Abstract
We have obtained a 1.55 Å crystal structure of a hammerhead ribozyme derived from Schistosoma mansoni in conditions that permit detailed observations of Na+ ion binding in the ribozyme's active site. At least two such Na+ ions are observed. The first Na+ ion binds to the N7 of G10.1 and the adjacent A9 phosphate in a manner identical to that previously observed for divalent cations. A second Na+ ion binds to the Hoogsteen face of G12, the general base in the hammerhead cleavage reaction, thereby potentially dissipating the negative charge of the catalytically active enolate form of the nucleotide base. A potential but more ambiguous third site bridges the A9 and scissile phosphates in a manner consistent with previous predictions. Hammerhead ribozymes have been observed to be active in the presence of high concentrations of monovalent cations, including Na+, but the mechanism by which monovalent cations substitute for divalent cations in hammerhead catalysis remains unclear. Our results enable us to suggest that Na+ directly and specifically substitutes for divalent cations in the hammerhead active site. The detailed geometry of the pre-catalytic active site complex is also revealed with a new level of precision, thanks to the quality of the electron density maps obtained from what is currently the highest resolution ribozyme structure in the protein data bank.
Introduction
The hammerhead ribozyme is found in satellite RNAs of various plant RNA virus genomes1,2, in the 3′-UTRs of mammals3, and within introns of many eukaryotes4,5. The ribozyme consists of a conserved core of about 15 mostly invariant residues 6 and, for optimal activity, requires the presence of sequences in stems I and II that interact to form tertiary contacts 7,8. (The optimal form has been termed the “natural” or “full-length” hammerhead.) The hammerhead ribozyme catalyzes an RNA self-cleavage phosphodiester isomerization reaction that involves nucleophilic attack of the C17 2′O upon the adjacent scissile phosphate, producing two RNA product strands.
Perhaps the most substantial of the controversies9-11 remaining subsequent to elucidation of the full-length hammerhead ribozyme structures12,13 is the mechanistic role that metal ions might play in the chemistry of catalysis. At one extreme, it has been proposed, based on observed cleavage in the presence of very high ionic-strength monovalent cations (such as Li+ and Na+) but in absence of divalent metal ions, that Mg2+, when present, plays a purely structural role, and is not a required participant in the chemical mechanism of catalysis14. At the other extreme are proposals in which one or more Mg2+ ions participate directly in the transition-state of the hammerhead self-cleavage reaction. The first and arguably most important of these hypotheses is one in which a single Mg2+ ion is proposed to coordinate directly two non-bridging phosphate oxygens simultaneously, one belonging to the A9 phosphate, and the other belonging to the scissile phosphate of the cleavage site 15. Many other mechanistic proposals involving participation of monovalent and divalent metal cations that reside between these mechanistic extremes have also been suggested16-19.
The first crystal structure of the natural, full-length hammerhead ribozyme was obtained from crystals grown in a high concentration of ammonium sulphate. This structure did not reveal any metal ions bound near the active site12, even while revealing the A9 and scissile phosphates, previously implicated in binding a single metal ion, to be only 4.2 Å apart, a distance that would be easily bridged by a single Mg2+ ion. Even more puzzling is the mode of Mn2+ binding in the same crystals after Mn2+ is introduced. (Mn2+ is known to substitute for Mg2+ in many RNAs, and it has the advantage of possessing a distinct X-ray absorption signature enabling its unambiguous identification even at moderate resolution). Despite the A9 and scissile phosphates forming an apparently near-perfect potential divalent metal ion binding-site, Mn2+ is nevertheless observed to bind exclusively to the A9 phosphate and the adjacent N7 of G10.120, in a manner essentially identical to that observed for the minimal hammerhead 21, in which the scissile phosphate is 18 Å away. This in turn has lead to proposals that include migration of the metal ion from the A9-only position to the bridging position, and binding of an additional bridging metal ion during formation of the transition-state17,18.
Here we report the highest-resolution ribozyme structure to date. New crystallization conditions were obtained in the presence of a high concentration (over 3 M) of Na+ ions, in addition to producing better-diffracting crystals. The new structure reveals several well-ordered bound Na+ ions, including ones bound at the active site whose catalytic relevance is quite suggestive.
Results
New crystallization conditions permit observation of bound Na+ ions at near-atomic resolution
Hammerhead ribozyme crystals prepared using 1.7 M sodium malonate, buffered to pH 7.5 as a precipitating agent (replacing ammonium sulphate), and 35% PEG 3350, possessed the same space group and unit cell dimensions as our previous crystals 12, but yielded significantly improved diffraction (1.55 Å resolution vs. 2.2 Å). These crystals permit visualization of specifically-bound Na+ ions under nearly ideal conditions. An overall view of the hammerhead ribozyme (PDB 3ZP8) with associated Na+ ion sites is depicted in Figure 1. Data collection and refinement statistics are reported in Table I.
Figure 1.
