Abstract
Voltage-gated calcium (CaV) ion channels convert neuronal activity into rapid intracellular calcium signals to trigger a myriad of cellular responses. Their involvement in major neurological and psychiatric diseases, and importance as therapeutic targets, has propelled interest in subcellular-specific mechanisms that align CaV channel activity to specific tasks. Here we highlight recent studies that delineate mechanisms controlling the expression of CaV channels at the level of RNA and protein. We discuss the roles of RNA editing and alternative pre-mRNA splicing in generating CaV channel isoforms with activities specific to the demands of individual cells; the roles of ubiquitination and accessory proteins in regulating CaV channel expression; and the specific binding partners which contribute to both pre- and post- synaptic CaV channel function.
Keywords: voltage-gated calcium channels, alternative splicing, splicing factors, synaptic transmission, synaptic proteins, ubiquitination
Ten CaV Genes, Thousands of Different CaV mRNAs, and Many More Functionally Different Proteins
“...different Ca currents show so many differences in fundamental properties that we find it easier to assume that there are more than one type.” – Hagiwara & Byerly (1981)1
Voltage-gated calcium ion channels are present at every critical step of information transfer in the nervous system from signal detection to perception, and from neuronal development to programmed apoptosis. Strategically located at points of sensory detection and on both sides of the synapse, CaV channels have a defining role in integrating signals and influencing synaptic strength. All but one of the ten mammalian genes that encode the main α1 subunit of CaV channels are expressed in the nervous system (Box 1). Each CaV gene has a distinct expression profile reflecting its functional specialization to support specific cellular tasks.
For more than 50 years the functional diversity of CaV currents among different cell-types has been attributed to the expression of multiple channel types [1]. Indeed, individual neurons can express multiple CaV genes, consistent with the varied subcellular distribution of calcium channel subtypes controlling a range of neuronal functions. The value of highly selective toxins and drugs used to tease out the relative contributions of CaV channels within single cells is hard to overstate. Yet this is not always straightforward: for example, until recently, the absence of pharmacological tools to distinguish CaV1.2 and CaV1.3 channels [2], which collectively give rise to most dihydropyridine-sensitive Ca currents in neurons, has stalled progress in defining their individual contributions. Even when specific toxins are available, the challenges associated with isolating the functional importance of closely related CaV channels are magnified several-fold when considering the thousands of channel isoforms potentially generated from single genes by alternative pre-mRNA splicing and RNA editing.
All ten mammalian CACNA1 genes contain multiple sites of alternative splicing; each gene has the potential to generate thousands of unique splice-isoforms controlled by the expression and relative activities of cell-specific splicing factors (Figs. 1, 2). Typically these sites are located in coding regions that tolerate sequence variation and that are not essential for structural integrity: N-termini, cytoplasmic loops I-II and II-III, extracellular linkers that connect transmembrane α-helices S3 and S4 in domains III and IV, and the C-termini (Figs. 2A, 2B, 3). Consequently, these hyper-variable-like regions are hotspots for cell-specific posttranslational modification, second messenger action on, and protein binding to CaV channels (Figs. 3, 4). The tethering of appropriate CaV channels to presynaptic active zones or to postsynaptic specializations depends on specific nanodomain-level protein interactions (see Figs. 5, 6); as discussed below for the most recently discovered of these interactions. A common feature across these studies is the ability to resolve molecular-level detail of specific RNAs and proteins within subpopulations of neurons, and within functionally distinct subcellular compartments. Novel techniques have been developed and applied to identify: splicing factor binding sites on CaV channel pre-mRNAs, CaV channel RNAs in specific populations of neurons, and CaV proteins at active zones. These methodologies marry disciplines to yield a more comprehensive, multi-dimensional view of CaV channels and their regulation from RNA to Protein. We highlight these and other recent studies elucidating cell-specific control of CaV RNA processing, posttranslational CaV protein modifications, and CaV protein-protein interactions. Collectively this work demonstrates exciting progress made in defining the molecular mechanisms underlying the expression of CaV channel isoforms in specific neurons, and in linking their expression to critical cell functions.
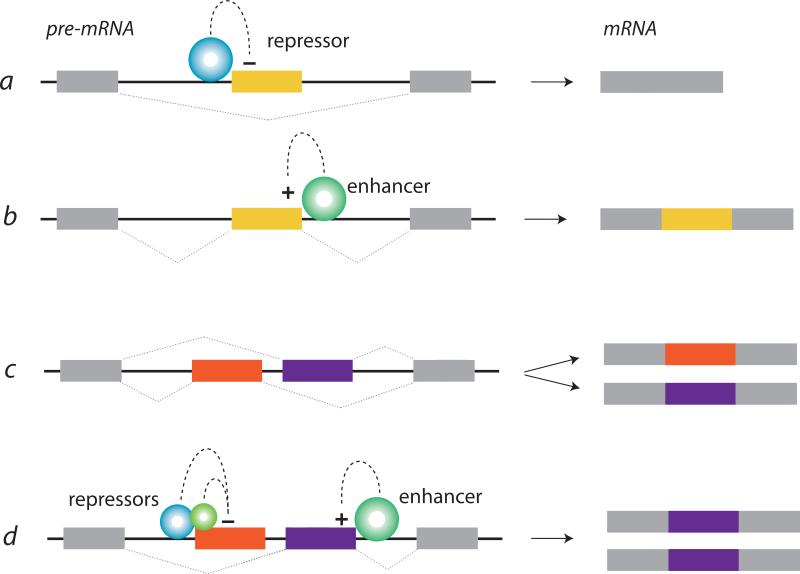
Figure 1. Splicing factors repress and enhance alternative exon inclusion during pre-mRNA processing.
Splicing factors bind to consensus nucleotide sequences in introns or target exons of pre-mRNAs to enhance or repress (silence) recognition of the exon by the spliceosome during pre-mRNA processing. Alternatively spliced exons (colored), constitutive exons (gray), and introns (black connecting lines) are illustrated. The position of splicing factor binding, relative to the target exon, is often predictive of splicing factor action. For example, members of the Nova, rbFox, and PTB splicing factor protein families tend to repress or silence exon inclusion when they bind their respective nucleotide sequence binding motifs upstream (a) or within (b) the target exon, and enhance exon inclusion when they bind their respective binding motifs downstream of the target exon (b). Alternatively spliced cassette exons are excluded or included during pre-mRNA splicing (a, b). Mutually exclusive alternative splicing involves two or more exons, only one of which is selected during pre-mRNA processing (c, d). Mutually exclusive exons often start or end with an incomplete codon. In this case, there is a shift in the reading frame in mRNAs that either lack both mutually exclusive exons or contain both, and early protein termination (not shown). Often exon selection is influenced by the concerted action of several splicing factors (d). The expression levels and activities of individual splicing factors depend on many cellular factors including those involved in development, neuronal activity, and defining neuronal subtype.
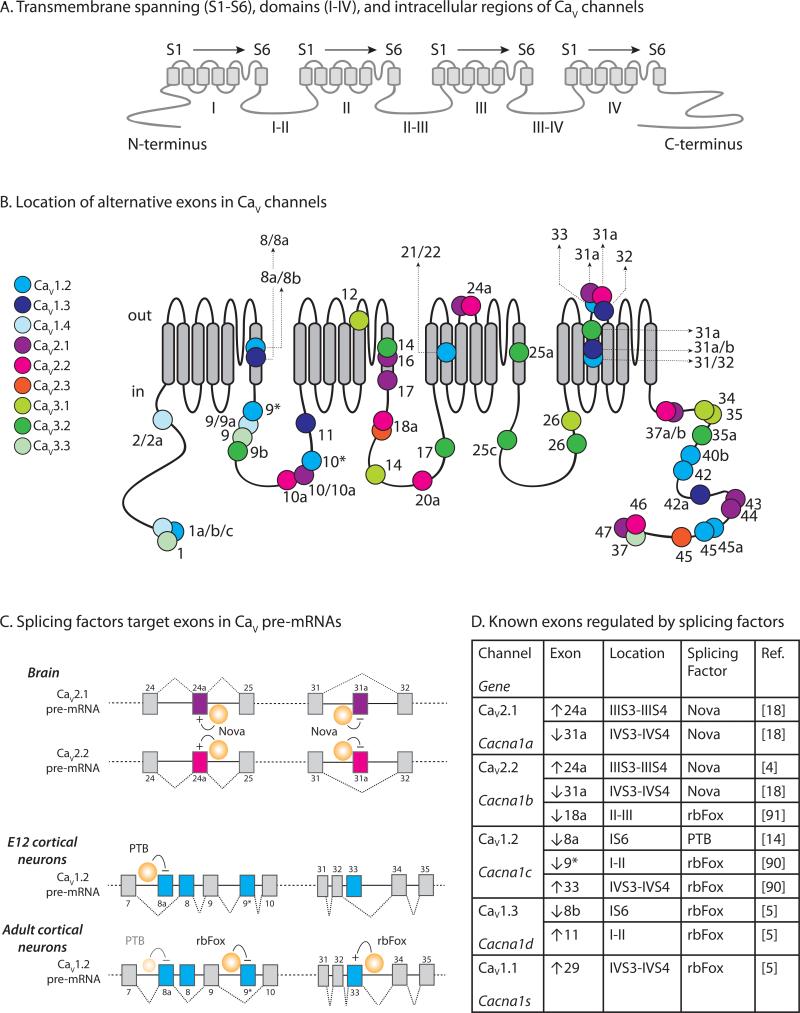
Figure 2. Alternative splicing is extensive in CaV channels and is regulated by the action of cell-specific splicing factors.
