Abstract
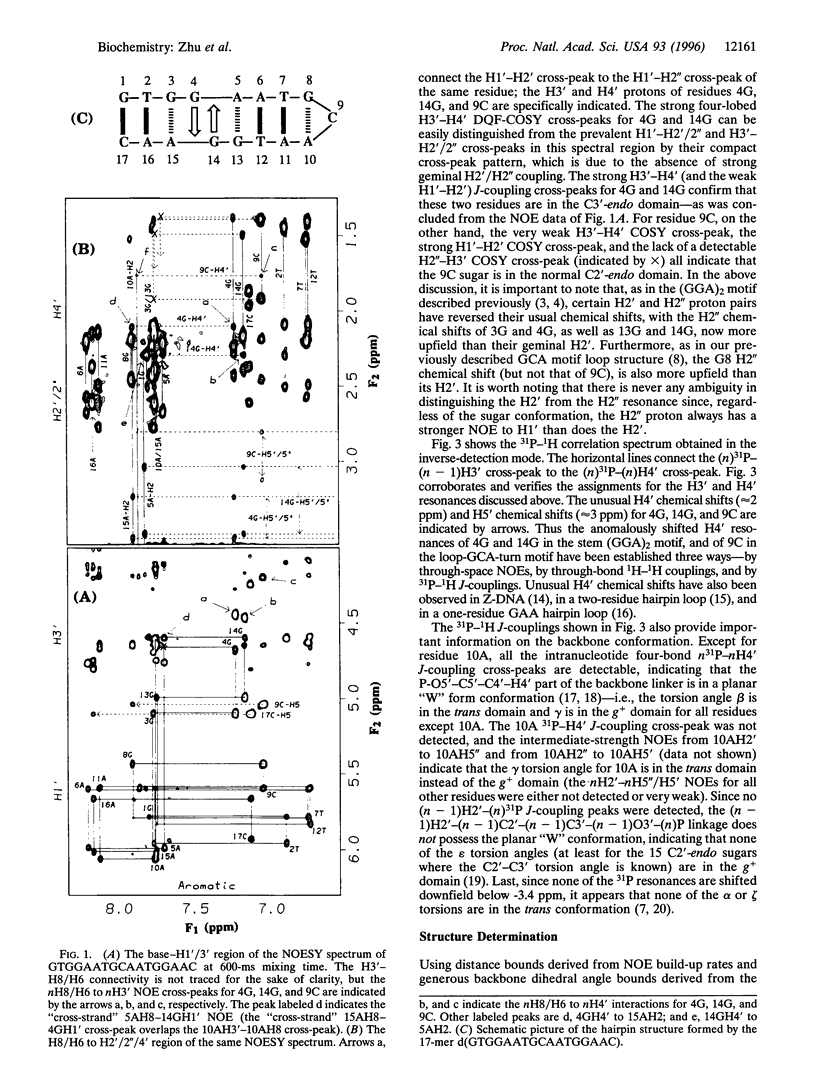
Recently, we established that satellite III (TGGAA)n tandem repeats, which occur at the centromeres of human chromosomes, pair with themselves to form an unusual "self-complementary" antiparallel duplex containing (GGA)2 motifs in which two unpaired guanines from opposite strands intercalate between sheared G.A base pairs. In separate studies, we have also established that the GCA triplet does not form bimolecular (GCA)2 motifs but instead promotes the formation of hairpins containing a GCA-turn motif in which the loop contains a single cytidine closed by a sheared G.A pair. Since TGCAA is the most frequent variant of TGGAA found in satellite III repeats, we reasoned that the potential of this variant to form GCA-turn miniloop fold-back structures might be an important factor in modulating the local structure in natural (TGGAA)n repeats. We report here the NMR-derived solution structure of the heptadecadeoxynucleotide (G)TGGAATGCAATGGAA(C) in which a central TGCAA pentamer is flanked by two TGGAA pentamers. This 17-mer forms a rather unusual and very stable hairpin structure containing eight base pairs in the stem, only four of which are Watson-Crick pairs, and a loop consisting of a single cytidine residue. The stem contains a (GGA)2 motif with intercalative 14G/4G stacking between two sheared G.A base pairs; the loop end of the stem consists of a sheared 8G.10A closing pair with the cytosine base of the 9C loop stacked on 8G. The remarkable stability of this unusual hairpin structure (Tm = 63 degrees C) suggests that it probably plays an important role in modulating the folding of satellite III (TGGAA)n repeats at the centromere.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blommers M. J., Haasnoot C. A., Walters J. A., van der Marel G. A., van Boom J. H., Hilbers C. W. Solution structure of the 3'-5' cyclic dinucleotide d(pApA). A combined NMR, UV melting, and molecular mechanics study. Biochemistry. 1988 Nov 1;27(22):8361–8369. doi: 10.1021/bi00422a011. [DOI] [PubMed] [Google Scholar]
- Blommers M. J., van de Ven F. J., van der Marel G. A., van Boom J. H., Hilbers C. W. The three-dimensional structure of a DNA hairpin in solution two-dimensional NMR studies and structural analysis of d(ATCCTATTTATAGGAT). Eur J Biochem. 1991 Oct 1;201(1):33–51. doi: 10.1111/j.1432-1033.1991.tb16253.x. [DOI] [PubMed] [Google Scholar]
- Boulard Y., Gabarro-Arpa J., Cognet J. A., Le Bret M., Guy A., Téoule R., Guschlbauer W., Fazakerley G. V. The solution structure of a DNA hairpin containing a loop of three thymidines determined by nuclear magnetic resonance and molecular mechanics. Nucleic Acids Res. 1991 Oct 11;19(19):5159–5167. doi: 10.1093/nar/19.19.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S. H., Cheng J. W., Fedoroff O., Reid B. R. DNA sequence GCGAATGAGC containing the human centromere core sequence GAAT forms a self-complementary duplex with sheared G.A pairs in solution. J Mol Biol. 1994 Aug 19;241(3):467–479. doi: 10.1006/jmbi.1994.1521. [DOI] [PubMed] [Google Scholar]
- Chou S. H., Cheng J. W., Reid B. R. Solution structure of [d(ATGAGCGAATA)]2. Adjacent G:A mismatches stabilized by cross-strand base-stacking and BII phosphate groups. J Mol Biol. 1992 Nov 5;228(1):138–155. doi: 10.1016/0022-2836(92)90497-8. [DOI] [PubMed] [Google Scholar]
