figure 3.
Structure of eMERGE Network. The Steering Committee, composed of the principal investigators from each institution and the NIH Project Scientist, is the governing body for the consortium. The External Scientific Panel provides input to the NHGRI director about the progress and direction of the network. The Coordinating Center provides centralized support and infrastructure. Genotyping Centers provide genotyping under CLIA certification for clinically actionable genetic variants. For details on the activities by the workgroups listed at the bottom of the figure, please see the main text. CLIA, Clinical Laboratory Improvement Amendments; eMERGE, Electronic Medical Records and Genomics; NHGRI, National Human Genome Research Institute; NIH, National Institutes of Health.
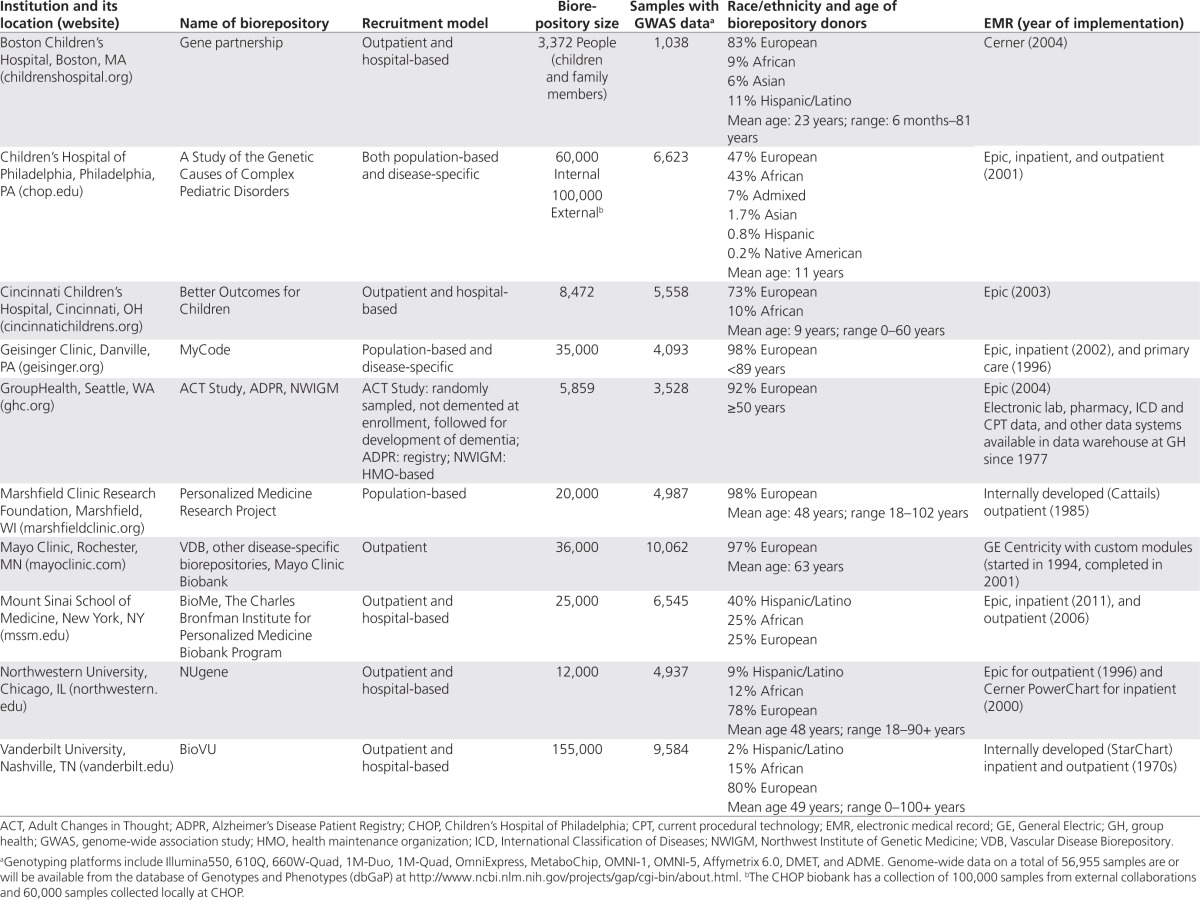
Table 1. Summary of biorepositories, electronic medical records, and available genome-wide data at 10 eMERGE institutions.
Summary of Phase I Scope and Aims
A request for applications from the NHGRI for eMERGE was released in March 20071 and was intended “to provide support for investigative groups affiliated with existing biorepositories to develop … methods and procedures for genome-wide studies in participants with phenotypes … derived from EMR.” In September 2007, grants were awarded to five sites (hereafter referred to as eMERGE-I)—Group Health Cooperative/University of Washington, Marshfield Clinic, Mayo Clinic, Northwestern University, and Vanderbilt University, which also served as the network's coordinating center.