A. Major domains and regions located on a schematic of the α1 subunit of a CaV channel. There are four structurally homologous domains I, II, III and IV. Each domain is comprised of 6 transmembrane spanning α-helices (S1-S6). The intracellular regions that link the domains are labeled I-II, II-III, and III-IV. The N-terminus and C-terminus are intracellular. This naming system is used to identify the major regions of CaV channels. For example, the third transmembrane spanning α-helix in domain IV is referred to as IVS3.
B. Location of alternative exons mapped on to a schematic of the α1 subunit of a CaV channel. The 24 transmembrane spanning domains are shown (gray) as well as extracellular and intracellular regions. The approximate locations of alternative exons are shown (circles) and color-coded to indicate specific CaV channel subtypes. Exon numbers are indicated and the numbering system used follows mouse Cacna1 gene numbering. Alternate first exons are shown at the start of the N-termini, and mutually exclusive alternative exons are designated as X/Y. The following references were used to compile information for CaV1.2 [14]; CaV1.3 [83]; CaV1.4 [84]; CaV2.1 [85]; CaV2.2 [83], accession # FJ609386; CaV2.3[83,86]; CaV3.1 [87]; CaV3.2 [88]; CaV3.3 [89].
C. Splicing factor target exons in CaV pre-mRNAs. Two splicing patterns are shown for three splicing factor RNA-binding protein families that have similar mechanisms of action: Nova, Fox and PTB. The first example (top) illustrates the action of Nova on alternatively spliced exon cassettes, e24a and e31a, in CaV2.1 and CaV2.2 pre-mRNAs. Nova enhances inclusion of e24a during pre-mRNA splicing of CaV2.1 and CaV2.2 by binding to elements in the respective introns downstream of the target exons (Intronic Splicing Enhancer; ISE), whereas Nova represses (or silences) inclusion of e31a during pre-mRNA splicing by binding elements upstream and within or overlapping the target exon (Intronic Splicing Silencer: ISS; Exonic Splicing Silencer: ESS). Nova is expressed in brain where CaV2.1 and CaV2.2 mRNAs containing e24a and lacking e31a dominate [18]. In the second example (lower), PTB represses inclusion of mutually exclusive e8a during splicing of CaV1.2 pre-mRNA. PTB is expressed in embryonic brain and therefore CaV1.2 mRNAs lacking e8a and containing e8 dominate early during development. In adult cortical neurons, PTB levels are reduced and it no longer represses e8a. By contrast, rbFox is upregulated during development and it represses inclusion of e9* and enhances inclusion of e33 [14,90]. In adult cortical neurons, CaV1.2 mRNA containing e8a, lacking e9*, and containing e33 dominate [14].
D. Alternatively spliced exons of CaV channel genes are shown together with splicing factors known to regulate their expression. Cassette exons are either included or excluded. E8a/e8 of CaV1.2, and e8b/e8a of CaV1.3 are mutually exclusive (see A and B). Therefore, repression of e8a during pre-mRNA splicing of CaV1.2 will promote inclusion of e8, and repression of e8b during pre-mRNA splicing of CaV1.3 will promote inclusion of e8a. CaV1.2 e8a is strongly repressed by PTB but more weakly repressed by the neuronal homologue, nPTB, despite their similar RNA binding motifs [14]. Based upon references [4,5,14,18,90,91].
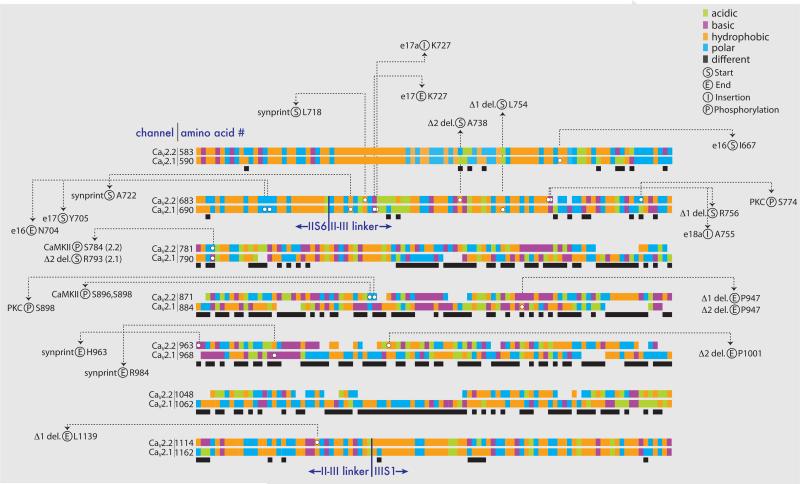
Figure 3. The II-III intracellular loop regions of CaV2.1 and CaV2.2 have divergent sequences.
Amino acid alignments for CaV2.1 (rat sequence: NM_012918) and CaV2.2 (rat sequence: AF055477) are shown for about 700 amino acids encoding the II-III intracellular loops as well as sequence in IIS6 and IIIS1 (Clustal Omega software [92]). The approximate locations of boundaries between transmembrane and intracellular domains are indicated. The chemical nature of each amino acid is coded according to the following color scheme: acidic (green) D, E; basic (purple) K, R, H; hydrophobic (orange) A, F, I, L, M, P, V, W; polar (blue) C, G, N, Q, S, T, Y. The black solid bars below the alignments indicate regions of differences in amino acid type between CaV2.1 and CaV2.2. White circles indicate amino acids of interest, the amino acid numbers are shown and they are coded according to the following categories: (S) alternative exon, deletion (del.), or synprint area start; (E) alternative exon del., or synprint area end; (I) alternative exon insertion; (P) phosphorylation. References: Alternative e16, e17, and e17a of CaV2.1 [85]; CaV2.2 e18a [93] [94]; CaV2.2 synprint [95]; CaV2.1 synprint [96]; PKC and CaMKII phosphorylation [97]; CaV2.1 Δ1 and Δ2 [98]; CaV2.
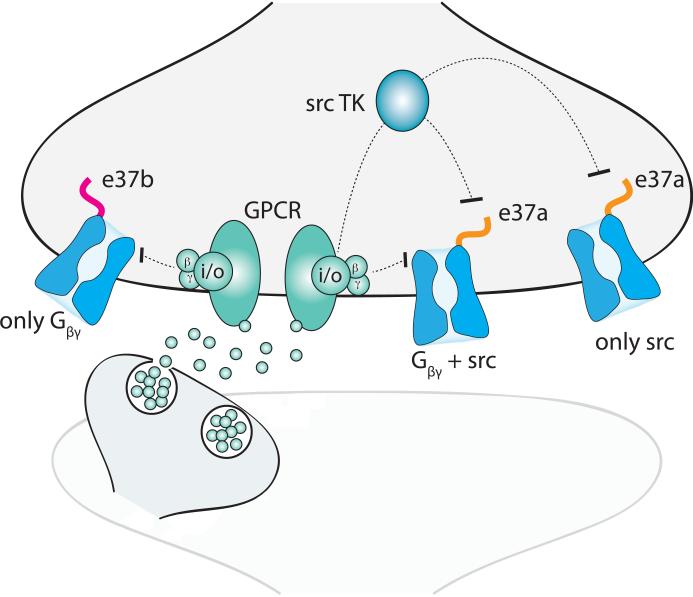
Figure 4. Alternative splicing influences G protein coupled receptor inhibition of CaV2.2 channels.
Illustration showing a Gi/o protein coupled receptor (GPCR; green) inhibiting presynaptic CaV2.2 channels (blue) by different signaling mechanisms. The GPCR is activated by neurotransmitter released from a neighboring neuron. Following activation, the G dimer dissociates from Gi/o, binds to and directly inhibits both CaV2.2 channel splice isoforms (e37a (orange) and e37b (pink)). A second G protein-dependent pathway, that requires src tyrosine kinase (TK) activation but that is independent of G, inhibits e37a-CaV2.2. This schematic is based on data published in [99] [23]. A prediction that follows from these studies is that e37a-CaV2.2 channels, that are more distant from GPCRs, will not be inhibited by membrane delimited G but may still be inhibited by src TK. This is because the latter mechanism involves a diffusible second messenger.
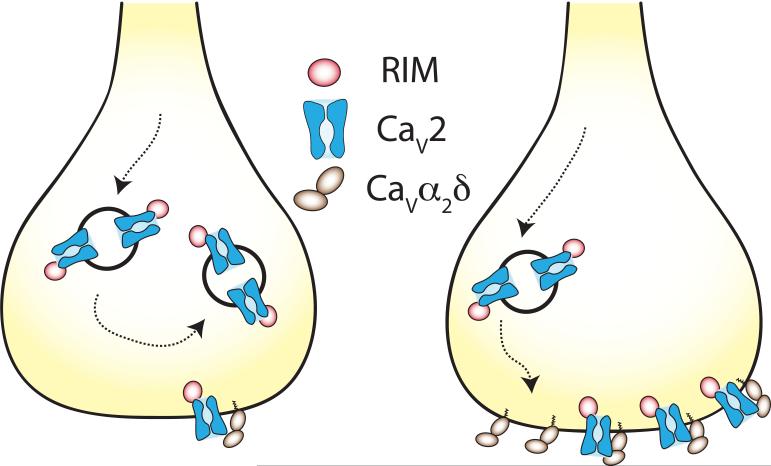
Figure 5. The density of CaV2 channels at presynaptic nerve terminals is controlled by RIM and the CaVα2δ-1 subunit.
Illustration shows that CaV2 channel trafficking to, or retention at presynaptic nerve terminals depends on association of CaV2 proteins with both RIM and CaVα2δ-1[45] [46] [38,49] [30]. The details of where RIM and CaVα2δ-1 interact with CaV2, in which intracellular compartment, are not known but the figure is intended to capture the finding that overexpression of CaV2 channels does not lead to an increase in functional presynaptic CaV2 channels (left) unless CaV21 is also over-expressed (right) [45] [46]. These data suggest that CaVα2δ-1 limits the insertion into or the stabilization of CaV2 channels at the active zones of the presynaptic nerve terminal [45].