- Chou S. H., Zhu L., Reid B. R. On the relative ability of centromeric GNA triplets to form hairpins versus self-paired duplexes. J Mol Biol. 1996 Jun 14;259(3):445–457. doi: 10.1006/jmbi.1996.0331. [DOI] [PubMed] [Google Scholar]
- Chou S. H., Zhu L., Reid B. R. The unusual structure of the human centromere (GGA)2 motif. Unpaired guanosine residues stacked between sheared G.A pairs. J Mol Biol. 1994 Dec 2;244(3):259–268. doi: 10.1006/jmbi.1994.1727. [DOI] [PubMed] [Google Scholar]
- Egli M., Gessner R. V. Stereoelectronic effects of deoxyribose O4' on DNA conformation. Proc Natl Acad Sci U S A. 1995 Jan 3;92(1):180–184. doi: 10.1073/pnas.92.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer N., Azorín F., Villasante A., Gutiérrez C., Abad J. P. Centromeric dodeca-satellite DNA sequences form fold-back structures. J Mol Biol. 1995 Jan 6;245(1):8–21. doi: 10.1016/s0022-2836(95)80034-4. [DOI] [PubMed] [Google Scholar]
- Frederick C. A., Coll M., van der Marel G. A., van Boom J. H., Wang A. H. Molecular structure of cyclic deoxydiadenylic acid at atomic resolution. Biochemistry. 1988 Nov 1;27(22):8350–8361. doi: 10.1021/bi00422a010. [DOI] [PubMed] [Google Scholar]
- Giessner-Prettre C., Pullman B., Tran-Dinh S., Neumann J. M., Huynh-Dinh T., Igolen J. Proton NMR study of the B----Z transition of d(CGm5CG)2 and d(CGm5CGCG)2: theory and experiment. Nucleic Acids Res. 1984 Apr 11;12(7):3271–3281. doi: 10.1093/nar/12.7.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorenstein D. G., Schroeder S. A., Fu J. M., Metz J. T., Roongta V., Jones C. R. Assignments of 31P NMR resonances in oligodeoxyribonucleotides: origin of sequence-specific variations in the deoxyribose phosphate backbone conformation and the 31P chemical shifts of double-helical nucleic acids. Biochemistry. 1988 Sep 20;27(19):7223–7237. doi: 10.1021/bi00419a009. [DOI] [PubMed] [Google Scholar]
- Grady D. L., Ratliff R. L., Robinson D. L., McCanlies E. C., Meyne J., Moyzis R. K. Highly conserved repetitive DNA sequences are present at human centromeres. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1695–1699. doi: 10.1073/pnas.89.5.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare D. R., Reid B. R. Three-dimensional structure of a DNA hairpin in solution: two-dimensional NMR studies and distance geometry calculations on d(CGCGTTTTCGCG). Biochemistry. 1986 Sep 9;25(18):5341–5350. doi: 10.1021/bi00366a053. [DOI] [PubMed] [Google Scholar]
- Hare D. R., Wemmer D. E., Chou S. H., Drobny G., Reid B. R. Assignment of the non-exchangeable proton resonances of d(C-G-C-G-A-A-T-T-C-G-C-G) using two-dimensional nuclear magnetic resonance methods. J Mol Biol. 1983 Dec 15;171(3):319–336. doi: 10.1016/0022-2836(83)90096-7. [DOI] [PubMed] [Google Scholar]
- Hirao I., Kawai G., Yoshizawa S., Nishimura Y., Ishido Y., Watanabe K., Miura K. Most compact hairpin-turn structure exerted by a short DNA fragment, d(GCGAAGC) in solution: an extraordinarily stable structure resistant to nucleases and heat. Nucleic Acids Res. 1994 Feb 25;22(4):576–582. doi: 10.1093/nar/22.4.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman A. A., Middleton K., Centola M., Mitchison T. J., Carbon J. Microtubule-motor activity of a yeast centromere-binding protein complex. Nature. 1992 Oct 8;359(6395):533–536. doi: 10.1038/359533a0. [DOI] [PubMed] [Google Scholar]