eMERGE-I had three major aims: (i) use EMR data for robust electronic phenotyping, (ii) conduct genome-wide association studies (GWAS) using the phenotypes derived in the above-mentioned first aim, and (iii) explore the ethical, legal, and social implications associated with EMR-based GWAS and wide-scale data sharing. The network formed workgroups that became the main drivers of progress in the key subject areas. In eMERGE-I, the workgroups included an informatics group, a genomics group, and a consent and community consultation group. Besides numerous publications (for a complete listing, see http://www.gwas.org), the workgroups had several accomplishments that were fundamental to the aims of phase I. The consent and community consultation group published model consent language for EMR-linked biorepositories, intended to harmonize the consent process for the collection and storage of human biospecimens and data for future research, particularly those collections that have an EMR component.2 The genomics workgroup created a unified data set of genotyped samples across all sites and published a “how to” paper that outlined the procedures and lessons learned from combining genotype data across a research network. The documented pitfalls of merging data from different genotyping facilities (even when generated on the same genotyping platform), such as inconsistencies in strand orientation, sample relatedness and population stratification across sites, site-specific batch effects, and errors introduced in the merging process, are of relevance to any group attempting to merge data from multiple sites.3 The informatics workgroup created and published a library of EMR-based phenotyping algorithms accrued throughout phase I that is available to investigators outside of the eMERGE Network.4
Lessons Learned from Phase I
Much of the success of eMERGE-I resulted from utilizing the full capacity of the network, and several key lessons learned were used to augment its structure.5
Although the founding sites initially focused on projects relating to phenotypes of local interest as well as joint projects, it became clear that projects had better outcomes when deployed across the network. Development of a phenotype algorithm was generally led by one site and then deployed at a second site. The issues encountered as the second site implemented the algorithm led to revisions that made it more robust and generalizable when deployed across the network. In addition, there was increased statistical power when cases and controls were shared. The eMERGE Network has played a major role in validating the concept that phenotypes derived from EMRs can be used successfully for GWAS and has disseminated its methods and findings extensively.6,7,8,9,10,11,12,13
Most eMERGE participants have consented to contributing their data to health research of any kind. However, whenever combining large data sets pertaining to individual-level information such as health or genomic data, even when fully deidentified, there exists the potential risk for the identification of individuals. Through network-wide projects, eMERGE-I was compelled to develop best practices for the sharing of genomic data and EMR-derived phenotypes while protecting the privacy of participants, and these have been published to aid other investigators engaged in the field.14,15,16,17,18,19
The issue of returning research results to participants emerged as another key point for discussion as network analyses identified individual-level chromosomal anomalies such as Klinefelter and Turner syndromes. In response, the network convened a return of results (RoR) oversight committee to provide ongoing support and clinical information on incidental findings from GWAS. These discussions were also brought to local constituencies for final decision making. The process is outlined and published and may form the basis for a deliberative model for adoption by other collaborative research groups faced with similar challenges.20
Transition to Phase II (eMERGE-II)
The key advances and challenges encountered in phase I were instrumental in shaping the goals of eMERGE as the network transitioned to phase II in August 2011 following a second request for applications.21 The memberships of the five initial sites were renewed and two new sites, Geisinger Clinic and Mount Sinai School of Medicine, were added. A separate award for the network coordinating center was granted to Vanderbilt University. In August 2012, following a request for applications22 for pediatric sites, eMERGE membership was extended to Children's Hospital of Philadelphia and a joint membership for Cincinnati Children's Hospital and Boston Children's Hospital (Figure 1). In particular, the new, larger network was interested in broadening its scope from using EMR data for discovery of genotype–phenotype associations all the way through to incorporation of genotype data into the EMR (Figure 2). This would allow the network to assess the utility of these results in clinical decision making such as informing clinicians of relevant pharmacogenomic (PGx) variants before a drug is prescribed or identifying persons at high genomic risk for a given condition.
This new focus required restructuring of the eMERGE-I workgroups for phase II. eMERGE-II introduced workgroups on EMR integration of genomic information, return of genomic results, and PGx, which was designed to address the complexities of linking genetic variation data with EMRs for effective clinical use as well as to address the difficulties in determining which results to use and how to return these results to participants and providers. The consent and community consultation group was restructured to include focus on clinician and patient education, and the informatics workgroup was restructured to become the phenotyping workgroup. As in phase I, an External Scientific Panel was formed to meet annually with eMERGE-II investigators in order to challenge the focus of the network and to encourage appropriate dissemination of products and lessons learned (Figures 2 and 3).
Major Goals and Activities of eMERGE Phase II
The eMERGE Network continues to discover genomic variants associated with clinical conditions identified using EMRs and to develop algorithms for electronic phenotyping. Building on this success, the network is now extending its focus to pilot studies for implementing genomic medicine through the EMR.23 Critical goals include determining the optimal methods and infrastructure needed for aspects such as patient consent, laboratory assays, RoR, integrating findings into the EMR, and providing sufficient decision support and patient/clinician education to use them effectively (Figure 2). These components are essential to facilitating the translation of genomic medicine from bench to bedside. To illustrate the regular activities of the eMERGE-II workgroups, case studies detailing a typical project have been authored by each group.
Phenotyping workgroup: phenotype algorithm development and PheKB
The phenotyping workgroup has as its goal the creation, validation, and execution of phenotype algorithms across the network and beyond. To aid in this process, investigators have developed Phenotype KnowledgeBase (PheKB),4 a repository for phenotype algorithms. Users can read, upload, search, and provide feedback on the algorithms and upload a variety of documents and metadata. Algorithms can be published and shared publicly or restricted to a particular collaborative group within a social networking framework to facilitate development and revising of the phenotypes. Users can comment and ask questions on phenotypes, receive e-mail notification when updates are made, and create “implementation” records, which capture site-specific validation of a phenotype algorithm. In eMERGE, phenotype algorithms on PheKB are validated at the creating site as well as at least 1–2 other institutions. PheKB is currently searchable by metadata fields.
Genomics workgroup: genotype imputation to facilitate network-wide genetic studies
To allow for the aggregation of genomic and phenotype data across all eMERGE sites, a genotype imputation pipeline was implemented to create a single and uniform data set for all individuals genotyped across the network. Genotype imputation is the process of inferring unobserved genotypes in a sample based on the haplotypes observed in a more densely genotyped reference sample. Imputation is computationally intensive and involves several steps including phasing the haplotypes, filling in the missing genotypes, and finally assembling and assessing the accuracy of the data. Version 1.0 of the eMERGE imputed data set includes more than 13 million single-nucleotide polymorphisms in more than 42,000 samples that have been imputed using the BEAGLE reference panel24 and the 1000 Genomes25 cosmopolitan reference panel, October 2011 release. The imputation process for eMERGE-II consumed ~1.1 × 106 CPU h.