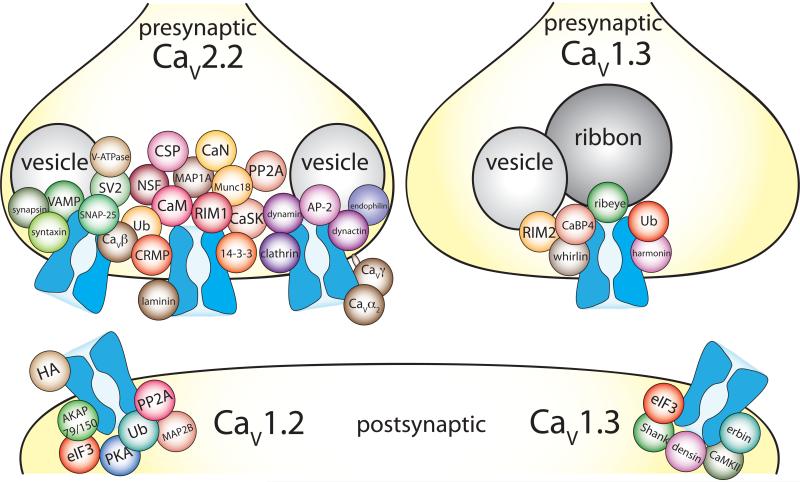
Figure 6. Presynaptic and postsynaptic CaV channels are associated with a number membrane anchoring and signaling proteins that define their function.
Illustrated are a subset of proteins shown to bind presynaptic CaV2.2, postsynaptic CaV1.2, and pre- and postsynaptic CaV1.3 channels. Not all proteins reported to bind to these CaV channels are shown, but those illustrated were confirmed by more than one proteomics screen and/or by a functional assay. In addition, some G proteins, chaperone, and cytoskeletal proteins are not included. The proteins shown are approximately grouped by function, but beyond that there is no significance to location in the synapse or relative to each other. Data were collected using the following references: CaV2.2: CSP [100], CRMP [101], laminin [102], MAP1A [103], Ub [36] [35], other proteins were identified by [54,55,104]. CaV1.2: PP2A, [105], PKA [106], AKAP79/150 [107] [108] [109], HA [110], Ub [34,111], eIF3 [112], MAP2B [108] [113], dynamin [112]. CaV1.3 presynapse: whirlin [114], RIM2 [48], harmonin [42], Ub [42], ribeye [115] [60], CaBP4 [116] [117]. CaV1.3 postsynapse: Shank [118], erbin [81], densin [64], CaMKII [64], eIF3 [112].
Cell-specific alternative pre-mRNA splicing and RNA editing regulate CaV channel function
“...the more you look, the more you see.” (Robert M. Pirsig, 1974)
Alternative pre-mRNA splicing is particularly prevalent in the mammalian brain, and is essential for normal neuronal development, axon targeting, neuronal excitability, and neural circuit formation. This form of pre-mRNA processing occurs in the cell's nucleus, and it is controlled by the concerted actions of a set of cell-specific, RNA binding proteins called splicing factors. Trans-acting splicing factors bind to consensus cis-sequence motifs in pre-mRNAs to influence the action of the spliceosome, promoting or repressing inclusion of alternatively spliced exons, and promoting or repressing the use of alternative splice acceptor or donor sites at intron/exon boundaries (Fig. 1). Nova, rbFox and polypyrimidine track binding (PTB) families of splicing factors direct inclusion or skipping of alternatively spliced exons in a number of neural gene pre-mRNAs including CACNA1 genes (Fig. 2). These splicing factor protein families have the capacity to either repress (silence) or enhance exon inclusion, in different target pre-mRNAs or within the same pre-mRNA, depending on the position of their consensus binding motifs, either upstream (5’) or downstream (3’) relative to the target exon [3-7] (Figs. 1, 2C). Genome-wide maps of splicing factor binding sites reveal certain common features in the mechanism of action of certain splicing factors and bring us closer to defining a working splicing code[8]. These splicing factor genomic maps have advanced our understanding of cell-specific regulation of channel structure and function. In addition, several studies have shown that alternative splicing can be controlled by epigenetic factors [9], signaling factors including cellular protein kinases and phosphatases[10], and by cell-specific alternative splicing of splicing factors themselves[5,6].
Splicing factors that target CACNA1 pre-mRNAs
CACNA1 pre-mRNAs are targets of splicing factors from the Nova, rbFox, PTB, and Muscleblind-like 2 (Mbnl2) RNA-binding protein families [4-7](Fig. 2D). Moreover, multiple rbFox splice isoforms act on the same CACNA1 pre-mRNA exon, but with different consequences [11]. Importantly, developmental changes in the patterns of CaV splice isoform expression can parallel the changes in expression levels of key splicing factors. For example, as PTB expression levels fall in the developing brain, levels of neuronal PTB (nPTB) increase [12,13]. PTB controls the choice between a pair of mutually exclusive exons in CACNA1C (CaV1.2), e8 and e8a, via binding to motifs in CACNA1C pre-mRNA upstream of e8a [14] (Fig. 2C). As PTB levels decrease during development, the ratio of e8a- to e8-containing CaV2.2 mRNAs increases because PTB no longer represses e8a. In this case, nPTB does not substitute for PTB as a repressor of e8a [14]. E8a and e8 encode alternate forms of transmembrane S6 in domain I of CaV1.2 in a region that is critical for normal channel function (Fig. 2B). E8a and e8 are of special interest because certain mutations in either exon were shown to cause the severe multi-organ hereditary disorder, Timothy Syndrome [15]. The severity of neurological symptoms in individuals with Type I and Type II Timothy Syndrome depends on which exon carries the mutation, presumably reflecting the different expression patterns of e8 and e8a in the brain [16].
CACNA1B (CaV2.2) pre-mRNAs are targets of Nova2, and several Nova2 binding sites align with previously identified alternatively spliced exons. For example, high-throughput sequencing combined with cross-linking immunoprecipitation methods (HiTS-CLIP) showed that Nova2 binding sites are located within the intron immediately upstream, and overlapping, a short six-nucleotide exon e31a in CACNA1B pre mRNAs [17] (Fig. 2C). The positioning of RNA binding motifs for Nova2 upstream of e31a predicts Nova2 repression of e31a. The HiTS-CLIP data are consistent with earlier reports that e31a-containing CaV2.2 mRNAs are found at very low levels in the rodent brain where Nova2 is expressed. By contrast, e31a-containing CaV2.2 mRNAs dominate in peripheral ganglia, which do not express Nova2 [18]. The same splicing factor can act differently on multiple alternatively spliced exons within the same pre-mRNA. This is the case for Nova2; in contrast to its silencing action on e31a, it binds motifs downstream of e24a to augment e24a expression in brain (Fig. 2C, 2D; [18]). Intriguingly, there are clusters of Nova2 binding sites in other regions of CACNA1B that do not map to known alternatively spliced exons, and the functional significance of these is not yet known.
RbFox and Nova splicing factor protein families have overlapping networks of synaptic protein targets, consistent with the collective action of multiple splicing factors on several alternative CACNA1 exons [17] (Fig. 1, 2C, 2D). Loss of rbFox2 results in reduced expression of Nova1, perhaps reflecting regulation of Nova1 pre-mRNA splicing by rbFox2 [17]. Several important studies have shown profound consequences—including gross neurodevelopmental abnormalities—of disrupting or eliminating splicing factors which regulate alternative splicing of multiple targets [5,7,19]. However, because the splicing of hundreds of neuronal pre-mRNAs—including those that encode other splicing factors—are disrupted in these studies, the relative contribution of splicing events within a single gene family, as as CACNA1, cannot be distinguished.
Individual Alternatively Spliced Exons Have Measurable Cellular and Behavioral Consequences
Many individual splice sites in CACNA1 genes are evolutionarily conserved, and it is often assumed that each adds functional value and fitness to the cells in which they are expressed. Another less considered possibility is that the functional impact of any individual splicing change may not be discernible at the cellular or behavioral level. Alternative pre-mRNA splicing of genes may have evolved as essential proteins operate over a broader dynamic range than would be possible from the activity of a single protein. This feature of alternative splicing could be critical during development, particularly in neurons that adapt to and continue to function in the face of dramatic changes in morphology and activity [20]. Yet there are several notable examples showing that exon choice in an influential gene can indeed modulate behavior. For example, reproductive dominance in honeybees is controlled by alternative splicing of exon 5 in a gene homologous to the gemini transcription factor of Drosophila [21], and cell-specific expression of a TRPV1 splice isoform confers unique infrared sensing capabilities to vampire bats [22].
Recently, the cellular and behavioral consequences of an alternatively spliced exon in the mouse Cacna1b gene were demonstrated. Enhanced expression of the nociceptor-specific exon 37a splice isoform of CaV2.2 increases both cellular sensitivity to inhibition by activated -opioid receptors and behavioral sensitivity to spinal morphine-induced analgesia [23] (Fig. 4). Cell-specific expression of e37a in wild-type mice may help explain why CaV2.2 channels in nociceptors are particularly sensitive to inhibition by drugs, transmitters, and hormones that act through G protein-coupled receptors. Additional alternatively spliced exons in Cacna1b and in the closely related Cacna1a (CaV2.1) and Cacna1e (CaV2.3) genes modify amino acid sequences in the cytoplasmic domains of CaV channels. The actions of G proteins and second messengers on other ion channels may also be influenced by cell-specific alternative splicing of ion channel pre-mRNAs in other parts of the nervous system.