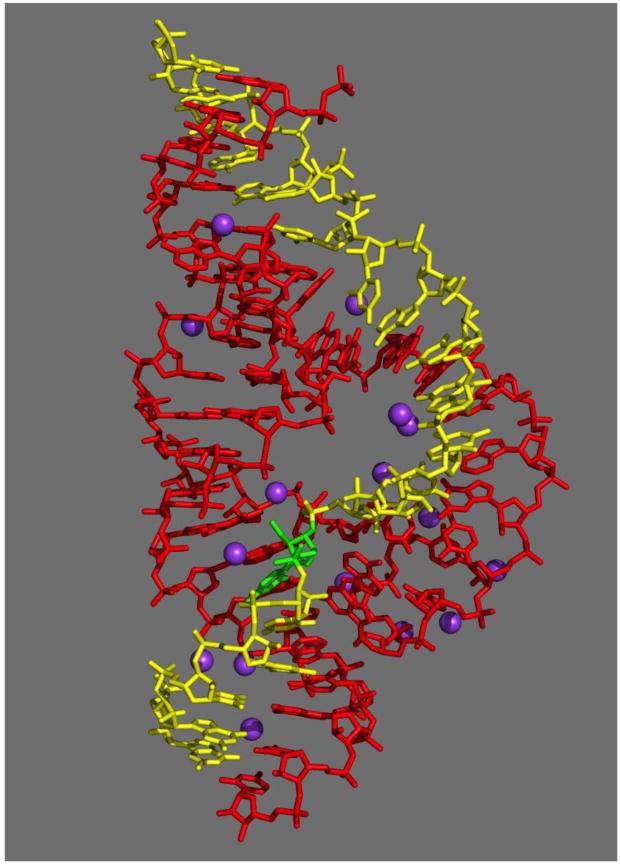
An all-bond representation of the 1.55 Å refined crystal structure of the full-length hammerhead ribozyme with bound Na+ ions. The enzyme strand is depicted in red, the substrate strand in yellow, the cleavage-site nucleotide (C17) in green, and the various Na+ ions are represented as purple spheres. Coordinates are available in the PDB (3ZP8).
Table I. Crystallographic data and refinement statistics.
| Data collection | ||
| Data Processing Software | iMosflm32, CCP4 suite, phenix.xtriage | |
| Space group | Monoclinic, C2 | |
| Unit cell parameters | a=50.82Å b=68.53Å c=58.90Å β = 112.35° | |
| Solvent Content, Vs (%) | 49.2 | |
| Matthews Coef., Vm (Å3/Mr) | 2.42 | |
| Overall | Highest res. shell | |
| Resolution range (Å) | 20.39 - 1.55 | 1.63-1.55 |
| No. unique reflections | 26462 | 3855 |
| Redundancy | 3.0 | 3.0 |
| I/σ | 13.90 | 1.80 |
| Completeness (%) | 97.6 | 99.6 |
| Rmergea | 0.03 | 0.63 |
| Structure refinement | ||
| Model building softwareb | COOT | |
| Refinement softwarec | phenix.refine | |
| Target | Maximum Likelihood (ML) | |
| R-factors | ||
| Rcrysd | 0.1867 | 0.2999 |
| Rfreee | 0.2157 | 0.3244 |
| Test set size (%) | 7.57 | 7.34 |
| Geometry | ||
| r.m.s.d. bond lengths (Å) | 0.004 | |
| r.m.s.d. bond angles (°) | 0.945 | |
| r.m.s.d. planarity (°) | 0.008 | |
| r.m.s.d. torsion angles (°) | 14.414 | |
| ML coordinate error (Å) | 0.21 | |
| ML phase error (°) | 23.41 | |
| β-factor from Wilson plot (Å2) | 24.76 | |
| No. TLS groupsf | 10 | |
Rmerge =©|I□ 〈I〉 | / ©(I), where I is the intensity of measured reflection and 〈I〉 is the mean intensity of all symmetry-related reflections.
Model-building and validation, and identification of Na+, was performed using COOT.35
Refinement and analysis was carried out using Phenix (phenix.refine and phenix.xtriage).34
Rcryst =Σ|Fcalc −Fobs|/ΣFobs, where Fobs and Fcalc are observed and calculated structure factors, respectively.
Rfree = ΣT|Fcalc − Fobs|/Σ Fobs, where T is a test data set of about 7.5% of the total unique reflections randomly chosen and set aside prior to refinement.
The quality of the near-atomic resolution electron density is shown in Figure 2a (a close-up of G12, the general base in the hammerhead cleavage reactions), and Figure 3 (a network of RNA-water-Na+ ion interactions near the active site). This is currently the highest resolution ribozyme structure available in the PDB database, and thus provides the best opportunity to identify monovalent metal ion interactions relevant to hammerhead ribozyme catalysis. Sixteen Na+ ion binding sites were identified based on coordination number and distances to adjacent water molecules, making reference to standard values obtained from large-angle X-ray scattering and double difference infrared spectroscopy experiments on hydrated Na+ ions in aqueous solution 22 and the MESPEUS data base23 of previously-determined macromolecular X-ray crystallographic structures in complex with metal ions in the PDB.
Figure 2.