RoR workgroup: penetrance of hemochromatosis mutations
The genetic and EMR data available in the eMERGE Network provide an opportunity to estimate the penetrance of genetic diseases, such as hemochromatosis, a common autosomal-recessive disorder of increased iron absorption, and subsequent adult-onset iron overload. Most individuals have C282Y or H63D mutations in the HFE gene but are asymptomatic. Homozygous and compound heterozygous adults for these HFE mutations will be identified from the eMERGE cohort, and a chart review will be carried out to establish the prevalence of hemochromatosis as well as the penetrance of related phenotypes. Because iron overload can be easily screened for and treated by phlebotomy, the cost–benefit of genetic screening is dependent on penetrance. The RoR workgroup is collaborating with the consent, education, regulation, and consultation workgroup on issues related to the process of returning clinically relevant HFE variants.
Consent, education, regulation, and consultation workgroup: evaluating the impact of returning hemochromatosis results
The consent, education, regulation, and consultation workgroup is working closely with the RoR workgroup on issues relating to the return of hemochromatosis results. Although there is compelling evidence that medical management of hemochromatosis can provide benefit to those with penetrant disease, a number of issues relating to the penetrance of HFE variants remain when making the decision to return results: is it possible to safely return low-penetrant results without unduly alarming participants and health-care providers? Do patients and their health-care providers find this information valuable? How do these decisions impact health-care costs? To answer some of these questions, the workgroup is developing a protocol to deliver HFE results and assess their impact. Education of research participants and health-care providers about low-risk genetic test results before the results are returned is critical. The effectiveness of educational tools, including those used within the EMR will be evaluated, and the amount of pre- and postreturn education required will be studied.
EMR integration workgroup: PGx pilot project
A major challenge in implementing genomic medicine is presenting relevant information to clinicians at the point of care. The increase of actionable genomic information needs to be matched with development and implementation of knowledge-based clinical decision support (CDS) systems deployed through EMRs. The eMERGE PGx project (also discussed in the next section) will preemptively genotype drug-naive patients who have an increased probability of receiving target drugs, primarily clopidogrel, warfarin, or simvastatin, in the next 3 years. The network consensus is that there is sufficient evidence and guidelines for preliminary clinical implementation of genotype-guided prescribing for these medications.26 For study patients, prescription of any one of these three drugs placed in computerized order entry systems will automatically trigger processing of clinical and genomic data. If predefined rules are met, information will be presented to the ordering clinician that could inform dosing or medication choice. Clinicians' decisions to use or disregard the information will be analyzed along with feedback to identify factors that promote or impede implementation. The outcomes measured in eMERGE-PGx will be primarily process outcomes (e.g., number of patients identified with an actionable pharmaceutical genotype, number of times a CDS rule fires, percentage of clinicians who follow recommendation, and appropriate changes in medication or dose based on recommendation). However, sites that are farther along the translation spectrum plan to include measurement of some health outcomes, including documented adverse drug reaction within 24 h of initiation of opioid medication, development of myopathy, and adherence to medication.
Collaborations with external groups
Of the lessons learned through the eMERGE experience, none is more prominent than that of collaboration. The many individuals and groups with diverse geography, experience, and expertise that constitute eMERGE have undoubtedly increased both the yield and quality of our work. The tools created by eMERGE investigators, as well as the genomic and clinical databases within the network, provide valuable resources for collaborations. In addition to collaborations within and between the eMERGE sites and workgroups, the network is also working closely with other groups focused on similar goals and activities.
The NHGRI's 2011 Strategic Plan emphasized implementation of genomic medicine, leading to the formation of the genomic medicine working group27 with members from more than 40 eMERGE and non-eMERGE institutions.28 The genomic medicine working group provides guidance to NHGRI and organizes meetings to discuss diverse implementation issues and develop pilot implementation projects.
Another key example of successful external collaboration is the eMERGE-PGx project, developed with the Pharmacogenetics Research Network.29 eMERGE-PGx will deploy targeted next-generation sequencing of 84 very important pharmacogenes. The activities of eMERGE-PGx include (i) clinical reporting restricted to very important pharmacogenes with evidence for “actionability” such as those included in guidelines promulgated by the Pharmacogenetics Research Network's Clinical Pharmacogenomics Implementation Consortium;26 (ii) preemptive testing and presentation of “actionable” variants in the EMR with CDS at the point of care; and (iii) creating a repository of the other very important pharmacogene variants that will enable future genotype–phenotype studies.
The eMERGE Network has also forged successful links with other NHGRI-funded consortia including the Population Architecture Using Genomics and Epidemiology Consortium,30 the Return of Results Consortium,31 and the Clinical Sequencing Exploratory Research Program.32 These links have allowed the network to exchange expertise with other groups doing complementary and often synergistic work in the genomic medicine domain.
The eMERGE Steering Committee has established guidelines on how external institutions can apply for affiliate membership to the eMERGE Network (http://www.gwas.org), and this is strongly encouraged.
eMERGE Phase II Network Opportunities, Challenges, and Lessons Learned
The combined resources of the eMERGE Network provide opportunities accompanied by some significant challenges, which the workgroups are addressing. Some notable examples are highlighted below.
Portability of electronic phenotypes within and outside eMERGE
There is currently no formal “phenotyping language” for the purpose of building EMR phenotyping algorithms nor is there a common approach to their implementation. Developing portable phenotyping algorithms is an area of high priority in eMERGE, with a view to easing implementation within and outside the network. One potential solution is the National Quality Forum's Quality Data Model, an XML-based information model for representing EMR-based quality measures to support meaningful-use reporting requirements.33,34,35 Nine algorithms have been implemented using the Quality Data Model, and eMERGE investigators are testing Drools36 and Konstanz Information Miner37 as common execution engines. The network's experiences will be formally documented and disseminated to the community.