Cell-specific inclusion of exons in Cacna1 genes can also impact disease states. Already mentioned are the different symptoms in Type I and Type II Timothy Syndrome determined by which exon, e8a or e8 in CACNA1C carries the mutation [16]. Other examples include the different consequences of Cacna1a (CaV2.1) mutations associated with familial hemiplegic migraine on the short (Δ47) and long (+47) splice isoforms of CaV2.1 [24], and an epilepsy causing mutation in mouse Cacna1h (CaV3.2) that results in different channel phenotypes depending on whether alternative exon 25 is expressed [25].
RNA editing of CACNA1D mRNA
Additional cell-specific alterations in amino acid sequence can arise from RNA editing, introducing potentially even finer specialization of CaV activities. In RNA editing, posttranscriptional deamination of adenosine to inosine (read as guanosine during translation) is catalyzed in RNA by the ADAR (adenosine deaminase acting on RNA) family of enzymes. Evidence for different RNA edited versions of CaV channel mRNAs was first reported for the Drosophila CaV2 homologue cacophony (Dmca1A) (Table 1) [26]. Recently, central nervous system (CNS)-specific editing of mammalian CaV1.3 RNAs by the ADAR protein ADR2 has been carefully documented and shown to generate four distinct sequences within the C-terminus IQ domain: IQDY, MQDY, IRDY, and MRDY [27]. By contrast, only one IQ sequence, IQDY, is found in CaV1.3 channels expressed outside of the CNS [27]. The IQ domain forms part of a critical calmodulin-binding site in CaV channels that mediates calcium-dependent inhibition. Calcium-dependent inhibition functions as an important feedback control on subsequent calcium entry; cytoplasmic calcium inhibits the further gating of the ion channel through which it flowed. Editing of the IQ site (e.g. to MQ) reduces calcium-dependent inactivation and as a consequence is thought to participate in shaping rhythmicity of action potential firing in neurons of the suprachiasmatic nucleus [27].
Table I.
| Voltage-gated Calcium Channel Nomenclature | ||||||||
|---|---|---|---|---|---|---|---|---|
| Protein name | Current type | Gene name | ||||||
| old | Human | Mouse, Rat | Zebrafish | Pufferfish | Drosophila | C.elegans | ||
| CaV1.1 | α 1S | L | CACNA1S | Cacna1s | cacna1sa,b | 1S-a,b,c | Ca-α1D | eql-19 |
| CaV1.2 | α 1C | CACNA1C | Cacna1c | cacna1c | 1C | |||
| CaV1.3 | α 1D | CACNA1D | Cacna1d | cacna1da,b | 1D-a,b,c,d | |||
| CaV1.4 | α 1F | CACNA1F | Cacna1f | cacna1f | 1F-a,b,c | |||
| CaV2.1 | α 1A | P/Q | CACNA1A | Cacna1a | cacna1aa,b | 1A-a,b | cac | unc-2 |
| CaV2.2 | α 1B | N | CACNA1B | Cacna1b | cacna1ba,b | 1B | ||
| CaV2.3 | α 1E | R | CACNA1E | Cacna1e | -- | 1E-a,b | ||
| CaV3.1 | α 1G | T | CACNA1G | Cacna1g | cacna1g | 1G-a,b | Ca-α1T | cca-1 |
| CaV3.2 | α 1H | CACNA1H | Cacna1h | -- | 1H-a,b | |||
| CaV3.3 | α 1I | CACNA1I | Cacna1i | cacna1i | 1I | |||
Cell-specific RNA editing could exert similar control over CaV channels in other populations of neurons, and could alter amino acid sequences of other critical domains that regulate specific functions. Because of the potential importance of CaV1.3 channels in promoting calcium-dependent cell death in dopaminergic neurons in the substantia nigra pars compacta and the proposed connection between CaV1.3 activity and Parkinson Disease [28], it will be interesting to know which edited versions of CaV1.3 RNAs dominate in these neurons. Mapping the cell-specific expression patterns of ADAR2 protein and mapping ADAR2 binding sites to specific RNAs would provide valuable information in this regard. However, as is the case for splicing factors, ADAR2 activity depends on several factors including phosphorylation-dependent prolyl-isomerase Pin1 that controls its nuclear localization [29], the ubiquitin E3 ligase WWP2 that promotes its proteasomal degradation [29], as well as other second messengers.
Mechanisms that Control Numbers of CaV Channels at the Cell Surface
CaV channel activity depends not only on the pattern of expression of functionally different splice and RNA edited isoforms but also on the overall expression level of CaV channel proteins in specific subcellular compartments. Counting CaV2 channels at active zones of different synapses by quantitative molecular and ultrastructural analyses recently demonstrated a tight correlation between presynaptic CaV2.1 and CaV2.2 channel number and vesicle release probability [30,31]. Whereas the third member of the CaV2 gene family, CaV2.3, was shown (with the exception of the interpeduncular nucleus) to be mostly restricted to postsynaptic structures, particularly dendritic shaft plasma membrane [32]. Thus, the overall activities of presynaptic CaV2.1 and CaV2.2 channels have a major influence on the efficacy of transmission at mammalian synapses. We know a great deal about G protein-coupled receptor mediated control of CaV2 channels, particularly CaV2.2. These well-studied signaling pathways terminate on CaV2.2 channels and account for short-term presynaptic effects of many neurotransmitters, hormones and drugs that modulate synaptic transmission. By contrast, the cellular mechanisms that control the overall number of CaV channels at active zones—which likely contribute to long-term changes in synaptic plasticity—were only recently elucidated [31].
Not surprisingly, cellular mechanisms analogous to those critical in setting the overall expression levels of postsynaptic receptors, including the ubiquitin proteasome system (UPS) [33], control surface expression of presynaptic CaV2.1 and CaV2.2 channels [34-36]. In addition, protein-CaV2 channel interactions described recently appear critical in establishing the number of CaV2 channels specifically at presynaptic terminals [30,37,38] (Fig. 5). Such molecular interactions are dynamic and subject to modulation, thereby regulating the density and number of presynaptic CaV2 channels during synaptic plasticity [30,31].
Ubiquitin Regulates CaV1 and CaV2 channels
Ubiquitination of several neuronal proteins is a critical posttranslational mechanism to modify the trafficking, endocytosis and activity of synaptic receptors and ion channels to adjust synaptic strength [34,39]. Ubiquitin covalently attaches to intracellular lysines of target proteins and, depending on the type of conjugate (i.e., monoubiquitination or polyubiquitination), it can promote internalization, modify protein function, or target protein for degradation via the UPS [40]. Activity-dependent ubiquitination of postsynaptic AMPA receptors is described and known to regulate synaptic plasticity. Until recently, CaV channels were rarely considered important targets of ubiquitin despite functional evidence that ubiquitin-dependent changes in synaptic efficacy involve presynaptic components [41]. Four reports now show that CaV1.3, CaV1.2, and CaV2.2 channels are all targets of posttranslational modification by ubiquitin [34-36,42]. These reports implicate the UPS in controlling activity-dependent pre- and postsynaptic calcium levels. Ubiquitination of CaVα1 subunits is influenced by their association with CaVβ subunits—known to regulate plasma membrane targeting of CaV—and by cell-specific alternative splicing. CaVβ subunit binding to CaV1.2 and CaV2.2 reduces CaVα ubiquitination and protects channels from proteasomal degradation [34,36]. Similarly, CaV2.2 channel splice isoforms with restricted expression (e37a-containing) have reduced ubiquitination compared to other broadly expressed isoforms (e37b-containing) and are associated with increased channel current densities [35]. Thus, the influence of the UPS on CaV2.2 channels is cell-specific, reflecting the particular expression profile of certain alternative splice forms. Cav2.2 was recently identified as a target of ubiquitin based on a large-scale proteomics analysis of diGly-modified lysine residues of proteins expressed in a human colorectal carcinoma cell line [43]. Modified lysines in the region of Cav2.2 encoded by e37a and e37b were not identified in this screen, suggesting that ubiquitination at this site is cell-specific. Future large-scale proteomics analyses that enrich for neuronal ion channels, and other membrane proteins, hold promise for mapping major sites of posttranslational modifications of critical synaptic proteins.
CaVα2δ and RIM regulate CaV2 expression
Two protein families have recently grabbed the limelight as essential for trafficking or tethering CaV2 channels to nerve terminals: CaVα2δ and RIM (Fig. 5, Fig. 6). CaVα2δ-1, first shown to interact with CaV channels ~25 years ago, is a glycosylphosphatidylinositol (GPI)-anchored extracellular protein [44]. This auxiliary subunit of CaV2 channels promotes membrane trafficking, speeds channel activation and inactivation, and binds the analgesics gabapentin and pregabalin. CaVα2δ-1 has now been shown to act as a rate-limiting factor controlling the number of CaV2.1 channels at presynaptic terminals [45]. Evidence for a limiting factor in CaV2 channel trafficking to presynaptic nerve terminals emerged from studies in cultured hippocampal neurons. Whereas somatic Ca currents were reliably enhanced in cultured neurons transiently expressing exogenous CaV2.1 or CaV2.2 channels, synaptic transmission was not [45,46]. However, co-expressing CaVα2δ-1 along with CaV2.1 or CaV2.2 channels in these neurons augmented channel function, facilitated CaV2 coupling to exocytosis, and enhanced synaptic transmission [45] (Fig. 5).
RIM proteins are also critical CaV2 channel partners at nerve terminals. The reported actions of RIM proteins on CaV2 channels are diverse: they augment CaV2.1 currents in presynaptic calyx of Held terminals [37], decrease voltage-dependent inactivation (VDI) of CaV2.2 channels, interfere with the inhibitory actions of μ-opioid receptors on CaV2.2 channels expressed in HEK-293 cells [47], and slow CaV1.3 channel inactivation in inner hair cells [48]. But their importance in tethering CaV2 channels to active zones is central to explaining their in vivo function [38,49]. Meticulous high-resolution analyses of the molecular and morphological architecture of glutaminergic synapses show that RIM protein numbers scale proportionately with presynaptic CaV2.1 channel numbers at active zone areas [30]. Moreover, the numbers of these complexes scale with and predict vesicle release probability, consistent with critical function [30] (Fig. 6).