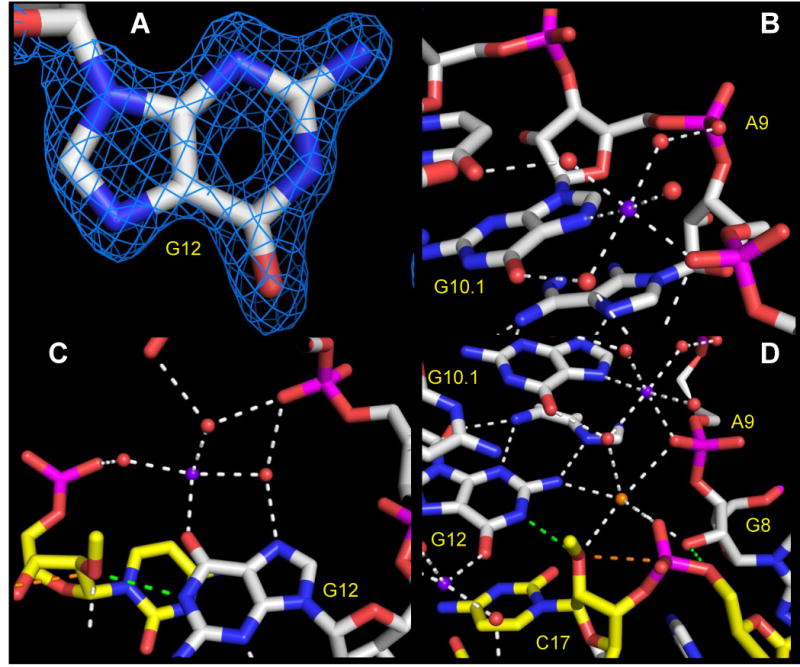
Atomic structure of the hammerhead ribozyme active site, with oxygen atoms depicted in red, nitrogen atoms in blue, phosphori in magenta, enzyme-strand carbon atoms in white, and substrate-strand carbon atoms in yellow. Na+ ions are shown as purple spheres. (a) A representative 1.55 Å resolution sigmaA-weighted 2Fo-Fc electron density map contoured at 2.0 r.m.s.d. reveals a hole in the six-membered aromatic ring of the general base, G12, in the active site. (b) The canonical A9 divalent metal ion binding site is occupied by Na+, (purple sphere), which forms inner-sphere interactions with the pro-R phosphate oxygen of A9, the N7 of G10.1, and four water molecules, three of which in turn form bridging interactions with other RNA atoms.(c) The Hoogsteen face of the general base, G12, within the active site, forms a second potentially catalytically relevant Na+ binding site (Na+ ion in purple). The O6 of G12 forms an inner-sphere interaction with the Na+, and one of the three unambiguously resolved water molecules it coordinates bridges to the N7 of G12. The other observed water molecules bridge to various phosphates near the active site. (d) A third potential ion or water molecule (orange sphere, see discussion in text) coordinates the pro-R oxygens of A9 and scissile phosphates, the 2′-O nucleophile of the cleavage site base, the 2′-O of G8 implicated in general acid catalysis, and a water molecule in a distorted octahedral complex.
Figure 3.

Wall-eyed stereo diagrams illustrating the network of hydrogen bonding interactions and other close contacts within the active site, as well as Na+ ions, shown as purple spheres, and well-resolved water molecules, whose oxygen atoms are shown as red spheres. The octahedrally coordinated water molecule or ion is shown in orange and labeled with a question mark. The carbon atoms within the enzyme strand are white, and those of the substrate strand are yellow. Green dotted lines correspond to the potentially active hydrogen bonds of the general base (N1 of G12 to 2′O of C17) and general acid (2′OH of G8 to 5′O of C1.1). The orange dotted line is the presumed trajectory of bond formation between the 2′O of C17 and the adjacent phosphorus atom of C1.1. (The blue mesh of the top figure is a 1.55 Å resolution sigmaA-weighted 2Fo-Fc electron density contoured at 1.2 r.m.s.d. The bottom figure is the same view, with the electron density map removed for clarity.
While most of the Na+ ions are in non-conserved regions of the ribozyme, two Na+ ion binding sites reside within the hammerhead ribozyme active site and are comprised of invariant residues. A third potential site is consistent with either Na+ ion or a water molecule. We focus on these sites primarily due to their potential mechanistic relevance to the chemistry of hammerhead ribozyme catalysis.
A Na+ ion binds to the A9 phosphate in a manner similar to divalent cations
A single strong divalent metal ion binding site has been observed in every minimal hammerhead ribozyme structure containing divalent metal ions24, including the first structure reported21. Because of the modest resolution of these initial structures, employment of Mn2+ or other divalent metal ions with a characteristic X-ray absorption signature greatly facilitated unambiguous identification 21,25. This divalent metal ion binding site, which has come to be known as the “McKay site,” is consistently characterized by direct coordination of the pro-R non-bridging phosphate oxygen atom of A9 with the metal ion, and direct coordination of the N7 of G10.1, the conserved nucleotide 3′-adjacent to the A9 phosphate at the proximal end of helical Stem II. The remaining four ligand positions in the octahedral complex are occupied by water molecules. This same mode of Mn2+ binding is also observed in the full-length hammerhead structure20.