Approaches to EMR integration of genomic information
EMRs and CDS systems can improve the quality of care and reduce adverse drug events,38,39,40,41 but no commercial EMR integrates pharmacogenetic information systematically even though the US Food and Drug Administration drug labels include pharmacogenetic variants for 105 drugs in 117 contexts.42 Nomenclatures and ontologies,43 such as SNOMED-CT and LOINC, reasonably represent concepts related to genetic tests, but mechanisms for long-term storage of genomic data as well as secure, generalizable, and interoperable data exchange between health-care settings are needed to ensure continuity of care.44 Given that most of the genomic data gained through high-density genotyping arrays or whole-exome/whole-genome sequencing are not actionable at this time, and that knowledge and interpretation are changing rapidly, the data will likely be stored external to the EMR.45 eMERGE is investigating external CDS, but there is no standard for external CDS and subsequent user actions (e.g., placing an order). An external CDS engine cannot specify choices for what happens next, whereas integrated CDS can specify a litany of options. eMERGE is collaborating with the Clinical Decision Support Consortium46 and participating in other national efforts to address these issues. These interactions are expected to lead to the establishment of a standard for genome-informed CDS.
Integration of pediatric sites
The addition of pediatric eMERGE sites affords opportunities to explore new phenotypes and data sets while posing several challenges. Integration of pediatric and adult projects into one eMERGE Network is nontrivial but could provide valuable information about heritable diseases that present early in life and continue to adulthood. In theory, identifying genetic contributions to complex diseases should be easier in children because environmental exposures have less time to take effect. A study of childhood obesity47 in which in addition to replicating adult obesity loci, novel loci were identified, supports this hypothesis. The network's experiences in combining adult and pediatric data will produce insights that are useful beyond the genomics community to large, heterogeneous collaborative research endeavors in general.
Longitudinal cost-effective genomic medicine discovery and implementation
The size and diversity of the collective eMERGE biobank and the rich EMR-linked phenotypic data provide a unique opportunity for cost-effective longitudinal studies in genomic medicine, permitting study of incident disease, age, and period biases,48 as well as reducing prevalence and incidence bias.49 Continued collection of data in the clinical setting at no additional cost to the research program not only increases its value and utility over time but may also necessitate informing participants about new interpretations of the results, either because knowledge about significant health impacts of identified variants50 is accruing rapidly or because new conditions or use of new medications change the risk profile context for the individual. The burden, ethics, and costs of revisiting genomic variation in a given person, as knowledge evolves about that person and the variation he/she carries, will continue to be a significant focus of the eMERGE Network. Any lessons learned are likely to be of great importance to the genomic medicine community as we near the possibility of comprehensive genomic information being the norm in clinical care.
Generalizable framework for the return of genomic results
The opportunities gained through longitudinal genomic discovery are strongly correlated with the challenges of returning results. It is generally accepted that results with an immediate impact on a person's health should be returned to the research participant.50,51,52,53 There is, however, far less consensus on how “medically actionable” or the related concept of “clinical utility” should be defined.53,54 Returning genomic research results raises practical, financial, psychosocial, and ethical challenges for both investigators and patients.53 The network is investigating models that allow patients to make choices about their results, evaluating the benefits and costs of returning results,50 and has also initiated consultation about returning research results with stakeholders, including physicians, patients, advisory committees, laboratory directors, and health plans.
The eMERGE network in the context of a translational framework
Implementing genomic medicine in the clinic is part of the strategic vision of the NHGRI and has been discussed recently.28,55,56 Five phases of moving genomic research into practice and policy have been defined,57,58,59 with the early phases focusing on biologic discoveries (T0), development of candidate health applications (T1), and assessing outcomes of interventions (T2). eMERGE-I focused largely on the T0 discovery phase through GWAS. eMERGE-II is developing T1 applications such as genomic risk prediction algorithms and clinically validated PGx assays, while continuing T0 discovery research through GWAS and phenome-wide association studies.10 eMERGE is not powered to assess outcomes directly (T2) but is building upon available literature and expert opinion to investigate how best to move genomic findings into health practice (T3) in its pilot implementation projects. The continued need for T2 research is expected to be greatly facilitated by the infrastructure for genomic research in biorepositories that eMERGE is developing and freely disseminating—especially its methods for electronic phenotyping and mining of EMRs, consent, returning results, patient education, and providing education and decision support to clinicians. eMERGE-II resources and findings will also facilitate the conduct of future T3 implementation research and potentially provide the foundation for comparative effectiveness research and public health surveillance (T4).
Conclusions
In the nearly 6 years since its inception, eMERGE has made great strides in the fields of genomics and informatics, contributing significantly to the now-established notion that the EMR is a powerful and cost-effective tool for genomics research. The network has developed tools and best practices that are being shared and utilized by the genomics and informatics communities and beyond. Building on its success, eMERGE is poised to lead the implementation of genomic medicine in clinical care through the EMR. It is hoped that this will result in improvements in health care, through safer and more effective prescribing, augmentation of primary and secondary prevention strategies, and enhanced understanding of the biology of disease. With the passage of the Patient Protection and Affordable Care Act and major changes to health-care delivery now upon us, there has never been a greater need and opportunity to improve safety and efficiency while reducing costs.
Disclosure
The authors declare no conflict of interests.
Acknowledgments
The eMERGE Network is funded by the NHGRI, with additional funding from the National Institute of General Medical Sciences through the following grants: U01HG004438 to Johns Hopkins University; U01HG004424 to the Broad Institute; U01HG004438 to Center for Inherited Disease Research; U01HG004610 and U01HG006375 to Group Health Cooperative; U01HG004608 to Marshfield Clinic; U01HG006389 to Essentia Institute of Rural Health; U01HG04599 and U01HG006379 to Mayo Clinic; U01HG004609 and U01HG006388 to Northwestern University; U01HG04603 and U01HG006378 to Vanderbilt University; U01HG006385 to the Coordinating Center; U01HG006382 to Geisinger Clinic; U01HG006380 to Mount Sinai School of Medicine; U01HG006830 to The Children's Hospital of Philadelphia; and U01HG006828 to Cincinnati Children's Hospital and Boston Children's Hospital.