Controlling Subcellular Targeting of CaV channels
Synapse-specific location of CaV2 channels
CaV2.1 and CaV2.2 channels couple differentially to neurotransmitter vesicle release machinery according to synapse type. In hippocampal interneurons, CaV2.1 channels mediate release of neurotransmitter from parvalbumin (PV)-expressing fast spiking basket cells while CaV2.2 channels mediate release of neurotransmitter from cholesystokinin (CCK)-expressing basket cells [50,51]. The physical distance between presynaptic channels and calcium sensors of exocytosis are predicted to be different between CaV2.1 and CaV2.2. These predictions as based on the differential ability of BAPTA and EGTA – that are fast and slow calcium chelators, respectively – to inhibit synaptic events mediated by CaV2.1 and CaV2.2 channels [50,51]. CaV2.1 channels at PV nerve terminals are predicted to be within nanometers of the calcium sensor that leads to synchronous GABA release, whereas CaV2.2 channels at CCK terminals are predicted to be within micrometers of the Ca sensor and are thought to underlie the highly asynchronous GABA release of CCK neurons [50-52]. However, the precise molecular identity of CaV2.2 and CaV2.1 splice isoforms at these synapses is not yet determined. It is therefore possible that different splice isoforms of CaV2, that are known to influence binding to release machinery, contribute to synapse-specific differences in calcium-dependent exocytosis.
Alternative splicing of auxiliary CaV subunits, as well as other interacting proteins, is also likely to contribute to the functional diversity among different synapses. For example, the four CACNA2D1-4 genes that encode mammalian CaVα2δ1-4 subunits are each subject to alternative pre-mRNA processing. Similarly, the Rims1 and Rims2 genes generate five principal RIM proteins by the use of independent promoters (RIM1α, RIM1β, RIM2α, RIM2β, and RIM2γ) and both Rims genes contain sites of alternative splicing [49]. This provides substantial capacity for synapse-specific differences in Ca-dependent transmitter release. Finally, asynchronous transmission can occur as a consequence of prolonged CaV2 channel openings, a phenomenon that is significantly augmented in CaV2.1 channels associated with CaVβ2a subunits [53]. CaVβ subunits strongly influence the rate of CaV2 channel inactivation, and CaVβ2a subunits in particular slow channel inactivation significantly compared to CaVβ1, CaVβ3, and CaVβ4 subunits (e.g. see [53]).
Proteins in Nanodomains with Synaptic CaV2 Channels
Although lacking resolution at the level of specific synapses, a proteomics screen combined with affinity purification and high-resolution quantitative mass spectroscopy has identified over 200 proteins that interact closely with CaV2 proteins in rodent brain [54]. This valuable data set includes a subset of proteins isolated from brain synaptosomes that were previously identified to interact with CaV2.2 channels [55] as well as cytoskeletal and chaperone proteins and a complex of novel proteins involved in both exocytosis and endocytosis (Fig. 6). Functional analyses, validating the significance of several of these protein-protein interactions, suggest the interactions are critical to the sub-cellular specialization of CaV2 channels. For example, the collapsin response mediator protein 2 (CRMP2)-CaV2.2 interaction controls CaV2.2 channel current densities in sensory neurons [56,57]. Disrupting this interaction using a cell permeable blocking peptide reduced CaV2.2 trafficking to presynaptic terminals, spontaneous excitatory postsynaptic currents in the spinal dorsal horn, and inflammatory and neuropathic pain [56]. The absence of gross behavioral or motor effects in these mice supports a unique role for CRMP2-CaV2.2 in maladaptive responses in sensory neurons to noxious stimuli. Other protein interactions that, like CRMP2, are mediated via C-termini of CaV2 channels may be important in stabilizing the presynaptic molecular architecture (Fig. 6).
Perhaps the best-studied region of CaV2 channels is the synprint site located in the II-III intracellular linkers of CaV2.1 and CaV2.2 (Fig. 3). The synprint region binds synaptic protein SNAREs (Syntaxin 1A and SNAP-25) and synaptotagmin (Fig. 6). This tripartite interaction, CaV2/synprint/synaptotagmin, so critical to the release process, is reviewed in detail elsewhere [58]. Less studied is the interaction between endocytotic proteins and presynaptic CaV2 channels, most notably AP-2 (adaptor protein for clathrin-mediated endocytosis) [54]. AP-2 was shown to associate with CaV2.2 and CaV2.1, but not CaV1.2, via the synprint region to control endocytosis, but not exocytosis, at calyx of Held nerve terminals [59]. The II-III linker sequence of CaV channels is highly variable among CACNA1 genes (Fig. 3) and it contains several sites of alternative pre-mRNA splicing (Fig. 3). Alternative splicing of CaV2.2 and CaV2.1 pre-mRNAs therefore generates II-III splice isoforms that have different capacities to interact with SNAREs [58] and potentially with AP-2.
Proteins that bind and modulate pre and post synaptic CaV1.3
CaV1.3 channels are found both pre- and post- synaptically in cochlear inner hair cells (IHCs). Presynaptic CaV1.3 channels regulate transmitter release, interact with ribeye (the ribbon synapse protein that promotes channel clustering), and negatively regulate the size of the ribbon body [60] [61] (Fig. 6). Harmonin also associates with presynaptic CaV1.3 channels at the onset of hearing in 2 week-old mice. This interaction promotes CaV1.3 ubiquitination and leads to a decrease in channel surface expression. In the deaf circler mouse, dfcr, a mutant allele of the harmonin gene disrupts the harmonin interaction with CaV1.3, leading to abnormally high CaV1.3 currents in IHCs [42]. Postsynaptic CaV1.3 channels also play a critical role in the auditory brainstem where they are required for the normal development of the inhibitory medial nucleus of the trapezoid body (MNTB) to the lateral superior olive (LSO) projection [62,63]. There is substantial reduction in the number of MNTB-LSO axons over the first two weeks of life concomitant with strengthening of the remaining synapses. Thus, postsynaptic CaV1.3 channels in the LSO neurons are thought to be vital for the development and maturation of inhibitory MNTB–LSO projections [62]. Postsynaptic CaV1.3 channels isolated from brain are also found in a complex with densin and CaMKII where they localize to dendritic spines [64]. Densin promotes calcium-dependent facilitation of CaV1.3 channels [64]. Alternative pre-mRNA splicing of CACNA1D pre-mRNA must generate functionally distinct presynaptic and postsynaptic CaV1.3 channels. Defining the major splicing patterns of CaV1.3 isoforms, their differential expression patterns, and functional properties should provide valuable insight into structural domains essential for presynaptic and postsynaptic function.
Challenges
The number of molecularly distinct CaV proteins that can be generated from CACNA1 genes is stunning. Cell-specific gene expression, alternative pre-mRNA splicing, RNA editing, posttranslational modifications including ubiquitination, miRNA targeting, and subcellular specific protein-protein interactions are all employed according to the demands of the cell.
At the level of RNA, we still lack cell-specific information about which mRNA is expressed and when. Several cell-type specific transcriptome data sets have been generated from the combined application of methods including high throughput mRNA sequencing, pooling of genetically homogenous cells, and enriching for polysomal RNAs [13,65]. These methods are cataloguing CACNA1 transcripts according to neuronal sub-type and developmental stage, although substantial variability in the composition of certain transcripts has been observed across different studies and stochastic variations in transcriptomes are evident even in genetically similar cell types [13]. Genetically targeted and affinity-based miRNA profiling (miRAP) has also generated the first comparative analysis of cell-specific miRNAs in glutamatergic and GABAergic neurons of neocortex and cerebellum [66], giving insights into miRNA-dependent control of mRNA translation and stability in different classes of neurons. In addition, recent genome-wide mapping of splicing factor binding-sites by CLIP-seq, as already discussed, locates sites of splicing factor binding relative to target exons in pre-mRNAs which, combined with other information, could eventually lead to a splicing code [8]. These, and other sequence-based data sets not mentioned here, are publicly accessible (http://genome.ucsc.edu/) and recently integrated with the encyclopedia of DNA elements project (ENCODE, http://www.genome.gov/10005107).
At the protein level, major challenges remain but exciting technological advances are being made that combine high resolution imaging with proteomic analyses of single synapses [67]. It should soon be possible to visualize and distinguish among CaV isoforms with the spatial resolution needed to place them at individual active zones and synapses. Similarly, it may be possible to define unique patterns of posttranslational modifications, and characterize unique protein-CaV interactions that occur in highly localized, specialized subcellular regions of neurons. Such information should reveal the molecular mechanisms that underlie functional specialization at the level of individual synapses, and may suggest new therapeutic strategies to target specific regions or neural circuits within the brain.
At the gene level, several hereditary diseases and disorders originate from point mutations or tri-nucleotide expansions in CACNA1 genes but a major, as yet unexplained finding implicates CACNA1C (CaV1.2) in determining an individual's susceptibility to a range of psychiatric illnesses. In the most recent and largest genome wide association study of its kind, four risk loci were identified with significant and overlapping links to autism spectrum disorder, attention-deficit/hyperactivity disorder, bipolar disorder, major depressive disorder, and schizophrenia [68]. Two of these risk SNPs are located in calcium channel genes, CACNA1C (CaV1.2) and CACNB2 (CaVβ2) [69] [70,71] [72]. The widespread expression of CaV1.2 channels in the body including muscle cells of the cardiovascular system, endocrine cells, and smooth muscle adds to the intrigue. The cellular mechanisms described here that individualize CaV channel function according to cell-type, and in particular alternative pre-mRNA splicing, could be selectively disrupted in individuals exhibiting psychiatric illness without involvement of non-CNS systems.