Our 1.55 Å hammerhead ribozyme structure reveals that Na+ binds to the A9 phosphate and the N7 of G10.1 in the same way that Mn2+ binds (Figure 2b). The distance between Na+ and the pro-R phosphate oxygen of A9 is 2.36 Å, and the distance between Na+ and the N7 of G10.1 is 2.56 Å. The angle between these three atoms is 109°, indicating that the octahedral complex is somewhat distorted. Bond distances to the four water molecules range between 2.4 Å and 2.6 Å, with the other bond angles rather more close to 90°. (See Table II). Three of the coordinated water molecules form bridging interactions with other RNA atoms, including a hydrogen bond between water and the pro-R phosphate oxygen atom of G10.1, a hydrogen bond between water and the O6 of G10.1, and a hydrogen bond between water and O4 of U10.2. The remaining water molecule, does not appear to make additional hydrogen bonding interactions.
Table II. Metal Ion Binding Site Bond Distances (Å).
| G10.1/A9 phosphate site | current structure 3ZP8 | Mähler & Persson f | MESPEUS db(Na) g | MESPEUS db(Mg) g | Mn(II) structure 2OEU h | MESPEUS db(Mn) g |
|---|---|---|---|---|---|---|
|
| ||||||
| Me-N7 | 2.61 | --- | 2.64a | 2.77a | 2.08 | 2.42a |
| 3.06b | 2.11b | |||||
|
| ||||||
| Me-OPR | 2.33 | --- | 2.77c | 2.04c | 2.01 | 2.03 |
| 2.59d | 2.20d | |||||
|
| ||||||
| Me-OH2(3) | 2.36 | 2.43f | 2.52e | 2.18e | 2.41 | 2.26e |
|
| ||||||
| Me-OH2(4) | 2.51 | 2.43 | 2.52 | 2.18 | 2.24 | 2.26 |
|
| ||||||
| Me-OH2(5) | 2.33 | 2.43 | 2.52 | 2.18 | 2.02 | 2.26 |
|
| ||||||
| Me-OH2(6) | 2.56 | 2.43 | 2.52 | 2.18 | 1.70 | 2.26 |
Me-N7 Gua contact in DNA and RNA
All Me-N contacts
Me-O for non-bridging phosphate oxygens
Me-O for all oxygens
Me-O for water only
Solution values obtained from large angle X-ray scattering and double difference infrared spectroscopy. 22
MESPEUS data base23
Mn identified with unambiguous anomalous X-ray scattering signature, 20.
Numbers in italics are average values from solution scattering experiments (f) and MESPEUS database (g). Non-italics numbers are from hammerhead ribozyme crystal structures (3ZP8 for Na+ and 20EU for Mn2+(h)).
The G10.1/A9 phosphate metal binding site thus serves as an important positive control in the current structural analysis. Table II reports the coordination distances to Na+ in the current structure and to Mn2+ in the previous full-length hammerhead structure, as well as average coordination distance values for these and Mg2+ from the literature and databases cited. Based upon the chemical identities of the ligands in the octahedral complex, the bond distances and angles, identification of the ion as Na+ is reasonably certain. Hence Na+ is observed to bind to the A9 phosphate in a manner very similar to that of previously observed divalent cations, including Mn2+ and Mg2+.
A Na+ ion binds to the Hoogsteen face of G12, the general base in the cleavage reaction
Based upon proximity to the attacking nucleophile 12, in addition to compelling biochemical evidence 26, G12 has previously been identified as the general base in the hammerhead ribozyme cleavage reaction. Deprotonation of the N1 of G12 is thought to lead to the transient formation of a negatively charged enolate form of guanine, in which the negative charge becomes dispersed between N1 and O6. Upon initiation of the cleavage reaction by G12's abstraction of the 2′-proton from C17, the uncharged keto form of guanosine is restored.
A Na+ ion (Figure 2c) directly coordinates the O6 of G12 in the plane of the base, with a bond distance of 2.4 Å, and directly coordinates a water molecule, with a distance of 2.6 Å. This latter water molecule bridges to the N7 of G12, with a hydrogen bonding distance of 2.8 Å. Two additional water molecules are well-resolved, with bond distances of 2.3 Å and 2.5 Å, and angles consistent with an octahedral complex. These also form hydrogen bonding bridges to phosphate oxygen atoms. Weaker electron density corresponds to the two remaining ligand sites in the presumed octahedral complex; water molecules were not assigned to these positions in the PDB coordinates due to their rather high temperature factors.
This is the first crystallographic observation of a metal ion interaction with G12, although a similar interaction has been predicted previously, based upon biochemical evidence27,28.