Members of the External Scientific Panel: Eta Berner, University of Alabama; Jeffrey Botkin, MD, MPH, University of Utah; Charis Eng, MD, Cleveland Clinic; Gerardo Heiss, MD, PhD, University of North Carolina; Stan Huff, MD, InterMountain Healthcare; Howard McLeod, PhD (chair), University of North Carolina; and Lisa Parker, PhD, University of Pittsburgh.
External advisors for the eMERGE PGx project: Deborah A. Nickerson, PhD, University of Washington; and Steven E. Scherer, PhD, Baylor College of Medicine.
eMERGE Network teams and site-specific acknowledgements:
Cincinnati Children's Hospital and Boston Children's Hospital. John Harley, MD, PhD, principal investigator; Isaac Kohane, MD, PhD, principal investigator; Ingrid Holm, MD, MPH, gene partnership and return of results; John Hutton, MD, bioinformatics; Beth L. Cobb, MBA, and Cassandra Perry, MS, project managers; Bahram Namjou, MD, phenotyping and data analysis; Julie Bickel, MD, phenotyping; Cindy Prows, RN, return of results and pharmacogenomics; Imre Solti, MD, PhD, Guergana Savova, PhD, Pei Chen, BS, and Todd Lindgren, MS, phenotyping and natural-language processing; Keith Marsolo, PhD, John Bickel, MD, and Michael Wagner, PhD, bioinformatics; Alexander Vinks, PhD, and Wendy Wolf, PhD, pharmacogenomics.
The Children's Hospital of Philadelphia. Hakon Hakonarson, MD, PhD, principal investigator; Brendan Keating, PhD, genome sciences; Patrick Sleiman, PhD, statistical genetics; John Connolly, PhD, integrative genomics; Rosetta Chiavacci, RN, clinical research; Frank Mentch, PhD, database manager; Haijun Qiu, PhD, bioinformatics; Meckenzie Behr, clinical coordinator.
Geisinger Clinic. David J. Carey, PhD, principal investigator; Marc S. Williams, MD, coprincipal investigator; Gerard Tromp, PhD, bioinformatics and genome science; Helena Kuivaniemi, MD, PhD, genome science; W. Andrew Faucett, MS, genetic counseling; David H. Ledbetter, PhD, genome science; Glenn S. Gerhard, MD, genome science; Diane T. Smelser, PhD, statistical genetics; Kenneth Borthwick, programmer/natural-language processing; Ryan Colonie, data analyst; Jonathan Bock, data analyst; Samantha Fetterolf, project manager; G. Craig Wood, MS, statistician; Janet L. Williams, MS, CGC, genetic counseling; Laura Rogers, MS, CGC, genetic counseling; Bethanny Packard-Smith, MS, CGC, genetic counseling; Xin Chu, PhD, obesity study investigator; Evan J. Ryer, MD, clinical expert; James R. Elmore, MD, clinical expert; Christopher D. Still, DO, clinical expert; Tamara R. Vrabec, MD, clinical expert; M. Joshua Shellenberger, DO, clinical expert; Steven R. Steinhubl, MD, clinical expert; Agnes Sundaresan, MD, clinical expert; Robert C. Elston, PhD, Case Western Reserve University, site principal investigator, statistical genetics; Alan R. Shuldiner, MD, University of Maryland, site principal investigator; Braxton D. Mitchell, PhD, University of Maryland, genome science.
The biobanking and genotyping at Geisinger Clinic was funded by the Pennsylvania Commonwealth Universal Research Enhancement Program, the Ben Franklin Technology Development Fund of PA, grants from the National Institutes of Health (P30DK072488, R01DK088231, and R01DK091601), the Geisinger Clinical Research Fund, and a grant-in-aid from the American Heart Association.
Group Health and University of Washington: Eric B. Larson, MD, MPH, principal investigator; Gail Jarvik, MD, PhD, coprincipal investigator, genetic analysis, phenotyping and bioethics; James Ralson, MD, MPH, medical informatics; Andrea Hartzler, PhD, medical informatics; David S. Carrell, PhD, programmer/natural-language processing, phenotyping; Paul Crane, MD, MPH, general internist, phenotyping; David Crosslin, PhD, research genetic analysis, genomics; Daniel S. Kim, genetic analysis, phenotyping; Carlos J. Gallego, MD, genetic analysis, outcomes, phenotyping; Shubhabrata Mukherjee, PhD, genetic analysis, statistics; Stephanie Malia Fullerton, PhD, bioethics; Susan Brown Trinidad, MA, bioethics; Kathleen A. Leppig, MD, clinical implementation; Christopher S. Carlson, PhD, Fred Hutchinson Cancer Research Center, genetic analysis.
The Group Health subject collection was part of an ongoing National Institute on Aging project, Adult Changes in Thoughts (ACT) study (AG06781) and was also supported in part by the Northwest Institute of Genetic Medicine with funds from the Washington State Life Sciences Discovery funds (grant 265508).
Marshfield Clinic, Essentia Institute of Rural Health and Pennsylvania State University: Catherine A. McCarty, PhD, MPH, principal investigator; Murray Brilliant, PhD, site principal investigator; Simon Lin, PhD, biomedical informatics; Ariel S. Brautbar, MD, medical genetics; Richard Patchett, MD, ophthalmologist; Peggy Peissig, informatics; Richard Berg, MS, statistician; Rob Strenn, database/programmer/analyst; James Linneman, programmer/analyst; Carla Rottscheit, programmer/analyst; Terrie Kitchner, senior research coordinator; Marylyn Ritchie, PhD, Penn State University, site principal investigator, computational genetics; Shefali Setia Verma, bioinformatics; Gretta D. Armstrong, project manager.