Box 1. By Any Other Name.
Voltage-gated calcium (CaV) ion channels are highly specialized plasma membrane proteins that convert small changes in the membrane potential into rapid, localized increases in intracellular calcium. Only a limited number of ion channels, including several ligand-gated ion channels, allow the passage of calcium. However, CaV channels are exquisitely selective for calcium over other cations as long as extracellular calcium concentrations exceed ~ 10 M. In the absence of extracellular calcium, CaV channels lose their high selectively to calcium and readily conduct a range of cations.
Most CaV channels have a dual function: they support ionic current that changes the membrane potential, and they permit the flow of calcium across the plasma membrane that serves as an intracellular second messenger. In mammals, ten genes encode the ten major 1 subunits of CaV channels (CACNA1A through CACNA1I, and CACNA1S). They have unique expression profiles, cellular function, pharmacology, and are associated with various diseases. The numerous aliases used to identify and distinguish the different CaV channel genes, proteins, and currents are shown for seven different species (Table I).
The major 1 subunits of CaV channels fall into three classes based on sequence homology CaV1 (1.1, 1.2, 1.3, and 1.4), CaV2 (2.1, 2.2, and 2.3) and CaV3 (3.1, 3.2, and 3.3). The classes are functionally distinguished by a combination of pharmacological tools and biophysical properties, although pharmacological differences among CaV1 (dihydropyridine-sensitive), CaV2 (ω-agatoxin-IVA inhibits CaV2.1; ω-conotoxin GIVA inhibits CaV2.2), and CaV3 channels more reliably distinguish among CaV sub-types. Pharmacological, genetic and functional studies have helped delineate the different cellular roles controlled by CaV channels. Presynaptic calcium entry, and subsequent transmitter release is mediated by CaV2 (mainly CaV2.1 and CaV2.2) channels throughout the nervous system, by CaV1.3 in inner hair cells [73] and by CaV1.4 in retina [74,75]. Dendritic postsynaptic CaV1 (mainly CaV1.2) channels regulate calcium entry that controls gene expression [76,77]. Dendritic CaV2.3 channels [32] are implicated in acquisition of dendritic phenotype [78] and oscillatory burst discharge in the reticular thalamus[79]. Postsynaptic CaV1.3 channels underlie pacemaking in certain cells [80], are important for brain stem neuron development [63], and are implicated in calcium-dependent death of dopaminergic neurons [2]. The roles of presynaptic CaV2 channels in synaptic transmission and short-term plasticity [58] and the role of postsynaptic CaV1 channels in activity-dependent gene expression in neurons have been reviewed elsewhere [81]. A functional, correctly targeted CaV channel depends on its association with other calcium channel subunits including CaVβ, CaVα2β, and other proteins – some of these aspects of CaV channel regulation are detailed in other recent reviews [82].
Highlights.
Highlights for “Control of Neuronal Voltage-Gated Calcium Ion Channels From RNA to Protein”
How many different voltage-gated calcium channels are there and should you care?
All mammalian Cacna1 genes have the potential to generate hundreds of CaV channels
Cell-specific mechanisms control CaV channel function at RNA and Protein levels according to cell type
Cell-specific protein-protein interactions control sub-cellular CaV channel trafficking and function
Cell-specific and subcellular expression patterns of CaV isoforms is important for disease and treatment developement
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hagiwara S, Byerly L. Membrane biophysics of calcium currents. Fed Proc. 1981;40:2220–2225. [PubMed] [Google Scholar]
- 2.Kang S, et al. CaV1.3-selective L-type calcium channel antagonists as potential new therapeutics for Parkinson's disease. Nat Commun. 2012;3:1146. doi: 10.1038/ncomms2149. [DOI] [PubMed] [Google Scholar]
- 3.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ule J, et al. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 5.Gehman LT, et al. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet. 2011;43:706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeo GW, et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charizanis K, et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron. 2012;75:437–450. doi: 10.1016/j.neuron.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irimia M, Blencowe BJ. Alternative splicing: decoding an expansive regulatory layer. Curr Opin Cell Biol. 2012;24:323–332. doi: 10.1016/j.ceb.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Z, et al. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol Cell. 2012;47:422–433. doi: 10.1016/j.molcel.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dredge BK, Jensen KB. NeuN/Rbfox3 nuclear and cytoplasmic isoforms differentially regulate alternative splicing and nonsense-mediated decay of Rbfox2. PLoS One. 2011;6:e21585. doi: 10.1371/journal.pone.0021585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boutz PL, et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang F, Lao K, Surani MA. Development and applications of single-cell transcriptome analysis. Nat Methods. 2011;8:S6–11. doi: 10.1038/nmeth.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang ZZ, et al. Regulation of the mutually exclusive exons 8a and 8 in the CaV1.2 calcium channel transcript by polypyrimidine tract-binding protein. J Biol Chem. 2011;286:10007–10016. doi: 10.1074/jbc.M110.208116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Splawski I, et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 16.Splawski I, et al. Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations. Proc Natl Acad Sci U S A. 2005;102:8089–96. doi: 10.1073/pnas.0502506102. discussion 8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang C, et al. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science. 2010;329:439–443. doi: 10.1126/science.1191150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen SE, Darnell RB, Lipscombe D. The neuronal splicing factor Nova controls alternative splicing in N-type and P-type CaV2 calcium channels. Channels (Austin) 2010;4:483–489. doi: 10.4161/chan.4.6.12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gehman LT, et al. The splicing regulator Rbfox2 is required for both cerebellar development and mature motor function. Genes Dev. 2012;26:445–460. doi: 10.1101/gad.182477.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turrigiano G. Homeostatic synaptic plasticity: local and global mechanisms for stabilizing neuronal function. Cold Spring Harb Perspect Biol. 2012;4:a005736. doi: 10.1101/cshperspect.a005736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jarosch A, Stolle E, Crewe RM, Moritz RF. Alternative splicing of a single transcription factor drives selfish reproductive behavior in honeybee workers (Apis mellifera). Proc Natl Acad Sci U S A. 2011;108:15282–15287. doi: 10.1073/pnas.1109343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gracheva EO, et al. Ganglion-specific splicing of TRPV1 underlies infrared sensation in vampire bats. Nature. 2011;476:88–91. doi: 10.1038/nature10245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andrade A, Denome S, Jiang YQ, Marangoudakis S, Lipscombe D. Opioid inhibition of N-type Ca2+ channels and spinal analgesia couple to alternative splicing. Nat Neurosci. 2010;13:1249–1256. doi: 10.1038/nn.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams PJ, et al. Ca(V)2.1 P/Q-type calcium channel alternative splicing affects the functional impact of familial hemiplegic migraine mutations: implications for calcium channelopathies. Channels (Austin) 2009;3:110–121. doi: 10.4161/chan.3.2.7932. [DOI] [PubMed] [Google Scholar]
- 25.Powell KL, et al. A Cav3.2 T-type calcium channel point mutation has splice-variant-specific effects on function and segregates with seizure expression in a polygenic rat model of absence epilepsy. J Neurosci. 2009;29:371–380. doi: 10.1523/JNEUROSCI.5295-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith LA, et al. A Drosophila calcium channel alpha1 subunit gene maps to a genetic locus associated with behavioral and visual defects. J Neurosci. 1996;16:7868–7879. doi: 10.1523/JNEUROSCI.16-24-07868.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang H, et al. RNA editing of the IQ domain in Ca(v)1.3 channels modulates their Ca(2)(+)-dependent inactivation. Neuron. 2012;73:304–316. doi: 10.1016/j.neuron.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ilijic E, Guzman JN, Surmeier DJ. The L-type channel antagonist isradipine is neuroprotective in a mouse model of Parkinson's disease. Neurobiol Dis. 2011;43:364–371. doi: 10.1016/j.nbd.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marcucci R, et al. Pin1 and WWP2 regulate GluR2 Q/R site RNA editing by ADAR2 with opposing effects. EMBO J. 2011;30:4211–4222. doi: 10.1038/emboj.2011.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holderith N, et al. Release probability of hippocampal glutamatergic terminals scales with the size of the active zone. Nat Neurosci. 2012;15:988–997. doi: 10.1038/nn.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheng J, et al. Calcium-channel number critically influences synaptic strength and plasticity at the active zone. Nat Neurosci. 2012;15:998–1006. doi: 10.1038/nn.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parajuli LK, et al. Quantitative regional and ultrastructural localization of the Ca(v)2.3 subunit of R-type calcium channel in mouse brain. J Neurosci. 2012;32:13555–13567. doi: 10.1523/JNEUROSCI.1142-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henley JM, Barker EA, Glebov OO. Routes, destinations and delays: recent advances in AMPA receptor trafficking. Trends Neurosci. 2011;34:258–268. doi: 10.1016/j.tins.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altier C, et al. The Cavbeta subunit prevents RFP2-mediated ubiquitination and proteasomal degradation of L-type channels. Nat Neurosci. 2011;14:173–180. doi: 10.1038/nn.2712. [DOI] [PubMed] [Google Scholar]