An additional potential ion binding site bridging the A9 and scissile phosphates
Another potential ion binding site, observed in the hammerhead ribozyme active site, corresponds to a well-ordered electron density peak at water position 1, bridging the pro-R non-bridging phosphate oxygen atoms of the A9 and scissile phosphates with distances of 3.1 Å and 2.5 Å, respectively, and a bond angle of 85.6°. Additional contacts include the 2′O of C17, the cleavage-site nucleotide, at 3.2 Å; the 2′-O of G8 implicated in general acid catalysis, at 3.3 Å, and water molecule (2131 in the 3zp8 PDB file), at 3 Å, which is also coordinated by the Na+ ion bound at the canonical A9 phosphate site. The final position in the slightly distorted octahedral complex is occupied by the exocyclic amine of G12, forming a close-contact at 2.9 Å.
We have chosen to model this position as a water molecule rather than as a sodium ion in the PDB file 3ZP8 as we believe it is the more conservative interpretation of the data. Although the observed octahedral coordination of the molecule or ion at position 1 is consistent with assignment of a Na+ ion rather than water, the close contact with the exocyclic amine of G12 and the longer bond distances are more consistent with a hydrogen bond to a water molecule. The position was refined assuming the identity of water with an occupancy of 1.0 and having 10 electrons. (Na+ also has 10 electrons, and therefore an experimentally indistinguishable X-ray scattering factor.) The identity of this feature is addressed further in the discussion section.
Further details regarding the active site geometry
The high resolution diffraction data permit us to measure the most critical non-bonded inter-atomic distances and associated angles in the hammerhead ribozyme active site with unique precision. The ribozyme-substrate complex includes a 2′-OMe C17 at the active site; this functions as a substrate analogue-inhibitor in that the active 2′-H is replaced with an inert methyl ether linkage that can only function as a hydrogen bond acceptor. When G12 is protonated at N1, the N1 proton forms a 3.1 Å hydrogen bond with the 2′-O of C17. The corresponding bond angles with respect to C2 and C6 in G12 are 106° and 129°. (An ideal symmetric hydrogen bond would possess angles of about 117°, given the internal ring angle of guanine.) The interatomic distance between the attacking nucleophile, i.e., the 2′O of C17, and the adjacent scissile phosphorus atom is 3.3 Å, and the accompanying in-line attack angle is 156.8°. The ideal transition-state (or reaction intermediate) in-line attack angle will approach 180° as the scissile phosphate approaches a pentacoordinated trigonal-bipyramidal oxyphosphorane geometry, and the final bond distance between the 2′O and the phosphorus atom in the final reaction product will be about 1.59 Å. The 2′-OH of G8 donates a 3.2 Å hydrogen bond to the 5′O of C1.1, the leaving group in the cleavage reaction, belonging to the nucleotide immediately 3′ to the cleavage site. These and several additional contacts are illustrated in Figure 3.
Discussion
Although the hammerhead ribozyme is catalytically active in the absence of divalent metal ions14, it does require a high concentration of positive charge in the form of molar quantities of monovalent cations such as Na+, or exchange-inert trivalent complexes such as cobalt hexaamine, i.e., Co(NH3)63+, for activity28. Under physiological conditions, the most likely sources of high cationic strength are Mg2+ ions or monovalent ions such as Na+ or K+. It is thus of considerable importance to understand how Na+ may substitute for Mg2+ in hammerhead ribozyme catalysis, in terms of its structural interactions as well as catalytic potential. Crystallographic identification of Na+ binding sites is not always straightforward, especially when dealing with moderate diffraction resolutions typically associated with RNA crystals. Na+, like Mg2+ and water, has only 10 electrons, and unlike divalent metal ions such as the more electron-rich Mn2+ often used to identify metal binding sites indirectly, Na+ does not possess a useful X-ray absorption edge that would facilitate unambiguous identification. Therefore, a combination of coordination geometry, biochemistry, and known propensities to form nucleotide complexes must be used together with high-resolution X-ray diffraction data to identify Na+ binding sites unambiguously. Obtaining hammerhead ribozyme crystals that diffract to 1.55 Å resolution in crystallization conditions that include a high concentration of Na+ ions presents a unique opportunity to identify monovalent cation binding sites that may have mechanistically significant implications.
The single most prominent divalent metal ion binding site in both the minimal and full-length hammerhead structures, the “McKay Site,” involves inner-sphere coordination of the pro-R phosphate oxygen of A9, and inner-sphere coordination of the N7 of G10.1, the adjacent nucleotide base. The binding mode appears identical for Mn2+ in both the minimal and full-length structures, despite their very different active site conformations21,27. Because of its proximity to the scissile phosphate in the full-length hammerhead structure, and because of biochemical evidence suggesting that the A9 and scissile phosphate are bridged by a single metal ion in the transition-state of the cleavage reaction15, the McKay Site metal ion has received considerable attention.
The A9 and scissile phosphates are separated by 4.2 Å in the full-length hammerhead ribozyme structure, and form a nearly geometrically ideal potential divalent metal ion binding site for a metal ion favoring formation of an octahedral complex. Thus it is quite puzzling that A9 phosphate metal ion binding favors coordination with N7 of G10.1 rather than the pro-R oxygen of the scissile phosphate in the full-length hammerhead. This has lead to suggestions that this metal ion migrates and bridges the two phosphates only as the reaction approaches the transition-state, or that the metal ion's binding mode is somehow disrupted by the presence of the 2′-OMe at the cleavage site, or that the corresponding Mg2+ ion simply binds differently compared to the observed mode of Mn2+ binding, as has been witnessed in the context of tRNA29. These proposals have been tested computationally17,20, and in the case of unmodified substrate structures, crystallographically, for both the minimal30 and full-length13 hammerhead ribozyme structures. We are now able to add direct high-resolution crystallographic observations.