Mayo Clinic: Iftikhar J. Kullo, MD, principal investigator; Christopher G. Chute, MD, PhD, coprincipal investigator; Barbara A. Koenig, PhD, bioethics; Mariza de Andrade, PhD, statistical genetics; Suzette Bielinski, PhD, epidemiology; Jyotishman Pathak, PhD, informatics; John A. Heit, MD, clinical expert.
Mount Sinai School of Medicine: Erwin Bottinger, MD, principal investigator; Omri Gottesman, MD, lead physician, genomics, informatics, EMR, CDS; Stuart Scott, PhD, clinical and molecular genetics; Jean-Sebastien Hulot, MD, PhD, pharmacogenomics; Joseph Kannry, MD, informatics, EMR, CDS; Steve Ellis, informatics, EMR, CDS; Yolanda Keppel, program manager; Shaun Purcell, PhD, quantitative analysis, GWAS; Weijia Zhang, PhD, quantitative analysis, GWAS; Inga Peter, PhD, quantitative analysis, GWAS; Rajiv Nadukuru, programmer; Vaneet Lotay, MS, genomics, informatics, programmer; Michael Parides, PhD, quantitative analysis; Carol Horowitz, MD, MPH, community-based participation; Rosamond Rhodes, PhD, community-based participation; Saskia Sanderson, PhD, communication aides; Randi Zinberg, MS, ethical and social implications; Jennifer Lin, MD, clinician leader and educator; Thomas Ullman, MD, MSc, clinician leader and educator; Douglas Dieterich, MD, clinician leader and educator; Scott Friedman, MD, clinician leader and educator; Tanisha Brown, MBA, biobank manager; Ana Mejia, clinical research coordinator; Richard Cooper, MD, consultant; Sekar Kathiresan, MD, consultant. From Columbia University Department of Biomedical Informatics, George Hripsak, MD, PhD, bioinformatics; Carol Friedman, PhD, natural-language processing; Chunhua Weng, PhD, bioinformatics; Casey Lynette Overby, PhD, bioinformatics; The Mount Sinai BioMe Biobank Program is supported by the Andrea and Charles Bronfman Philanthropies.
Northwestern University: Rex L. Chisholm, PhD, coprincipal investigator; Maureen E. Smith, MS, CGC, coprincipal investigator; Abel Kho, MD, MS, internist and medical informatics; M. Geoffrey Hayes, PhD, statistical geneticist; Laura Rasmussen-Torvik, PhD, genetic epidemiologist; Justin Starren, MD, PhD, biomedical informatics; Carl Christensen, informatics; Stephen Persell, MD, MPH, internist, decision support; Sharon Aufox, MS, CGC, genetic counselor; Jennifer Pacheco, programmer analyst; Luke Rasmussen, programmer analyst; William L. Thompson, PhD, programmer, natural-language processing; Vivian Pan, MS, CGC, genetic counselor; Catherine Wicklund, genetic counselor, genomics policy. Institutional support for the NUgene biorepository was received from the Feinberg School of Medicine and the Center for Genetic Medicine at Northwestern University.
Vanderbilt University: Dan M. Roden, MD, principal investigator; Ellen Clayton, MD, JD, bioethics; Dana Crawford, PhD, statistical genetics/genetic epidemiology; Joshua C. Denny, MD, MS, informatics/phenotyping; Bradley A Malin, PhD, informatics/privacy protection; Josh F. Peterson, MD, MPH, implementation/clinical outcomes for role/specialty with VGER/PREDICT; Jonathan S. Schildcrout, PhD, biostatistics; Russ Wilke, MD, pharmacogenomics/genome science; Lisa Bastarache, MS, informatics; Ioana Danciu, MS, informatics/clinical outcomes; Jessica Delaney, MD, clinical phenotyping; Logan Dumitrescu, PhD, statistical genetics/genetic epidemiology; Robert Goodloe, MS, statistical genetics/genetic epidemiology; Raymond Heatherly, informatics/privacy protection; Eugenia McPeek Hinz, MD, informatics/clinical phenotyping; Janina Jeff, PhD, statistical genetics/genetic epidemiology; Jason Karnes, PharmD, PhD, clinical phenotyping; Jennifer Malinowski, statistical genetics/genetic epidemiology; A. Scott McCall, clinical phenotyping; Jonathan Mosley, MD, PhD, clinical phenotyping; Alexander Saip, PhD, informatics; Sarah Stallings, PhD, project management; Sara Van Driest, MD, PhD, clinical phenotyping; Xiaoming Wang, MS, informatics; Matthew Westbrook, bioethics.
Coordinating Center: Jonathan L. Haines, PhD, principal investigator; Joshua C. Denny, MD, MS, informatics; Bradley A. Malin, PhD, informatics/privacy protection; Marylyn D. Ritchie, PhD, Penn State University, statistical genetics/genome science; Melissa Basford, MBA, program management; Gretta Armstrong, MA, genomics project management; Yuki Bradford, MS, statistical genetics; James Cowan, informatics; Jacqueline Kirby, MS, project management; Lauren Melancon, program coordination; Brandy Mapes, MLIS, program coordination; Peter Speltz, informatics; Anurag Verma, MS, informatics; Shefali Verma, MS, statistical genetics; Weiyi Xia, informatics.
Supplementary Material
References
- National Human Genome Research Institute eMERGE Phase 1 RFA. 2007. http://grants.nih.gov/ grants/guide/rfa-files/RFA-HG-07-005.html . Accessed January 2013.