- 35.Marangoudakis S, et al. Differential ubiquitination and proteasome regulation of Ca(V)2.2 N-type channel splice isoforms. J Neurosci. 2012;32:10365–10369. doi: 10.1523/JNEUROSCI.0851-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waithe D, Ferron L, Page KM, Chaggar K, Dolphin AC. Beta-subunits promote the expression of Ca(V)2.2 channels by reducing their proteasomal degradation. J Biol Chem. 2011;286:9598–9611. doi: 10.1074/jbc.M110.195909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han Y, Kaeser PS, Sudhof TC, Schneggenburger R. RIM determines Ca(2)+ channel density and vesicle docking at the presynaptic active zone. Neuron. 2011;69:304–316. doi: 10.1016/j.neuron.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaeser PS, et al. RIM proteins tether Ca2+ channels to presynaptic active zones via a direct PDZ-domain interaction. Cell. 2011;144:282–295. doi: 10.1016/j.cell.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rotin D, Staub O. Role of the ubiquitin system in regulating ion transport. Pflugers Arch. 2011;461:1–21. doi: 10.1007/s00424-010-0893-2. [DOI] [PubMed] [Google Scholar]
- 40.MacGurn JA, Hsu PC, Emr SD. Ubiquitin and membrane protein turnover: from cradle to grave. Annu Rev Biochem. 2012;81:231–259. doi: 10.1146/annurev-biochem-060210-093619. [DOI] [PubMed] [Google Scholar]
- 41.Rinetti GV, Schweizer FE. Ubiquitination acutely regulates presynaptic neurotransmitter release in mammalian neurons. J Neurosci. 2010;30:3157–3166. doi: 10.1523/JNEUROSCI.3712-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gregory FD, et al. Harmonin inhibits presynaptic Cav1.3 Ca(2)(+) channels in mouse inner hair cells. Nat Neurosci. 2011;14:1109–1111. doi: 10.1038/nn.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davies A, et al. The alpha2delta subunits of voltage-gated calcium channels form GPI-anchored proteins, a posttranslational modification essential for function. Proc Natl Acad Sci U S A. 2010;107:1654–1659. doi: 10.1073/pnas.0908735107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoppa MB, Lana B, Margas W, Dolphin AC, Ryan TA. alpha2delta expression sets presynaptic calcium channel abundance and release probability. Nature. 2012;486:122–125. doi: 10.1038/nature11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao YQ, Tsien RW. Different relationship of N- and P/Q-type Ca2+ channels to channel-interacting slots in controlling neurotransmission at cultured hippocampal synapses. J Neurosci. 2010;30:4536–4546. doi: 10.1523/JNEUROSCI.5161-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weiss N, et al. Rim1 modulates direct G-protein regulation of Ca(v)2.2 channels. Pflugers Arch. 2011;461:447–459. doi: 10.1007/s00424-011-0926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gebhart M, et al. Modulation of Cav1.3 Ca2+ channel gating by Rab3 interacting molecule. Mol Cell Neurosci. 2010;44:246–259. doi: 10.1016/j.mcn.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 49.Kaeser PS, Deng L, Fan M, Sudhof TC. RIM genes differentially contribute to organizing presynaptic release sites. Proc Natl Acad Sci U S A. 2012;109:11830–11835. doi: 10.1073/pnas.1209318109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goswami SP, Bucurenciu I, Jonas P. Miniature IPSCs in hippocampal granule cells are triggered by voltage-gated Ca2+ channels via microdomain coupling. J Neurosci. 2012;32:14294–14304. doi: 10.1523/JNEUROSCI.6104-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hefft S, Jonas P. Asynchronous GABA release generates long-lasting inhibition at a hippocampal interneuron-principal neuron synapse. Nat Neurosci. 2005;8:1319–1328. doi: 10.1038/nn1542. [DOI] [PubMed] [Google Scholar]
- 52.Daw MI, Tricoire L, Erdelyi F, Szabo G, McBain CJ. Asynchronous transmitter release from cholecystokinin-containing inhibitory interneurons is widespread and target-cell independent. J Neurosci. 2009;29:11112–11122. doi: 10.1523/JNEUROSCI.5760-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Few AP, et al. Asynchronous Ca2+ current conducted by voltage-gated Ca2+ (CaV)-2.1 and CaV2.2 channels and its implications for asynchronous neurotransmitter release. Proc Natl Acad Sci U S A. 2012;109:E452–E460. doi: 10.1073/pnas.1121103109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muller CS, et al. Quantitative proteomics of the Cav2 channel nano-environments in the mammalian brain. Proc Natl Acad Sci U S A. 2010;107:14950–14957. doi: 10.1073/pnas.1005940107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khanna R, Li Q, Bewersdorf J, Stanley EF. The presynaptic CaV2.2 channel-transmitter release site core complex. Eur J Neurosci. 2007;26:547–559. doi: 10.1111/j.1460-9568.2007.05680.x. [DOI] [PubMed] [Google Scholar]
- 56.Brittain JM, et al. Suppression of inflammatory and neuropathic pain by uncoupling CRMP-2 from the presynaptic Ca(2)(+) channel complex. Nat Med. 2011;17:822–829. doi: 10.1038/nm.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson SM, et al. Further insights into the antinociceptive potential of a peptide disrupting the N-type calcium channel-CRMP-2 signaling complex. Channels (Austin) 2011;5:449–456. doi: 10.4161/chan.5.5.17363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Catterall WA, Few AP. Calcium channel regulation and presynaptic plasticity. Neuron. 2008;59:882–901. doi: 10.1016/j.neuron.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 59.Watanabe H, et al. Involvement of Ca2+ channel synprint site in synaptic vesicle endocytosis. J Neurosci. 2010;30:655–660. doi: 10.1523/JNEUROSCI.3214-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sheets L, Trapani JG, Mo W, Obholzer N, Nicolson T. Ribeye is required for presynaptic Ca(V)1.3a channel localization and afferent innervation of sensory hair cells. Development. 2011;138:1309–1319. doi: 10.1242/dev.059451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sheets L, Kindt KS, Nicolson T. Presynaptic CaV1.3 channels regulate synaptic ribbon size and are required for synaptic maintenance in sensory hair cells. J Neurosci. 2012;32:17273–17286. doi: 10.1523/JNEUROSCI.3005-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hirtz JJ, et al. Synaptic refinement of an inhibitory topographic map in the auditory brainstem requires functional Cav1.3 calcium channels. J Neurosci. 2012;32:14602–14616. doi: 10.1523/JNEUROSCI.0765-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hirtz JJ, et al. Cav1.3 calcium channels are required for normal development of the auditory brainstem. J Neurosci. 2011;31:8280–8294. doi: 10.1523/JNEUROSCI.5098-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jenkins MA, et al. Ca2+-dependent facilitation of Cav1.3 Ca2+ channels by densin and Ca2+/calmodulin-dependent protein kinase II. J Neurosci. 2010;30:5125–5135. doi: 10.1523/JNEUROSCI.4367-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okaty BW, Sugino K, Nelson SB. Cell type-specific transcriptomics in the brain. J Neurosci. 2011;31:6939–6943. doi: 10.1523/JNEUROSCI.0626-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He M, et al. Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron. 2012;73:35–48. doi: 10.1016/j.neuron.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Micheva KD, Busse B, Weiler NC, O'Rourke N, Smith SJ. Single-synapse analysis of a diverse synapse population: proteomic imaging methods and markers. Neuron. 2010;68:639–653. doi: 10.1016/j.neuron.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smoller JW, et al. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nyegaard M, et al. CACNA1C (rs1006737) is associated with schizophrenia. Mol Psychiatry. 2010;15:119–121. doi: 10.1038/mp.2009.69. [DOI] [PubMed] [Google Scholar]
- 71.Ferreira MA, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee MT, et al. Genome-wide association study of bipolar I disorder in the Han Chinese population. Mol Psychiatry. 2011;16:548–556. doi: 10.1038/mp.2010.43. [DOI] [PubMed] [Google Scholar]
- 73.Johnson SL, Marcotti W. Biophysical properties of CaV1.3 calcium channels in gerbil inner hair cells. J Physiol. 2008;586:1029–1042. doi: 10.1113/jphysiol.2007.145219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bech-Hansen NT, et al. Loss-of-function mutations in a calcium-channel alpha1-subunit gene in Xp11.23 cause incomplete X-linked congenital stationary night blindness. Nat Genet. 1998;19:264–267. doi: 10.1038/947. [DOI] [PubMed] [Google Scholar]
- 75.Morgans CW. Localization of the alpha(1F) calcium channel subunit in the rat retina. Invest Ophthalmol Vis Sci. 2001;42:2414–2418. [PubMed] [Google Scholar]
- 76.Dolmetsch RE, Pajvani U, Fife K, Spotts JM, Greenberg ME. Signaling to the nucleus by an L-type calcium channel-calmodulin complex through the MAP kinase pathway. Science. 2001;294:333–339. doi: 10.1126/science.1063395. [DOI] [PubMed] [Google Scholar]
- 77.Wheeler DG, Barrett CF, Groth RD, Safa P, Tsien RW. CaMKII locally encodes L-type channel activity to signal to nuclear CREB in excitation-transcription coupling. J Cell Biol. 2008;183:849–863. doi: 10.1083/jcb.200805048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nishiyama M, et al. Semaphorin 3A induces CaV2.3 channel-dependent conversion of axons to dendrites. Nat Cell Biol. 2011;13:676–685. doi: 10.1038/ncb2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zaman T, et al. Cav2.3 channels are critical for oscillatory burst discharges in the reticular thalamus and absence epilepsy. Neuron. 2011;70:95–108. doi: 10.1016/j.neuron.2011.02.042. [DOI] [PubMed] [Google Scholar]