We observe that a Na+ ion binds to the McKay site via exactly the same mode as observed in all previous hammerhead crystal structures with more electron-rich divalent cations. This mode involves inner-sphere coordination of the pro-R phosphate oxygen of A9, and inner-sphere coordination of the N7 of G10.1, the adjacent nucleotide base (Figure 2b). This mode of binding is preserved, despite the fact that the uncharged N7 is a softer ligand than oxygen, a more typical ligand for a hard divalent metal ion such as Mg2+ ion. In other words, this Na+ ion prefers a coordination environment consisting of one hard and one soft ligand (in addition to four bound water molecules, as one might propose for a softer divalent metal ion such as Mn2+), rather than a coordination environment consisting instead of two hard ligands (the pro-R oxygens of the A9 and scissile phosphates).
Table II lists the coordination bond distances observed for Na+ and Mn2+ in the G10.1/A9 phosphate site, as well as average coordination distances for these metals and Mg2+. Overall, the coordination distances are consistent with Na+ and clearly indicate that Na+ can substitute for divalent metal ions in the most prominent metal binding site in the hammerhead ribozyme.
A second active-site Na+ ion forms an inner-sphere interaction with the exocyclic O6 keto oxygen of G12, the nucleotide implicated as the general base in the hammerhead cleavage reaction (Figure 2c). This Na+ ion is ideally positioned on the Hoogsteen face of G12 to counter-balance (or disperse) the transient negative charge accumulated at O6 as an enolate ion accompanying deprotonation of the N1 of G12, in turn required to initiate the cleavage reaction via abstraction of the 2′-H of C17. This Na+ ion, in other words, may favorably perturb the pKa of G12 to enhance general base catalysis. This sort of Na+ ion interaction with G12 has in fact been suggested previously, based upon biochemical evidence27,28.
A third, but more ambiguous, potential ion site forms a distorted octahedral complex that bridges the pro-R oxygens of the A9 and scissile phosphates, in addition to forming interactions with the 2′-O attacking nucleophile of C17, and the 5′-O leaving group of C1.1, and a bridging water molecule that spans between it and the McKay Site Na+ ion (Figure 2d).
We have conservatively modeled the bridging entity as a water molecule because its identity is somewhat ambiguous. Specifically, it forms a close-contact with the exocyclic amine of G12 at the sixth and final octahedral coordination position. The coordination distance is rather long, and the chemical identity of the ligand is unexpected, as noted in the results section. It is therefore unlikely to be a very stable interaction, even by the standards of Na+. The distance is consistent with a hydrogen bond to water, but modeling as water does not permit us to account for more than one of the other five observed coordination interactions. The scattering center in question refines well to 10 electrons, equally consistent with either Na+ ion or water. We therefore chose the more cautious interpretation of the data, and suggest that if indeed this is the predicted bridging metal ion site, the coordination environment may change somewhat as the transition-state geometry is approached, or it may prefer to bind K+ under physiological conditions, as K+ is arguably a more physiologically likely monovalent cation, and its binding would in fact be more consistent with the observed potential coordination distances (Figure 4).
Figure 4.
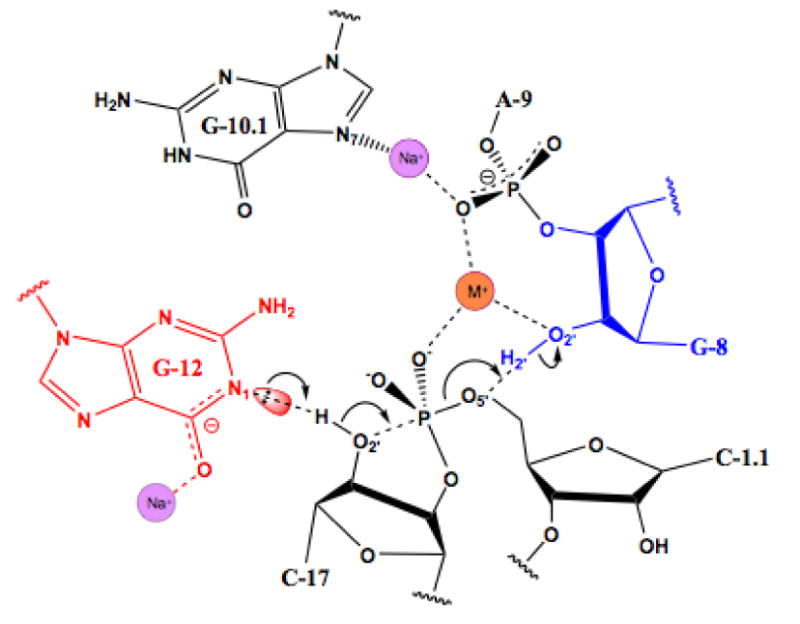
Schematic diagram of a possible hammerhead ribozyme reaction mechanism, corresponding to the structural representation in Figure 2D. Na+ ions are indicated, as in the previous figures, in purple, and the potential Na+ or K+ ion binding site (M+) is indicated in orange. The Na+ ion associated with G12 may help to stabilize the negative charge associated with deprotonation of the N1 of G12, and the bridging potential Na+ or K+ ion may help to stabilize the close approach of two negatively-charged phosphates.