- Li Y., Zon G., Wilson W. D. NMR and molecular modeling evidence for a G.A mismatch base pair in a purine-rich DNA duplex. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):26–30. doi: 10.1073/pnas.88.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrew J., Diehl B., Fitzgerald-Hayes M. Single base-pair mutations in centromere element III cause aberrant chromosome segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Feb;6(2):530–538. doi: 10.1128/mcb.6.2.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooren M. M., Pulleyblank D. E., Wijmenga S. S., van de Ven F. J., Hilbers C. W. The solution structure of the hairpin formed by d(TCTCTC-TTT-GAGAGA). Biochemistry. 1994 Jun 14;33(23):7315–7325. doi: 10.1021/bi00189a037. [DOI] [PubMed] [Google Scholar]
- Ng R., Carbon J. Mutational and in vitro protein-binding studies on centromere DNA from Saccharomyces cerevisiae. Mol Cell Biol. 1987 Dec;7(12):4522–4534. doi: 10.1128/mcb.7.12.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbons L. P., van der Marel G. A., van Boom J. H., Altona C. An NMR study of polymorphous behaviour of the mismatched DNA octamer d(m5C-G-m5C-G-A-G-m5C-G) in solution. The B-duplex and hairpin forms. Eur J Biochem. 1987 Dec 30;170(1-2):225–239. doi: 10.1111/j.1432-1033.1987.tb13690.x. [DOI] [PubMed] [Google Scholar]
- Prosser J., Frommer M., Paul C., Vincent P. C. Sequence relationships of three human satellite DNAs. J Mol Biol. 1986 Jan 20;187(2):145–155. doi: 10.1016/0022-2836(86)90224-x. [DOI] [PubMed] [Google Scholar]
- Sarma R. H., Mynott R. T., Wood D. J., Hruska F. E. Determination of the preferred conformations constrained along the C-4'-C-5' and C-5'-O-5' bonds of beta-5'-nucleotides in solution. Four-bond 31P-1H coupling. J Am Chem Soc. 1973 Sep 19;95(19):6457–6459. doi: 10.1021/ja00800a055. [DOI] [PubMed] [Google Scholar]
- Sklenár V., Miyashiro H., Zon G., Miles H. T., Bax A. Assignment of the 31P and 1H resonances in oligonucleotides by two-dimensional NMR spectroscopy. FEBS Lett. 1986 Nov 10;208(1):94–98. doi: 10.1016/0014-5793(86)81539-3. [DOI] [PubMed] [Google Scholar]
- Sussman J. L., Seeman N. C., Kim S. H., Berman H. M. Crystal structure of a naturally occurring dinucleoside phoaphate: uridylyl 3',5'-adenosine phosphate model for RNA chain folding. J Mol Biol. 1972 May 28;66(3):403–421. doi: 10.1016/0022-2836(72)90423-8. [DOI] [PubMed] [Google Scholar]
- Vissel B., Nagy A., Choo K. H. A satellite III sequence shared by human chromosomes 13, 14, and 21 that is contiguous with alpha satellite DNA. Cytogenet Cell Genet. 1992;61(2):81–86. doi: 10.1159/000133374. [DOI] [PubMed] [Google Scholar]
- Zhu L., Chou S. H., Reid B. R. The structure of a novel DNA duplex formed by human centromere d(TGGAA) repeats with possible implications for chromosome attachment during mitosis. J Mol Biol. 1995 Dec 8;254(4):623–637. doi: 10.1006/jmbi.1995.0643. [DOI] [PubMed] [Google Scholar]
- Zhu L., Chou S. H., Xu J., Reid B. R. Structure of a single-cytidine hairpin loop formed by the DNA triplet GCA. Nat Struct Biol. 1995 Nov;2(11):1012–1017. doi: 10.1038/nsb1195-1012. [DOI] [PubMed] [Google Scholar]
- Zhu L., Reid B. R. An improved NOESY simulation program for partially relaxed spectra: BIRDER. J Magn Reson B. 1995 Mar;106(3):227–235. doi: 10.1006/jmrb.1995.1038. [DOI] [PubMed] [Google Scholar]