- Electronic Medical Records and Genomics (eMERGE) Network Model consent language for EMR-linked bioepositories. 2009. http://www.genome.gov/Pages/ PolicyEthics/InformedConsent/eMERGEModelLanguage2009-12–15.pdf . Accessed January 2013.
- Zuvich RL, Armstrong LL, Bielinski SJ, et al. Pitfalls of merging GWAS data: lessons learned in the eMERGE network and quality control procedures to maintain high data quality. Genet Epidemiol. 2011;35:887–898. doi: 10.1002/gepi.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- eMERGE PheKb: Phenotype KnowledgeBase. 2012. http://www.PheKb.org . Accessed January 2013.
- McCarty CA, Chisholm RL, Chute CG, eMERGE Team et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullo IJ, Ding K, Shameer K, et al. Complement receptor 1 gene variants are associated with erythrocyte sedimentation rate. Am J Hum Genet. 2011;89:131–138. doi: 10.1016/j.ajhg.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosslin DR, McDavid A, Weston N, Electronic Medical Records and Genomics (eMERGE) Network et al. Genetic variants associated with the white blood cell count in 13,923 subjects in the eMERGE Network. Hum Genet. 2012;131:639–652. doi: 10.1007/s00439-011-1103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, Ritchie MD, Crawford DC, et al. Identification of genomic predictors of atrioventricular conduction: using electronic medical records as a tool for genome science. Circulation. 2010;122:2016–2021. doi: 10.1161/CIRCULATIONAHA.110.948828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, Crawford DC, Ritchie MD, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet. 2011;89:529–542. doi: 10.1016/j.ajhg.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, Denny JC, Crawford DC, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010;86:560–572. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho AN, Hayes MG, Rasmussen-Torvik L, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Assoc. 2012;19:212–218. doi: 10.1136/amiajnl-2011-000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullo IJ, Ding K, Jouni H, Smith CY, Chute CG. A genome-wide association study of red blood cell traits using the electronic medical record. PLoS One. 2010;5(9):e13011. doi:10.1371/journal.pone.0013011 doi: 10.1371/journal.pone.0013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukides G, Denny JC, Malin B. The disclosure of diagnosis codes can breach research participants' privacy. J Am Med Inform Assoc. 2010;17:322–327. doi: 10.1136/jamia.2009.002725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukides G, Gkoulalas-Divanis A, Malin B. Anonymization of electronic medical records for validating genome-wide association studies. Proc Natl Acad Sci USA. 2010;107:7898–7903. doi: 10.1073/pnas.0911686107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malin B. Secure construction of k-unlinkable patient records from distributed providers. Artif Intell Med. 2010;48:29–41. doi: 10.1016/j.artmed.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Malin B, Benitez K, Masys D. Never too old for anonymity: a statistical standard for demographic data sharing via the HIPAA Privacy Rule. J Am Med Inform Assoc. 2011;18:3–10. doi: 10.1136/jamia.2010.004622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamersoy A, Loukides G, Denny JC, Malin B. Anonymization of administrative billing codes with repeated diagnoses through censoring. AMIA Annu Symp Proc. 2010;2010:782–786. [PMC free article] [PubMed] [Google Scholar]
- McGuire AL, Basford M, Dressler LG, et al. Ethical and practical challenges of sharing data from genome-wide association studies: the eMERGE Consortium experience. Genome Res. 2011;21:1001–1007. doi: 10.1101/gr.120329.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton SM, Wolf WA, Brothers KB, et al. Return of individual research results from genome-wide association studies: experience of the Electronic Medical Records and Genomics (eMERGE) Network. Genet Med. 2012;14:424–431. doi: 10.1038/gim.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Human Genome Research Institute eMERGE Phase II RFA. 2011. http://grants.nih.gov/ grants/guide/rfa-files/RFA-HG-10-009.html . Accessed January 2013.
- National Human Genome Research Institute eMERGE phase II pediatric RFA. 2011. http://grants.nih.gov/grants/ guide/rfa-files/RFA-HG-11–022.html . Accessed January 2013.
- Kullo IJ, Jarvik GP, Manolio TA, Williams MS, Roden DM. Leveraging the electronic health record to implement genomic medicine. Genet Med. 2013;15:270–271. doi: 10.1038/gim.2012.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical Pharmacogenetics Implementation Consortium Clinical Pharmacogenetics Implementation Consortium. 2009. http://www.pharmgkb.org/page/cpic . Accessed January 2013.
- National Human Genome Research Institute Genomic Medicine Working Group (GMWG). 2011. http://www.genome.gov/27549220 . Accessed January 2013.
- Manolio TA, Chisholm RL, Ozenberger B, et al. Implementing genomic medicine in the clinic: the future is here. Genet Med. 2013;15:258–267. doi: 10.1038/gim.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharmacogenetics Research Network Pharmacogenetics Research Network. 2006. http://www.pgrn.org . Accessed January 2013.
- National Human Genome Research Institute Population Architecture using Genomics and Epidemiology (PAGE) Consortium. 2008. http://www.genome.gov/27541456 . Accessed January 2013.
- National Human Genome Research Institute Return of Results Consortium. 2011. http://www.genome.gov/27545526 . Accessed January 2013.
- National Human Genome Research Institute Clinical Sequencing Exploratory Research (CSER) Program. 2011. http://www.genome.gov/27546194 . Accessed January 2013.