Previous molecular simulations studies successfully predict many of the features of Na+ ions were predicted to bind at the cleavage site. The monovalent cation site #1 of Figure 1 of this study 31 exactly corresponds to the putative Na+/K+ ion binding we have observed experimentally in the hammerhead ribozyme structure 31. Specifically, a minimum or threshold occupancy of cations was found to be necessary for proper folding of the active site into the pre-catalytic conformation. This requirement is predicted to be fulfilled by either divalent metal ions or monovalent metal ions in high concentration. In particular, one or more Na+ ions were predicted to bind at the cleavage site. The monovalent cation site #1 of Figure 1 of this study 31 exactly corresponds to the putative Na/K site identified crystallographically, both in terms of spatial position and coordination geometry. Two additional monovalent metal ion sites are also predicted at the active site that have not as yet been observed crystallographically. Nonetheless, our structural results test what is perhaps the single most important prediction of the simulation studies, and thus serve to validate this computational approach as well as corroborate its primary result.
Experimental Methods
Full-length hammerhead ribozyme crystals were obtained via vapor diffusion, as described previously12,20, except the crystallization conditions were modified as follows: The reservoir contained 1.7 M sodium malonate, buffered to pH 7.5, and 1 mM MgCl2. The hanging drop contained 1/2 concentration of the reservoir solution mixed with the RNA solution prepared as described previously12. Crystals were stabilized in a mother liquor containing 1.7 M sodium malonate, pH 7.5, and 10 mM MgCl2, and flash-frozen, using the sodium malonate as a cryoprotectant. The data collection is summarized in Table I. The data were processed using iMosflm32 and CCP433, and refined using Phenix34, beginning with rigid-body refinement using 2GOZ (now 3ZD5) after substituting ribouridine for 5′-bromouridine. This was followed by simulated annealing and TLS refinement using default phenix.refine protocols. Model building and adjustment was performed within COOT35, including identification and rejection of water molecules and Na+ ions. The refined structural coordinates and accompanying Fobs are currently available in the Protein Data Bank as 3ZP8.
Supplementary Material
Hammerhead Ribozymes are catalytically active in the presence of monovalent cations such as Na+
Highest resolution ribozyme structure
Na+ binding sites revealed
Potential catalytic relevance examined
Acknowledgments
This work was supported by the National Institutes of Health grant R01GM087721 to WGS. We thank the members of the Center for Molecular Biology of RNA for helpful discussions, advice and shared facilities, Donald Burke for discussing metal interactions with the Hoogsteen face of G12, Norm Pace for pointing out that solvent site #1 may be predisposed to binding K+, Darrin York and his research group for discussions regarding monovalent and divalent metal ion binding modes and their molecular simulation, and the reviewers for many helpful suggestions, including the accurate identification of metal ion binding sites.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Prody GA, Bakos JT, Buzayan JM, Schneider IR, Breuning G. Autolytic processing of dimeric plant virus satellite RNA. Science. 1986;231:1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- 2.Symons RH. Plant pathogenic RNAs and RNA catalysis. Nucleic Acids Res. 1997;25:2683–2689. doi: 10.1093/nar/25.14.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martick M, Horan LH, Noller HF, Scott WG. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature. 2008;454:899–902. doi: 10.1038/nature07117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.García-Robles I, Sánchez-Navarro J, de la Peña M. Intronic hammerhead ribozymes in mRNA biogenesis. Biol Chem. 2012;393:1317–1326. doi: 10.1515/hsz-2012-0223. [DOI] [PubMed] [Google Scholar]
- 5.Hammann C, Luptak A, Perreault J, de la Peña M. The ubiquitous hammerhead ribozyme. RNA. 2012;18:871–885. doi: 10.1261/rna.031401.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruffner DE, Stormo GD, Uhlenbeck OC. Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry. 1990;29:10695–10702. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]
- 7.De la Peña M, Gago S, Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- 9.McKay DB. Structure and function of the hammerhead ribozyme: an unfinished story. Rna. 1996;2:395–403. [PMC free article] [PubMed] [Google Scholar]
- 10.Blount KF, Uhlenbeck OC. The structure-function dilemma of the hammerhead ribozyme. Annu Rev Biophys Biomol Struct. 2005;34:415–440. doi: 10.1146/annurev.biophys.34.122004.184428. [DOI] [PubMed] [Google Scholar]