- 80.Vandael DH, Mahapatra S, Calorio C, Marcantoni A, Carbone E. Cav1.3 and Cav1.2 channels of adrenal chromaffin cells: Emerging views on cAMP/cGMP-mediated phosphorylation and role in pacemaking. Biochim Biophys Acta. 2013;1828:1608–1618. doi: 10.1016/j.bbamem.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 81.Calin-Jageman I, Lee A. Ca(v)1 L-type Ca2+ channel signaling complexes in neurons. J Neurochem. 2008;105:573–583. doi: 10.1111/j.1471-4159.2008.05286.x. [DOI] [PubMed] [Google Scholar]
- 82.Dolphin AC. Calcium channel auxiliary alpha2delta and beta subunits: trafficking and one step beyond. Nat Rev Neurosci. 2012;13:542–555. doi: 10.1038/nrn3311. [DOI] [PubMed] [Google Scholar]
- 83.Lipscombe D, Castiglioni AJ. Calcium Channel Pharmacology. Springer; 2004. pp. 369–409. [Google Scholar]
- 84.Boycott K, Maybaum T, Naylor M, Weleber R. A summary of 20 CACNA1F mutations identified in 36 families with incomplete X-linked congenital stationary night blindness, and characterization of splice variants. Human genetics. 2001 doi: 10.1007/s004390100461. [DOI] [PubMed] [Google Scholar]
- 85.Soong TW, et al. Systematic identification of splice variants in human P/Q-type channel alpha1(2.1) subunits: implications for current density and Ca2+-dependent inactivation. J Neurosci. 2002;22:10142–10152. doi: 10.1523/JNEUROSCI.22-23-10142.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pereverzev A, et al. Alternate splicing in the cytosolic II-III loop and the carboxy terminus of human E-type voltage-gated Ca(2+) channels: electrophysiological characterization of isoforms. Mol Cell Neurosci. 2002;21:352–365. doi: 10.1006/mcne.2002.1179. [DOI] [PubMed] [Google Scholar]
- 87.Ernst WL, Noebels JL. Expanded alternative splice isoform profiling of the mouse Cav3.1/alpha1G T-type calcium channel. BMC Mol Biol. 2009;10:53. doi: 10.1186/1471-2199-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhong X, Liu JR, Kyle JW, Hanck DA, Agnew WS. A profile of alternative RNA splicing and transcript variation of CACNA1H, a human T-channel gene candidate for idiopathic generalized epilepsies. Hum Mol Genet. 2006;15:1497–1512. doi: 10.1093/hmg/ddl068. [DOI] [PubMed] [Google Scholar]
- 89.Gomora JC, Murbartian J, Arias JM, Lee JH, Perez-Reyes E. Cloning and expression of the human T-type channel Ca(v)3.3: insights into prepulse facilitation. Biophys J. 2002;83:229–241. doi: 10.1016/s0006-3495(02)75164-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tang ZZ, Zheng S, Nikolic J, Black DL. Developmental control of CaV1.2 L-type calcium channel splicing by Fox proteins. Mol Cell Biol. 2009;29:4757–4765. doi: 10.1128/MCB.00608-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Minovitsky S, Gee SL, Schokrpur S, Dubchak I, Conboy JG. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic Acids Res. 2005;33:714–724. doi: 10.1093/nar/gki210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Coppola T, et al. Molecular cloning of a murine N-type calcium channel alpha 1 subunit. Evidence for isoforms, brain distribution, and chromosomal localization. FEBS Lett. 1994;338:1–5. doi: 10.1016/0014-5793(94)80105-3. [DOI] [PubMed] [Google Scholar]
- 94.Pan JQ, Lipscombe D. Alternative splicing in the cytoplasmic II-III loop of the N-type Ca channel alpha 1B subunit: functional differences are beta subunit-specific. J Neurosci. 2000;20:4769–4775. doi: 10.1523/JNEUROSCI.20-13-04769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sheng ZH, Rettig J, Cook T, Catterall WA. Calcium-dependent interaction of N-type calcium channels with the synaptic core complex. Nature. 1996;379:451–454. doi: 10.1038/379451a0. [DOI] [PubMed] [Google Scholar]
- 96.Rettig J, et al. Isoform-specific interaction of the alpha1A subunits of brain Ca2+ channels with the presynaptic proteins syntaxin and SNAP-25. Proc Natl Acad Sci U S A. 1996;93:7363–7368. doi: 10.1073/pnas.93.14.7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yokoyama CT, et al. Mechanism of SNARE protein binding and regulation of Cav2 channels by phosphorylation of the synaptic protein interaction site. Mol Cell Neurosci. 2005;28:1–17. doi: 10.1016/j.mcn.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 98.Rajapaksha WR, et al. Novel splice variants of rat CaV2.1 that lack much of the synaptic protein interaction site are expressed in neuroendocrine cells. J Biol Chem. 2008;283:15997–16003. doi: 10.1074/jbc.M710544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Raingo J, Castiglioni AJ, Lipscombe D. Alternative splicing controls G protein-dependent inhibition of N-type calcium channels in nociceptors. Nat Neurosci. 2007;10:285–292. doi: 10.1038/nn1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Swayne LA, Beck KE, Braun JE. The cysteine string protein multimeric complex. Biochem Biophys Res Commun. 2006;348:83–91. doi: 10.1016/j.bbrc.2006.07.033. [DOI] [PubMed] [Google Scholar]
- 101.Chi XX, et al. Regulation of N-type voltage-gated calcium channels (Cav2.2) and transmitter release by collapsin response mediator protein-2 (CRMP-2) in sensory neurons. J Cell Sci. 2009;122:4351–4362. doi: 10.1242/jcs.053280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nishimune H, Sanes JR, Carlson SS. A synaptic laminin-calcium channel interaction organizes active zones in motor nerve terminals. Nature. 2004;432:580–587. doi: 10.1038/nature03112. [DOI] [PubMed] [Google Scholar]
- 103.Leenders AG, et al. The role of MAP1A light chain 2 in synaptic surface retention of Cav2.2 channels in hippocampal neurons. J Neurosci. 2008;28:11333–11346. doi: 10.1523/JNEUROSCI.3078-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Khanna R, Zougman A, Stanley EF. A proteomic screen for presynaptic terminal N-type calcium channel (CaV2.2) binding partners. J Biochem Mol Biol. 2007;40:302–314. doi: 10.5483/bmbrep.2007.40.3.302. [DOI] [PubMed] [Google Scholar]
- 105.Davare MA, Horne MC, Hell JW. Protein phosphatase 2A is associated with class C L-type calcium channels (Cav1.2) and antagonizes channel phosphorylation by cAMP-dependent protein kinase. J Biol Chem. 2000;275:39710–39717. doi: 10.1074/jbc.M005462200. [DOI] [PubMed] [Google Scholar]
- 106.Johnson BD, Scheuer T, Catterall WA. Voltage-dependent potentiation of L-type Ca2+ channels in skeletal muscle cells requires anchored cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 1994;91:11492–11496. doi: 10.1073/pnas.91.24.11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Oliveria SF, Dell'Acqua ML, Sather WA. AKAP79/150 anchoring of calcineurin controls neuronal L-type Ca2+ channel activity and nuclear signaling. Neuron. 2007;55:261–275. doi: 10.1016/j.neuron.2007.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Davare MA, Dong F, Rubin CS, Hell JW. The A-kinase anchor protein MAP2B and cAMP-dependent protein kinase are associated with class C L-type calcium channels in neurons. J Biol Chem. 1999;274:30280–30287. doi: 10.1074/jbc.274.42.30280. [DOI] [PubMed] [Google Scholar]
- 109.Hall DD, et al. Critical role of cAMP-dependent protein kinase anchoring to the L-type calcium channel Cav1.2 via A-kinase anchor protein 150 in neurons. Biochemistry. 2007;46:1635–1646. doi: 10.1021/bi062217x. [DOI] [PubMed] [Google Scholar]
- 110.Kochlamazashvili G, et al. The extracellular matrix molecule hyaluronic acid regulates hippocampal synaptic plasticity by modulating postsynaptic L-type Ca(2+) channels. Neuron. 2010;67:116–128. doi: 10.1016/j.neuron.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kawaguchi M, Minami K, Nagashima K, Seino S. Essential role of ubiquitin-proteasome system in normal regulation of insulin secretion. J Biol Chem. 2006;281:13015–13020. doi: 10.1074/jbc.M601228200. [DOI] [PubMed] [Google Scholar]
- 112.Green EM, Barrett CF, Bultynck G, Shamah SM, Dolmetsch RE. The tumor suppressor eIF3e mediates calcium-dependent internalization of the L-type calcium channel CaV1.2. Neuron. 2007;55:615–632. doi: 10.1016/j.neuron.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Davare MA, Hell JW. Increased phosphorylation of the neuronal L-type Ca(2+) channel Ca(v)1.2 during aging. Proc Natl Acad Sci U S A. 2003;100:16018–16023. doi: 10.1073/pnas.2236970100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kersten FF, et al. Association of whirlin with Cav1.3 (alpha1D) channels in photoreceptors, defining a novel member of the usher protein network. Invest Ophthalmol Vis Sci. 2010;51:2338–2346. doi: 10.1167/iovs.09-4650. [DOI] [PubMed] [Google Scholar]
- 115.Knirsch M, et al. Persistence of Ca(v)1.3 Ca2+ channels in mature outer hair cells supports outer hair cell afferent signaling. J Neurosci. 2007;27:6442–6451. doi: 10.1523/JNEUROSCI.5364-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yang PS, et al. Switching of Ca2+-dependent inactivation of Ca(v)1.3 channels by calcium binding proteins of auditory hair cells. J Neurosci. 2006;26:10677–10689. doi: 10.1523/JNEUROSCI.3236-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cui G, et al. Ca2+-binding proteins tune Ca2+-feedback to Cav1.3 channels in mouse auditory hair cells. J Physiol. 2007;585:791–803. doi: 10.1113/jphysiol.2007.142307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang H, et al. Association of CaV1.3 L-type calcium channels with Shank. J Neurosci. 2005;25:1037–1049. doi: 10.1523/JNEUROSCI.4554-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]