- Blumenthal D, Tavenner M. The “meaningful use” regulation for electronic health records. N Engl J Med. 2010;363:501–504. doi: 10.1056/NEJMp1006114. [DOI] [PubMed] [Google Scholar]
- Kallem C. Transforming clinical quality measures for EHR use. NQF refines emeasures for use in EHRs and meaningful use program. J AHIMA. 2011;82:52–53. [PubMed] [Google Scholar]
- Thompson WK, Rasmussen LV, Pacheco JA, et al. An evaluation of the NQF Quality Data Model for representing Electronic Health Record driven phenotyping algorithms. AMIA Annu Symp Proc. 2012;2012:911–920. [PMC free article] [PubMed] [Google Scholar]
- Rea S, Pathak J, Savova G, et al. Building a robust, scalable and standards-driven infrastructure for secondary use of EHR data: the SHARPn project. J Biomed Inform. 2012;45:763–771. doi: 10.1016/j.jbi.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Shrestha G, Murthy S, et al. Modeling and executing electronic health records driven phenotyping algorithms using the NQF Quality Data Model and JBoss® Drools engine. AMIA Annu Symp Proc. 2012;2012:532–541. [PMC free article] [PubMed] [Google Scholar]
- Bright TJ, Wong A, Dhurjati R, et al. Effect of clinical decision-support systems: a systematic review. Ann Intern Med. 2012;157:29–43. doi: 10.7326/0003-4819-157-1-201207030-00450. [DOI] [PubMed] [Google Scholar]
- Haynes RB, Wilczynski NL, Computerized Clinical Decision Support System (CCDSS) Systematic Review Team Effects of computerized clinical decision support systems on practitioner performance and patient outcomes: methods of a decision-maker-researcher partnership systematic review. Implement Sci. 2010;5:12. doi: 10.1186/1748-5908-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers MW, Smeulers M, Vermeulen H, Peute LW. Effects of clinical decision-support systems on practitioner performance and patient outcomes: a synthesis of high-quality systematic review findings. J Am Med Inform Assoc. 2011;18:327–334. doi: 10.1136/amiajnl-2011-000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph AG, Haynes RB, Wyatt JC, Cook DJ, Guyatt GH. Users' Guides to the Medical Literature: XVIII. How to use an article evaluating the clinical impact of a computer-based clinical decision support system. JAMA. 1999;282:67–74. doi: 10.1001/jama.282.1.67. [DOI] [PubMed] [Google Scholar]
- US Food and Drug Administration Table of pharmacogenomic biomarkers in drug labels. 2012. http://www.fda.gov/drugs/ scienceresearch/researchareas/pharmacogenetics/ucm083378.htm . Accessed January 2013.
- Hoffman M, Arnoldi C, Chuang I. The clinical bioinformatics ontology: a curated semantic network utilizing RefSeq information. Pac Symp Biocomput. 2005. pp. 139–150. [DOI] [PubMed]
- Mitchell DR, Mitchell JA. Status of clinical gene sequencing data reporting and associated risks for information loss. J Biomed Inform. 2007;40:47–54. doi: 10.1016/j.jbi.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Starren J, Williams MS, Bottinger EP. Crossing the omic chasm: a time for omic ancillary systems. JAMA. 2013;309:1237–1238. doi: 10.1001/jama.2013.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical Decision Support Consortium Clinical Decision Support Consortium. 2008. http://www.partners.org/cird/cdsc/ . Accessed January 2013.
- Bradfield JP, Taal HR, Timpson NJ, Early Growth Genetics Consortium et al. A genome-wide association meta-analysis identifies new childhood obesity loci. Nat Genet. 2012;44:526–531. doi: 10.1038/ng.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilienfeld DE, Stolley PD.Foundation of Epidemiology3rd edn. Oxford University Press: New York; 1994 [Google Scholar]
- Patry JL. Methodological consequences of situation specificity: biases in assessments. Front Psychol. 2011;2:18. doi: 10.3389/fpsyg.2011.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabsitz RR, McGuire A, Sharp RR, et al. Ethical and practical guidelines for reporting genetic research results to study participants: updated guidelines from a National Heart, Lung, and Blood Institute working group. Circ Cardiovasc Genet. 2010;3:574–580. doi: 10.1161/CIRCGENETICS.110.958827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beskow LM, Burke W. Offering individual genetic research results: context matters. Sci Transl Med. 2010;2:38cm20. doi: 10.1126/scitranslmed.3000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman D, Murphy J, Scott J, Hudson K. Subjects matter: a survey of public opinions about a large genetic cohort study. Genet Med. 2008;10:831–839. doi: 10.1097/GIM.0b013e31818bb3ab. [DOI] [PubMed] [Google Scholar]
- Berg JS, Khoury MJ, Evans JP. Deploying whole genome sequencing in clinical practice and public health: meeting the challenge one bin at a time. Genet Med. 2011;13:499–504. doi: 10.1097/GIM.0b013e318220aaba. [DOI] [PubMed] [Google Scholar]
- Burke W, Laberge AM, Press N. Debating clinical utility. Public Health Genomics. 2010;13:215–223. doi: 10.1159/000279623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green ED, Guyer MS, National Human Genome Research Institute Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Green ED. Genomics reaches the clinic: from basic discoveries to clinical impact. Cell. 2011;147:14–16. doi: 10.1016/j.cell.2011.09.012. [DOI] [PubMed] [Google Scholar]
- Khoury MJ, Gwinn M, Yoon PW, Dowling N, Moore CA, Bradley L. The continuum of translation research in genomic medicine: how can we accelerate the appropriate integration of human genome discoveries into health care and disease prevention. Genet Med. 2007;9:665–674. doi: 10.1097/GIM.0b013e31815699d0. [DOI] [PubMed] [Google Scholar]
- Khoury MJ, Gwinn M, Bowen MS, Dotson WD. Beyond base pairs to bedside: a population perspective on how genomics can improve health. Am J Public Health. 2012;102:34–37. doi: 10.2105/AJPH.2011.300299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow RE, Vinson C, Chambers D, Khoury MJ, Kaplan RM, Hunter C. National Institutes of Health approaches to dissemination and implementation science: current and future directions. Am J Public Health. 2012;102:1274–1281. doi: 10.2105/AJPH.2012.300755. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.