- 11.Nelson JA, Uhlenbeck OC. Hammerhead redux: does the new structure fit the old biochemical data? RNA. 2008;14:605–615. doi: 10.1261/rna.912608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi YI, Martick M, Lares M, Kim R, Scott WG, Kim SH. Capturing hammerhead ribozyme structures in action by modulating general base catalysis. PLoS Biol. 2008;6:e234. doi: 10.1371/journal.pbio.0060234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray JB, Seyhan AA, Walter NG, Burke JM, Scott WG. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, Karbstein K, Peracchi A, Beigelman L, Herschlag D. Identification of the hammerhead ribozyme metal ion binding site responsible for rescue of the deleterious effect of a cleavage site phosphorothioate. Biochemistry. 1999;38:14363–14378. doi: 10.1021/bi9913202. [DOI] [PubMed] [Google Scholar]
- 16.Vogt M, Lahiri S, Hoogstraten CG, Britt RD, DeRose VJ. Coordination environment of a site-bound metal ion in the hammerhead ribozyme determined by 15N and 2H ESEEM spectroscopy. J Am Chem Soc. 2006;128:16764–16770. doi: 10.1021/ja057035p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee TS, Silva López C, Giambasu GM, Martick M, Scott WG, York DM. Role of Mg2+ in hammerhead ribozyme catalysis from molecular simulation. J Am Chem Soc. 2008;130:3053–3064. doi: 10.1021/ja076529e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong KY, Lee TS, York DM. Active participation of Mg ion in the reaction coordinate of RNA self-cleavage catalyzed by the hammerhead ribozyme. J Chem Theory Comput. 2011;7:1–3. doi: 10.1021/ct100467t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward WL, Derose VJ. Ground-state coordination of a catalytic metal to the scissile phosphate of a tertiary-stabilized Hammerhead ribozyme. RNA. 2012;18:16–23. doi: 10.1261/rna.030239.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martick M, Lee TS, York DM, Scott WG. Solvent structure and hammerhead ribozyme catalysis. Chem Biol. 2008;15:332–342. doi: 10.1016/j.chembiol.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pley HW, Flaherty KM, McKay DB. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- 22.Mähler J, Persson I. A study of the hydration of the alkali metal ions in aqueous solution. Inorg Chem. 2012;51:425–438. doi: 10.1021/ic2018693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsin K, Sheng Y, Harding M, Taylor P, Walkinshaw D. MESPEUS: a database of the geometry of metal sites in proteins. Journal of Applied Crystallography. 2008;41:963–968. [Google Scholar]
- 24.Scott WG. RNA structure, metal ions, and catalysis. Curr Opin Chem Biol. 1999;3:705–709. doi: 10.1016/s1367-5931(99)00029-0. [DOI] [PubMed] [Google Scholar]
- 25.Murray JB, Terwey DP, Maloney L, Karpeisky A, Usman N, Beigelman L, Scott WG. The structural basis of hammerhead ribozyme self-cleavage. Cell. 1998;92:665–673. doi: 10.1016/s0092-8674(00)81134-4. [DOI] [PubMed] [Google Scholar]
- 26.Han J, Burke JM. Model for general acid-base catalysis by the hammerhead ribozyme: pH-activity relationships of G8 and G12 variants at the putative active site. Biochemistry. 2005;44:7864–7870. doi: 10.1021/bi047941z. [DOI] [PubMed] [Google Scholar]
- 27.Roychowdhury-Saha M, Burke DH. Extraordinary rates of transition metal ion-mediated ribozyme catalysis. RNA. 2006;12:1846–1852. doi: 10.1261/rna.128906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roychowdhury-Saha M, Burke DH. Distinct reaction pathway promoted by non-divalent-metal cations in a tertiary stabilized hammerhead ribozyme. RNA. 2007;13:841–848. doi: 10.1261/rna.339207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jovine L, Djordjevic S, Rhodes D. The crystal structure of yeast phenylalanine tRNA at 2.0 A resolution: cleavage by Mg(2+) in 15-year old crystals. J Mol Biol. 2000;301:401–414. doi: 10.1006/jmbi.2000.3950. [DOI] [PubMed] [Google Scholar]
- 30.Scott WG, Murray JB, Arnold JR, Stoddard BL, Klug A. Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme. Science. 1996;274:2065–2069. doi: 10.1126/science.274.5295.2065. [DOI] [PubMed] [Google Scholar]
- 31.Lee TS, Giambaşu GM, Sosa CP, Martick M, Scott WG, York DM. Threshold occupancy and specific cation binding modes in the hammerhead ribozyme active site are required for active conformation. J Mol Biol. 2009;388:195–206. doi: 10.1016/j.jmb.2009.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winn MD. An overview of the CCP4 project in protein crystallography: an example of a collaborative project. J Synchrotron Radiat. 2003;10:23–25. doi: 10.1107/s0909049502017235. [DOI] [PubMed] [Google Scholar]
- 34.Adams PD, Grosse-Kunstleve RW, Hung L, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallographica. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 37.Painter J, Merritt EA. TLSMD web server for the generation of multigroup TLS models. Journal of Applied Crystallography. 2006;39:109–111. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


